Abstract
Background
To date, no genome-wide association study (GWAS) has considered the combined phenotype of asthma with hay fever. Previous analyses of family data from the Tasmanian Longitudinal Health Study provide evidence that this phenotype has a stronger genetic cause than asthma without hay fever.
Objective
We sought to perform a GWAS of asthma with hay fever to identify variants associated with having both diseases.
Methods
We performed a meta-analysis of GWASs comparing persons with both physician-diagnosed asthma and hay fever (n = 6,685) with persons with neither disease (n = 14,091).
Results
At genome-wide significance, we identified 11 independent variants associated with the risk of having asthma with hay fever, including 2 associations reaching this level of significance with allergic disease for the first time: ZBTB10 (rs7009110; odds ratio [OR], 1.14; P = 4 × 10−9) and CLEC16A (rs62026376; OR, 1.17; P = 1 × 10−8). The rs62026376:C allele associated with increased asthma with hay fever risk has been found to be associated also with decreased expression of the nearby DEXI gene in monocytes. The 11 variants were associated with the risk of asthma and hay fever separately, but the estimated associations with the individual phenotypes were weaker than with the combined asthma with hay fever phenotype. A variant near LRRC32 was a stronger risk factor for hay fever than for asthma, whereas the reverse was observed for variants in/near GSDMA and TSLP. Single nucleotide polymorphisms with suggestive evidence for association with asthma with hay fever risk included rs41295115 near IL2RA (OR, 1.28; P = 5 × 10−7) and rs76043829 in TNS1 (OR, 1.23; P = 2 × 10−6).
Conclusion
By focusing on the combined phenotype of asthma with hay fever, variants associated with the risk of allergic disease can be identified with greater efficiency.
Keywords: Rhinitis, atopy, selection, genetic correlation, bivariate, single nucleotide polymorphism
The comorbidity between asthma and rhinitis (or hay fever) is well established1,2 and might be a consequence of a causal relationship3,4 or a shared cause.5,6 Consistent with the latter hypothesis, results from twin and family studies have estimated that both diseases share 50% to 90% of their genetic susceptibility and 20% to 50% of their environmental susceptibility.7,8
Genome-wide association studies (GWASs) are a proved approach to discover genetic variants associated with asthma and hay fever. To date, GWASs have identified variants in 18 loci for which there are genome-wide significant associations with asthma,9 including ORMDL3,10–15 PDE4D,16 DENND1B,17 HLA,11,15,18–21 IL33,11,12,15 IL1RL1,11,12,15,20 SMAD3,11,15 IL2RB,11 SLC30A8,18 PYHIN1,12 TSLP,12,15 GAB1,19 IKZF4,19 10p14,19 LRRC32,14,15 IL6R,14 RORA,20 and TLR1.15 Two GWASs of hay fever have been published recently,22,23 with one variant in the LRRC32 region being genome-wide significant.22 In addition, Hinds et al15 reported that 11 of 23 variants discovered in a GWAS of self-reported allergy were also associated with hay fever risk at the genome-wide significance level, including those in or near TLR1, TSLP, LRRC32, IL1RL1, HLA-DQA1, HLA-C, PLCL1, LPP, IL33, SMAD3, and ETS1. Therefore, to date, variants in 7 loci (TLR1, TSLP, LRRC32, IL1RL1, HLA, IL33, and SMAD3) have been established as risk factors for both diseases, but many more shared risk variants remain to be identified. On the basis of analyses of family data from the population-based Tasmanian Longitudinal Health Study,24 we hypothesized that by performing a GWAS that explicitly considers the combined phenotype of asthma with hay fever we can substantially improve power to detect genetic risk factors shared between both diseases.
We found that considering asthma and hay fever as 3 mutually exclusive phenotypes was informative; the strengths and the familial aspects of their relationship with eczema in infancy differed across the 3 outcomes.24 Specifically, the childhood disease associations with infantile eczema were more pronounced and more likely to have a causal component for asthma with hay fever (A+H+) than for hay fever without asthma (A−H+) and asthma without hay fever (A+H−).
