Abstract
A total of 300 gastric biopsy samples and 50 Helicobacter pylori isolates were collected from Ethiopian adult dyspeptic patients. The vacA and cagA genes were detected in 90 and 79% of biopsy specimens, respectively, and in 100 and 87% of clinical isolates, respectively. Both genes were detected in 84% of the gastric biopsy samples and in 87% of the clinical isolates. Among vacA genotypes, the s1/m1 genotype was the most common in gastric biopsy samples (48%). The vacA and cagA positive H. pylori strains were detected to a higher degree in patients with chronic active gastritis (71%) than patients with other histopathological findings (29%) (P < 0.05).
Several Helicobacter pylori virulence genes related to the risk of gastroduodenal diseases have been proposed. The vacuolating cytotoxin (vacA) gene is present in virtually all H. pylori strains and contains at least two variable regions, the signal (s) region, which encodes the signal peptide, and the middle (m) region (4). The s region has been divided into two subtypes, s1 and s2, and the m region has been divided into two subtypes, m1 and m2 (19). The amount of cytotoxin produced is highest with the s1/m1 allele, followed by the s1/m2 allele, while no cytotoxin activity is found when s2/m2 is present (19). The cytotoxin-associated gene (cagA) is a marker for a genomic pathogenicity island of 40 kb (6). A significant association between the presence of ulcers or gastric carcinoma and the presence of vacA type s1 and cagA gene (5, 19). The present study represents the first in Ethiopia to detect H. pylori vacA and cagA genotypes from gastric biopsy samples and clinical isolates using PCR-based methods.
MATERIALS AND METHODS
Study subjects.
A total of 300 consecutive informed and consenting adult patients with dyspeptic symptoms from the gastrointestinal referral and follow-up clinics of Department of Internal Medicine, Tikur Anbassa University Hospital, Addis Ababa, Ethiopia, were investigated for H. pylori between November 2000 and August 2002. The mean age of the patients was 36.5 years (standard deviation, 13.8 years; range, 15 to 90 years). The majority of patients (76%) were between the ages of 15 and 44 years. Of the 300 patients, 186 (62%) were males and 114 (38%) were females (resulting in an overall male to female ratio of 1.6:1).
The study was approved by the Department Graduate Committee, the Faculty Research Publications Committee and endorsed by the Faculty Academic Commission and has been ethically cleared.
Culture and identification.
Antral gastric biopsy samples were taken from each dyspeptic patient. The biopsy specimens were put into sterile phosphate-buffered saline containing 15% glycerol and immediately transported to laboratory for culture. Biopsy samples for molecular analysis were kept frozen in 15% tryptone soy broth (Oxoid Ltd., Basingstoke, England) and stored at −70°C until analyzed.
H. pylori was cultured from antral biopsy specimens using a standard method (17). H. pylori identification was based on morphology, Gram staining, oxidase, catalase, and urease tests. All the isolated H. pylori strains were kept frozen at −70°C in the tryptone soy broth medium containing 15% (vol/vol) glycerol until genotyping was performed. The H. pylori reference strain (CCUG 17874) (Culture Collection, University of Gothenburg, Gothenburg, Sweden) was cultured throughout the study for quality control.
Histopathology.
Gastric biopsy specimens were fixed in 10% formalin and embedded in paraffin. The sections (4 to 5 μm thick) were cut and stained with hematoxylin and eosin (2). The histological findings from the sections stained with hematoxylin and eosin were scored according to the updated Sydney system of classification and grading of gastritis (7).
Genomic DNA extraction.
Biopsy specimens and isolates were centrifuged at 10,000 × g for 5 min. The DNA was extracted from the pellets by use of the QIAamp DNA kit (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations and DNA stored at −20°C until analysis. DNA extraction negative controls were performed in parallel by including sterile tubes without samples to check for contamination of the DNA extraction reagents.
PCR-DGGE.
