Abstract
The current study investigates whether microRNA (miRNA) regulators of epithelial-mesenchymal transition (EMT), tissue fibrosis, and angiogenesis are differentially expressed in human primary pterygium. Genome-wide miRNA and mRNA expression profiling of paired pterygium and normal conjunctiva was performed in the context of conventional excision of pterygium with autotransplantation of conjunctiva (n=8). Quantitative real time polymerase chain reaction (qRT-PCR) was used to validate the expression of key molecules previously detected by microarray. In pterygium, 25 miRNAs and 31 mRNAs were significantly differentially expressed by more than two-fold compared to normal conjunctiva. 14 miRNAs were up-regulated (miR-1246, −486, −451, −3172, −3175, −1308, −1972, −143, −211, −665, −1973, −18a, 143, and −663b), whereas 11 were down-regulated (miR-675, −200b-star, −200a-star, −29b, −200b, −210, −141, −31, −200a, −934, and −375). Unsupervised hierarchical cluster analysis demonstrated that members of the miR-200 family were coexpressed and down-regulated in pterygium. The molecular and cellular functions that were most significant to the miRNA data sets were cellular development, cellular growth and proliferation, and cellular movement. qRT-PCR confirmed the expression of 15 of the 16 genes tested and revealed that miR-429 was down-regulated by more than two-fold in pterygium. The concerted down-regulation of four members from both clusters of the miR-200 family (miR-200a/−200b/−429 and miR-200c/−141), which are known to regulate EMT, and up-regulation of the predicted target and mesenchymal marker fibronectin (FN1), suggest that EMT could potentially play a role in the pathogenesis of pterygium and might constitute promising new targets for therapeutic intervention in pterygium.
Keywords: Pterygium, gene microarray, microRNA, mRNA, epithelial mesenchymal transition, miR-200 family
1. Introduction
Pterygium is a common degenerative disease of the ocular surface in which wedge-shaped ingrowth of conjunctival tissue invades the peripheral cornea. The condition is associated with chronic ultraviolet radiation exposure and is characterized by induction of cell proliferation, squamous metaplasia, goblet cell hyperplasia, inflammation, fibrosis, angiogenesis, and extracellular matrix breakdown (Chui et al., 2008). Recent studies also provide evidence that pterygium is a stem cell disorder with premalignant features (Chui et al., 2011; Hirst et al., 2009), and that epithelial-mesenchymal transition (EMT) may play a key role in the pathogenesis (Ando et al., 2011; Kato et al., 2007b). Although considerable progress has been made towards understanding the etiology of the disease, the pathogenesis of pterygium is not completely understood (Bradley et al., 2008; Chui et al., 2008).
EMT is critical in both developmental processes (Bolender and Markwald, 1979; Duband et al., 1995; Griffith and Hay, 1992; Viebahn, 1995), wound healing and tissue remodeling (Weber et al., 2012), and tumor metastasis (Thiery et al., 2009) and describes a reversible series of events during which epithelial cells lose cell-cell contacts and acquire mesenchymal characteristics (Gregory et al., 2008b). These events involve molecular reprogramming of the cell, including loss or redistribution of epithelial-specific cell-cell adhesion molecules such as E-cadherin, and turning on of mesenchymal markers including fibronectin, vimentin and N-cadherin (Thiery and Sleeman, 2006). Studies of tumor biology suggest that signaling pathways involving TGFβ, Wnt, Notch and growth factors such as PDGF and FGF may induce EMT (Moustakas and Heldin, 2007). Upon stimulation by TGFβ, ZEB transcription factors (ZEB1/ZEB2) are up-regulated, resulting in their binding to E-box elements and repression of E-cadherin and polarity factor genes, which ultimately leads to EMT (Gregory et al., 2008b).
MicroRNAs (miRNA) are a class of noncoding RNAs of 18–24 nucleotides that post-transcriptionally down-regulate gene expression and modulate the expression of 20% or more of the human genome (Lewis et al., 2005; Xie et al., 2005). miRNAs down-regulate gene expression by binding to the 3’-untranslated region (UTR) of protein coding transcripts, resulting in either mRNA cleavage or translational repression (Ambros, 2004; Bartel, 2004; Lai, 2003). In recent years, the miRNA transcriptomes of the mammalian retina (Arora et al., 2007; Karali et al., 2007; Lagos-Quintana et al., 2003; Loscher et al., 2007; Ryan et al., 2006; Xu et al., 2007), lens (Frederikse et al., 2006; Ryan et al., 2006) and cornea (Ryan et al., 2006) have been identified and characterized, and similarities in the miRNA expression profiles have called into question whether ocular tissues may have common miRNA signatures (Xu, 2009). In spite of these progresses, the function and pathophysiological role of miRNAs in ophthalmology are still largely elusive. To date, only a few studies have investigated the gene expression profile of human pterygia, reporting differentially expressed extracellular matrix-related, fibrogenic, angiogenic, proinflammatory, and oncogenic genes (John-Aryankalayil et al., 2006; Tong et al., 2009). However, Chen et al. are the only ones to report data on the role of miRNAs in pterygium demonstrating that miR-766 and miR-215 may cause changes in genes that regulate wound healing processes (Chen S, et al. IOVS 2010;51: ARVO E-Abstract 2403).
We hypothesized that miRNAs are involved in the molecular pathogenesis of pterygium, and aimed to identify miRNAs in pterygium in order to improve our understanding of the pterygium pathogenesis. Specifically, the current study investigates whether miRNA regulators of EMT (Burk et al., 2008; Gregory et al., 2008a; Korpal et al., 2008; Park et al., 2008), tissue fibrosis (Kato et al., 2007a; Kwiecinski et al., 2011; Pottier et al., 2009; Thum et al., 2008; van Rooij et al., 2008; Wang et al., 2008) and ocular angiogenesis (Shen et al., 2008) are differentially expressed in human primary pterygium compared to normal conjunctiva.
