Abstract
Purpose of Review
The frequency and severity of asthma differs between different racial and ethnic groups. An understanding of the genetic basis for these differences could constitute future genetic biomarker panels for predicting asthma risk and progression in individuals from different ethnic groups.
Recent Themes
The recent mixing of different ancestries during the European colonization of the Americas and the African slave trade has resulted in the complex population structures identified in different ethnic groups. These population structures represent varying degrees of genetic diversity which impacts the allele frequency of individual variants and, thus, how gene variation is utilized in genetic association studies. In this review, we will discuss the basis for the complex population structures of modern human genomes and the impact of genetic diversity on genetic studies in different ethnic groups. We will also highlight the potential for admixture and rare variant-based genetic studies to identify novel genetic loci for asthma susceptibility and severity.
Summary
The ability to account for the consequences of genetic diversity in different racial and ethnic groups will be critical in developing genetic profiles for personalized or precision medicine approaches tailored to asthmatics from different ethnic groups.
Keywords: asthma, genetics, genome-wide association study, admixture, ancestry
Introduction
The frequency and severity of asthma differs between different racial and ethnic groups. In the United States (U.S.), African Americans have a greater frequency of asthma and experience greater asthma-related morbidity and mortality compared to non-Hispanic Whites [1]. In addition, Puerto Ricans have the highest frequency of asthma and the highest morbidity and mortality among U.S. ethnic subgroups, while asthma is much less common and severe in Mexican Americans [2–4]. An understanding of the genetic basis for inter-racial and inter-ethnic differences in disease expression could provide the basis for genetic biomarker panels for predicting asthma risk, disease progression, and responsiveness to pharmacological therapies for individuals from different ethnic or racial groups.
High-throughput genotyping technologies have allowed for the analysis 100,000’s to millions of single nucleotide polymorphisms (SNPs) to test for associations with asthma using genome-wide association studies (GWAS). The most recent advances of next-generation DNA sequencing has led to an expanding catalogue of human genetic diversity in populations representing different ancestral populations. In this review, we will discuss the basis for the genetic diversity and population structure of modern human genomes. We will summarize how the genetic diversity of different human populations has impacted genetic studies and led to the identification of unique susceptibility loci in different ethnic groups. Finally, we will highlight the potential for admixture and rare variant-based genetic studies to identify genetic loci for asthma susceptibility and severity.
Genetic Diversity and Population Structure in Recently Admixed Populations
The Human Genome Project primarily catalogued common gene variants, particularly SNPs, the most common form of gene variation in human genomes [5]. SNPs are heritable landmarks for population structure and genetic diversity which have provided clues about the demographic history of our species and the basis for the genetic diversity of different ancestral populations [6, 7]. Based on recombination models, the first modern humans migrated to Europe from sub-Saharan African nearly 40,000 years ago resulting in a collapse or loss of genetic diversity followed by the population growth of the Upper Paleolithic period [7].
The loss of genetic diversity during this “bottleneck” resulted in descendant populations from a European ancestry with gene variants highly co-inherited or correlated over longer genomic regions through (linkage disequilibrium). Very simply defined, the inheritance of genes together, often due to physical proximity on a chromosome, is labeled linkage disequilibrium. This was primarily due to fewer number of recombination events over fewer generations since the last “collapse.” In comparison, descendant populations from an ancient African ancestry have greater genetic diversity resulting from fewer co-inherited gene variants in shorter genomic regions originating from a greater number of recombination events over more generations (i.e. shorter regions of linkage disequilibrium) [7]. Thus, population structures from European or African ancestries represent varying degrees of genetic diversity which impacts linkage disequilibrium throughout the genome and how individual genetic variants can be used to “tag” gene variation in genetic association studies in different ethnic groups (Figure 1).
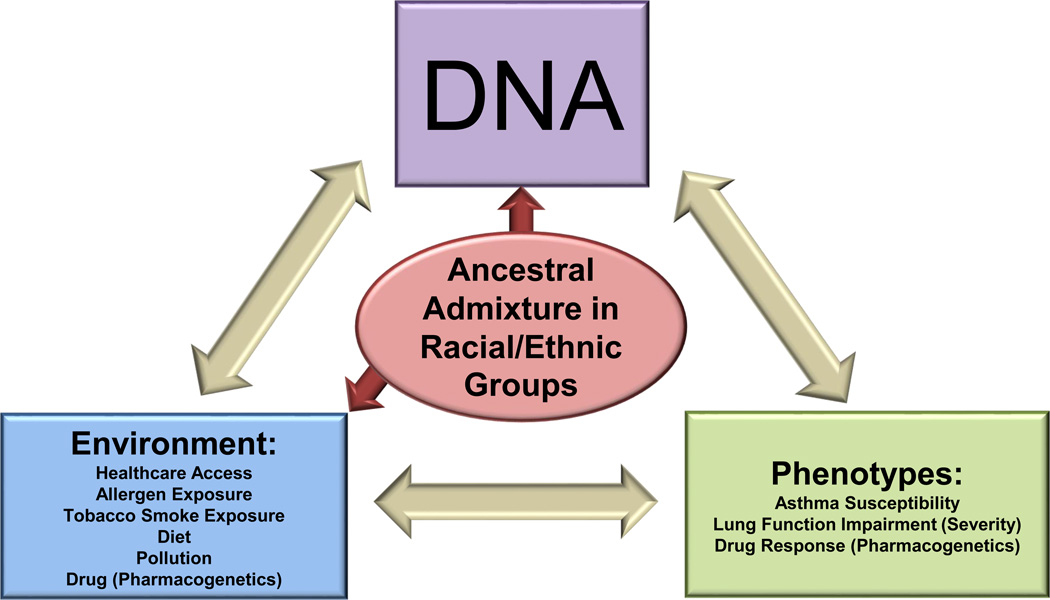
Figure 1. Ancestral Admixture and Genetic Research in Complex Diseases.