Therefore we estimated familial associations separately for the 3 phenotypes. For phenotype A+H+, we found strong associations between first-degree relatives, with odds ratios (ORs) for sibling, mother-offspring, and father-offspring pairs of approximately 6, 5, and 3, respectively (all P < .0001). For phenotypes A−H+ and A+H−, the associations between first-degree relatives were also highly statistically significant but attenuated. For A−H+, they were approximately 4.5, 2.5, and 2, respectively, whereas for A+H−, they were approximately 3, 3, and 2, respectively. However, of potentially more importance for informing GWASs of the genetic causes of these phenotypes, there were also spousal associations for phenotypes A−H+ and A+H−, with the ORs for parent pairs being 1.7 and 2.5, respectively (both P < .001). For phenotype A+H+, however, the parents were not associated (OR, 1.2; P = .6); that is, one cannot exclude at least part of the familial associations for phenotypes A−H+ and A+H− being caused by nongenetic factors, but this cannot be said of phenotype A+H+.
The lack of a spousal correlation for phenotype A+H+ and the strong associations between first-degree relatives for this phenotype led us to focus our efforts on the discovery of genetic causes of asthma and hay fever by studying GWAS data based on the combined phenotype of asthma with hay fever. Specifically, we performed a meta-analysis of GWASs considering persons with both physician-diagnosed asthma and hay fever to be cases and persons with neither disease to be control subjects.
METHODS
Studies included in the GWAS of asthma with hay fever
Participants (n = 20,776) for this study were from 4 studies (see Table E1 in this article’s Online Repository at www.jacionline.org), as summarized below. Informed consent was obtained from all participants, and the study protocols were reviewed and approved by the appropriate ethics committees.
Australian Asthma Genetics Consortium (n = 2,137)
A total of 2,669 case subjects of European ancestry with physician-diagnosed asthma and 4,528 control subjects of European ancestry without asthma were genotyped for a recent GWAS of asthma described in detail elsewhere.14 For the present study, we selected the subset of participants for whom hay fever information was also available, including 1,505 asthmatic subjects who had a history of hay fever (as cases) and 632 nonasthmatic subjects without a history of hay fever (as control subjects). Cases and control subjects were drawn from 4 studies: the Queensland Institute of Medical Research (n = 921), the Lung Institute of Western Australia (n = 475), the Busselton Health Study (n = 445), and the Tasmanian Longitudinal Health Study (n = 296). Asthmatic subjects (mean age, 41 years; 61% female) were considered to also have a history of hay fever if they answered affirmatively to a hay fever screening question included in epidemiologic questionnaires (see Table E2 in this article’s Online Repository at www.jacionline.org). Control subjects (mean age, 37 years; 60% female) reported never having had hay fever.
23andMe (n = 15,072)
Participants were customers of 23andMe (Mountain View, Calif) who had been genotyped as part of the 23andMe Personal Genome Service. We selected cases and control subjects from participants who had completed a survey about asthma. Cases (n = 4,230; mean age, 48 years; 54% female) responded yes to the question “Have you ever been diagnosed by a doctor with asthma or bronchial asthma?” and also responded affirmatively to the question “Have you ever had allergic rhinitis (stuffed or dripping nose caused by allergies)?” Control subjects (n = 10,842; mean age, 49 years; 39% female) answered no to both questions. Only persons of European ancestry were included in the analysis.
Avon Longitudinal Study of Parents and Their Children (n = 2800)
The Avon Longitudinal Study of Parents and Their Children (ALSPAC) is based on a longitudinal population-based birth cohort recruited through pregnant women residing in Avon, United Kingdom, with an expected delivery date between April 1, 1991, and December 31, 1992.25 Fourteen thousand five hundred forty-one pregnant women were initially enrolled, with 14,062 children born. Cases (n = 668; mean age, 14 years; 42% female) were reported by their mothers to have a lifetime doctor’s diagnosis of asthma on questionnaires completed when the children were age 7½ or 14 years and to have hay fever (ever or current) on questionnaires completed when the children were age 11 or 14 years. Control subjects (n = 2,132; mean age, 14 years; 52% females) were reported by their mothers to never have had asthma or hay fever on the same questionnaires. Only unrelated persons of European ancestry were included in the analysis.