2. Materials and methods
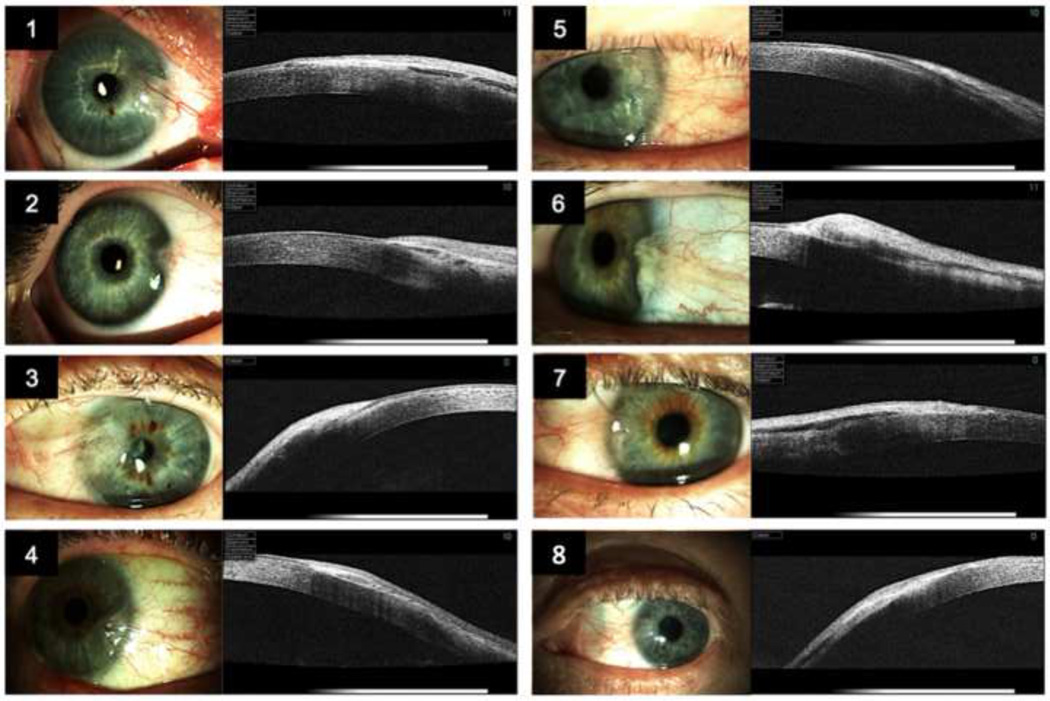
2.1 Patients and specimens
The study was approved by the South-Eastern Norway Regional Committee for Research Ethics, and was conducted in accordance with the official regulations for clinical research and the Declaration of Helsinki. Inclusion criteria were the presence of untreated primary nasal pterygium, clinical indication for conventional excision of pterygium with auto-transplantation of conjunctiva, and Caucasian origin to remove the potential confounding effect of ethnicity on the microarray analysis. Eight patients referred for elective pterygium surgery, six males (aged 47 to 82 years old, mean age 65 years), and two females (aged 27 to 49 years old, mean age 38 years), provided written informed consent and were included in the study. Preoperative slit lamp photography (Haag-Streit BQ900/IM900, Koeniz, Switzerland) and anterior segment optical coherence tomography (Topcon 3D OCT 1000, Topcon Corp., Tokyo, Japan) were performed in all study participants to demonstrate the extension of the pterygium onto the cornea (Fig.1). The corneal ingrowth in patients 1–8 was 4 mm, 2 mm, 4.5 mm, 3 mm, 3 mm, 2 mm, 3 mm, and 4 mm, respectively. Conventional excision of pterygium with autotransplantation of a superotemporal conjunctival graft and the use of fibrin tissue adhesive (Suppl. Fig. 1) was carried out under local anesthesia by a single surgeon (DHE). A small rectangular piece of normal conjunctival tissue was excised from the autograft and served as control tissue. The pterygium tissue was immediately trimmed under a Nikon ZMZ800 stereoscopic microscope (Nikon corp., Chiyoda-ku, Tokyo, Japan) using aseptic technique, and normal conjunctival tissue was eliminated from the resection edges of the pterygium. Next, the tissues were placed in individual culture dishes and washed three times with sterile, physiological NaCl-solution. Finally, the tissues were transferred to 2 ml Eppendorf Safe-Lock microcentrifuge tubes (Eppendorf, Hamburg, Germany) and stored at −86°C.
Figure 1. Patients.
Anterior photography and anterior segment optical coherent tomography of the 8 study patients demonstrating the hyperreflective pterygium extending onto the cornea destructing the Bowman’s membrane.
2.2 RNA isolation
Total RNA was extracted from biopsies using Qiagen miRNeasy Mini Kit (Qiagen Inc. Inc., Venlo, Netherlands), according to the manufacturer’s protocol. The frozen biopsies were treated with 700 µl of QIAzol Lysis Reagent (Qiagen Inc.) and shaken for two minutes by a Qiagen TissueLyser (Qiagen Inc.) with a frequency of 30/min to assure sample disruption. In addition, Phase Lock Gel Heavy 2 ml (5 PRIME, Hamburg, Germany) was used to increase yield and eliminate interphase contamination of nucleic acid solution. Concentration was determined by measurement with the Nano Drop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed by the Agilent BioAnalyzer 2100 System and RNA 6000 Nano Assay (Agilent Technologies, Santa Clara, CA). RNA samples were immediately frozen and stored at −80°C.
2.3 RNA labeling, microarray hybridization, preprocessing and normalization of raw microarray data miRNA expression analysis
200 ng of total RNA was used for labeling of microRNA by the Genisphere FlashTag HSR kit following the manufacturer's recommendations (Genisphere, Hatfield, PA). Labeled RNAs were hybridized to the GeneChip miRNA 2.0 Array (Affymetrix, Santa Clara, CA) as recommended by the manufacturer. The arrays were washed and stained using FS-450 fluidics station (Affymetrix). Signal intensities were detected by Hewlett Packard Gene Array Scanner 3000 7G (Hewlett Packard, Palo Alto, CA). The scanned images were processed using the AGCC (Affymetrix GeneChip Command Console) software, and the CEL files were imported into Partek Genomics Suite software (Partek, Inc. MO, USA). The Robust Multichip Analysis algorithm was applied for generation of signal values and normalization. Gene transcripts with maximal signal values that were less than six across all arrays were removed to filter for low and non-expressed genes, reducing the number of gene transcripts to 428.
2.4 mRNA expression analysis
100 ng of total RNA were subjected to Ambion WT Expression Kit (Ambion, Austin, TX) and GeneChip WT Terminal Labeling Kit (Affymetrix), following the manufacturers’ protocols for whole genome gene expression analysis. Microarray analyses were performed using the Affymetrix GeneChip Human Gene 1.1 ST Array strips (Affymetrix), which contain more than 28,000 gene transcripts. The arrays were hybridized, washed, stained and scanned using the GeneAtlas™ Personal Microarray System (Affymetrix). The scanned images were processed using the GeneAtlas Instrument Control Software, and the CEL files were imported into Partek Genomics Suite software (Partek). The RMA algorithm was applied for generation of signal values and normalization. Gene transcripts with maximal signal values of less than 16 across all arrays were removed to filter for low and non-expressed genes, reducing the number of gene transcripts to 18,690.
2.5 Microarray data analysis
The expression profiles were compared using a two-way ANOVA model, and the expression level was given as average normalized (log transformed) signal intensity. Correction for multiple testing was performed using false discovery rate (FDR) controlling procedures (Benjamini and Hochberg, 1995). Lists with differentially expressed genes were generated using both unadjusted values and FDR<10%. A p-value <0.05 was considered statistically significant. To further reduce the complexity of data and identify co-expressed genes or the similarity of samples, hierarchical clustering analysis was made using Partek Genomics Suite Software (Partek). Biological functional analysis, pathway analysis, and prediction of miRNA-targeted genes were performed using Ingenuity Pathways Analysis (IPA, Ingenuity Systems, Redwood City, CA, USA, www.ingenuity.com). Briefly, the data sets containing gene identifiers, corresponding fold changes, and p-values were uploaded into the web-delivered application. Then, each gene identifier was mapped to its corresponding gene object in the Ingenuity Knowledge Base (IKB, Ingenuity Systems). The biological functional analysis identified the biological functions and/or diseases that were most significant to the data sets. Fisher’s exact test was performed to calculate the probability that each biological function and/or disease assigned to the data set was due to chance alone. A network based upon the IKB was generated by connecting protein nodes to represent direct and indirect biological relationships. To simplify the view, only genes of proteins related to EMT, tissue fibrosis, and ocular angiogenesis were included, and unconnected nodes were removed. Predicted miRNA targets were determined using the Target Filter of IPA. The Target Filter is using the miRNA-mRNA target databases TargetScan, TarBase, miRecords, and Ingenuity expert findings in order to identify mRNA targets of miRNAs.