Genetic research studies the role of genetic variability in determining risk for complex airway diseases, related phenotypes, and disease severity. Genetic risk can be directly altered by gene variation that impacts biological function directly or through gene-by-environment interactions. The population structures of modern human genomes represent varying degrees of genetic diversity due to recent admixture which impacts the allele frequency of individual variants which determine phenotype in different racial or ethnic groups. In addition, individuals from these different populations share similarities in languages, social experiences, and cultural behaviors which could result in differences in environmental exposures and gene-by-environment interactions which confound gene variant associations.
In addition, the human population has experienced a nearly three-fold expansion in growth over the past 400 generations which resulted in an increased frequency of recent mutations and, thus, rare variants [8, 9]. Rare variants with an allele frequency less than 0.005 were found to be enriched in whole-genomes representing 14 different populations from the 1,000 Genomes Project [9, 10]. The 1,000 Genomes Project also demonstrated that the frequency of individual rare variants (allele frequency <0.05) was greater than more common variants and included hundreds of rare coding variants and variation in conserved, regulatory regions. Thus, these rare variants could exhibit strong biologic effects as reflected in the “common disease-rare allele” hypothesis [10–12].
In the 1,000 Genomes Project, rare variants were three times more frequent in populations from an African ancestry compared to those from a primarily European and Asian ancestry consistent with the “bottleneck” history of ancestral non-African populations [7, 10]. Thus, populations from European or African ancestries have differing frequencies of individual gene variants, particularly rare variants which might be unique to a single ancestry (Table 1) [13, 14]. The variability in allelic frequencies of more common polymorphisms has the potential to alter the genetic diversity captured by individual SNPs in populations from differing ancestral backgrounds (Figure 1, Table 1).
Table 1.
Variant Allele Frequencies of Asthma Susceptibility Loci in Different Ancestral Populations and United States Ethnic Groups.
| Gene Names | ChrPos | Gene ID | Associated SNP Locusa |
Alleles Major/Minor |
Minor Allele Frequency by Racial Groupb | References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CEU | YRI | ASW | MEX | CHB | JPT | ||||||
| Interleukin-6 Receptor | 1q21 | IL6R | rs4129267 | C/T | 0.35 | 0.07 | 0.15 | 0.52 | 0.39 | 0.36 | [17] |
| Pyrin and HIN Domain Family Member 1 | 1q23 | PYHIN1 | rs1102000 | A/G | 0.00 | 0.35 | NA | NA | 0 | 0 | [18] |
| Interleukin-1 Receptor | 2q12 | IL1RL1 | rs1420101 | C/T | 0.35 | 0.32 | 0.43 | 0.27 | 0.39 | 0.46 | [15, 16] |
| Interleukin-18 Receptor | 2q12 | IL18R1 | rs3771166 | G/A | 0.41 | 0.72 | 0.65 | 0.28 | 0.13 | 0.18 | [15] |
| Dipeptidyl Peptidase-10 | 2q14 | DPP10 | rs1435879 | A/G | 0.10 | 0.03 | 0.04 | 0.19 | 0.31 | 0.30 | [19] |
| GRB2-Associated Binding Protein 1 | 4q31 | GAB1 | rs1397527 | G/T | 0.45 | 0.84 | 0.74 | NA | 0.31 | 0.30 | [20] |
| Ubiquitin Specific Peptidase 38 | 4q31 | USP38 | rs7686660 | T/G | 0.21 | 0.47 | 0.44 | 0.52 | 0.74 | 0.72 | [20] |
| cAMP-Specific Phosphodiesterase 4D | 5q11 | PDE4D | rs1588265 | A/G | 0.36 | 0.16 | 0.22 | 0.22 | 0.70 | 0.73 | [21] |
| WD Repeat Domain 36 | 5q22 | WDR36 | rs2416257 | C/T | 0.14 | 0.14 | 0.10 | 0.06 | 0.07 | 0.04 | [22] |
| Thymic Stromal Lymphopoietin | 5q22 | TSLP | rs1837253 | C/T | 0.28 | 0.34 | 0.29 | 0.30 | 0.62 | 0.66 | [15, 16, 19, 20] |
