Abstract
AR signaling is essential for the growth and survival of prostate cancer (PCa), including most of the lethal castration-resistant PCa (CRPC). We previously reported that TGF-β signaling in prostate stroma promotes prostate tumor angiogenesis and growth. By using a PCa/stroma co-culture model, here we show that stromal TGF-β signaling induces comprehensive morphology changes of PCa LNCaP cells. Furthermore, it induces AR activation in LNCaP cells in the absence of significant levels of androgen, as evidenced by induction of several AR target genes including PSA, TMPRSS2, and KLK4. SD-208, a TGF-β receptor 1 specific inhibitor, blocks this TGF-β induced biology. Importantly, stromal TGF-β signaling together with DHT induce robust activation of AR. MDV3100 effectively blocks DHT-induced, but not stromal TGF-β signaling induced AR activation in LNCaP cells, indicating that stromal TGF-β signaling induces both ligand-dependent and ligand-independent AR activation in PCa. TGF-β induces the expression of several growth factors and cytokines in prostate stromal cells, including IL-6, and BMP-6. Interestingly, BMP-6 and IL-6 together induces robust AR activation in these co-cultures, and neutralizing antibodies against BMP-6 and IL-6 attenuate this action. Altogether, our study strongly suggests tumor stromal microenvironment induced AR activation as a direct mechanism of CRPC.
Keywords: TGF-β, AR, tumor microenvironment, prostate stroma, prostate cancer, co-culture
INTRODUCTION
Prostate cancer (PCa) is the most commonly diagnosed cancer and the second leading cause of cancer death in American men. AR signaling is essentially required for most human prostate cancers. Therefore, ADT is the standard treatment for advanced and metastatic PCa. Most prostate tumors initially respond to ADT therapy. However, most tumors will relapse and become the castration-resistant PCa (CRPC). Currently, there is no cure for CRPC. Interestingly, AR signaling is re-activated in most CRPC tumors in the presence of castration level of androgen ligands. Hence, AR remains as a major therapeutic target for CRPC.
Several molecular mechanisms may count for this AR re-activation in CRPC, including AR gene amplification/overexpression, AR mutation, the presence of AR splice variants, enhanced AR co-regulators signaling, alterations in steroid metabolism, growth factor and/or cytokine induced AR activation etc [1]. Tumor microenvironment also plays critical roles in regulating prostate cancer progression [2]. Prostate cancer is enriched in reactive stromal microenvironment, including reactive myofibroblasts that are uniquely presented in wound repair and tumor microenvironment [3–5]. Transforming Growth Factor β (TGF-β) is generally overexpressed in most carcinomas associated with a reactive stroma, including breast, colon, and prostate [3, 6–9]. Overexpression of TGF-β in carcinoma cells is usually associated with a down-regulation of functional TGF-β receptors in carcinoma cells but not in stromal cells [9–12]. Subcutaneous injection of TGF-β1 is sufficient to induce a stromal reaction with differentiation to myofibroblasts, enhanced collagen production and stimulated angiogenesis [13, 14]. Therefore, TGF-β1 may be a key factor inducing a reactive stroma in wound repair and cancer. By using the differential reactive stroma (DRS) xenograft model [15–18], we have shown that human prostate stromal cells differentially promote rate of PCa progression [15]. By conditional knockout of TGF-β Receptor II (TβRII) and overexpression of a dominant negative Smad3 in prostate stromal cells in LNCaP DRS xenograft model, we have demonstrated that Smad3-mediated TGF-β signaling in prostate stroma promotes prostate tumor growth and angiogenesis [16, 18] and this stromal TGF-β action is partially mediated by Connective Tissue Growth Factor (CTGF) and Fibroblast Growth Factor 2 (FGF-2) signaling [17, 18]. Therefore, TGF-β signaling in prostate stroma regulates PCa progression.
Interleukin-6 (IL-6) is a pleiotropic cytokine that play important roles in regulating immune system and inflammation. It is also a key cytokine in regulating human cancers, including PCa [19]. Serum IL-6 level is associated with PCa progression and metastasis [20, 21]. Functionally, IL-6 can induce AR expression and AR activation, and promote PCa cell growth [22–28]. IL-6 has also been shown to promote castration-resistance of PCa including that to enzalutamide (MDV3100, a second-generation antiandrogen) [25, 28, 29]. Interestingly, the circulating levels of both IL-6 and TGF-β1 were elevated in patients with metastatic PCa [30]. Bone morphogenetic proteins (BMPs) play important roles in inducing bone formation. BMP-6 expression is frequently elevated in PCa [31]. It can promote PCa bone metastases and its expression is associated with a more invasive phenotype [32, 33]. Interestingly, two most recent studies revealed a role of BMP-6 in promoting castration-resistance of PCa [34, 35].
In this report, we used direct co-cultures of LNCaP cells with three different human prostate stromal cell lines to show that prostate stroma-specific TGF-β signaling induces AR activation in LNCaP cells in the absence of significant amount of androgens, and that treatment of MDV3100, a second-generation antiandrogen [36], only partially attenuates this AR activation. Our study also revealed robust cooperative activity between stromal TGF-β signaling and DHT ligand in inducing AR activation in PCa cells. Finally, we showed that IL-6 and BMP-6 together induces robust AR activation, and that they partially mediate this prostate stromal TGF-β signaling induced AR activation in PCa cells.
RESULTS
Prostate stroma – specific TGF-β signaling induces morphological changes in LNCaP cells
We have previously shown that stromal TGF-β signaling promotes prostate tumor growth [18]. To further delineate the underlying mechanisms, we generated LNCaP cells overexpressing an HA-tagged constitutively activated TGF-β1 ligand (LNCaP-TGF-β1(a)) and control LNCaP cells (LNCaP-Ctrl) as described before [37]. We then performed in vitro PCa/stroma co-cultures by plating LNCaP-TGF-β1(a) cells or LNCaP-Ctrl cells on top of the mostly confluent HPS-19I cells, a previously generated human prostate stromal cell line [38]. Since LNCaP cells are defective in TGF-β receptor I (TβRI/ALK-5) that is essential for mediating TGF-β signaling [39], only HPS19I cells can respond to TGF-β ligand in these co-cultures. This provides a unique opportunity to study how prostate stromal cell-specific TGF-β signaling regulates PCa biology. We performed these co-cultures in 0.2% FBS containing minimal amount of growth factors and cytokines for up to 28 days. We found that while the LNCaP-Ctrl cells remained relatively flat, LNCaP-TGF-β1(a) cells formed sphere-like structures in these co-cultures (Figure 1a & b, Supplementary Figure 1), indicating that prostate stromal cell-specific TGF-β signaling induces profound biological changes in LNCaP cells.
Figure 1. Prostate stromal TGF-β signaling induces profound changes in the co-cultured LNCaP cells.