2.6 Quantitative real-time reverse transcription polymerase chain reaction (Q-RT-PCR)
Q-RT-PCR reactions were performed using ViiA 7 Real-Time PCR System (Applied Biosystems, Carlsbad, CA) to validate a selected panel of miRNAs and mRNAs primarily related to epithelial-mesenchymal transition, tissue fibrosis, and angiogenesis. 10 ng of totalRNA from pterygium and normal conjunctival tissue were reverse transcribed using the TaqMan MicroRNA RT Kit (Applied Biosystems) and specific primer/probes for each microRNA. Thereafter, 1.3 µL product from the RT reaction (diluted 1:15), 7.7 µL H2O, and 1 µL primer/probes (TaqMan MicroRNA Assay 20X, Applied Biosystems) were added to 10 µL universal PCR Master Mix (TaqMan, Applied Biosystems). MIR23b, showing a variance of 0.019 in the microarray experiments, served as endogenous control. Assays used were MIR200A-502, MIR200B-2251, MIR200C-2300, MIR141-463, MIR429-1024, MIR29A-2112, MIR29B-413, MIR29C-587, and MIR23B-400 (Applied Biosystems).
From the pterygium and normal conjunctival tissue, 200 nanograms of totalRNA were reverse transcribed using the Omniscript RT Kit (Qiagen Inc.). Nine µL cDNA (diluted 1:10 in H2O) and 1 µL of primer/probes (TaqMan Gene Expression Assays, Applied Biosystems) were added to 10 µL universal PCR master mix (TaqMan, Applied Biosystems). Each gene was run in triplicates. EXOSC10 (exosome component 10), showing a variance of 0.004 in the microarray study, was used as endogenous control. The following assays were used: FN1-Hs00365052_m1, ZEB1-Hs00232783_m1,EB2-Hs00207691_m1, VIM-Hs00185584_m1, LTBP2-Hs00166367_m1, PECAM1-Hs00169777_m1, VWF-Hs00169795_m1, CCL19-Hs00171149_m1, and EXOSC10-Hs00160216_m1 (Applied Biosystems).
The relative changes of each transcript were calculated by the cycle threshold method (ΔΔCt method) (Livak and Schmittgen, 2001). Statistical comparison of Q-RT-PCR data was performed with the paired Student’s t-test (Excel, Microsoft, Redmond, WA). P<0.05 was considered significant.
3. Results
3.1 Global miRNA and gene expression profiling
Inter-individual variability in gene expression was the major source of variation in the genome-wide miRNA and mRNA expression analysis (Suppl. Fig. 2). Using filtering criteria of a 1.5 or greater fold change in expression, and a p-value of < 0.05, 70 miRNAs out of 15.644 probe sets were differentially expressed in human primary pterygium samples relative to normal conjunctiva. 25 miRNAs were significantly differentially expressed by more than two-fold. 14 of these genes (56%) showed increased expression (Table 1), while 11 (44%) showed decreased expression (Table 2). With regard to miRNAs involved in EMT, the miR-200 family members miR-200a, miR-200b, and miR-141 were down-regulated in pterygium, whereas miR-200c (FC=−1.36, P=0.007) and miR-205 (FC=1.19, P=0.23) exhibited less than a 1.5fold change. miR-429 was not detected either in pterygium or normal conjunctiva. The tissue fibrosis regulators miR-29b and miR-192 were down-regulated in pterygium, whereas mir-29a and mir-29c demonstrated less than a 1.5-fold change. Among the putative regulators of ocular angiogenesis, miR-146, −199a, and −451 were up-regulated, and miR-31 was down-regulated in pterygium. None of the 70 miRNAs remained significant after correction for multiple testing using a FDR of 10%.
TABLE 1.
Up-regulated miRNAs in human primary pterygium (fold change ≥ 2, P<0.05)
| Gene symbol | Official name provided by HGNCa | Fold changeb | P-value |
|---|---|---|---|
| MIR1246 | miRNA1246 | 4.5 | 0.001 |
| MIR486 | miRNA486 | 4.4 | 0.004 |
| MIR451 | miRNA451 | 4.1 | 0.010 |
| MIR3172 | miRNA3172 | 3.4 | 0.009 |
| MIR3175 | miRNA3175 | 3.3 | <0.001 |
| MIR1308 | miRNA1308 | 3.2 | 0.02 |
| MIR1972 | miRNA1972 | 3.0 | <0.001 |
| MIR143c | miRNA143 | 2.7 | 0.008 |
| MIR211 | miRNA211 | 2.7 | 0.03 |
| MIR665 | miRNA665 | 2.3 | 0.01 |
| MIR1973 | miRNA1973 | 2.2 | 0.04 |
| MIR18A | miRNA18A | 2.1 | 0.004 |
| MIR143 | miRNA143 | 2.0 | 0.006 |
| MIR663B | miRNA663b | 2.0 | 0.02 |
Up-regulated miRNAs in human primary pterygium versus normal conjunctival tissue.
HGNC: HUGO Gene Nomenclature Committee.
Expression level is given as average normalized (log transformed) signal intensity.
Antisense miRNA
TABLE 2.
Down-regulated miRNAs in human primary pterygium (fold change ≥ 2, P<0.05)
| Gene symbol | Official name provided by HGNCa | Fold changea | P-value |
|---|---|---|---|
| MIR675 | miRNA675 | −2.0 | 0.005 |
| MIR200Bc | miRNA200b | −2.1 | 0.002 |
| MIR200Ac | miRNA200a | −2.3 | 0.002 |
| MIR29B | miRNA29b | −2.3 | 0.005 |
| MIR200B | miRNA200b | −2.3 | <0.001 |
| MIR210 | miRNA210 | −2.4 | <0.001 |
| MIR141 | miRNA141 | −2.5 | <0.001 |
| MIR31 | miRNA31 | −2.6 | 0.02 |
| MIR200A | miRNA200a | −2.7 | <0.001 |
| MIR934 | miRNA934 | −3.0 | <0.001 |
| MIR375 | miRNA375 | −3.7 | 0.03 |
Down-regulated miRNAs in human primary pterygium versus normal conjunctival tissue.