| RAD50 Homolog | 5q31 | RAD50 | rs2244012 | A/G | 0.20 | 0.73 | 0.51 | 0.18 | 0.19 | 0.19 | [23] |
| Interleukin-13 | 5q31 | IL13 | rs1295686 | C/T | 0.22 | 0.73 | 0.59 | 0.48 | 0.34 | 0.30 | [23, 24] |
| α-1B-Adrenergic Receptor | 5q33 | ADRA1B | rs10515807 | G/A | 0.16 | 0.03 | NA | NA | 0.32 | 0.34 | [25] |
| TNFAIP3 Interacting Protein 1 | 5q33 | TNIP1 | rs1422673 | C/T | 0.19 | 0.43 | 0.37 | 0.47 | 0.56 | 0.51 | [24] |
| Psoriasis Susceptibility 1 Candidate 1 | 6p21 | PSORS1C1 | rs3094663 | C/T | 0.27 | 0.24 | 0.36 | 0.28 | 0.31 | 0.35 | [16] |
| Human Leukocyte Antigen Complex DQB1 | 6p21 | HLA-DQB1 | rs9273349 | C/T | 0.42 | 0.48 | 0.47 | 0.23 | 0.39 | 0.44 | [15, 16, 23] |
| Human Leukocyte Antigen Complex DRA | 6p21 | HLA-DRA | rs2395185 | G/T | 0.43 | 0.19 | 0.20 | 0.35 | 0.37 | 0.39 | [24] |
| Interleukin-33 | 9p24 | IL33 | rs1342326 | A/C | 0.17 | 0.35 | 0.32 | 0.15 | 0.00 | 0.00 | [15, 16, 19] |
| GATA Binding Protein 3 | 10p14 | GATA3 | rs10508372 | G/A | 0.04 | 0.22 | 0.21 | 0.35 | 0.59 | 0.56 | [20] |
| Ikaros Family Zinc Finger 4 | 12q13 | IKZF4 | rs1701704 | T/G | 0.32 | 0.07 | 0.15 | 0.21 | 0.23 | 0.22 | [20] |
| SMAD Family Member 3 | 15q22 | SMAD3 | rs744910 | A/G | 0.45 | 0.68 | 0.67 | 0.54 | 0.58 | 0.56 | [15] |
| RAR-Related Orphan Receptor A | 15q22 | RORA | rs11071559 | C/T | 0.15 | 0.56 | 0.43 | 0.11 | 0.14 | 0.23 | [15] |
| ORM1-Like 3 | 17q12 | ORMDL3 | rs7216389 | C/T | 0.49 | 0.88 | 0.70 | 0.60 | 0.66 | 0.72 | [15, 18, 19, 21, 26–29] |
| Gasderminlike B | 17q12 | GSDMB | rs2305480 | G/A | 0.47 | 0.05 | 0.25 | 0.40 | 0.33 | 0.28 | [15, 16, 24, 29] |
| Ikaros Family Zinc Finger 3 | 17q12 | IKZF3 | rs907092 | G/A | 0.49 | 0.07 | 0.28 | 0.39 | 0.33 | 0.34 | [16] |
| Prion-Related Protein | 20p12 | PRNP | rs6052761 | T/C | 0.10 | 0.35 | 0.39 | 0.17 | 0 | 0.03 | [25] |
| Interleukin-2 Receptor, β Subunit | 22q12 | IL2RB | rs2284033 | G/A | 0.42 | 0.39 | 0.45 | 0.32 | 0.65 | 0.58 | [15] |
Asthma susceptibility loci selected among the first identified by genome-wide association studies or admixture mapping and denoted by reference sequence number (rs) [15–29].
Minor or less common, variant allele frequencies are based on data from the International HapMap Project Genome Browser release 28 accessed on June 23, 2014 (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/).
Abbreviations from each group are as follows: CEU=Utah residents with ancestry from northern and western Europe; YRI=Individuals from Yoruba in Ibadan, Nigeria; ASW=African Americans from the southwest United States; MEX=Mexican Americans from Los Angeles, CA; CHB=Han Chinese from Beijing, China; JPT=Japanese from Tokyo, Japan.
The recent mixing of different ancestries over the past 500 years during the European colonization of the Americas and the African slave trade has resulted in the complex population structures identified in modern U.S. ethnic groups. As a consequence, recently admixed populations such as African Americans, Puerto Ricans, and Mexican Americans have varying percentages of African, Amerindian, and European ancestries due to different demographic histories which can even significantly vary between individuals of the same ethnicity (Figure 2) [30–32]. The complex population structures of these recently admixed populations have not been covered by most whole-genome genotyping platforms. Large-scale next-generation sequencing projects such as the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project (GO ESP); and the NIH NHLBI Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) Consortium provide insight into the genetic diversity of these populations [33]. In addition, whole-genome sequencing data from CAAPA has been used to design a genotyping platform which captures the population structure of these populations, including coverage for rare variants.