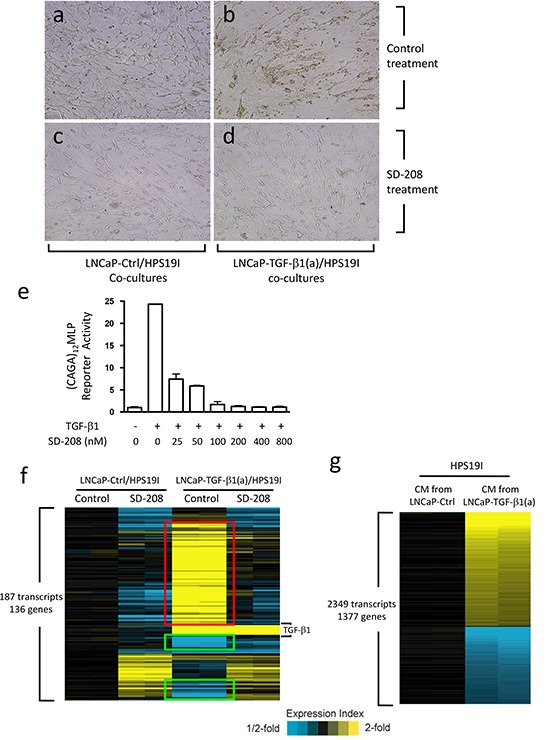
(a-d) LNCaP-TGF-β1(a) cells and LNCaP-Ctrl cells were co-cultured with HPS19I human prostate stromal cells in RPMI1640 supplemented with 0.2% FBS, and treated with 400 nM of SD-208 or control for 15 days. Representative photographs were shown for (a) control and (c) SD208 treated LNCaP-Ctrl/HPS19I co-cultures, and (b) control and (d) SD208 treated LNCaP-TGF-β1(a)/HPS19I co-cultures. HPS19I cells are at the bottom layer. (e) HPS19I cells were co-transfected with (CAGA)12MLP and pRL-null vectors, and treated with 50 pM of TGF-β1 and different dosages of SD-208 compound for 24 hours. Cell lysates were prepared and assayed for luciferase activity. (f) LNCaP-TGF-β1(a) cells or LNCaP-Ctrl cells were co-cultured with HPS19I cells in RPMI1640 supplemented with 0.2% FBS and treated with 400 nM of SD-208 or vehicle control for 6 days. Total RNA was extracted and microarrays were performed to compare differential gene expression among these differentially treated co-cultures. Two independent experiments were performed. (g) HPS19I cells were treated for 6 days with conditioned media (CM) from LNCaP-TGF-β1(a) cells or LNCaP-Ctrl cells in RPMI1640 supplemented with 0.2% FBS. Microarrays were performed to compare differential gene expression between these two differentially treated groups. Two independent experiments were performed.
SD-208 is a TβRI (ALK-5) protein kinase-specific inhibitor [40]. To determine its efficacy in inhibiting TGF-β signaling in HPS19I cells, we transfected HPS19I cells with a (CAGA)12MLP (Smad binding sequence) reporter construct [41] and a pRL-null vector (Promega, control for transfection efficiency). We then treated these cells with 50 pM of TGF-β1 along with different dosages of SD-208. TGF-β1 induced a 25-fold increase in (CAGA)12MLP reporter activity in the transfected HPS19I cells, and this induction was inhibited by SD-208 compound in a dose-dependent manner (Figure 1e). 400 nM of SD-208 abolished TGF-β signaling in HPS19I cells, thus was used in the co-culture experiments (Figure 1c and 1d). Consistent with its TGF-β signaling inhibition activity, 400 nM of SD-208 treatment blocked the LNCaP-TGF-β1(a) cells from forming sphere-like structures in the LNCaP-TGF-β1(a)/HPS19I co-cultures (Figure 1d).
TGF-β induces profound gene expression changes in LNCaP/HPS19I co-cultures
To explore how prostate stroma-specific TGF-β signaling regulates LNCaP cell biology in these co-cultures, we carried out cDNA microarray study to compare differential gene expression among (1) control treated LNCaP-Ctrl/HPS19I co-cultures, (2) 400 nM of SD-208 treated LNCaP-Ctrl/HPS19I co-cultures, (3) control treated LNCaP-TGF-β1(a)/HPS19I co-cultures, and (4) 400 nM of SD-208 treated LNCaP-TGF-β1(a)/HPS19I co-cultures. After 6-day treatment, total RNAs were directly extracted from these co-cultures from two independent studies, reverse transcribed, and submitted for microarray analysis using the 4×44K Whole Human Genome Oligo Microarray chip (Agilent Technologies). We found that 187 transcripts representing 136 unique genes were differentially expressed among these groups (ANOVA P<0.001, standard deviation > 0.2), including 79 unique genes (in red) that were induced by TGF-β, but blocked or reversed by SD-208 treatment, as well as 23 genes (in green) that were repressed by TGF-β, but blocked or reversed by SD-208 treatment (Figure 1f). The most significantly upregulated genes in these LNCaP-TGF-β1(a)/HPS19I co-cultures include TIMP3, COMP, FN1, TSPAN2, CILP, TNFAIP6, ENC1, CDKN2B, MRAS, LTBP2, LOX, POSTN, LRRC32 etc., as well as notably KLK3 (PSA), a prostate epithelial cell specific marker and an AR target gene (Table 1a). The most significantly downregulated genes include UGT2B17, FAM111A, ZNF294, ANK3, MAPRE2 etc. (Table 1b). Control and SD-208 treated LNCaP-TGF-β1(a)/HPS19I co-cultures all exhibited overexpression of nine transcripts, all of which represent TGF-β1, the ectopically overexpressed gene in LNCaP-TGF-β1(a) cells.
Table 1A. Most up-regulated genes in LNCaP-TGF-β1(a)/HPS19I co-cultures (microarray).
| RefSeq Nuc | Gene | TGF-β1+Ctrl vs. Ctrl+Ctrl | TGF-β1+SD208 vs.TGF-β1+Ctrl | p-value (ANOVA) |
|---|---|---|---|---|
| NM_000362 | TIMP3 | 21.80112275 | 0.035626662 | 0.000969463 |
| NM_000095 | COMP | 18.43130951 | 0.063331859 | 3.72E-05 |
| NM_212482 | FN1 | 11.94239303 | 0.12931702 | 0.000623914 |
| NM_005725 | TSPAN2 | 10.28542467 | 0.101405721 | 9.73E-05 |
| NM_003613 | CILP | 10.21670312 | 0.091015909 | 0.000805663 |
| NM_007115 | TNFAIP6 | 8.549476011 | 0.117397514 | 1.33E-06 |
| NM_003633 | ENC1 | 8.46863116 | 0.132486872 | 0.000143214 |
| NM_078487 | CDKN2B | 7.857379819 | 0.142359863 | 0.000247918 |
| NM_012219 | MRAS | 6.906686977 | 0.120416016 | 3.32E-05 |
| NM_000428 | LTBP2 | 6.530185903 | 0.17879509 | 0.00021206 |
| NM_002317 | LOX | 6.253405306 | 0.162652276 | 0.000493453 |
| NM_006475 | POSTN | 6.131065054 | 0.268041215 | 0.000381746 |
| NM_005512 | LRRC32 | 6.027300939 | 0.155609222 | 0.000244534 |
| NM_001013398 | IGFBP3 | 5.892165506 | 0.181799757 | 0.000340864 |
| NM_000660 | TGFB1 | 5.448264587 | 0.78251392 | 0.000170351 |
| NM_002667 | PLN | 5.105920007 | 0.214734252 | 0.000696765 |
| NM_014632 | MICAL2 | 4.972581483 | 0.17152412 | 0.000559198 |
| NM_016931 | NOX4 | 4.849811685 | 0.225379795 | 0.000265966 |
| NM_014631 | SH3PXD2A | 4.341337763 | 0.24444475 | 0.000103435 |
| NM_000393 | COL5A2 | 4.280097929 | 0.165513816 | 5.77E-05 |
| NM_003474 | ADAM12 | 4.189365259 | 0.274394503 | 0.000101548 |
| NM_014467 | SRPX2 | 4.147952958 | 0.257998723 | 4.06E-05 |
| NM_006851 | GLIPR1 | 4.092470638 | 0.201018413 | 4.62E-05 |
| NM_005613 | RGS4 | 4.074850693 | 0.251673169 | 4.92E-05 |
| NM_001018004 | TPM1 | 3.947191935 | 0.195719983 | 0.000839361 |
| NM_006329 | FBLN5 | 3.701593985 | 0.245752501 | 0.000232936 |
| BC014203 | FAM101B | 3.623415448 | 0.309548366 | 0.000792052 |
| NM_000399 | EGR2 | 3.490980445 | 0.261212761 | 0.00023454 |
| NM_001648 | KLK3 | 3.481810093 | 0.288453944 | 0.000272881 |
| NM_000096 | CP | 3.324909574 | 0.332527772 | 0.000485262 |
| NM_001898 | CST1 | 3.287001556 | 0.335459083 | 0.00068421 |
| NM_181847 | AMIGO2 | 3.249577889 | 0.261229048 | 0.000858199 |
| NM_138455 | CTHRC1 | 3.217756975 | 0.313332784 | 0.000253826 |
| NM_153026 | PRICKLE1 | 3.179655624 | 0.38858927 | 0.000640482 |
| NM_012293 | PXDN | 3.039816136 | 0.257492324 | 0.000777339 |
Table 1B. Most down-regulated genes in LNCaP-TGF-β1(a)/HPS19I co-cultures (microarray).