HGNC: HUGO Gene Nomenclature Committee
Expression level is given as average normalized (log transformed) signal intensity
Antisense miRNA
Of the mRNAs, 390 out of 28.869 probe sets were differentially expressed in human primary pterygium samples relative to normal conjunctiva using filtering criteria of a 1.5 or greater fold change in expression and a p-value less than 0.05. Table 3 presents a selection of mRNAs related to EMT, tissue fibrosis, and angiogenesis. Key molecules in EMT including collagen, type III, alpha 1 (COL3A1), fibronectin 1 (FN1), latent transforming growth factor beta binding protein 2 (LTBP2), matrix metallopeptidase 2 (MMP2), notch 1 (NOTCH1), secreted protein acidic cysteine-rich (osteonectin /SPARC), transcription factor 4 (TCF4), transforming growth factor beta 2 (TGB2), and vimentin (VIM) were up-regulated in pterygium. On the other hand, E-Cadherin (CDH1, FC=−1.0, P=0.58), catenin beta-1 (CTNNB1, FC=−1.0, P=0.46), snail homolog 1 (Drosophila) (SNAI1) and 2 (SNAI2, FC=−1.1, P=0.42), and zinc finger E-box binding homeobox 1 (ZEB1, FC=1.4, P=0.02) and 2 (ZEB2, FC=1.2, P=0.03) demonstrated less than a 1.5-fold change or were not detected. Regarding tissue fibrosis and angiogenesis, actin, alpha 2, smooth muscle, aorta (ACTA), aquaporin 1 (Colton blood group) (AQP1), chemokine (C-C motif) ligand 19 (CCL19), COL3A1, connective tissue growth factor (CTGF), platelet/endothelial cell adhesion molecule 1 (PECAM1), and von Willebrand factor (VWF) were up-regulated in pterygium. 31 genes were differentially expressed genes using a FDR of 10%. Among those, 17 probe sets (55%) were up-regulated (Suppl. Table 1), whereas 14 probe sets (45%) were down-regulated (Suppl. Table 2). LTBP1, FN1, AQP1, CCL19, PECAM1, and VWF remained significant after correction for multiple testing.
TABLE 3.
mRNAs related to epithelial-mesenchymal transition, tissue fibrosis, and angiogenesis in human primary pterygium
| Gene symbol | Official name provided by HGNCa | Fold changeb |
|---|---|---|
| Epithelial-mesenchymal transition | ||
| CALD1 | Caldesmon 1 | 1.6 |
| CAMK2N1 | Calcium/calmodulin-dependent protein kinase II inhibitor 1 | −2.2 |
| COL3A1 | Collagen, type III, alpha 1 | 1.7 |
| FN1 | Fibronectin 1 | 2.6 |
| IGFBP4 | Insulin-like growth factor binding protein 4 | 1.5 |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 1.8 |
| MSN | Moesin | 1.5 |
| NOTCH 1 | Notch 1 | 1.5 |
| PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | 1.6 |
| SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 1.6 |
| TGFB2 | Transforming growth factor, beta 2 | 1.6 |
| TFPI2 | Tissue factor pathway inhibitor 2 | −2.3 |
| VIM | Vimentin | 1.5 |
| Tissue fibrosis | ||
| ACTA2 | Actin, alpha 2, smooth muscle, aorta | 2.7 |
| AQP1 | Aquaporin 1 (Colton blood group) | 2.7 |
| CCL19 | Chemokine (C-C motif) ligand 19 | 2.0 |
| COL3A1 | Collagen, type III, alpha 1 | 1.7 |
| CTGF | Connective tissue growth factor | 2.0 |
3.2 Clustering analysis
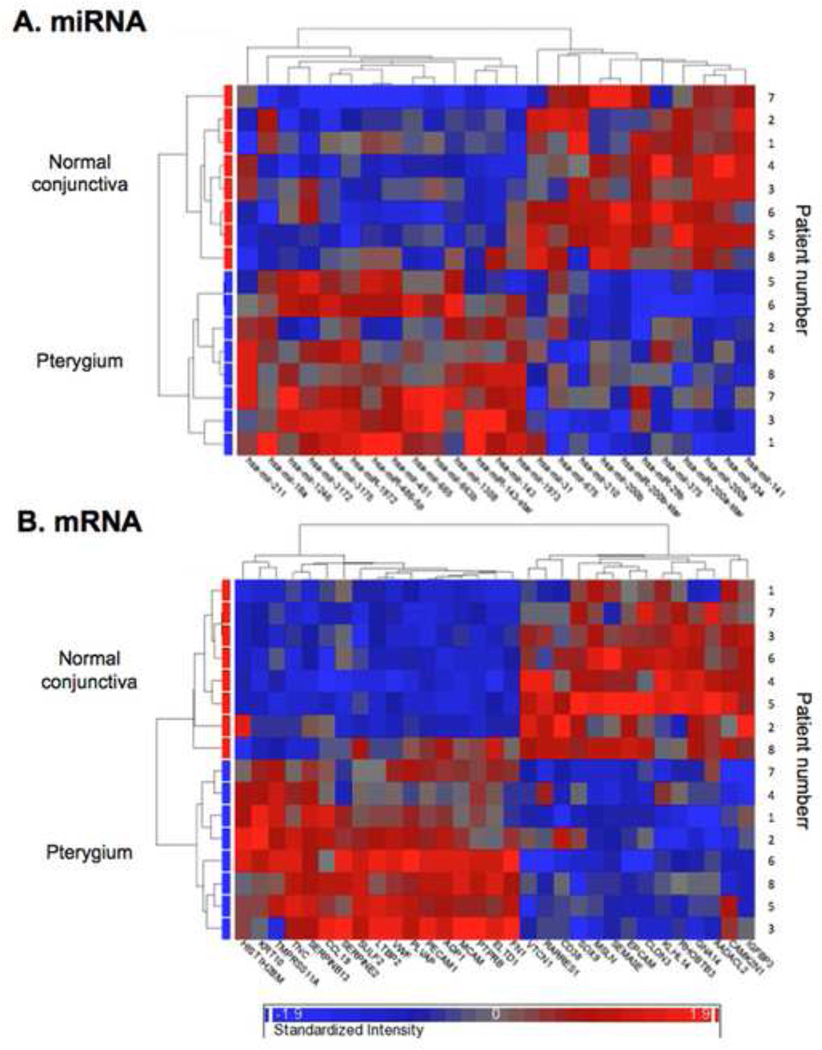
The euclidean, unsupervised, hierarchical clustering of the 25 miRNA probe sets and 31 mRNA probe sets is visualized in the form of heat maps (Fig. 2A and B). A cluster of down-regulated miRNAs, containing the miR-200 family members miR-200a, miR-200b, and miR-141 with a similar expression profile across all the samples was detected in pterygium. Furthermore, miR-29b was coexpressed with members of the miR-200 family. No specific cluster was found among the differentially expressed mRNAs.
Figure 2. Hierarchical clustering analysis.
Two-dimensional euclidean unsupervised hierarchical clustering analysis of the (A) 25 miRNA probe sets and (B) 31 mRNA probe sets visualized in the form of heat maps. Expression levels are indicated on a color scale where red represents the up-regulated genes and blue the down-regulated genes. The shorter the length of the branches, the more co-expressed the members of the genes are.