The PCR amplification was carried out using a GeneAmp 2700 Thermal cycler (Applied Biosystems, Foster City, Calif.). A seminested Helicobacter genus-specific PCR assay targeting the 16S rDNA was used to amplify Helicobacter DNA (9). Denaturing gradient gel electrophoresis (DGGE) analysis of the PCR products was performed in a DCode system (Bio-Rad, Hercules, Calif.) as recently described (1). Migration ladder containing PCR products of reference Helicobacter strains (H. muridarum [CCUG 29262], H. bilis [CCUG 38995], H. pullorum [NCTC 12825], H. pylori [CCUG 17874], “Flexispira rappini” [CCUG 23435], H. hepaticus [CCUG 33637], and H. bizzozeronii [AF 53]) was run in parallel as a mobility ladder.
vacA and cagA genotyping.
Detection of H. pylori vacA and cagA genes was performed on gastric biopsy specimens and isolates positive for H. pylori by PCR-DGGE as previously described (15, 19). As a positive control, H. pylori (CCUG 17874) DNA (∼0.1 ng) was added to the reaction mixture, while 5 μl of sterile deionized Millipore-filtered water was added to the reaction mixture as a negative control. Estimation of size of the PCR products was done by using Gene ruler 100-bp DNA ladder (Fermentas, Vilnius, Lithuania). The products of each PCR assay were visualized by electrophoresis in a 1.5% agarose gel containing ethidium bromide (15).
Statistical analysis.
Epi info version 2000 (Centers for Disease Control and Prevention, Atlanta, Ga.) was used for statistical analysis. Chi-square or Fisher's exact test was applied to test whether differences between values were significant. P values <0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Histopathological examinations were performed on 276 (92%) of the gastric biopsy specimens, whereas the remaining 24 (8%) were not adequate (too small). Abnormal findings were observed in all examined specimens. Chronic gastritis was found in 48 (17.4%) patients, chronic active gastritis in 185 (67%), chronic atrophic gastritis in 24 (9%); chronic atrophic gastritis with intestinal metaplasia in 17 (6%) and malignant lesions in 2 (0.6%) patients. The overall prevalence of chronic gastritis was 99.3%. The most common histopathological findings in the present study were similar to those reported from other parts of Africa (10).
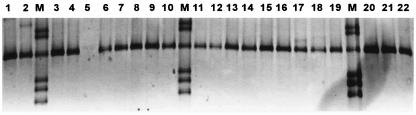
The seminested Helicobacter genus-specific PCR assay detected Helicobacter DNA in 273 of 300 (91%) of the biopsy specimens. DGGE analysis showed that all PCR products have mobility pattern similar to the H. pylori reference strain (Fig. 1).
FIG. 1.
DGGE analysis of PCR products amplified by using Helicobacter genus-specific primers. Lanes, 1 to 4 and 6 to 22, H. pylori-positive samples; 5, H. pylori-negative sample; M, mobility marker (PCR products of reference Helicobacter spp. [from the top: H. bilis, H. pullorum, H. pylori, “F. rappini,” H. hepaticus, and H. bizozzeroni]).
The vacA gene was detected in 246 of 273 (90%) of H. pylori-positive gastric biopsy specimens, which is similar to reported results from The Netherlands (93%) and Hong Kong (95.8%) (14, 21), emphasizing high sensitivity of the PCR method employed in the present study. The vacA genotypes s1/m1, s1/m2, s2/m2, and s2/m1 were found in 48, 28, 9, and 2% of the specimens, respectively (Table 1), whereas, 15 biopsy specimens (6%) were incomplete and thus did not yield a detectable PCR product for the vacA s or m regions. The pattern of vacA alleles in this study is in agreement with those reported in other studies (3, 8, 16, 18, 20). However, the frequency of vacA s1/m1 allelic type in this study is higher than figures reported from The Netherlands (36%), Hong Kong (26 to 31%), and Nigeria (24%), but lower than figures reported from Brazil (80%) and Korea (78%) (3, 11, 16, 20, 21). In the present investigation, the rare vacA s2/m1 allele was detected in 4 (2%) of the 246 gastric biopsy specimens examined, also reported in studies in South Africa and Chile (12, 13). Multiple vacA genotypes were found in 18 (7%) of the 246-biopsy specimens examined. The most frequent multiple vacA genotypes were s1/m1m2 (11 of 18; 61%).