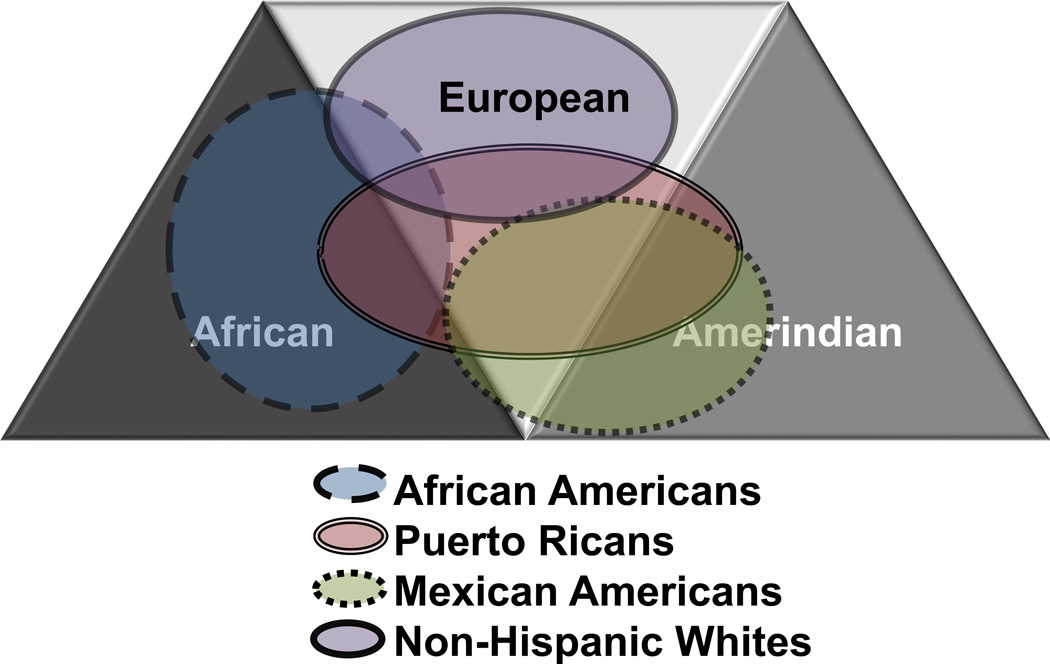
Figure 2. Ancestral Backgrounds of Different United States Ethnic Groups.
The recent mixing of different ancestries during the European colonization of the Americas and the African slave trade has resulted in the complex population structures of recently admixed populations such as African Americans, Puerto Ricans, and Mexican Americans. Individuals from each ethnic group have varying percentages of African (dark triangle), Amerindian (gray triangle), and European (light triangle) ancestries due to different demographic histories; however, between individuals from the same ethnic group there can also be significant variability in ancestral backgrounds. This Venn diagram shows that, on average, non-Hispanic Whites (solid circle) have undergone little recent admixture while African Americans (long-dashed circle), Puerto Ricans (double circle), and Mexican Americans (dotted circle) have varying proportions of different ancestries due to recent mixing [30–32].
Population Structure and Genetic Diversity in Early Genetic Studies
Before the first whole-genomes were sequenced, genetic studies for asthma susceptibility were performed with family-based approaches studies avoiding issues of ancestry since analysis was performed by family unit. Family-based genetic studies used genetic variants equally spaced throughout the genome to identify susceptibility regions or loci which co-segregated or were co-inherited with a trait such as asthma. These early studies demonstrated that risk for asthma and related phenotypes were determined by multiple loci throughout the genome [34–36]. These family-based studies contributed to our initial understanding of the complex, polygenic nature of asthma susceptibility determined by loci which varied between ethnic groups from varying ancestral backgrounds, gene-gene interactions, and gene-by-environment interactions (Figure 1) [35, 37–39].
Family-based studies were followed by an increasing number of biologic candidate gene association studies which compared the frequency of a common SNP allele between unrelated cases and controls. Association studies have been useful for genetic studies of common diseases where alleles may have a weak or modest effect on disease risk (the “common allele-common allele” hypothesis). Over 100 genes have been studied based on chromosomal regions identified in family-based linkage studies or biologic plausibility. For each candidate gene that has been discovered and replicated as an asthma susceptibility locus, there are additional studies which have not shown significant associations. The most replicated candidate genes belong to the broad categories of innate immunity (HLA-DRB1, HLA-DQB1), TH2 inflammatory pathway signaling (IL4, IL13, IL4R), cellular inflammation (TNF), and lung development (ADAM33) [40]. In many instances, inconsistencies in replication may have been due to small, underpowered sample sizes. However, the variability in allele frequencies of associated SNPs between populations could have resulted in different variants within the same gene similarly associated with asthma in different populations (i.e. “loose” replication, Figure 1) [41, 42].
More importantly, association testing without accounting for population structure or genetic ancestry (i.e. population stratification) could result in spurious differences (i.e., false positives). If allele frequencies differences between unrelated cases and controls frequencies are due to ethnic differences, this could lead to false positive results. Most candidate gene studies accounted for ancestry by separating subjects by self-reported ethnic group; however, whole-genome genotyping provides information about population structure and ancestral admixture. In addition, individuals from different ethnic groups share similarities in languages, social experiences, cultural behaviors, and geography which could result in common environmental exposures or gene-by-environment interactions which could confound genetic studies (Figure 1) [43].