| RefSeq Nuc | Gene | TGF-β1+Ctrl vs. Ctrl+Ctrl | TGF-β1+SD208 vs.TGF-β1+Ctrl | p-value (ANOVA) |
|---|---|---|---|---|
| NM_001077 | UGT2B17 | 0.196197102 | 4.554852029 | 0.000695519 |
| NM_022074 | FAM111A | 0.33585709 | 2.325687499 | 0.000978972 |
| NM_015565 | ZNF294 | 0.356016518 | 2.631507725 | 0.000769972 |
| AK126851 | ANK3 | 0.413492187 | 3.394052268 | 9.89916E-05 |
| NM_014268 | MAPRE2 | 0.447337037 | 2.56792461 | 0.000371084 |
| NM_002467 | MYC | 0.484325292 | 2.653538875 | 0.000233651 |
| NM_003759 | SLC4A4 | 0.489633359 | 0.985494206 | 0.00077358 |
| NM_021190 | PTBP2 | 0.490358498 | 1.585425699 | 0.000671709 |
| AK075235 | SVEP1 | 0.521242168 | 1.139842928 | 5.82E-04 |
| NM_018689 | KIAA1199 | 0.550188967 | 1.452227923 | 6.60136E-05 |
| NM_000214 | JAG1 | 0.572787648 | 1.261023084 | 0.000639491 |
| NM_001634 | AMD1 | 0.633893667 | 1.61911846 | 9.31E-05 |
| NM_006358 | SLC25A17 | 0.651353495 | 1.882434831 | 0.000460944 |
| NM_002113 | CFHR1 | 0.672707916 | 1.258311297 | 7.55E-04 |
| NM_017686 | GDAP2 | 0.692205019 | 1.111019264 | 0.000774186 |
| NM_058191 | C21orf66 | 0.694078519 | 1.479905697 | 7.82E-05 |
| NM_004687 | MTMR4 | 0.697685088 | 1.504510216 | 0.000276199 |
In order to identify the prostate epithelia-specific gene that was regulated by prostate stromal TGF-β signaling, we also treated HPS19I cells alone using conditioned media collected from LNCaP-TGF-β1(a) cells or LNCaP-Ctrl cells cultured in RPMI1640 supplemented with 0.2% FBS. Conditional media, but not TGF-β1 ligand, were used here to more closely simulate the co-culture conditions. After 6 days of treatment, we extracted total RNA from these HPS19I cells and performed microarray. 2349 transcripts representing 1377 genes were differentially expressed among these two treatment groups (fold change>1.5, Figure 1g). The most significantly upregulated genes in HPS19I cells include ELN, EGR2, COMP, NOX4, CILP, CDKN2B, ENC1, AGT, PMEPA1, TNFAIP6, TIMP3 etc. (Table 2a), and most significantly downregulated genes include ADH1C, ADH1A, ALDH1A1, VCAM1, HSD17β2, SMPDL3A, ZFP36L2, SECTM1, ADAMTS5, NR2F1 etc. (Table 2b).
Table 2A. Most up-regulated genes in LNCaP-TGF-β1(a) cell conditioned media treated HPS19I cells (microarray).
| RefSeq Nuc | Gene | TGF-β1 CM vs. Ctrl CM |
|---|---|---|
| BC065566 | ELN | 16.67449355 |
| NM_000399 | EGR2 | 14.05299758 |
| NM_000095 | COMP | 12.98900057 |
| NM_016931 | NOX4 | 12.32605115 |
| NM_003613 | CILP | 9.059147489 |
| NM_078487 | CDKN2B | 8.803843969 |
| NM_003633 | ENC1 | 8.260572524 |
| NM_000029 | AGT | 7.191604093 |
| NM_020182 | PMEPA1 | 7.070540703 |
| NM_007115 | TNFAIP6 | 6.750100868 |
| X77690 | TIMP3 | 6.366782695 |
| NM_004385 | VCAN | 6.149750603 |
| NM_012219 | MRAS | 5.992620419 |
| NM_000501 | ELN | 5.807745301 |
| NM_006216 | SERPINE2 | 5.755856917 |
| NM_005725 | TSPAN2 | 5.661870275 |
| NM_005725 | TSPAN2 | 5.352542656 |
| NM_031866 | FZD8 | 5.312916891 |
| NM_002318 | LOXL2 | 5.290455686 |
| NM_020152 | C21orf7 | 5.266205737 |
| NM_002667 | PLN | 4.972344608 |
| NM_054034 | FN1 | 4.877547028 |
| NM_014631 | SH3PXD2A | 4.589092327 |
| NM_014399 | TSPAN13 | 4.524283014 |
| NM_003474 | ADAM12 | 4.349125589 |
| NM_182943 | PLOD2 | 4.30726254 |
| NM_014632 | MICAL2 | 4.05405353 |
| NM_001013398 | IGFBP3 | 4.052272457 |
| NM_006475 | POSTN | 4.052068264 |
| NM_000846 | GSTA2 | 4.041328015 |
| NM_000428 | LTBP2 | 4.023595863 |
| NM_002317 | LOX | 3.946875107 |
| NM_004750 | CRLF1 | 3.89952373 |
| NM_012261 | C20orf103 | 3.871335965 |
| NM_014840 | NUAK1 | 3.628631621 |
| NM_005613 | RGS4 | 3.583530694 |
| NM_001845 | COL4A1 | 3.569510901 |
| NM_138409 | MRAP2 | 3.500158747 |
Table 2B. Most down-regulated genes in LNCaP-TGF-β1(a) cell conditioned media treated HPS19I cells (microarray).
| RefSeq Nuc | Gene | TGF-β1 CM vs. Ctrl CM |
|---|---|---|
| NM_000669 | ADH1C | 0.048337642 |
| NM_000667 | ADH1A | 0.049204652 |
| NM_018689 | KIAA1199 | 0.055600352 |
| NM_000689 | ALDH1A1 | 0.090425225 |
| NM_001078 | VCAM1 | 0.100168911 |
| NM_002153 | HSD17B2 | 0.123017582 |
| NM_006714 | SMPDL3A | 0.124668133 |
| NM_006887 | ZFP36L2 | 0.1669457 |
| NM_003004 | SECTM1 | 0.181835508 |
| NM_007038 | ADAMTS5 | 0.184125165 |
| NM_007021 | C10orf10 | 0.184605811 |
| NM_005654 | NR2F1 | 0.206777199 |
| NM_004753 | DHRS3 | 0.207888106 |
| NM_014585 | SLC40A1 | 0.208805616 |
| NM_012342 | BAMBI | 0.21522776 |
| NM_000891 | KCNJ2 | 0.218105292 |
| NM_001031716 | OBFC2A | 0.222516674 |
| NM_019105 | TNXB | 0.22609769 |
| NM_020379 | MAN1C1 | 0.228067582 |
| NM_005329 | HAS3 | 0.228406025 |
| NM_001430 | EPAS1 | 0.232550842 |
| NM_030781 | COLEC12 | 0.239762468 |
| NM_005602 | CLDN11 | 0.239772493 |
| NM_005410 | SEPP1 | 0.245152972 |
| NM_006287 | TFPI | 0.246145446 |
| NM_018043 | ANO1 | 0.247547036 |
| NM_000076 | CDKN1C | 0.247573166 |
| NM_001083 | PDE5A | 0.248350261 |
| NM_018043 | ANO1 | 0.250724637 |
| NM_198194 | STOM | 0.251440257 |
| NM_002825 | PTN | 0.255859758 |
| NM_153225 | C8orf84 | 0.256217479 |
| NM_018487 | TMEM176A | 0.258119323 |
| NM_001146 | ANGPT1 | 0.25867152 |
| NM_000599 | IGFBP5 | 0.261560301 |
| NM_014020 | TMEM176B | 0.262431295 |
| NM_175861 | TMTC1 | 0.270304699 |
| NM_006988 | ADAMTS1 | 0.275587107 |
We compared the obtained TGF-β regulated gene lists in the LNCaP/HPS19I co-cultures and those in the HSP19I cells cultured alone. As expected, most of the TGF-β regulated genes in the LNCaP/HPS19I co-cultures were also presented in the TGF-β treated HSP19I cells, indicating that these genes might also be TGF-β regulated genes in HPS19I cells of these co-cultures. Again, PSA (KLK3), a prostate epithelial cell specific and AR regulated gene, was induced in the LNCaP-TGF-β1 (a) / HPS19I cell co-cultures, but not expressed in HPS19I cells when cultured alone. Furthermore, SD-208 treatment blocked this induction (Table 1a).