Western Australian Pregnancy Cohort (Raine, n = 767)
Recruitment for the Raine cohort has previously been described in detail.26–28 In brief, between 1989 and 1991, 2,900 pregnant women were recruited before 18 weeks’ gestation into a randomized controlled trial to evaluate the effects of repeated ultrasound in pregnancy. The resulting children have since been comprehensively phenotyped from birth to 21 years of age by trained members of the Raine research team. Data collection has included questionnaires completed by the child’s primary caregiver and by the adolescent from age 14 years, physical assessments by trained assessors at all follow-up years, and DNA collection from the year 14 follow-up. Cases (N = 282; mean age, 17 years; 45% female) were defined as children who were reported by their mothers on any available survey (year 5, 8, 10, 14, or 17 follow-ups) to have received a diagnosis of both asthma and hay fever by a pediatrician/specialist or general practitioner. Control subjects (n = 485; mean age, 17 years; 51% female) were reportedly free of asthma and hay fever diagnosis across all available time points. Only persons of European ancestry were included in the analysis.
Individual GWASs
Genotyping, single nucleotide polymorphism (SNP) imputation, and association testing procedures for each of the 4 GWASs (Australian Asthma Genetics Consortium [AAGC], 23andMe, ALSPAC, and Raine) are described in detail in the Methods section in this article’s Online Repository at www.jacionline.org. Briefly, 20,776 samples were genotyped with Illumina (550K, 610K, 660K or OmniExpress) arrays, and standard quality control (QC) filters were applied before imputation. Genomic coverage in each study was then expanded by imputing unmeasured variants against the HapMap and/or 1000 Genomes reference haplotypes, including the X chromosome. Autosomal SNPs directly genotyped or imputed with high confidence were tested for association with case-control status in each study while adjusting for significant covariates, such as age and sex.
Meta-analysis of GWASs
Before the meta-analysis, we excluded SNPs from each GWAS with (1) a minor allele frequency (MAF) of less than 0.01; (2) an imputation information score or r2 value of less than 0.3; (3) outlier SE for the allelic association estimate; (4) more than 2 distinct alleles across cohorts; and/or (5) an MAF difference of greater than 0.05 between any 2 studies. After QC, results were available for 4,972,397 SNPs, of which 57.9% were present in both the AAGC and 23andMe cohorts, 42.0% in all 4 cohorts, and less than 0.1% in 3 cohorts. No correction for genomic inflation of test statistics was applied before (to individual GWASs) or after (overall results) the meta-analysis, which was performed with METAL29 by using a fixed-effects model. The Cochran Q test was used to identify SNPs for which there was significant heterogeneity in associations across studies.
Choice of threshold for genome-wide significance testing
Through imputation based on reference panels from the 1000 Genomes Project, we were able to test 4.9 million variants for association with asthma with hay fever, including 4,180,629 common (84%, MAF ≥ 0.05) and 791,768 less common (16%, 0.01 ≤ MAF < 0.05) variants. As such, correction for multiple SNP testing required a more stringent genome-wide significance level threshold than the commonly used threshold of 5 × 10−8 based on approximately 1 million effectively independent common variants for Europeans, as estimated from HapMap project data.30 Instead, we used a genome-wide significance threshold based on (1) the updated estimate of approximately 1.63 million effectively independent common variants for Europeans estimated from 1000 Genomes Project data31 and (2) 257,403 less common variants tested, which excludes a redundant set of 534,365 highly correlated (r2 > 0.8) less common variants. Therefore we used a P value of 0.05/(1,634,900 + 257,403) = 3 × 10−8 as a threshold for genome-wide significance.