3.3 Biological functional analysis
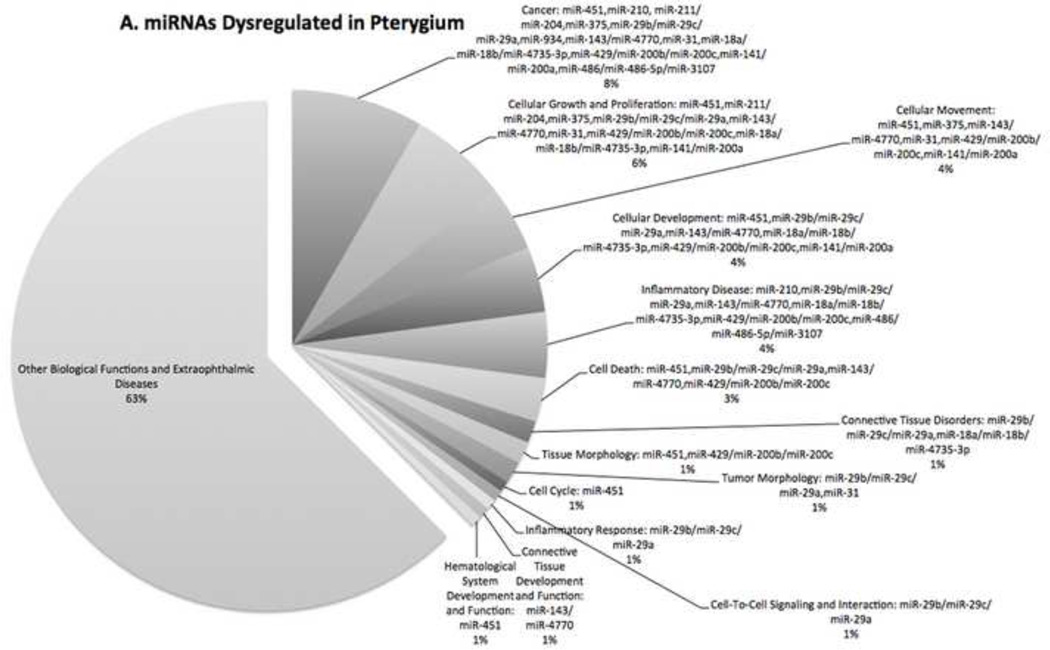
The 25 miRNA probe sets and 31 mRNA probe sets were subject to biological functional analysis. The molecular and cellular functions that were most significant to the miRNA data sets were cellular development, cellular growth and proliferation, and cellular movement. According to Ingenuity Pathway Analysis, cellular development describes functions associated with the development and differentiation of cells. This includes cellular functions that are involved in specific kinds of differentiation as well as developmental functions such as maturation and senescence of cells.
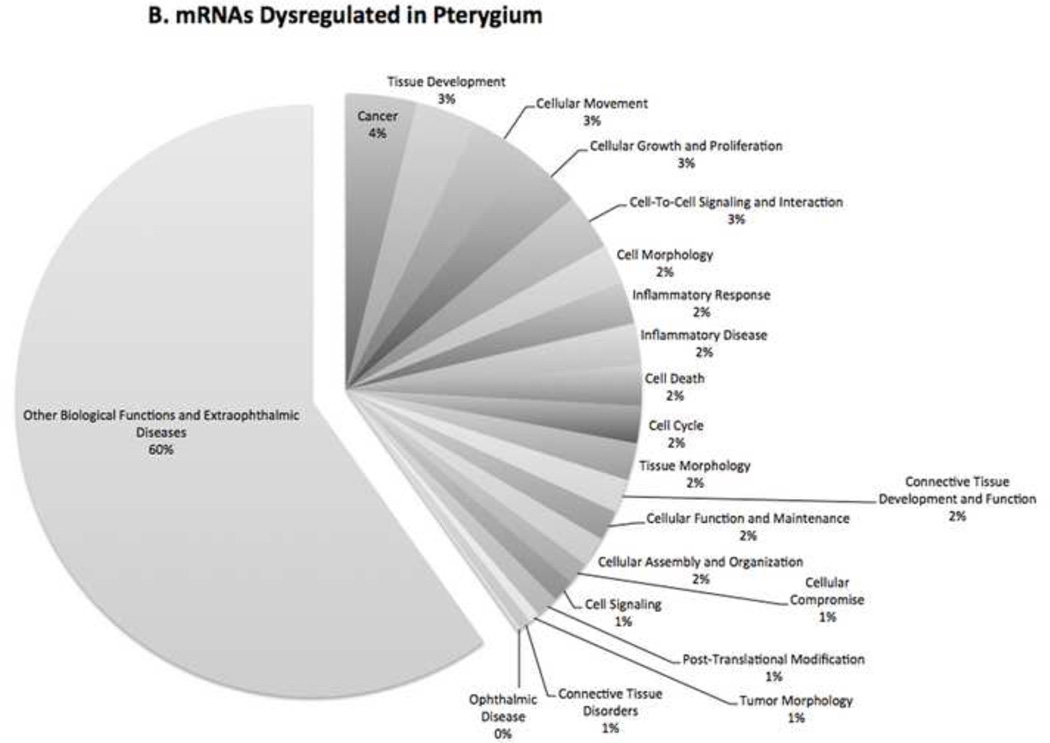
The top three molecular and cellular functions that were most significant to the mRNA data sets were cellular movement, cell-to-cell signaling and interaction, and cellular compromise. Cellular compromise describes functions associated with the damage or degeneration of cells or any process that might compromise the function of the cell.
Figure 3 demonstrates the percentage of miRNAs (Fig. 3A) and mRNAs (Fig. 3B) with regard to the number of molecules with different biological functions that were dysregulated in pterygium.
Figure 3. Biological Function of Dysfunctional Genes.
Pie charts showing the percentage of (A) miRNAs and (B) mRNAs with different biological functions that were dysregulated in pterygium. The biological functional analysis identified the biological functions and/or diseases that were most significant to the data sets.