Prostate stromal TGF-β signaling induces AR activation in PCa cells
Since PSA is a well-established AR target gene in human PCa, our above observation suggested that stroma-specific TGF-β signaling might induce AR activation in PCa cells. To test this, we performed qPCR on the expression of additional well-established PCa epithelia-specific AR target genes including KLK4 and TMPRSS2 in these LNCaP/HPS19I co-cultures. TGF-β induced the expression of all these three AR target genes in the LNCaP-TGF-β1(a)/HPS19I co-cultures, but not in LNCaP cells (data not shown) or HPS19I cells cultured alone (Figure 2a). Furthermore, these inductions were blocked by addition of 400 nM of SD-208, further confirming that TβRI (ALK5) protein kinase activity in the HPS19I cells was required for TGF-β ligand induced AR activation in LNCaP cells (Figure 2a).
Figure 2. Prostate stromal TGF-β signaling induces AR activation in the co-cultured LNCaP cells.
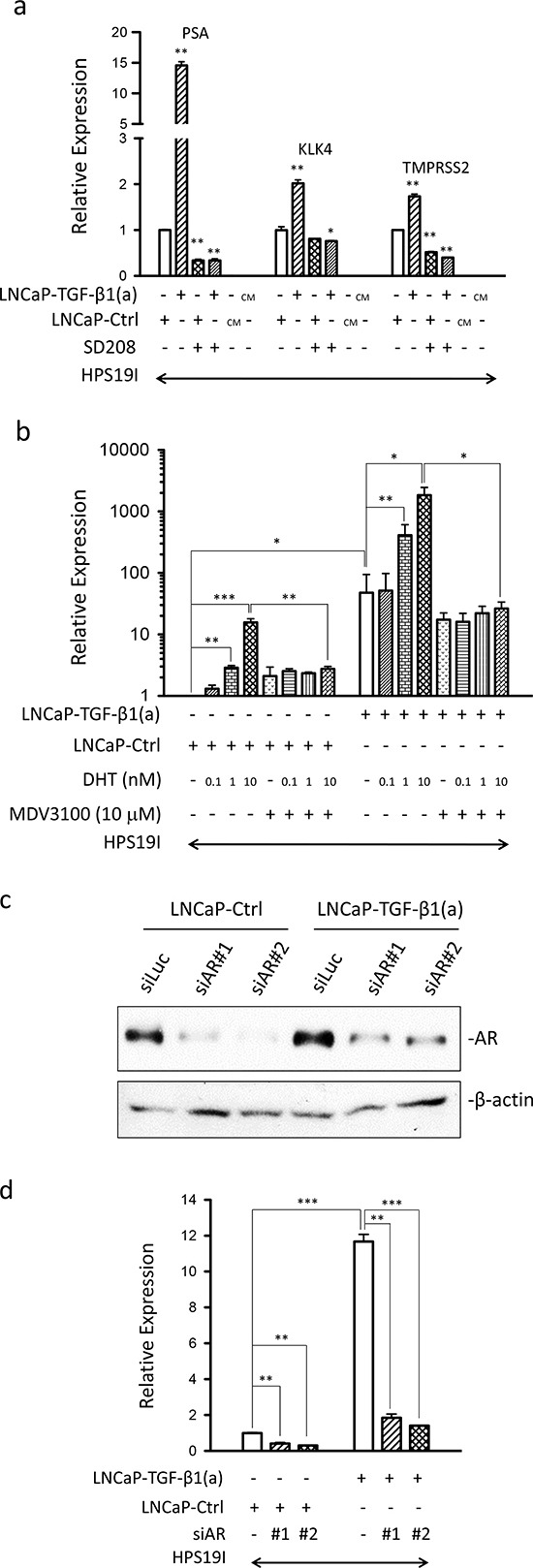
(a) qPCR were used to analyze the expression of PSA, KLK4, and TMPRSS2 in the differentially treated LNCaP-TGF-β1(a)/HPS19I, LNCaP-Ctrl/HPS19I, and HPS19I cells as described in Figure 1f & g. CM: conditioned media. Representative data is from two independent experiments. Gene expression data were normalized to GAPDH expression. *p<0.05, **p<0.01 (unpaired Student's t-test). (b) LNCaP-TGF- β1(a) cells and LNCaP-Ctrl cells were co-cultured with HPS19I cells in RPMI1640 supplemented with 0.2% charcoal-stripped FBS, and treated with different dosages of DHT in the presence (+) or absence (−) of 10 μM of MDV3100 for 6 days. Total RNA was extracted and reverse transcribed. qPCR were used to analyze the expression of PSA, KLK4, and TMPRSS2 in these co-cultures. Data is from three independent experiments. Gene expression data were normalized to GAPDH expression. *p<0.05, **p<0.01, ***p<0.001 (paired Student's t-test). (c, d) LNCaP-TGF-β1(a) cells and LNCaP-Ctrl cells were transfected with two independent siRNAs against human AR (siAR#1 and siAR#2), along with siRNA against luciferase (siLuc) as control. On the second day, the cells were trypsinized and collected. (c) Part of the cells were used for continuing culturing in regular media for 4 days, after which cell lysates were prepared and subjected to Western blot for AR and β-actin. (d) Part of the cells were used for co-cultures with HPS19I cells in RPMI1640 supplemented with 0.2% charcoal-stripped FBS for 4 days, after which total RNA was extracted and reverse transcribed. qPCR were used to analyze the expression of PSA in these co-cultures. Representative data is from two independent experiments. Gene expression data were normalized to GAPDH expression. **p<0.01, ***p<0.001 (unpaired Student's t-test).