TABLE 1.
H. pylori vacA genotypes in gastric biopsy samples and clinical isolates
| vacA genotype | No. (%) of samples or isolates
|
|
|---|---|---|
| Gastric biopsy | Clinical | |
| s1/m1 | 118 (48) | 31 (60) |
| s1/m2 | 68 (28) | 14 (27) |
| s2/m2 | 23 (9) | 4 (7) |
| s2/m1 | 4 (2) | 0 |
| s1/m1m2a | 11 (4) | 2 (4) |
| s2/m1m2a | 2 (0.7) | 0 |
| s1s2/m2a | 4 (2) | 1 (2) |
| s1s2/m1m2a | 1 (0.3) | 0 |
| Incomplete (vacA s or m)b | 15 (6) | 0 |
| Total vacA positive | 246 (100) | 52 (100) |
Multiple vacA genotypes.
For vacA s and m regions, n = 7 and n = 8, respectively.
The vacA was detected in all 52 H. pylori isolates tested (Table 1). The prevalence of the vacA subtypes s1/m1, s1/m2, and s2/m2 was 60, 27, and 7%, respectively. Three (6%) of the isolates contained mixed vacA subtypes; two s1/m1m2 and one s1s2/m2 from a single H. pylori isolate. The multiple vacA genotypes detected in this study are similar to results from Italy reported by Blaser and Berg (5). Surprisingly, the prevalence of multiple vacA genotypes in this study was much lower compared with results reported from Brazil (13%), Chile (32%), Korea (18%), and The Netherlands (11%) (3, 11, 13, 20). The low prevalence of multiple vacA genotypes in a country with a high prevalence of H. pylori infections in the general population may be the result of a low number of mosaics of any combination of signal (s) and mid-region (m) alleles of the bacteria circulating in the community.
Of the 273 H. pylori PCR-positive biopsy specimens, 217 (79%) were cagA positive. Four different genotypic combinations were recognized based on analysis of the positive and negative vacA and cagA results—vacA+ cagA+, vacA+ cagA, vacA cagA, and vacA cagA+, which were found in 76, 14, 6, and 4% of specimens, respectively (Table 2). Forty-five of the 52 (87%) H. pylori strains were positive for both vacA and cagA, whereas the remaining isolates 7 (13%) were only vacA positive (Table 2). Statistical analysis showed no difference in the detection of the vacA and cagA in gastric biopsy specimens and clinical isolates (P = 0.70 for vacA, P = 0.96 for cagA). In addition, no statistical differences in the frequency of detection of the different vacA allelic types from gastric biopsy specimens and clinical isolates were found (P > 0.05). The prevalence of cagA positive H. pylori strains varies from one geographic region to another, e.g., 38% in Chile, 48% in Sri Lanka, 67% in The Netherlands, 81% in the United Sates, 90% in Hong Kong, 97% in Korea, 93% in Nigeria and 94% in Brazil (3, 8, 11, 13, 16, 18, 20, 21). Correlation of histopathology results with vacA and cagA genotypes showed that vacA and cagA positive strains were detected to a higher degree in patients with chronic active gastritis (71%) compared with patients with other histopathological findings (29%) (P < 0.05) (Table 3).
TABLE 2.
Distribution of H. pylori vacA and cagA genotypes in 273 gastric biopsy samples and 52 clinical isolates
| Genotype status | No. (%) of samples or isolates
|
|
|---|---|---|
| Gastric biopsy | Clinical | |
| vacA+cagA+ | 207 (76) | 45 (87) |
| vacA+cagA | 39 (14) | 7 (13) |
| vacA cagA | 17 (6) | 0 |
| vacA cagA+ | 10 (4) | 0 |
TABLE 3.