Association analyses stratified by asthma and hay fever status
To estimate the associations between individual variants and asthma and hay fever risks separately, we performed the following 7 additional analyses, comparing (1) asthma cases (A+) with asthma-free (A−) control subjects (ie, estimating association with asthma while ignoring hay fever status); (2) hay fever cases (H+) with hay fever–free (H−) control subjects (ie, estimating association with hay fever while ignoring asthma status); (3) asthma cases (A+) with control subjects who were asthma free and hay fever free (A−H−); (4) hay fever cases (H+) with control subjects who were asthma free and hay fever free (A−H−); (5) asthma without hay fever cases (A+H−) with control subjects who were asthma free and hay fever free (A−H−); (6) hay fever without asthma cases (A−H+) with control subjects who were asthma free and hay fever free (A−H−); and (7) asthma without hay fever cases (A+H−) with hay fever without asthma (A−H+) cases. All 4 GWASs described above (AAGC, 23andMe, ALSPAC, and Raine) contributed to these analyses, which included an additional 2,320 (AAGC), 9,821 (23andMe), 2,513 (ALSPAC), and 508 (Raine) samples that were not part of the asthma with hay fever GWAS, as detailed in Table E3 in this article’s Online Repository at www.jacionline.org. These analyses were restricted to variants found to be associated with asthma with hay fever at the genome-wide significance level (P ≤ 3 × 10−8).
Replication study
Variants associated with asthma with hay fever at the suggestive significance level (3 × 10−8 < P ≤ 5 × 10−6) were followed up by using GWAS data from an additional 3,333 persons (878 cases and 2,455 control subjects, see Table E4 in this article’s Online Repository at www.jacionline.org) who completed the 23andMe asthma survey described above after the meta-analysis of GWASs was finalized. As such, data from these persons were not available to be included in the 23andMe discovery GWAS. Ascertainment of participants, determination of disease status, and genotyping were as described above for the 23andMe study. The 3,333 persons were confirmed to be unrelated (to each other and to the 23andMe discovery set) and of European ancestry through analysis of genome-wide allele sharing.
RESULTS
Eleven variants associated with risk of having asthma with hay fever
We performed a GWAS using 4 independent studies (see Table E1), with cases (N = 6,685) defined as persons who reported a lifetime doctor’s diagnosis of asthma and also hay fever (see Table E2). Control subjects (n = 14,219) were persons who had a diagnosis of neither asthma nor hay fever.
After QC, association results for 4.9 million SNPs were meta-analyzed by using a fixed-effects model. The ratio of the observed to the expected median association χ2 statistic (λ) was 1.061 (see Fig E1 in this article’s Online Repository at www.jacionline.org), corresponding to a rescaled32–34 λ1000 of 1.007, which might be consistent with the effects of residual technical artifacts, a polygenic component to disease risk, or both.35 Because these effects could not be distinguished and the inflation was modest, genomic control was not used to adjust the observed association results.
Eleven independent (r2 < 0.1) SNPs located in 10 chromosomal regions had P values exceeding our genome-wide significance threshold of 3 × 10−8 (Table I and see Fig E2 in this article’s Online Repository at www.jacionline.org). All 11 associations would have been genome-wide significant had genomic control been applied (see Table E5 in this article’s Online Repository at www.jacionline.org). Of these, 9 SNPs were located in or near established risk loci for allergic disease, namely HLA-DQB1 (P = 4 × 10−14), TLR1 (P = 5 × 10−12), WDR36 (P = 3 × 10−11), LRRC32 (P = 5 × 10−11), IL1RL1 (P = 4 × 10−11), GSDMA (P = 4 × 10−10), TSLP (P = 10−9), IL33 (P = 2 × 10−9), and SMAD3 (P = 4 × 10−9).
TABLE I.