To further reduce androgen levels, we also performed LNCaP-TGF-β1(a)/HPS19I co-cultures and LNCaP-Ctrl/HPS19I co-cultures in RPMI1640 media supplemented with 0.2% charcoal stripped FBS deprived of androgen, and treated them with increasing dosages of DHT and/or 10 μM of MDV3100, a second-generation antiandrogen [36], for six days. We similarly extracted total RNA directly from these co-cultures, and performed reverse transcription reactions and qPCR analysis for PSA expression. As expected, DHT induced PSA expression in the control LNCaP-Ctrl/HPS19I co-cultures in a dose-dependent manner. We also unexpectedly observed that MDV3100 treatment somehow modestly induced PSA expression in these LNCaP-Ctrl/HPS19I co-cultures maintained in 0.2% charcoal stripped FBS, whereas the AR antagonist activity of MDV3100 was confirmed by its strong inhibition of 10 nM DHT induced PSA expression in these LNCaP-Ctrl/HPS19I co-cultures (Figure 2b). Consistent with the data obtained in LNCaP/HPS19I co-cultures in 0.2% FBS (Figure 2a), PSA expression in the LNCaP-TGF-β1(a)/HPS19I co-cultures in 0.2% charcoal-stripped FBS was also greatly induced, to levels comparable to 10nM DHT induced PSA expression in LNCaP-Ctrl/HPS19I co-cultures. These observations further confirmed prostate stroma-specific TGF-β signaling induction of AR activation in the absence of significant amount of androgen. Addition of DHT dose-dependently promoted PSA expression in these LNCaP-TGF-β1(a)/HPS19I co-cultures, with robust induction observed in 1 nM and 10 nM of DHT treatments, indicating strong cooperative or additive actions between the stromal TGF-β signaling induced AR activation and the DHT ligand-induced AR activation. Consistent with the AR antagonist activity, 10 μM of MDV3100 robustly blocked 1nM and 10 nM DHT-induced PSA expression in the LNCaP-TGF-β1(a)/HPS19I co-cultures. In contrast, MDV3100 treatment only modestly reduced stromal TGF-β signaling induced PSA expression in the absence of externally added DHT ligand, indicating that a large fraction of stromal TGF-β signaling induced AR activation is ligand-independent (Figure 2b). Finally, LNCaP-TGF-β1(a) cells and LNCaP-Ctrl cells expressed comparable levels of basal and DHT-induced PSA (Supplementary Figure 2), further confirming that stroma-specific TGF-β signaling is essential for inducing AR activation in LNCaP cells in these co-cultures.
To confirm that our observed stromal TGF-β signaling induced PSA expression is AR dependent, we performed knockdown of AR in LNCaP-Ctrl and LNCaP-TGF-β1(a) cells using two independent siRNAs against human AR. Western blots confirmed significant knockdown of AR in both cell lines (Figure 2c). These AR-knockdown LNCaP cells were used for co-cultures with HPS19I cells in RPMI1640 media supplemented with 0.2% charcoal stripped FBS for four days. We extracted total RNA directly from these co-cultures, and performed reverse transcription reactions and qPCR analysis for PSA expression. Indeed, knockdown of AR in LNCaP cells greatly inhibited PSA expression in these co-cultures, confirming that stromal TGF-β signaling induced PSA expression is dependent on AR (Figure 2d).
To further expand our observation, we also performed co-cultures of LNCaP cells with HTS33B cells and HPS33Q cells, two additional human prostate stromal cell lines that we prepared following a previously described protocol [5, 38]. These LNCaP/HTS33B co-cultures and LNCaP/HPS33Q co-cultures were treated with 50 pM of TGF-β1 or vehicle control in RMPI1640 supplemented with 0.2% charcoal stripped FBS for six days. Total RNA was similarly directly extracted from these co-cultures, reverse transcribed, and used for qPCR analysis for gene expression. We showed that TGF-β treatment induced the expression of PSA, KLK4, and TMPRSS2 in all these co-cultures, further confirming that prostate stromal cell-specific TGF-β signaling induces AR activation in LNCaP cells (Figure 3b, c).
Figure 3. IL-6 and BMP-6 induces AR activation in LNCaP cells.
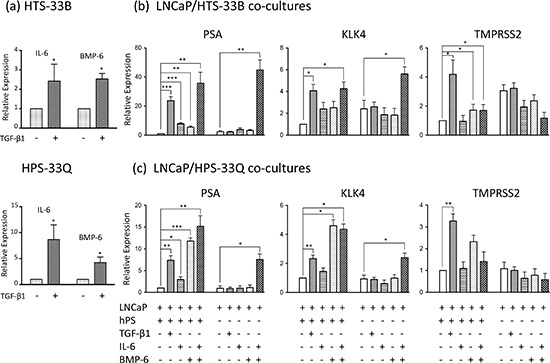
(a) HTS33B and HPS33Q human prostate stromal cells were cultured in RPMI1640 supplemented with 0.2% charcoal-stripped FBS and treated with TGF-β1 (50 pM) for 3 days. Total RNA was extracted and reverse transcribed. qPCR were used to analyze the expression of IL-6 and BMP-6. Data is from three independent experiments. (b) LNCaP cells were co-cultured with HTS33B cells in RPMI1640 supplemented with 0.2% charcoal-stripped FBS, and treated with TGF-β1 (50 pM), IL-6 (25 ng/ml), and/or BMP-6 (50 ng/ml) for 6 days. Total RNA was extracted and reverse transcribed. qPCR were used to analyze the expression of PSA, KLK4, TMPRSS2 expresssion in these co-cultures. Data is from three independent experiments. (c) LNCaP cells were similarly co-cultured with HPS33Q cells and treated with TGF-β1 (50 pM), IL-6 (25 ng/ml), and/or BMP-6 (50 ng/ml) for 6 days, and analyzed for the expression of PSA, KLK4, TMPRSS2 (qPCR). Data is from three independent experiments. All gene expression data were normalized to GAPDH expression. hPS (human prostate stromal cells) stands for either HTS33B (b) or HPS33Q cells (c). *p<0.05, **p<0.01, ***p<0.001 (paired Student's t-test).
IL-6 and BMP-6 activity partially mediates prostate stromal TGF-β signaling induced AR activation in LNCaP cells
IL-6 is a key cytokine regulating PCa biology, including inducing AR activity and promoting androgen-independent growth [22–25]. BMP-6 was recently reported to promote castration-resistant PCa [34, 35]. We found that TGF-β induces the expression of both IL-6 and BMP-6 in human prostate stromal cells cultured in 0.2% charcoal stripped FBS (Figure 3a). To examine whether IL-6 and/or BMP-6 mediate the stromal TGF-β signaling induced AR activation in LNCaP cells, we first treated LNCaP/HTS33B co-cultures and LNCaP/HPS33Q co-cultures in RMPI1640 supplemented with 0.2% charcoal stripped FBS with 25 ng/ml of IL-6 and/or 50 ng/ml of BMP-6 for six days. While IL-6 treatment and BMP-6 treatment each can induce PSA expression in all these co-cultures, co-treatments of both IL-6 and BMP-6 robustly induced PSA expression to levels exceeding those induced by TGF-β treatment. These results indicate that IL-6 and BMP-6 together strongly induces AR activation in LNCaP cells and that this action may greatly contribute to stromal TGF-β signaling induced AR activation in LNCaP cells.
We next examined whether the presence of prostate stromal cells is required for the IL-6/BMP-6 induced AR activation in LNCaP cells. In contrast to our above observations in the co-culture system, neither IL-6 treatment alone nor BMP-6 treatment alone can greatly enhance PSA expression in LNCaP cells cultured alone in RPMI 1640 media supplemented with 0.2% charcoal stripped FBS. However, co-treatments of both IL-6 and BMP-6 induced robust expression of PSA in the LNCaP-only cell cultures, further confirming the cooperative activities of IL-6 and BMP-6 in inducing AR activation in LNCaP cells (Figure 3b, c). We made similar observations in KLK4 expression, but not in TMPRSS2 expression, in these IL-6 and BMP-6 treated LNCaP/stromal co-cultures as well as in LNCaP cells cultured alone (Figure 3b, c). These data suggest that although IL-6 and BMP-6 may greatly contribute to stromal TGF-β signaling induced AR activities including inducing PSA and KLK4 expression, additional signaling molecules/pathways may be required for mediating stromal TGF-β signaling induced other AR activities such as inducing TMPRSS2 expression.
Finally, to further evaluate the functional roles of IL-6 and BMP-6 in mediating stromal TGF-β signaling induced AR activation in LNCaP cells, we treated the LNCaP/HTS33B and LNCaP/HPS33Q co-cultures with IL-6 and BMP-6 neutralizing antibodies. Use of IL-6 neutralizing antibody produced an apparent reduction in stromal TGF-β signaling induced AR activation in LNCaP cells, whereas BMP-6 neutralizing antibody attenuated this induction to a less extent (Figure 4a, b). Altogether, these data suggest that IL-6 and BMP-6 mediate prostate stromal TGF-β signaling induced AR activation.