Distribution of vacA and cagA allelic types according to histopathological findings in the antra of 276 dyspeptic patients
| Histopathological findingsb | No. (%) with vacA and cagA results
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
s1/m1
|
s1/m2
|
s2/m1
|
s2/m2
|
Mixed
|
Inca
|
Negative
|
||||||||
| cagA+ | cagA | cagA+ | cagA | cagA+ | cagA | cagA+ | cagA | cagA+ | cagA | cagA+ | cagA | cagA+ | cagA | |
| Nonatrophic gastritis | ||||||||||||||
| CG (n = 48) | 9 (19) | 2 (4) | 4 (8) | 6 (13) | 0 | 0 | 1 (2) | 0 | 4 (8) | 0 | 3 (6) | 0 | 1 (2) | 18 (38) |
| CAG (n = 185) | 80 (43) | 2 (1) | 36 (20) | 4 (2) | 2 (1) | 0 | 10 (6) | 10 (6) | 6 (3) | 4 (2) | 6 (3) | 4 (3) | 2 (1) | 19 (10) |
| Atrophic gastrits | ||||||||||||||
| CAAG (n = 24) | 9 (38) | 1 (4) | 10 (42) | 2 (8) | 0 | 0 | 0 | 0 | 1 (4) | 0 | 0 | 0 | 0 | 1 (4) |
| CAAGI (n = 17) | 9 (53) | 0 | 3 (18) | 0 | 0 | 0 | 0 | 0 | 1 (6) | 0 | 1 (6) | 0 | 0 | 3 (18) |
| Malignant lesions | ||||||||||||||
| Adenocarcinoma (n = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 |
| MALT lymphoma (n = 1) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (n = 276) | 108 (39) | 5 (2) | 53 (19) | 12 (4) | 2 (1) | 0 | 11 (4) | 10 (4) | 13 (5) | 4 (1) | 10 (4) | 4 (1.4) | 3 (1) | 41 (15) |
vacA gene was incomplete (Inc) to yield a detectable PCR product for the vacA s or m region.
Abbreviations: CG, chronic gastritis; CAG, chronic active gastritis; CAAG, chronic active atrophic gastritis; CAAGI, chronic active atrophic gastritis with intestinal metaplasia; MALT, mucosa-associated lymphatic tissue.
Molecular analyses demonstrated that more than 80% of the Ethiopian H. pylori strains (detected from dyspeptic patients) harbor both vacA and cagA genes. The presence of such combined genotypes in infected patients has been proposed to increase the risk for development of clinical complications such as peptic ulcerations and gastric cancer (4, 20). Further genetic analysis should be conducted to determine the homology of the H. pylori genome in members of the same family in order to study the transmission of H. pylori from person to person.
Acknowledgments
This research project was supported by grants from the Swedish International Development Cooperation Agency with developing countries (SIDA/SAREC) program for Bio-Medical Research and Training and a grant from the Swedish Research Council (16X04723 and 6X11229 to T.W.) and from the University Hospital of Lund (A.L.F.).
REFERENCES
- 1.Abu Al-Soud, W., M. Bennedsen, S. L. W. On, I.-S. Ouis, P. Vandamme, H.-O. Nilsson, A. Ljungh, and T. Wadström. 2003. Assessment of PCR-DGGE for the identification of diverse Helicobacter species, and application to faecal samples from zoo animals to determine helicobacter prevalence. J. Med. Microbiol. 52:765-771. [DOI] [PubMed] [Google Scholar]
- 2.Anim, J. T., N. Al-Sobkie, A. Prasad, B. John, P. N. Sharma, and I. Al-Hamar. 2000. Assessment of different methods for staining Helicobacter pylori in endoscopic gastric biopsies. Acta Histochem. 102:129-137. [DOI] [PubMed] [Google Scholar]
- 3.Ashour, A. A., P. P. Magalhaes, E. N. Mendes, G. B. Collares, V. R. de Gusmao, D. M. Queiroz, A. M. Nogueira, G. A. Rocha, and C. A. de Oliveira. 2002. Distribution of vacA genotypes in Helicobacter pylori strains isolated from Brazilian adult patients with gastritis, duodenal ulcer or gastric carcinoma. FEMS Immunol. Med. Microbiol. 33:173-178. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and D. E. Berg. 2001. Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Investig. 107:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon, M. F., R. M. Genta, J. H. Yardley, and P. Correa. 1996. Classification and grading of gastritis. The updated Sydney system. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg Pathol. 20:1161-1181. [DOI] [PubMed] [Google Scholar]