Genetic Diversity and Population Structure in GWAS
In most GWAS, the allele frequencies of a large numbers of SNPs representing the entire genome (500,000 to more than one million SNPs) are compared between unrelated cases and controls. Whole-genome genotyping with these chips also provides information about ancestral differences to adjust observed SNP associations and minimize confounding by population stratification. The first GWAS for asthma susceptibility were performed on European Non-Hispanic White asthma subjects and controls. Thus, it did not represent other racial or ethnic groups with different ancestries where unique susceptibility loci could have been identified. This is particularly important for ethnic groups such as African Americans and Puerto Ricans who experience a higher risk for asthma and greater asthma-related morbidity compared to other ethnic groups [1, 3, 4]. The first GWAS for asthma identified a novel locus on 17q12 which contained the ORM1-like 3 gene (ORMDL3) and the neighboring gasdermin-like genes (GSDMB) which have become the most replicated loci for asthma susceptibility (Table 1) [17, 18, 21, 26–28]. Subsequent GWAS in primarily European and Non-Hispanic White populations have also identified susceptibility loci at IL33 on chromosome 9p24, IL1RL1/IL18RL1 (between the IL-1 and IL-18 receptor genes) on 2q12, WDR36/TSLP on 5q22, HLA-DRA and HLA-DQB1 on 6p21, and IL13 on 5q31 (Table 1) [15, 18, 23, 24].
The EVE consortium performed a large GWAS meta-analysis of a combined, multi-ethnic population of European Americans, African Americans, African Caribbeans, Mexican Americans, and Puerto Ricans which confirmed asthma susceptibility loci on chromosome 17q12 (GSDMB/ORMDL3), IL1RL1, and TSLP, and IL33 in these ethnic groups (Table 1) [15, 19, 26]. These loci and an adjacent SNP on 17q12 (IKZF3) were also associated with asthma in a recent GWAS of Puerto Ricans and Mexican Americans from the Genes-environments and Admixture in Latino Americans study (GALA II); however, other loci identified in non-Hispanic Whites populations have not been associated with asthma in other ethnic groups [16, 19]. These inter-ethnic differences may have possibly been due to varying inter-ethnic allele frequencies of risk loci (Table 1), differences in the genomic regions tagged by selected SNPs, or the fact that other genes are important loci in these populations.
The EVE consortium GWAS meta-analysis identified one of the first asthma susceptibility loci (PYHIN1) unique to African Americans and African Caribbeans. The associated PYHIN1 SNPs occurred at an allele frequency of 0.26 to 0.35 in African ancestral populations, but were not polymorphic in European Whites and rare in Hispanics (Table 1) [19]. GWAS meta-analyses of additional populations from a primarily African ancestry also identified novel susceptibility loci in ADRA1B and PRNP while confirming dipeptidyl peptidase (DPP10) as a risk locus from prior linkage studies [25, 44]. These GWAS demonstrate how the complexity of population structure can lead to differences in genetic susceptibility loci in populations from different ancestries.
A major emphasis of genetic studies in asthma has been on susceptibility. However, there is emerging evidence that genes which influence disease severity or progression differ from those that influence disease risk [24, 45]. The identification of loci which determine disease severity requires well-phenotyped asthma cohorts and has the potential to expose important pathways which underlie inter-ethnic differences in asthma disease severity [1, 3].
Lung function is a fundamental determinant of asthma severity influenced by multiple gene loci that were identified in GWAS of primarily European White general populations [46–49]. Gene variation in one of these genes coding for the hedgehog interacting protein (HHIP), was associated with lung function in non-Hispanic White and African American asthmatics from the NHLBI-sponsored Severe Asthma Research Program (SARP). Each copy of the HHIP risk variant allele was associated with an 85 milliliter reduced FEV1 in non-Hispanic White asthmatics and a 252 milliliter reduced FEV1 in African Americans. SNPs in five additional genes associated with lung function in the general population (FAM13A, NOTCH4, THSD4, PTCH1, and PID1) were also associated with lung function and asthma severity in non-Hispanic Whites asthmatics with additive effects, but these associations were not found in African American asthmatics [50]. These inter-ethnic differences might be related to differences in linkage disequilibrium tagged by the selected SNPs; however, it is possible that distinct loci from an African ancestry determine lung function and asthma severity in admixed African descent populations in a similar, additive fashion [31, 50, 51].
Ancestry-Based Genetic Studies
Differences in asthma susceptibility and severity between recently admixed ethnic groups are a rationale for ancestry-based genetic studies to identify genetic loci. Whole-genome genotyping provides data on SNPs with marked variable allele frequencies between different ancestral populations which can estimate genetic ancestry for ancestry-based genetic studies [52]. Studies of whole-genome or global genetic ancestry have shown that African Americans have an estimated 80 percent African ancestry, on average, (20 percent European ancestry) while Puerto Ricans and Mexican Americans have a combination of three different ancestries: European, African Ancestry, and Amerindian (Figure 2) [30, 31]. Studies of global African ancestry have shown African ancestry is higher in African Americans with self-reported asthma compared to controls suggesting that genetic variation from an African ancestry could influence asthma susceptibility [53].
Ancestry-based genetic studies in different ethnic groups have suggested a role for gene variation related to African ancestry on lung function. Global African ancestry was inversely associated with baseline lung function measures in three independent African American general populations and Puerto Rican asthmatics, a trend predicted by race-based lung function predictive equations [31, 51, 54]. Global African ancestry was also associated with severe asthma exacerbations in a male subset of asthmatics from the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) cohort [55]. The relationship between African ancestry and asthma severity requires replication and evaluation using ancestry-based genetic approaches focused on identifying a causative locus.