Figure 4. IL-6 and BMP-6 neutralizing antibody treatment attenuates stromal TGF-β induced AR activation in the co-cultured LNCaP cells.
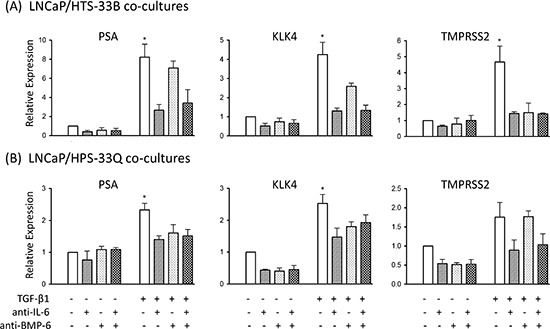
(a) LNCaP cells were cultured alone or co-cultured with HTS33B cells in RPMI1640 supplemented with 0.2% charcoal-stripped FBS, and treated with TGF-β1 (50 pM) along with neutralizing antibody against IL-6 and/or BMP-6 (both at 2 μg/ml) for 6 days. Total RNA was extracted and reverse transcribed. qPCR were used to analyze the expression of PSA, KLK4, TMPRSS2 expresssion in these co-cultures. Data is from two independent experiments. (b) LNCaP cells were similarly cultured alone or co-cultured with HPS33Q cells and treated with TGF-β1 (50 pM) along with neutralizing antibody against IL-6 and/or BMP-6 (both at 2 μg/ml) for 6 days, and analyzed for the expression of PSA, KLK4, TMPRSS2 (qPCR). Data is from two independent experiments. All gene expression data were normalized to GAPDH expression. *p<0.05 (unpaired Student's t-test).
DISCUSSION
Tumor microenvironment is important for prostate cancer progression. TGF-β is a key cytokine in regulating cancer progression, either by directly regulating tumor cells, such as inducing epithelial to mesenchymal transition (EMT), or indirectly by regulating tumor microenvironment, such as inducing extracellular matrix remodeling, regulating angiogenesis as well as immune responses. We showed here that TGF-β could indirectly induce AR activation in PCa cells through directly modulating stromal cells in tumor microenvironment. Since AR re-activation is requisite for the recurrence of most CRPC tumors, this stromal TGF-β signaling induced AR activation in PCa cells may provide a direct mechanism for CRPC. Importantly, stromal TGF-β signaling greatly potentiated and enhanced DHT-induced AR activation, suggesting that it might be a therapeutic target to inhibit AR activation in CRPC tumors in the presence of castration-levels of androgen.
Our current data suggest that IL-6 and BMP-6 might partially mediate stromal TGF-β signaling induced AR activation. Interestingly, although neither IL-6 nor BMP-6 could robustly induce AR activation in LNCaP cells when cultured alone, IL-6 and BMP-6 together greatly enhance AR activity in these cells. Notably, when prostate stromal cells are presented in the LNCaP/stromal cell co-cultures, IL-6 and BMP-6 can each significantly enhance AR activation in LNCaP cells, which may attribute to the basal activation of IL-6 and BMP-6 signaling pathway in these co-cultures. As the expression of IL-6 and BMP-6 are elevated in the tumor microenvironment, these may be important factors in the evolution of castration resistant disease.
Consistent with many previous reports, MDV3100 showed robust AR antagonist activities in most of our LNCaP/HPS19I co-cultures studies. However, MDV3100 also exhibited slight apparent AR agonist activity under certain experimental conditions, such as in LNCaP/HPS19I cell co-cultured in 0.2% of charcoal-stripped FBS as described in Figure 2. The nature and significance of this unexpected slight AR agonist activity of MDV3100 is still to be determined. Finally, stromal TGF-β signaling induced AR activation in LNCaP cells can only be partially attenuated by treatment of MDV3100 (Figure 2), indicating both ligand-dependent and ligand-independent AR activation. Interestingly, our microarray study and qPCR analysis revealed that stromal TGF-β signaling strongly repressed expression of UGT2B17 (Table 1b), as well as UGT2B15 (data not shown), two key epithelium-specific UDP-glucuronosyltransferases for glucuronidation and inactivation of testosterone and DHT ligands for their efficient clearance from normal prostate epithelial cells as well as PCa cells [42, 43]. These suggest a possibility that repression of UGT2B15 and UGT2B17 in PCa cells may contribute to stromal TGF-β signaling induced ligand-dependent AR activation in PCa cells, which warrants future studies on the functional roles of these two UDP-glucuronosyltransferases in regulating prostate stroma induced AR activation in PCa cells.
MATERIALS AND METHODS
Reagents
The Porcine TGF-β1(204-B-002), recombinant human BMP-6 (507-BP), recombinant human IL-6 (206-IL-010), antibodies against human BMP-6 (AF-507), human IL-6 (AF-206-NA), and normal goat IgG (AB-108-C) were purchased from R&D System (Minneapolis, MN, USA). MVD3100 was purchased from Selleckchem (Houston, TX, USA). SD-208 was obtained from Scios, Inc. (now Alza Corporation Vacaville, CA, USA; part of Johnson and Johnson).
Cell cultures
LNCaP cells were purchased from ATCC and cultured in RPMI1640 supplemented with 10% FBS (Hyclone, Logan, UT or Invitrogen, Carlsbad, CA, USA). LNCaP-TGF-β1(a) cells overexpressing an HA-tagged constitutively activated TGF-β1 ligand, HA-TGF-β1(a), and control LNCaP (LNCaP-Ctrl) cells were generated as previously described [37]. Human prostate stromal cell lines HPS19I, HTS33B, and HPS33Q were prepared following a previously described protocol and cultured in Bfs medium: DMEM supplemented with 5% FBS (Hyclone or Invitrogen), 5% Nu serum (Collaborative Research, Bedford, MA), 0.5 μg/mL testosterone, 5 μg/mL insulin, 100 units/mL penicillin, and 100 μg/mL streptomycin (Sigma, St. Louis, MO, USA) [5, 38].
Human prostate cancer cell and prostate stromal cell co-cultures
HPS19I, HTS33B, and HPS33Q cells were first allowed to grow in Bfs media to confluence on 6 well plates. LNCaP cells were then plated on top of the stromal cells at 1×105 cells per well and subjected to various treatments in RPMI1640 supplemented with 0.2% FBS or charcoal stripped FBS for 6 to 28 days. The treatments include 50 pM (1.25ng/ml) porcine TGF-β1 (R&D Systems), 400 nM of SD-208, 0.1, 1, and 10 nM of DHT (Sigma), 10 μM of MDV3100 (Selleckchem, Houston, TX, USA), 25 ng/ml of IL-6, 50 ng/ml of BMP-6, 2 μg/ml of IL-6 neutralizing antibody, 2 μg/ml of IL-6 neutralizing antibody (R&D system), and various vehicle controls. When applicable, LNCaP cells and prostate stromal cells were each similarly cultured and treated as those for the LNCaP/Stroma co-cultures. Total RNA was extracted from these co-cultures using the RNeasy Miniprep kit (Qiagen, Valencia, CA, USA) or TRIzol® reagent (Invitrogen).
Knockdown of AR in LNCaP cells
Two siRNAs from Invitrogen, AR/HSS179973 (siRNA#1) and AR/HSS179972 (siRNA#2), were used to knock down AR expression in LNCaP-TGF-β1(a) cells and LNCaP-Ctrl cells. The corresponding sequences are CCGGAAGCUGAAGAAACUUGGUAAU (sense), AUUACCAAGUUUCUUCAGCUUCCGG (antisence) for AR/HSS179973, and GAUGAAGCUUCUGGGUGUCACUAUG (sense), CAUAGUGACACCCAGAAGCUUCAUC (antisense) for AR/HSS179972. siRNAs were transfected into LNCaP cells using GenMute™ siRNA Transfection Reagent following manufacture's protocol (SignaGen, Rockville, MD, USA). Western blots were used to verify knockdown of AR. Antibodies used are anti-AR (sc-816, Santa Cruz Biotechnology, Dallas, TX, USA) and anti-β-actin (A1978, Sigma).