Eleven variants associated with the risk of having asthma with hay fever at the genome-wide significance level (P ≤ 3 × 10−8)
| Chromosome | Position (bp) | Nearest gene (kb distance) | SNP† (risk allele) | Risk allele frequency (range) | OR (95% CI) | Association P value | Heterogeneity test, P value (I2; 95% CI) |
|---|---|---|---|---|---|---|---|
| 6p21 | 32734579 | HLA-DQB1 (1) | rs9273373 (G) | 0.54–0.58 | 1.24 (1.17–1.30) | 4 × 10−14 | .97 (0; 0–89) |
| 4p14 | 38476105 | TLR1 (2)* | rs4833095 (T) | 0.74–0.76 | 1.20 (1.14–1.26) | 5 × 10−12 | .37 (5; 0–86) |
| 5q22 | 110495398 | WDR36 (1) | rs1438673 (C) | 0.49–0.52 | 1.16 (1.11–1.21) | 3 × 10−11 | .59 (0; 0–85) |
| 2q12 | 102332981 | IL1RL1 (2)* | rs10197862 (A) | 0.85–0.86 | 1.24 (1.16–1.32) | 4 × 10−11 | .16 (41; 0–80) |
| 11q13 | 75976842 | LRRC32 (69) | rs2155219 (T) | 0.48–0.52 | 1.16 (1.11–1.21) | 5 × 10−11 | .28 (22; 0–88) |
| 17q21 | 35376206 | GSDMA (3)* | rs7212938 (G) | 0.46–0.48 | 1.16 (1.11–1.20) | 4 × 10−10 | .84 (0; 0–85) |
| 5q22 | 110429771 | TSLP (6) | rs1837253 (C) | 0.71–0.75 | 1.17 (1.12–1.24) | 1 × 10−9 | .11 (50; 0–84) |
| 9p24 | 6165855 | IL33 (40) | rs72699186 (T) | 0.15–0.16 | 1.26 (1.16–1.35) | 2 × 10−9 | .45 (0; 0–89) |
| 8q21 | 81454434 | ZBTB10 (106) | rs7009110 (T) | 0.36–0.41 | 1.14 (1.09–1.19) | 4 × 10−9 | .77 (0; 0–85) |
| 15q22 | 65255339 | SMAD3 (19)* | rs17294280 (G) | 0.23–0.27 | 1.18 (1.11–1.25) | 4 × 10−9 | .59 (0; 0–85) |
| 16p13 | 11136213 | CLEC16A (47)* | rs62026376 (C) | 0.72–0.74 | 1.17 (1.11–1.24) | 1 × 10−8 | .76 (0; 0–89) |
SNP is located within reported gene.
SNPs rs9273373, rs72699186, and rs62026376 were not tested in ALSPAC (n = 2,800) and Raine (n = 767), and therefore results for these 3 SNPs are based on the AAGC (n = 2,137) and 23andMe (n = 15,072) studies. The most significantly associated SNPs in LD (r2 > 0.5) with each of these 3 SNPs and available in all 4 studies were rs6906021 (risk allele C; OR, 1.16; P = 3 × 10−11; heterogeneity test, P = .73), rs1342326 (risk allele C; OR, 1.17; P = 2 × 10−7; heterogeneity test, P = .73), and rs12935657 (risk allele G; OR, 1.15; P = 5 × 10−8; heterogeneity test, P = .33), respectively.
The remaining 2 SNPs were located in loci with variants previously reported to be associated with allergic disease at the suggestive (P ≤ 5 × 10−6) but not genome-wide level of significance. These include rs7009110 (OR per allele, 1.14; P = 4 × 10−9), which is 106 kb upstream of ZBTB10 on chromosome 8q21 (see Fig E3 in this article’s Online Repository at www.jacionline.org), and rs62026376 (OR, 1.17; P = 10−8), which is located in intron 21 of CLEC16A on chromosome 16p13 (see Fig E4 in this article’s Online Repository at www.jacionline.org). The direction of association for both variants was the same in all 4 studies (see Table E6 in this article’s Online Repository at www.jacionline.org).