3.4 Pathway analysis
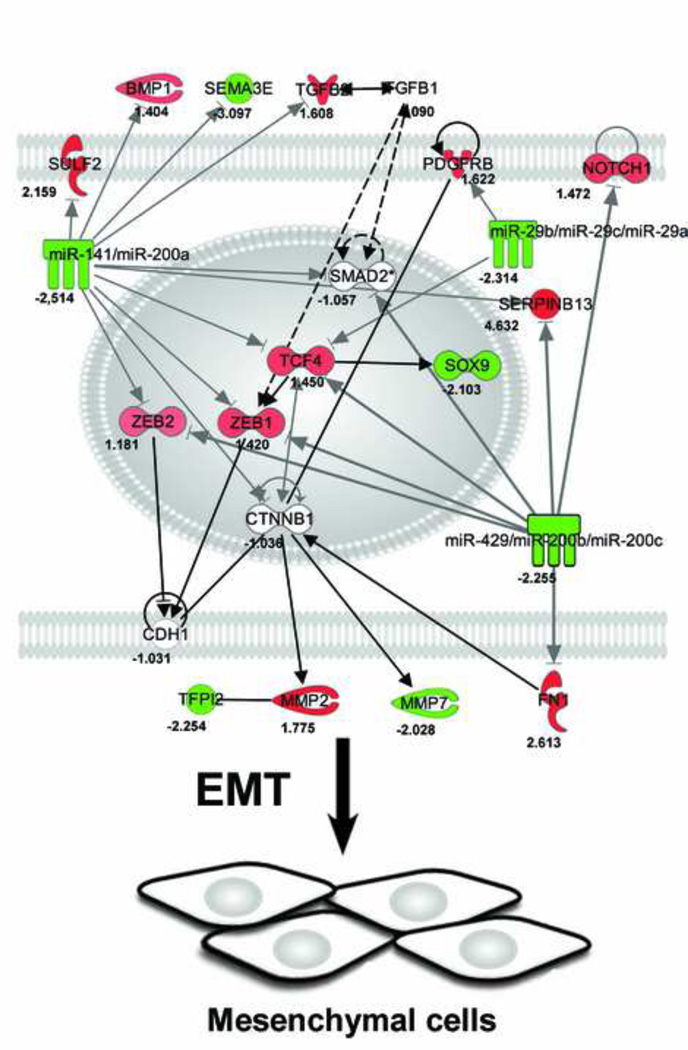
Pathway analysis demonstrated important consistencies between the observed down-regulation of members of the miR-200 family and up-regulation of targets associated with EMT including markers of EMT such as FN1 and MMP2 and activators of EMT such as NOTCH1, TCF4, ZEB1, and ZEB1 (Fig. 4).
Figure 4. Pathway analysis.
Network demonstrating direct and indirect biological relationships in human primary pterygium. Each node represents a molecule or cluster of molecules, and the intensity of node color in the networks indicates the degree of downregulation (green) or upregulation (red) of gene expression. Arrows indicate the direction of 25 intermolecular actions. The diagram shows important consistencies between the observed downregulation of members of the miR-29 and miR-200 families and targets associated with epithelial-mesenchymal transition (EMT) including activators of EMT such as NOTCH1, TCF4, ZEB1, and ZEB1, and markers of EMT such as MMP2 and FN.
3.5 Prediction of miRNA-targeted genes
Predicted targets among the 31 differentially expressed mRNAs (fold change ≥ 2, FDR of 10%) of miRNAs specifically involved in EMT (miR-200a, −200b, and −141), fibrosis (miR-29b), and angiogenesis (miR-31 and miR-451) are presented in Table 4. In particular, FN1 was identified as a target of miR-200b.
TABLE 4.
Prediction of miRNA-targeted genes
| Gene symbol | Predicted targets among the 31 differentially expressed mRNAs (fold change ≥ 2. false discovery rate of 10%) |
|---|---|
| Epithelial-mesenchymal transition | |
| MIR200A | SERPINB13, SEMA3E, SULF2 |
| MIR200B | FN1, SERPINB13, SEMA3E, SULF2, KLHL14 |
| MIR141 | SERPINB13, SEMA3E, SULF2 |
| Tissue fibrosis | |
| MIR29B | None |
| Angiogenesis | |
| MIR31 | None |
| MIR451 | None |
The miRNA-targeted genes were predicted by the miRNA-mRNA target databases Target Scan (www.targetscan.org/). TargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA.
3.6 Validation of gene microarray data
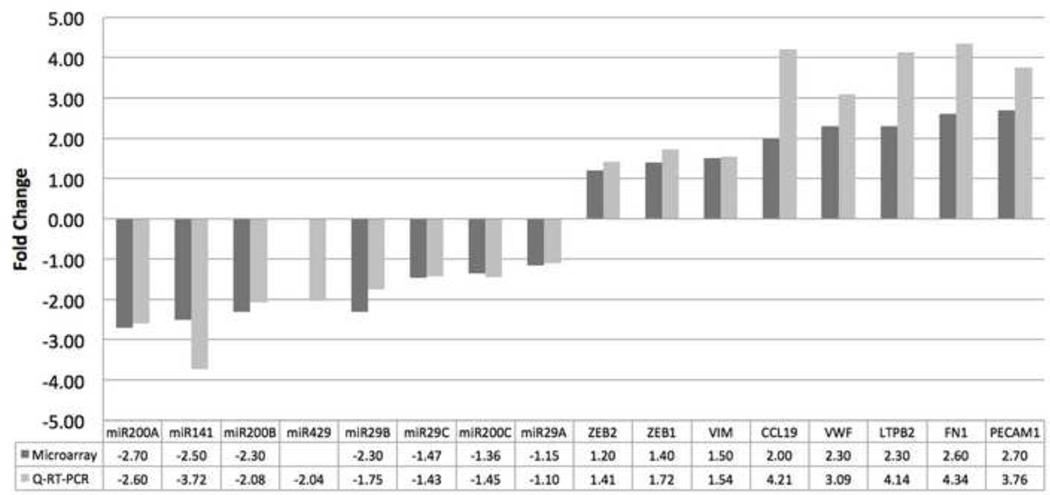
The qRT-PCR analysis demonstrated that the direction and magnitude of changes were consistent with the results obtained from the microarray analysis for all genes selected for validation except miR-429 which was down-regulated by more than two-fold in pterygium (FC=−2.04, P=0.002) (Fig. 5). All genes except miR-29a and miR-29c were significantly changed (P<0.05).
Figure 5. Validation of Microarray Data.
Bar graph showing the correlation of microarray data with quantitative real-time reverse transcription polymerase chain reaction (Q-RT-PCR) transcript levels. The X axis shows a selected panel of miRNAs and mRNAs primarily related to epithelial-mesenchymal transition, tissue fibrosis, and angiogenesis. The Y-axis features black and grey bars which represent the fold change by microarray and Q-RT-PCR experiment, respectively.
4. Discussion
This study represents the first comparative genome-wide miRNA and mRNA expression analysis of human primary pterygium. Our data demonstrated that pterygium is associated with significant changes in expression of multiple miRNAs that could potentially play a role in the pathogenesis of pterygium.
Down-regulation of the miR-200 family has been shown to be critical for TGF-β-induced EMT (Gregory et al., 2008a). In the present study, there were important consistencies between the concerted down-regulation of members of the miR-200 family and the up-regulation of target genes associated with EMT. These genes included markers of EMT such as FN (Thiery, 2003) and MMP2 (Park and Schwarzbauer, 2013), the NOTCH1 receptor which can promote myofibroblast differentiation (Liu et al., 2009; Saad et al., 2010), and transcription factors such as TCF4, ZEB1, and ZEB2 that are known to contribute to the induction of EMT (Sanchez-Tillo et al., 2011; Schmalhofer et al., 2009). FN has been shown to enhance EMT through different mechanisms (Camara and Jarai, 2010; Park and Schwarzbauer, 2013). The down-regulation of miR-200a, miR-200b, and miR-141 and up-regulation of the mesenchymal marker and predicted target FN1 (Hu et al., 2009) were confirmed by qRT-PCR. Furthermore, qRT-PCR revealed that miR-429, the fourth member of the miR-200 family, was significantly down-regulated.