Luciferase reporter assay
HPS19I cells were transfected with 0.4 μg of (CAGA)12MLP [41] and 0.1 μg of pRL-null (Promega, Madison, WI, USA) in 12 well plate using FuGENE 6 transfection reagent (Roche Applied Sciences, Penzberg, Germany) as described before [18]. 24 hours later, cells were treated with 50 pM TGF-β1 and different dosages of SD-208 compound for 24 hours. Cell lysates were prepared and measured for luciferase activity using the Dual-Luciferase® Reporter Assay system (Promega). The reporter firefly luciferase activity was normalized to the co-transfected pRL-null Renilla luciferase activity.
cDNA microarray
HPS19I cells were grown to mostly confluence in T75 flasks. 5×105 of LNCaP-TGF-β1(a) cells or LNCaP-Ctrl cells were then plated on top of the HPS19I cells. The co-cultures were maintained in RPMI1640 supplemented with 0.2% FBS and subjected to treatments with 400 nM of SD-208 or vehicle control for 6 days. Conditioned media from LNCaP-TGF-β1(a) cells and LNCaP-Ctrl cells maintained in RPMI1640 supplemented with 0.2% FBS were also collected and used to treat HPS19I cells for 6 days. Total RNA was extracted from these LNCaP-TGF-β1(a)/HPS19I and LNCaP-Ctrl/HPS19I co-cultures and from the differentially treated HPS19I cells using the RNeasy Miniprep kit (Qiagen). The cDNA reverse transcription and fluorescent labeling reactions were carried out using the SuperScript® Plus Direct cDNA Labeling System with Alexa Fluor® aha-dUTPs kit (Invitrogen). Briefly 3 μg of RNA and 3 μg of Universal Human Reference RNA (Stratagene, La Jolla, CA) were labeled in reverse transcription with Alexa647 and Alexa555 aha-dUTPs respectively for each sample. After labeling, each cDNA was mixed with reference cDNA and purified with SuperScript Plus Direct cDNA Labeling System purification module according to manufacturer's instructions. Eluted sample was mixed with 10xBlocking Solution and 2xHiRPM buffer (Agilent Technologies, Santa Clara, CA, USA), incubated for 2 min at 95–98° C and hybridized on 4×44K Whole Human Genome Oligo Microarray chip using SureHyb DNA Microarray Hybridization Chambers in DNA Microarray hybridization oven (Agilent Technologies) at 10 rpm, 65°C for 20 hrs. After hybridization, slides were washed in Gene Expression Wash Buffer 1 and 2 for 1 min and than dried by centrifugation at 2000 rpm for 2 min. Microarrays were scanned with a dynamic autofocus microarray scanner (Agilent Microarray Scanner- G2565BA, Agilent Technologies,) using Agilent-provided parameters (Red and Green PMT were each set at 100%, and scan resolution was set to 5 um). The Feature Extraction Software v9.1.3.1 (Agilent Technologies) was used to extract and analyze the signals.
The microarray data were processed using bioconductor (loess normalization), with Combat to adjust for batch effects [44]. Fold changes and ANOVA tests for each gene were computed using log-transformed values. Array data have been deposited into the Gene Expression Omnibus (GSE51624).
Quantitative PCR
1 μg total RNA was reverse transcribed with random hexamer primers using the SuperScript III First Strand Synthesis kit (Invitrogen). Reversed transcribed product was then analyzed by real-time PCR using Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) on the ABI Prism PE7700 sequence analyzer (Applied Biosystems, Foster City, CA, USA) or using the PerfeCTa CYBR Green FastMix (Quanta Biosciences, Gaithersburg, MD, USA) on the Light Cycler® 480II (Roche Applied Science). PCR primers used are PSA: 5′ ACCAGAGGAGTTCTTGACCCCAAA 3′ and 5′ CCCCAGAATCACCCGAGCAG 3′, KLK4: 5′ GGCACTGGTCATGGAAAACGA 3′ and 5′ TCAAGACTGTGCAGGCCCAGCC 3′ (Annealing temperature 56), TMPRSS2: 5′ CGCTGGCCTACTCTGGAA 3′ and 5′ CTGAGGAGTCGCACTCTATCC 3′, GAPDH: 5′ AGCCACATCGCTCAGACAC 3′ and 5′ GCCCAATACGACCAAATCC 3′, IL-6: 5′ CAAAGATGGCTGAAAAAGATGGA 3′ and 5′ CTGTTCTGGAGGTACTCTAGGT 3′, and BMP-6: 5′ GGACAGCGCCTTCCTCAAC 3′ and 5′ CGTACTCCACCAGGTTCACAAA 3′. Relative expression levels were determined by the ΔΔCt method, and normalized to GAPDH.
Statistics
The statistical analyses on microarray data were described above. Statistical analyses (Student's t-tests) for comparing gene expression were generated using Graphpad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). P < 0.05 was considered statistically significant.
SUPPLEMENTARY FIGURES
Acknowledgments
This work was supported by NIH grants R01-CA058093, R01-DK045909, U01-CA84296 (DRR), P30 CA125123 (CJC), Department of Defense grants W81XWH-04-1-0189 (DRR), W81XWH-07-1-0200, W81XWH-09-1-0651, W81XWH-13-1-0163 (FY), and Cancer Prevention Research Institute of Texas (CPRIT) grant RP130651 (FY). SD-208 compound was provided by Scios, Inc. (now Alza Corporation, Vacaville, CA, USA; part of Johnson and Johnson).
Footnotes
Conflict of interests
The authors declare no conflict of interest.