A variant in linkage disequilibrium (LD; r2 = 0.52) with the sentinel SNP in ZBTB10 was recently reported to be associated with atopic dermatitis (AD),36 with a consistent but weaker association (rs7000782; OR per allele, 1.09; P = 10−6). The association we observed with asthma with hay fever remained when analyzing a subset of eczema-free subjects (see Table E7 in this article’s Online Repository at www.jacionline.org), suggesting that it was not driven by eczema status.
Similarly, a recent GWAS of hay fever22 reported a suggestive association with rs887864 (OR, 1.16; P = 1 × 10−6), a variant in LD (r2 = 0.53) with the CLEC16A sentinel SNP. This region was also reported to be associated with AD at a near-suggestive level by a Japanese study (rs9923856; OR, 1.17; P = 6 × 10−6; r2 with sentinel SNP = 0.50 for Europeans and 0.28 for East Asians).37 The published direction of association with hay fever and AD was the same as that observed in our study.
Association with asthma and hay fever risks
The 11 variants above were also associated (P < .05) with the risk of both asthma (A+ vs A−) and hay fever (H+ vs H−) when each disease was considered as a separate phenotype. However, for all variants, the estimated associations with asthma and hay fever were weaker than with the combined asthma with hay fever phenotype (see Table E8 in this article’s Online Repository at www.jacionline.org). Three (ZBTB10, SMAD3, and CLEC16A) of the 11 associations did not reach the genome-wide significance threshold when asthma and hay fever were considered separately, even when restricting the analysis to asthma-free and hay fever–free control subjects (A+ vs A−H− and H+ vs A−H−, see Table E8).
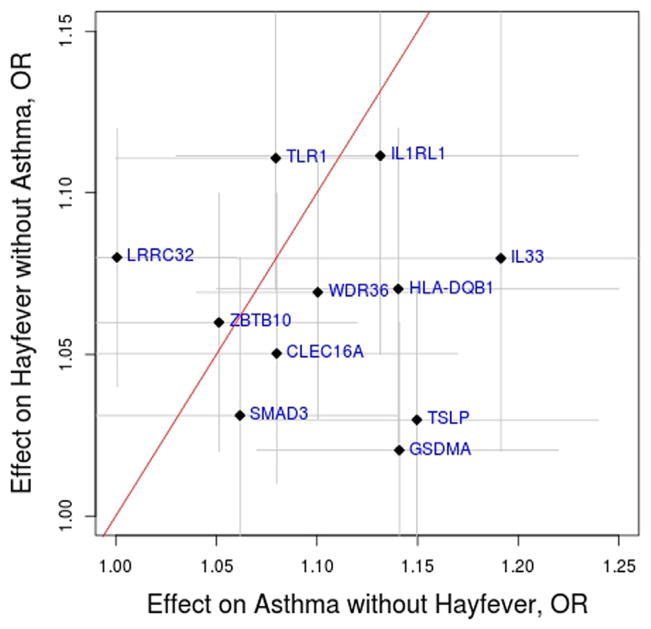
When considering the associations on the mutually exclusive phenotypes of asthma without hay fever (A+H− vs A−H−) and hay fever without asthma (A−H+vs A−H−), there were notable differences for 3 variants (Fig 1 and see Table E8). First, the LRRC32 variant was associated with an increased risk of hay fever without asthma (OR, 1.08; 95% CI, 1.04–1.12) but not asthma without hay fever (OR, 1.00; 95% CI, 0.94–1.06). The difference in risk allele frequency between A+H− cases and A−H+ cases was statistically significant (P = .014, see Table E9 in this article’s Online Repository at www.jacionline.org), indicating that the association with hay fever risk per se is stronger than that with asthma risk per se. In the AAGC study the prevalence of allergic sensitization to a common allergen (ie, atopy) was only modestly higher for A−H+ cases (112/160 [70%]) when compared with A+H− cases (277/440 [63%]). As such, differences in atopy status might contribute to but are unlikely to fully explain the weaker association observed in the A+H− group. Second, the GSDMA and TSLP variants were associated with risk of asthma without hay fever (OR of 1.14 and 1.15 and 95% CI of 1.07–1.22 and 1.07–1.24, respectively) but less so with risk of hay fever without asthma (OR of 1.02 and 1.03 and 95% CI of 0.98–1.06 and 0.98–1.07, respectively). In both situations the difference in risk allele frequency between A+H− cases and A−H+ cases was statistically significant (P = .0003 and P = .004, respectively; see Table E9). These results suggest that the GSDMA and TSLP variants are stronger risk factors for asthma than for hay fever.