Admixture mapping (i.e. mapping by admixture linkage disequilibrium, Figures 3A and 3B) is a whole-genome scanning approach which has mostly been used to identify asthma susceptibility loci. In contrast to GWAS, admixture mapping tests for associations between estimates of ancestry at each SNP and a phenotype of interest. Admixture mapping requires a substantially smaller number of genetic markers than GWAS and can identify regions with rare variants (Figures 3A and 3B) [56, 57]. An admixture mapping study of African Americans from SARP and the NHLBI-sponsored Collaborative Study on the Genetics of Asthma (CSGA) identified an admixture mapping association peak on chromosome 6q14. This peak contained a novel SNP locus only associated with asthma in African Americans with a local European ancestral background suggesting that the risk allele was inherited from recent European admixture. These novel SNP associations were replicated in Puerto Ricans from the Genetics of Asthma in Latino Americans (GALA) cohort demonstrating that genetic ancestry has the potential to interact with susceptibility loci in recently admixed populations [58].
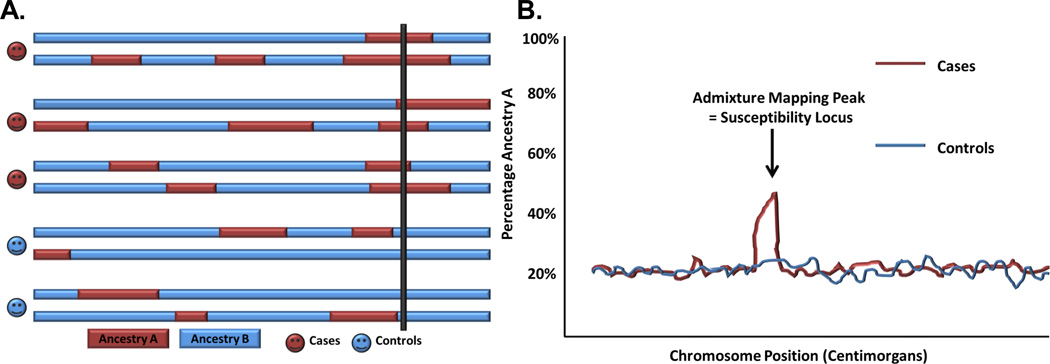
Figure 3. A and 3B: Illustration of Admixture Mapping.
A. The hypothesis behind mapping by admixture linkage disequilibrium or admixture mapping is that chromosomes from an admixed population (shown with darker and lighter colored genetic regions from different ancestries) contain a susceptibility allele which is more frequent in the darker ancestral regions versus the lighter regions. Admixture mapping identifies increased ancestry at a susceptibility locus in affected individuals (region intersected by thick black line).
B. Loci with significant associations between ancestry and disease risk are represented by admixture mapping peaks or chromosomal regions with an overrepresentation of ancestry from the ancestral population, with the highest proportion of risk alleles at the locus containing the risk-invoking variant. Reproduced from Montana G, et al. Am J Hum Genet 2004;75:771–789. and Patterson N, et al. Am J Hum Genet 2004. 74:979–1000 [56, 57].
Ancestry mapping studies in Puerto Rican and Mexican American asthma cases and controls from the GALA cohort also identified 62 admixture mapping peaks of which six contained loci previously associated with asthma. One peak contained the LYN gene which was associated with airways disease in a LYN-knockout murine model [59, 60]. More recently, admixture mapping of a larger Latino cohort from GALA II was combined with allelic association testing to identify a novel susceptibility locus in PSORS1C1 [16].
Another recent admixture-based study identified novel rare variation associated with bronchodilator lung function reversibility in response to the short-acting beta2-adrenergic receptor agonist (beta agonist or SABA) albuterol, a diagnostic asthma phenotype. In Puerto Ricans and Mexican Americans asthmatics from GALA II, admixture mapping in combination with GWAS identified rare variants within two solute carrier genes (SLC24A4 and SLC22A15) associated with SABA bronchodilation [61]. Admixture-based approaches should be employed in combination with allelic association testing to identify the genetic basis for inter-ethnic differences in asthma susceptibility, severity, and response to pharmacologic therapies [32, 62].
DNA Sequencing and Rare Variants
Most genetic studies of asthma susceptibility and severity have been limited to common “tagging” SNPs; however, large-scale next-generation sequencing projects from the NHLBI GO ESP, CAAPA, and the 1,000 Genomes Project now provide an increasingly diverse map of the genomes of different admixed populations including an expanding catalogue of rare variation [10, 33]. Rare variant studies of nine resequenced asthma candidate genes identified rare coding variants associated with asthma susceptibility in DPP10 and IL12RB1 in African Americans and non-Hispanic Whites, respectively [14]. Another study identified rare variants in the receptor target for beta agonists, the beta2-adrenergic receptor gene (ADRB2), associated with severe exacerbations requiring hospitalization in African Americans and non-Hispanic Whites treated with a long-acting beta agonist [13]. In both rare variant studies, associated rare variants differed between ethnic groups but resulted in similar effects, an example of allelic heterogeneity [13, 14, 63]. In these studies, African Americans had a greater frequency of rare variants compared to non-Hispanic Whites providing a potential genetic basis for inter-ethnic differences in asthma risk, severity, and therapeutic responsiveness [13, 14, 32, 62].