EMT is a process essential to wound healing and tissue remodeling after injuries like thermal burns (Weber et al., 2012) characterized by differentiation of epithelial cells into myofibroblasts that rebuild the extracellular matrix and facilitate wound contraction. The hypothesis of EMT being involved in the pathogenesis of pterygium which is associated with chronic ultraviolet radiation exposure, was first proposed by Kato et al. (Kato et al., 2007b). Histopathology and electron microscopy revealed aberrant, fibrotic proliferation beneath the pterygium epithelium and dissociated epithelial cells that were surrounded by activated fibroblast-like cells. Immunohistochemical analyses demonstrated down-regulated E-cadherin, intranuclear accumulation of β-catenin and lymphoid-enhancer-factor-1, and immunopositivity of MMP-7, SNAI1 and SNAI2 in pterygial epithelial cells. Down-regulation of E-cadherin and β-catenin were later confirmed by Kase et al. (Kase et al., 2007).
The miR-200 family of miRNAs is an important regulator of EMT (Burk et al., 2008; Gregory et al., 2008a; Korpal et al., 2008; Park et al., 2008) and consists of five members that in humans are found in two clusters located on chromosome 1 (miR-200a/−200b/−429) and chromosome 12 (miR-200c/−141). In brief, the miR-200 family promotes the epithelial phenotype through post-transcriptional repression of ZEB1/ZEB2 and TGFβ2, thereby enabling the expression of E-cadherin and polarity factors that are integral in forming cell-cell junctions (Gregory et al., 2008b). Although we did not detect a significant change in mRNA expression of the ZEB1 and ZEB1 regulated gene CDH1, it has to be taken in consideration that transcriptional regulation by ZEB1 and ZEB2 is complex and involves multiple co-factors (Camara and Jarai, 2010). Variations in the levels of different ZEB1 co-factors might modulate ZEB1 activity, thus preventing its inhibitory effect on the CDH1 promoter. In addition, the deregulation of other CDH1 modulators different from ZEB1 and ZEB2 could potentially contribute mask its repression by these transcription factors.
Among the proposed regulators of tissue fibrosis (miR-21, −29a, −29b, −29c, − 155, −192, and −377) (Kato et al., 2007a; Kwiecinski et al., 2011; Pottier et al., 2009; Thum et al., 2008; van Rooij et al., 2008; Wang et al., 2008) and ocular angiogenesis (miR-31, −106a, −146, −150, −181, −184, −199a, −214, −424, and −451) (Shen et al., 2008) miR-29b, −192, −31, −146, −199a, and −451 were differentially expressed using filtering criteria of a 1.5 or greater fold change in expression and a p-value of < 0.05. Potential targets of the dysregulated miRNAs were not identified in the present study, even though fibrosis related genes including ACTA2, AQP1, CCL19, COL3A1, and CTGF, and genes associated with angiogenesis, such as PECAM1 and VWF, were up-regulated.
Reports on the expression of miRNA and mRNA in human pterygium are limited. Recently, a study comparing paired human pterygium and conjunctival tissues using a miRNA array with 3100 miRNA probes found that miR-766 was significantly up-regulated, and the predicted down-regulation of miR-766 targets nuclear receptor subfamily 4, group A, member 1 (NR4A1) and EGF containing fibulin-like extracellular matrix protein 1 (EFEMP1) (Chen S, et al. IOVS 2010;51: ARVO E-Abstract 2403). Moreover, miR-215 was down-regulated, and the predicted targets collagen, type III, alpha 1 (COL3A1) and collagen, type IV, alpha 2 (COL4A2) were up-regulated. In summary, the authors concluded that the change in expression of miR-766 and miR-215 may cause changes in genes that regulate wound healing processes. Our results were partly in line with those of Chen et al. confirming the down-regulation of EFEMP1 and up-regulation of COL3A1, whereas neither miR-766, miR-215, or NR4A1 were differentially expressed in our study. The discrepancies may reflect the use of different microarrays and differences in normalization. Similarly to John-Aryankalayil et al. and Tong et al., we found FN1, PECAM1, and VFW to be up-regulated in pterygium compared to normal conjunctiva (Suppl. Table 3) (John-Aryankalayil et al., 2006; Tong et al., 2009). Tong also reported changes in the direction and magnitude of AQP1, LTBP2, SERPINB13, SERPINE2, RARRES1, and IGFBP3 expression that were comparable to our results (Tong et al., 2009).
Several diseases are characterized by global changes in a number of different regulatory systems, for example DNA repair, apoptosis, and cell adhesion (Garzon et al., 2010). Since miRNAs may alter a high number of genes, only one or a few miRNAs may therefore be sufficient to affect complex regulatory systems. By altering miRNA levels, these systems may, therefore, effectively be reverted to a normal and healthy state (Garzon et al., 2010). For up-regulation of miRNAs, there are two major strategies. miRNA mimics (duplexes) can be used in conjunction with different sorts of lipid formulations, or miRNAs can be delivered using a viral vector, encoding a miRNA hairpin (Garzon et al., 2010). Down-regulation of miRNAs can be obtained through administration of anti-sense nucleotides, which are often chemically modified to enable both stability and specificity. A clear advantage of sequence-based miRNA drugs is its simple and essentially one-dimensional nature. This contrasts sharply with de novo development of more traditional protein-based drugs with challenging three-dimensional protein structures and protein–drug interactions. Moreover, knowledge on miRNA sequence readily translates to the most appropriate design of potential associated drugs. This may boost progress in therapeutic drug development (Lanford et al., 2010). In 2005, only 12 years after miRNA was discovered, the first demonstration that miRNA antagonism has the potential to become a new efficient kind of therapy was published (Krutzfeldt et al., 2005). Since then, several therapies have been introduced clinically and one miRNA-based therapeutic has already entered Phase 2 clinical trials (Lanford et al., 2010; Vinther et al., 2012).
There are limitations to the study that should be acknowledged. First, the number of study patients was limited. Second, the relationship between gene expression and pterygium severity with regard to corneal ingrowth, pterygial thickness or pterygial vascularity was not studied. Third, fluctuations in mRNA translation and protein level were not investigated. Caution should therefore be exercised when generalizing our data to a wider population of patients with pterygium and making conclusions on the role of miRNAs in the pathogenesis of pterygium. Adequately powered studies are warranted applying immunofluorescence, proteomics or protein level quantification of the miRNA targets in patients with pterygium staged according to severity level.
In conclusion, our data demonstrated that pterygium is associated with significant changes in expression of multiple miRNAs. Some of the changes identified in this study suggest new mechanisms potentially relevant in the formation of pterygium. Future investigations should help to evaluate the relevance of these miRNAs in the pathogenesis of pterygium and their potential as therapeutic targets for this disease.
Supplementary Material
Highlights.
Members of the miR-200 family are coexpressed and down-regulated in pterygium.
The mesenchymal marker fibronectin is up-regulated in pterygium.
Epithelial-mesenchymal transition may play a role in the pathogenesis of pterygium.
miR-200 family members are potential targets for therapeutic intervention in pterygium.