- 8.Fernando, N., J. Holton, D. Vaira, M. DeSilva, and D. Fernando. 2002. Prevalence of Helicobacter pylori in Sri Lanka as determined by PCR. J. Clin. Microbiol. 40:2675-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto, K., H. Ohashi, A. Takakura, and T. Itoh. 2000. Current status of helicobacter contamination of laboratory mice, rats, gerbils, and house musk shrews in Japan. Curr. Microbiol. 41:161-166. [DOI] [PubMed] [Google Scholar]
- 10.Kidd, M., J. A. Louw, and I. N. Marks. 1999. Helicobacter pylori in Africa: observations on an ′enigma within an enigma'. J. Gastroenterol. Hepatol. 14:851-858. [DOI] [PubMed] [Google Scholar]
- 11.Kim, S. Y., C. W. Woo, Y. M. Lee, B. R. Son, J. W. Kim, H. B. Chae, S. J. Youn, and S. M. Park. 2001. Genotyping cagA, vacA subtype, iceA1, and babA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J. Korean Med Sci. 16:579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letley, D. P., A. Lastovica, J. A. Louw, C. J. Hawkey, and J. C. Atherton. 1999. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J. Clin. Microbiol. 37:1203-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez, A., C. Gonzalez, F. Kawaguchi, R. Montoya, A. Corvalan, J. Madariaga, J. Roa, A. Garcia, F. Salgado, H. Solar, and M. Palma. 2001. Helicobacter pylori: cagA analysis and vacA genotyping in Chile. Detection of a s2/m1 strain. Rev. Med. Chil. 129:1147-1153. [PubMed] [Google Scholar]
- 14.Scholte, G. H., L. J. van Doorn, W. G. Quint, and J. Linderman. 2001. Genotyping of Helicobacter pylori strains in formalin-fixed or formaldehyde-sublimate-fixed paraffin-embedded gastric biopsy specimens. Diagn. Mol. Pathol. 10:166-170. [DOI] [PubMed] [Google Scholar]
- 15.Sjunnesson, H., T. Falt, E. Sturegard, W. Abu Al-Soud, A. Ljungh, and T. Wadström. 2003. PCR-denaturing gradient gel electrophoresis and two feces antigen tests for detection of Helicobacter pylori in mice. Curr. Microbiol. 47:278-285. [DOI] [PubMed] [Google Scholar]
- 16.Smith, S. I., C. Kirsch, K. S. Oyedeji, A. O. Arigbabu, A. O. Coker, E. Bayerdoffer, and S. Miehlke. 2002. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in Nigerian patients with duodenal ulcer disease. J. Med. Microbiol. 51:851-854. [DOI] [PubMed] [Google Scholar]
- 17.Soltesz, V., B. Zeeberg, and T. Wadström. 1992. Optimal survival of Helicobacter pylori under various transport conditions. J. Clin. Microbiol. 30:1453-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straus, E. W., H. Patel, J. Chang, R. M. Gupta, V. Sottile, J. Scirica, G. Tarabay, S. Iyer, S. Samuel, and R. D. Raffaniello. 2002. H. pylori infection and genotyping in patients undergoing upper endoscopy at inner city hospitals. Dig. Dis. Sci. 47:1575-1581. [DOI] [PubMed] [Google Scholar]
- 19.van Doorn, L. J., C. Figueiredo, R. Rossau, G. Jannes, M. van Asbroeck, J. C. Sousa, F. Carneiro, and W. G. V. Quint. 1998. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J. Clin. Microbiol. 36:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 21.Wong, B. C., Y. Yin, D. E. Berg, H. H. Xia, J. Z. Zhang, W. H. Wang, W. M. Wong, X. R. Huang, V. S. Tang, and S. K. Lam. 2001. Distribution of distinct vacA, cagA and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter 6:317-324. [DOI] [PubMed] [Google Scholar]