Genetic Diversity and the Future of Precision Medicine
The ability to account for the consequences of genetic diversity in different racial and ethnic groups in genetic studies will be critical in developing genetic profiles for personalized or precision medicine approaches tailored to asthmatics from different ethnic groups [64]. These profiles will primarily be identified through multi-gene models which account for gene variation, both common and rare, specific to individuals from different ancestral populations. Future genetic studies in all racial and ethnic groups will require integration of more comprehensive genotyping and next-generation sequencing data with analytical methods which account for the varying diversity and population structure of different ancestral backgrounds. Future studies will also require the enrollment of a larger number of subjects from underrepresented ethnic groups into asthma susceptibility or severity studies as well as clinical trials for GWAS, admixture-based genetic studies, and candidate gene studies.
Conclusions
Genetic studies have the potential to identify loci important for asthma risk in individuals from different ancestries while also identifying gene pathways which influence asthma progression and severity. For the individual asthmatic, the development of genetic profiles would provide guidance on the risk for disease onset and progression and individualize an appropriate intervention or treatment [64]. For diverse asthma populations, these genetic profiles have the potential to define the genes for asthma susceptibility and severity unique to different ethnic and racial groups, which could account for the healthcare-related disparities we are challenged with today.
Key Points.
The more recent mixing of different ancestries during the European colonization of the Americas and the African slave trade has resulted in the complex population structures identified in modern U.S. ethnic groups.
The population structures of modern human genomes represent varying degrees of genetic diversity which impacts the allele frequency of individual variants, particularly rare variants.
Differences in genetic diversity between individuals from different ethnic groups impacts how individual genetic variants can be used to “tag” gene variation in genetic association studies and must be accounted for in all genetic association studies, particularly GWAS.
Admixture-based approaches and rare variant studies in different ethnic groups have suggested a role for gene variation related to ancestry on asthma susceptibility and severity and have the potential to elucidate the genetic basis for inter-ethnic differences when complimented by allelic association testing or GWAS.
The ability to account for the consequences of genetic diversity in different racial and ethnic groups will be critical in developing genetic profiles for personalized or precision medicine approaches tailored to asthmatics from different ethnic groups.
Acknowledgements
Funding Sources: NIH NHLBI Grants R01 NR013700, U10 HL109164, U01 HL 65899, and U10 HL098103
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Wenzel SE, Busse WW. National Heart L, Blood Institute's Severe Asthma Research P. Severe asthma: lessons from the Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(1):14–21. doi: 10.1016/j.jaci.2006.10.025. quiz 2–3. [DOI] [PubMed] [Google Scholar]
- 2.Rose D, Mannino DM, Leaderer BP. Asthma prevalence among US adults, 1998–2000: role of Puerto Rican ethnicity and behavioral and geographic factors. Am J Public Health. 2006;96(5):880–888. doi: 10.2105/AJPH.2004.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169(3):386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 4.Homa DM, Mannino DM, Lara M. Asthma mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am J Respir Crit Care Med. 2000;161(2 Pt 1):504–509. doi: 10.1164/ajrccm.161.2.9906025. [DOI] [PubMed] [Google Scholar]
- 5.Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407(6803):513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 6.Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 7.Marth G, Schuler G, Yeh R, Davenport R, Agarwala R, Church D, et al. Sequence variations in the public human genome data reflect a bottlenecked population history. Proc Natl Acad Sci U S A. 2003;100(1):376–381. doi: 10.1073/pnas.222673099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts L. 9 billion? Science. 2011;333(6042):540–543. doi: 10.1126/science.333.6042.540. [DOI] [PubMed] [Google Scholar]
- 9.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuji S. Genetics of neurodegenerative diseases: insights from high-throughput resequencing. Hum Mol Genet. 2010;19(R1):R65–R70. doi: 10.1093/hmg/ddq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 13.Ortega VE, Hawkins GA, Moore WC, Hastie AT, Ampleford EJ, Busse WW, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting beta agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir Med. 2014;2(3):204–213. doi: 10.1016/S2213-2600(13)70289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torgerson DG, Capurso D, Mathias RA, Graves PE, Hernandez RD, Beaty TH, et al. Resequencing candidate genes implicates rare variants in asthma susceptibility. Am J Hum Genet. 2012;90(2):273–281. doi: 10.1016/j.ajhg.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: The Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2013.08.055. [Epub ahead of print]. This article summarizes one of the largest GWAS and admixture mapping studies for asthma in Latino populations consisting of primarily Puerto Ricans and Mexican Americans from the Genes-environments and Admixture in Latino Americans study (GALA II). This GWAS identified a SNP adjacent to IKZF3 associated with asthma while admixture mapping was combined with allelic association testing to identify a novel susceptibility locus in PSORS1C1 which replicated in Latinos from the EVE consortium.