REFERENCES
- 1.Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–238. doi: 10.1016/j.pharmthera.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Barron DA, Rowley DR. The reactive stroma microenvironment and prostate cancer progression. Endocr Relat Cancer. 2012;19:R187–204. doi: 10.1530/ERC-12-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166:2472–2483. [PubMed] [Google Scholar]
- 4.Rowley DR. What might a stromal response mean to prostate cancer progression? Cancer Metastasis Rev. 1999;17:411–419. doi: 10.1023/a:1006129420005. [DOI] [PubMed] [Google Scholar]
- 5.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8:2912–2923. [PubMed] [Google Scholar]
- 6.Coffey RJ, Jr, Shipley GD, Moses HL. Production of transforming growth factors by human colon cancer lines. Cancer Res. 1986;46:1164–1169. [PubMed] [Google Scholar]
- 7.Barrett-Lee P, Travers M, Luqmani Y, Coombes RC. Transcripts for transforming growth factors in human breast cancer: clinical correlates. Br J Cancer. 1990;61:612–617. doi: 10.1038/bjc.1990.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastham JA, Truong LD, Rogers E, Kattan M, Flanders KC, Scardino PT, Thompson TC. Transforming growth factor-beta 1: comparative immunohistochemical localization in human primary and metastatic prostate cancer. Lab Invest. 1995;73:628–635. [PubMed] [Google Scholar]
- 9.Gerdes MJ, Larsen M, McBride L, Dang TD, Lu B, Rowley DR. Localization of transforming growth factor-beta1 and type II receptor in developing normal human prostate and carcinoma tissues. J Histochem Cytochem. 1998;46:379–388. doi: 10.1177/002215549804600312. [DOI] [PubMed] [Google Scholar]
- 10.Kim IY, Ahn HJ, Zelner DJ, Shaw JW, Lang S, Kato M, Oefelein MG, Miyazono K, Nemeth JA, Kozlowski JM, Lee C. Loss of expression of transforming growth factor beta type I and type II receptors correlates with tumor grade in human prostate cancer tissues. Clin Cancer Res. 1996;2:1255–1261. [PubMed] [Google Scholar]
- 11.Williams RH, Stapleton AM, Yang G, Truong LD, Rogers E, Timme TL, Wheeler TM, Scardino PT, Thompson TC. Reduced levels of transforming growthfactor beta receptor type II in human prostate cancer: an immunohistochemical study. Clin Cancer Res. 1996;2:635–640. [PubMed] [Google Scholar]
- 12.Guo Y, Jacobs SC, Kyprianou N. Down-regulation of protein and mRNA expression for transforming growth factor-beta (TGF-beta1) type I and type II receptors in human prostate cancer. Int J Cancer. 1997;71:573–579. doi: 10.1002/(sici)1097-0215(19970516)71:4<573::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 13.Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122:103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, Fauci AS. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci U S A. 1986;83:4167–4171. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuxhorn JA, McAlhany SJ, Dang TD, Ayala GE, Rowley DR. Stromal cells promote angiogenesis and growth of human prostate tumors in a differential reactive stroma (DRS) xenograft model. Cancer Res. 2002;62:3298–3307. [PubMed] [Google Scholar]
- 16.Tuxhorn JA, McAlhany SJ, Yang F, Dang TD, Rowley DR. Inhibition of TGF-b activity decreases angiogenesis in a human prostate cancer reactive stroma xenograft model. Cancer Res. 2002;62:6021–6025. [PubMed] [Google Scholar]
- 17.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Strand DW, Rowley DR. Fibroblast growth factor-2 mediates transforming growth factor-beta action in prostate cancer reactive stroma. Oncogene. 2008;27:450–459. doi: 10.1038/sj.onc.1210663. [DOI] [PubMed] [Google Scholar]
- 19.Culig Z, Puhr M. Interleukin-6: a multifunctional targetable cytokine in human prostate cancer. Mol Cell Endocrinol. 2012;360:52–58. doi: 10.1016/j.mce.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima J, Tachibana M, Horiguchi Y, Oya M, Ohigashi T, Asakura H, Murai M. Serum interleukin 6 as a prognostic factor in patients with prostate cancer. Clin Cancer Res. 2000;6:2702–2706. [PubMed] [Google Scholar]
- 21.Shariat SF, Andrews B, Kattan MW, Kim J, Wheeler TM, Slawin KM. Plasma levels of interleukin-6 and its soluble receptor are associated with prostate cancer progression and metastasis. Urology. 2001;58:1008–1015. doi: 10.1016/s0090-4295(01)01405-4. [DOI] [PubMed] [Google Scholar]
- 22.Hobisch A, Eder IE, Putz T, Horninger W, Bartsch G, Klocker H, Culig Z. Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor. Cancer Res. 1998;58:4640–4645. [PubMed] [Google Scholar]
- 23.Chen T, Wang LH, Farrar WL. Interleukin 6 activates androgen receptor-mediated gene expression through a signal transducer and activator of transcription 3-dependent pathway in LNCaP prostate cancer cells. Cancer Res. 2000;60:2132–2135. [PubMed] [Google Scholar]
- 24.Lin DL, Whitney MC, Yao Z, Keller ET. Interleukin-6 induces androgen responsiveness in prostate cancer cells through up-regulation of androgen receptor expression. Clin Cancer Res. 2001;7:1773–1781. [PubMed] [Google Scholar]
- 25.Lee SO, Lou W, Hou M, de Miguel F, Gerber L, Gao AC. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin Cancer Res. 2003;9:370–376. [PubMed] [Google Scholar]
- 26.Lou W, Ni Z, Dyer K, Tweardy DJ, Gao AC. Interleukin-6 induces prostate cancer cell growth accompanied by activation of stat3 signaling pathway. Prostate. 2000;42:239–242. doi: 10.1002/(sici)1097-0045(20000215)42:3<239::aid-pros10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Giri D, Ozen M, Ittmann M. Interleukin-6 is an autocrine growth factor in human prostate cancer. Am J Pathol. 2001;159:2159–2165. doi: 10.1016/S0002-9440(10)63067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malinowska K, Neuwirt H, Cavarretta IT, Bektic J, Steiner H, Dietrich H, Moser PL, Fuchs D, Hobisch A, Culig Z. Interleukin-6 stimulation of growth of prostate cancer in vitro and in vivo through activation of the androgen receptor. Endocr Relat Cancer. 2009;16:155–169. doi: 10.1677/ERC-08-0174. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Zhu Y, Lou W, Cui Y, Evans CP, Gao AC. Inhibition of constitutively active Stat3 reverses enzalutamide resistance in LNCaP derivative prostate cancer cells. Prostate. 2014;74:201–209. doi: 10.1002/pros.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler HL, McCurdy MA, Kattan MW, Timme TL, Scardino PT, Thompson TC. Elevated levels of circulating interleukin-6 and transforming growth factor-beta1 in patients with metastatic prostatic carcinoma. J Urol. 1999;161:182–187. [PubMed] [Google Scholar]
- 31.Hamdy FC, Autzen P, Robinson MC, Horne CH, Neal DE, Robson CN. Immunolocalization and messenger RNA expression of bone morphogenetic protein-6 in human benign and malignant prostatic tissue. Cancer Res. 1997;57:4427–4431. [PubMed] [Google Scholar]
- 32.Dai J, Keller J, Zhang J, Lu Y, Yao Z, Keller ET. Bone morphogenetic protein-6 promotes osteoblastic prostate cancer bone metastases through a dual mechanism. Cancer Res. 2005;65:8274–8285. doi: 10.1158/0008-5472.CAN-05-1891. [DOI] [PubMed] [Google Scholar]
- 33.Darby S, Cross SS, Brown NJ, Hamdy FC, Robson CN. BMP-6 over-expression in prostate cancer is associated with increased Id-1 protein and a more invasive phenotype. J Pathol. 2008;214:394–404. doi: 10.1002/path.2292. [DOI] [PubMed] [Google Scholar]
- 34.Lee GT, Jung YS, Ha YS, Kim JH, Kim WJ, Kim IY. Bone morphogenetic protein-6 induces castration resistance in prostate cancer cells through tumor infiltrating macrophages. Cancer Sci. 2013;104:1027–1032. doi: 10.1111/cas.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee GT, Kang DI, Ha YS, Jung YS, Chung J, Min K, Kim TH, Moon KH, Chung JM, Lee DH, Kim WJ, Kim IY. Prostate cancer bone metastases acquire resistance to androgen deprivation via WNT5A-mediated BMP-6 induction. Br J Cancer. 2014;110:1634–1644. doi: 10.1038/bjc.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barron DA, Strand DW, Ressler SJ, Dang TD, Hayward SW, Yang F, Ayala GE, Ittmann M, Rowley DR. TGF-beta1 induces an age-dependent inflammation of nerve ganglia and fibroplasia in the prostate gland stroma of a novel transgenic mouse. PLoS One. 5:e13751. doi: 10.1371/journal.pone.0013751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schauer IG, Ressler SJ, Tuxhorn JA, Dang TD, Rowley DR. Elevated Epithelial Expression of Interleukin-8 Correlates with Myofibroblast Reactive Stroma in Benign Prostatic Hyperplasia. Urology. 2008;72:205–213. doi: 10.1016/j.urology.2007.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim IY, Ahn HJ, Zelner DJ, Shaw JW, Sensibar JA, Kim JH, Kato M, Lee C. Genetic change in transforming growth factor beta (TGF-beta) receptor type I gene correlates with insensitivity to TGF-beta 1 in human prostate cancer cells. Cancer Res. 1996;56:44–48. [PubMed] [Google Scholar]
- 40.Uhl M, Aulwurm S, Wischhusen J, Weiler M, Ma JY, Almirez R, Mangadu R, Liu YW, Platten M, Herrlinger U, Murphy A, Wong DH, Wick W, et al. SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 2004;64:7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- 41.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. Embo J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab. 2003;14:473–479. doi: 10.1016/j.tem.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Chouinard S, Barbier O, Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J Biol Chem. 2007;282:33466–33474. doi: 10.1074/jbc.M703370200. [DOI] [PubMed] [Google Scholar]
- 44.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