Acknowledgements
The authors thank Helge Raeder, Astrid Østerud, and Harald Fredrik Ulltveit-Moe for their assistance and support.
Abbreviations
- EMT
epithelial-mesenchymal transition
- miRNA
microRNA
- UTR
untranslated region
- FDR
false discovery rate
- IKB
Ingenuity Knowledge Base
- Q-RT-PCR
Quantitative real-time reverse transcription polymerase chain reaction
- TGB2
transforming growth factor beta 2
- LTBP2
latent transforming growth factor beta binding protein 2
- NOTCH1
notch 1
- VIM
vimentin
- FN1
fibronectin 1
- MMP2
matrix metallopeptidase 2
- COL3A1
collagen, type III, alpha 1
- SPARC
secreted protein acidic cysteine-rich
- CDH1
E-Cadherin
- ZEB
Zinc finger E-box binding homeobox SNAI, snail homolog (Drosophila)
- ACTA
actin, alpha 2, smooth muscle, aorta
- AQP1
aquaporin 1 (Colton blood group)
- CCL19
chemokine (C-C motif) ligand 19
- CTGF
connective tissue growth factor
- PECAM1
platelet/endothelial cell adhesion molecule 1
- VWF
von Willebrand factor
- NR4A1
nuclear receptor subfamily 4, group A, member 1
- EFEMP1
EGF containing fibulin-like extracellular matrix protein 1
- COL3A1
collagen, type III, alpha 1
- COL4A2
collagen, type IV, alpha 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ando R, Kase S, Ohashi T, Dong Z, Fukuhara J, Kanda A, Murata M, Noda K, Kitaichi N, Ishida S. Tissue factor expression in human pterygium. Mol Vis. 2011;17:63–69. [PMC free article] [PubMed] [Google Scholar]
- Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48:3962–3967. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bolender DL, Markwald RR. Epithelial-mesenchymal transformation in chick atrioventricular cushion morphogenesis. Scanning electron microscopy. 1979:313–321. [PubMed] [Google Scholar]
- Bradley JC, Yang W, Bradley RH, Reid TW, Schwab IR. The science of pterygia. Br J Ophthalmol. 2008;94:815–820. doi: 10.1136/bjo.2008.151852. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara J, Jarai G. Epithelial-mesenchymal transition in primary human bronchial epithelial cells is Smad-dependent and enhanced by fibronectin and TNF-alpha. Fibrogenesis & tissue repair. 2010;3:2. doi: 10.1186/1755-1536-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui J, Coroneo MT, Tat LT, Crouch R, Wakefield D, Di Girolamo N. Ophthalmic pterygium: a stem cell disorder with premalignant features. The American journal of pathology. 2011;178:817–827. doi: 10.1016/j.ajpath.2010.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui J, Di Girolamo N, Wakefield D, Coroneo MT. The pathogenesis of pterygium: current concepts and their therapeutic implications. The ocular surface. 2008;6:24–43. doi: 10.1016/s1542-0124(12)70103-9. [DOI] [PubMed] [Google Scholar]
- Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Acta anatomica. 1995;154:63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- Frederikse PH, Donnelly R, Partyka LM. miRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochemistry and cell biology. 2006;126:1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews. Drug discovery. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008a;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell cycle (Georgetown, Tex.) 2008b;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Griffith CM, Hay ED. Epithelial-mesenchymal transformation during palatal fusion: carboxyfluorescein traces cells at light and electron microscopic levels. Development. 1992;116:1087–1099. doi: 10.1242/dev.116.4.1087. [DOI] [PubMed] [Google Scholar]
- Hirst LW, Axelsen RA, Schwab I. Pterygium and associated ocular surface squamous neoplasia. Arch Ophthalmol. 2009;127:31–32. doi: 10.1001/archophthalmol.2008.531. [DOI] [PubMed] [Google Scholar]
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecologic oncology. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- John-Aryankalayil M, Dushku N, Jaworski CJ, Cox CA, Schultz G, Smith JA, Ramsey KE, Stephan DA, Freedman KA, Reid TW, Carper DA. Microarray and protein analysis of human pterygium. Mol Vis. 2006;12:55–64. [PubMed] [Google Scholar]
- Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- Kase S, Osaki M, Sato I, Takahashi S, Nakanishi K, Yoshida K, Ito H, Ohno S. Immunolocalisation of E-cadherin and beta-catenin in human pterygium. Br J Ophthalmol. 2007;91:1209–1212. doi: 10.1136/bjo.2007.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007a;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Shimmura S, Kawakita T, Miyashita H, Ogawa Y, Yoshida S, Higa K, Okano H, Tsubota K. Beta-catenin activation and epithelial-mesenchymal transition in the pathogenesis of pterygium. Invest Ophthalmol Vis Sci. 2007b;48:1511–1517. doi: 10.1167/iovs.06-1060. [DOI] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kwiecinski M, Noetel A, Elfimova N, Trebicka J, Schievenbusch S, Strack I, Molnar L, von Brandenstein M, Tox U, Nischt R, Coutelle O, Dienes HP, Odenthal M. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS One. 2011;6:e24568. doi: 10.1371/journal.pone.0024568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. microRNAs: runts of the genome assert themselves. Current biology : CB. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu T, Hu B, Choi YY, Chung M, Ullenbruch M, Yu H, Lowe JB, Phan SH. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. The American journal of pathology. 2009;174:1745–1755. doi: 10.2353/ajpath.2009.080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome biology. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer science. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Schwarzbauer JE. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2013 doi: 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice JM, Marcet B, Cardinaud B, Barbry P, Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- Saad S, Stanners SR, Yong R, Tang O, Pollock CA. Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor. The international journal of biochemistry & cell biology. 2010;42:1115–1122. doi: 10.1016/j.biocel.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci U S A. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer metastasis reviews. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Current opinion in cell biology. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature reviews. Molecular cell biology. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Tong L, Chew J, Yang H, Ang LP, Tan DT, Beuerman RW. Distinct gene subsets in pterygia formation and recurrence: dissecting complex biological phenomenon using genome wide expression data. BMC Med Genomics. 2009;2:14. doi: 10.1186/1755-8794-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebahn C. Epithelio-mesenchymal transformation during formation of the mesoderm in the mammalian embryo. Acta anatomica. 1995;154:79–97. doi: 10.1159/000147753. [DOI] [PubMed] [Google Scholar]
- Vinther J, Rukov JL, Shomron N. MicroRNAs and their antagonists as novel therapeutics. In: Erdmann VA, Barciszewski J, editors. From nucleic acids sequences to molecular medicine. Springer-Verlag; 2012. pp. 504–520. [Google Scholar]
- Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CE, Li NY, Wai PY, Kuo PC. Epithelial-mesenchymal transition, TGF-beta, and osteopontin in wound healing and tissue remodeling after injury. Journal of burn care & research : official publication of the American Burn Association. 2012;33:311–318. doi: 10.1097/BCR.0b013e318240541e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. microRNA expression in the eyes and their significance in relation to functions. Progress in retinal and eye research. 2009;28:87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. The Journal of biological chemistry. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.