- 17.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43(9):893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41(3):342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125(2):328–335. e11. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol. 2012;130(4):861–868. e7. doi: 10.1016/j.jaci.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125(2):336–346. e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19(4):458–464. doi: 10.1038/ejhg.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5(8):e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Q, Zhao H, Wang A, Gong Y, Liu Q. Association of genetic variants in chromosome 17q21 and adult-onset asthma in a Chinese Han population. BMC Med Genet. 2011;12:133. doi: 10.1186/1471-2350-12-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29(1):76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- 31.Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, et al. Genetic ancestry in lung-function predictions. N Engl J Med. 2010;363(4):321–330. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1):16–26. doi: 10.1016/j.jaci.2013.10.040. This review summarizes the origins of genetic diversity in recently admixed ethnic groups and describes the implications of genetic admixture on pharmacogenetic studies in asthma.
- 33.Seattle, WA: NHLBI GO Exome Sequencing Project (ESP); [cited 2013 May 1]. Exome Variant Server. Available from: (URL: http://evs.gs.washington.edu/EVS/). [Google Scholar]
- 34.A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15(4):389–392. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, et al. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001;68(6):1437–1446. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koppelman GH, Meyers DA, Howard TD, Zheng SL, Hawkins GA, Ampleford EJ, et al. Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med. 2009;180(10):929–935. doi: 10.1164/rccm.200810-1621OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colilla S, Nicolae D, Pluzhnikov A, Blumenthal MN, Beaty TH, Bleecker ER, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol. 2003;111(4):840–846. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- 38.Meyers DA, Postma DS, Stine OC, Koppelman GH, Ampleford EJ, Jongepier H, et al. Genome screen for asthma and bronchial hyperresponsiveness: interactions with passive smoke exposure. J Allergy Clin Immunol. 2005;115(6):1169–1175. doi: 10.1016/j.jaci.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 39.Dizier MH, Bouzigon E, Guilloud-Bataille M, Siroux V, Lemainque A, Boland A, et al. Evidence for gene × smoking exposure interactions in a genome-wide linkage screen of asthma and bronchial hyper-responsiveness in EGEA families. Eur J Hum Genet. 2007;15(7):810–815. doi: 10.1038/sj.ejhg.5201830. [DOI] [PubMed] [Google Scholar]
- 40.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7(2):95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins GA, Tantisira K, Meyers DA, Ampleford EJ, Moore WC, Klanderman B, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006;174(10):1101–1109. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112(4):717–722. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013;188(3):309–318. doi: 10.1164/rccm.201302-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003;35(3):258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol. 2013;132(2):313–320. e15. doi: 10.1016/j.jaci.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42(1):45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42(1):36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127(6):1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brehm JM, Acosta-Perez E, Klei L, Roeder K, Barmada MM, Boutaoui N, et al. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012;129(6):1484–1490. e6. doi: 10.1016/j.jaci.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flores C, Ma SF, Pino-Yanes M, Wade MS, Perez-Mendez L, Kittles RA, et al. African ancestry is associated with asthma risk in African Americans. PLoS One. 2012;7(1):e26807. doi: 10.1371/journal.pone.0026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159(1):179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 55.Rumpel JA, Ahmedani BK, Peterson EL, Wells KE, Yang M, Levin AM, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol. 2012;130(6):1302–1306. doi: 10.1016/j.jaci.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet. 2004;75(5):771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74(5):979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, et al. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 2012;130(3):622–629. e9. doi: 10.1016/j.jaci.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beavitt SJ, Harder KW, Kemp JM, Jones J, Quilici C, Casagranda F, et al. Lyn-deficient mice develop severe, persistent asthma: Lyn is a critical negative regulator of Th2 immunity. J Immunol. 2005;175(3):1867–1875. doi: 10.4049/jimmunol.175.3.1867. [DOI] [PubMed] [Google Scholar]
- 60.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012;130(1):76–82. e12. doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drake KA, Torgerson DG, Gignoux CR, Galanter JM, Roth LA, Huntsman S, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133(2):370–378. doi: 10.1016/j.jaci.2013.06.043. This article was among the first to describe a role for rare variants in bronchodilator response to a short-acting beta2-adrenergic receptor agonist (SABA), a diagnostic asthma phenotype. In Puerto Ricans and Mexican Americans asthmatics from GALA II admixture mapping in combination with GWAS identified rare variants within two solute carrier genes (SLC24A4 and SLC22A15) associated with SABA bronchodilation.
- 62.Wechsler ME, Castro M, Lehman E, Chinchilli VM, Sutherland ER, Denlinger L, et al. Impact of race on asthma treatment failures in the asthma clinical research network. Am J Respir Crit Care Med. 2011;184(11):1247–1253. doi: 10.1164/rccm.201103-0514OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma L, Roman-Campos D, Austin ED, Eyries M, Sampson KS, Soubrier F, et al. A novel channelopathy in pulmonary arterial hypertension. N Engl J Med. 2013;369(4):351–361. doi: 10.1056/NEJMoa1211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir Med. 2014;2(5):405–415. doi: 10.1016/S2213-2600(14)70012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]