FIG. 1.
Effect (OR ±95% CI) of the top 11 loci on the mutually exclusive phenotypes of asthma without hay fever (A+H−) and hay fever without asthma (A−H+). Results are based on the comparison between 14,091 control subjects (no asthma and no hay fever, A−H−) and either 2,776 asthma without hay fever (A+H−) cases or 9,301 hay fever without asthma (A−H+) cases.
Association between asthma and variants in the region of ZBTB10 and CLEC16A in the GABRIEL GWAS
Next, we investigated whether publicly available results from the GABRIEL asthma GWAS11 could provide independent support for the association we observed for variants in the region of ZBTB10 and CLEC16A. After excluding 2 overlapping cohorts (ALSPAC and Busselton), leaving 9,360 cases and 14,692 control subjects, a consistent association was observed between asthma risk and 2 proxy SNPs (r2 > 0.8) in both loci: rs6473226 upstream of ZBTB10 (OR, 1.07; P = .003) and rs17673553 in CLEC16A (OR, 1.07; P = .004; see Table E10 in this article’s Online Repository at www.jacionline.org). These results, together with the genome-wide significant association observed in our meta-analysis and the previous reported associations with hay fever and AD, establish the ZBTB10 and CLEC16A regions as containing bona fide genetic risk factors for allergic disease.
LD with nearby coding, regulatory, or other disease-associated variants
To provide some insight into the potential molecular mechanisms underlying the associations with variants in the ZBTB10 and CLEC16A regions, we searched for nearby variants in LD (r2 ≥ 0.3) with the sentinel SNPs at each locus using data for 379 persons of European descent from the 1000 Genomes Project. We focused on potentially deleterious (ie, nonsynonymous, missense, and splice site) coding variants and SNPs reported to influence gene expression levels,38–42 immune cell levels,43,44 or other inflammatory or immune-related diseases.9
For the CLEC16A locus, we identified a variant in LD (r2 = 0.95) with the sentinel SNP that reportedly influences the expression of the nearby DEXI gene (rs12935657, P = 2 × 10−18) in monocytes.41 The rs62026376:C allele associated with an increased asthma with hay fever risk is in phase with the rs12935657:G allele, which decreases DEXI expression. We also identified 2 variants that were associated (P < .005) with the numbers of peripheral blood CD56+ natural killer T cells (rs3901386) and eosinophils (rs9652582, see Table E11 in this article’s Online Repository at www.jacionline.org). The rs62026376:C allele was in phase with the rs9652582:G allele, which is associated with increased eosinophil levels. Lastly, the sentinel SNP was found to be in moderate LD with established type 1 diabetes (rs12708716) and multiple sclerosis (rs7200786) risk-associated variants (see Table E12 in this article’s Online Repository at www.jacionline.org), with the same direction of association for both diseases. We did not identify any noteworthy variants in LD with the ZBTB10 sentinel SNP apart from the variant (rs7000782) previously reported to be associated with AD.36
Suggestive associations
An additional 19 independent SNPs were associated with asthma with hay fever risk at a suggestive significance level (3 × 10−8 < P ≤ 5 × 10−6) in our meta-analysis (see Table E13 in this article’s Online Repository at www.jacionline.org). Of these, 4 are located in regions in which other SNPs have been found to be associated with allergic disease, namely SNPs located in or near SLC25A46 (rs3853750), IL33 (rs343496), IKZF3 (rs12450323), and LRRC32 (rs1320644). We studied these 19 SNPs using a further 878 cases and 2,455 control subjects (see Table E4), but no single SNP was found to be associated with disease after taking into account multiple testing (see Table E14 in this article’s Online Repository at www.jacionline.org). We nonetheless highlight the following 2 loci as those with the strongest evidence for association (P < 10−6) from the combined analysis of discovery and replication cohorts: rs41295115 (MAF, 5%; OR per allele, 1.28; P = 5 × 10−7), a variant located between IL2RA and RBM17, and rs76043829, which is located in TNS1 (MAF, 13%; OR per allele, 1.23; P = 2 × 10−6), a locus previously reported to contain SNPs associated with lung function.45
DISCUSSION
By explicitly considering the combined phenotype of asthma with hay fever, we identified 11 independent variants with genome-wide significant associations with disease risk using a relatively modest sample size (20,776 samples). Among these were variants in the 8q21 and 16p13 regions, which we have now established as containing bona fide genetic risk factors for allergic disease.
The 8q21 region had recently been identified as containing variants associated with AD by means of a meta-analysis of GWASs, but this was not validated by a replication study.36 For this reason, variation in that locus was not designated as a confirmed risk factor for AD and therefore, more generally, for allergic disease. We extended this original observation by finding that a common variant (MAF of approximately 40%) in this region was associated with a per-allele 1.14-fold increased risk of having asthma with hay fever. This association was independently supported by results from the GABRIEL asthma GWAS.11 Taken together, these results confirm that there are genetic variants associated with allergic disease in the 8q21 region near ZBTB10. The association with asthma was still detectable when persons with eczema were excluded (OR per allele, 1.16), suggesting that it was not confounded by eczema status. Conversely, further studies are required to establish whether the reported association with eczema36,37 is independent of or confounded by asthma/hay fever status.
We did not identify any nearby potentially functional variants in LD with the sentinel SNP in the 8q21 locus that could provide insight into the molecular mechanisms underlying this association. The 2 closest genes are ZBTB10 (106 kb away) and TPD52 (216 kb); to our knowledge, there is no known biological evidence linking either gene with allergic disease. ZBTB10 is a putative repressor of the Sp1 transcription factor,46 which regulates multiple immune-related genes47–50; as such, ZBTB10 might underlie the observed association with rs7009110. Further studies are needed to clarify the cause of this SNP association.
The second region that our results suggest contains variants associated with risk of the combined asthma with hay fever phenotype is on chromosome 16p13, with the sentinel SNP located in intron 21 of the CLEC16A gene and associated with an increased risk of 1.17-fold per allele. This is an established susceptibility locus for autoimmune diseases, with different risk SNPs reported for type 1 diabetes (OR, 1.23; MAF, 0.32),51 multiple sclerosis (OR, 1.15; MAF, 0.49),52 and primary biliary cirrhosis (OR, 1.32; MAF, 0.32).53 Variants in this region have also been previously reported to be associated at the suggestive level of significance with hay fever (OR, 1.16; MAF, 0.37)22 and AD (OR, 1.17; MAF, 0.19).37 The different disease-associated variants are in moderate LD with each other (0.1 < r2 < 0.6), but the predisposing alleles are all in phase, suggesting that the underlying causal variant or variants have the same direction of effect on both autoimmune and allergic diseases.
Although being located in an intronic region of the CLEC16A gene, variants in LD with our sentinel SNP have previously been found to be associated with the expression of the nearby DEXI gene in monocytes.41 The allele associated with an increased risk of asthma with hay fever decreases DEXI expression, which is consistent with recent reports for autoimmune disease–associated variants.54,55 This suggests that DEXI is a plausible causal gene underlying the observed association with allergic disease. DEXI is expressed in the lung, as well as in relevant immune cell types, such as CD4+ T cells and B cells.54 However, its function is unknown, and therefore studying its role in the immune system might provide new insights into the cause of allergic disease. In this respect it is noteworthy that the allele associated with an increased risk of asthma with hay fever and decreased DEXI expression is also associated with increased peripheral blood eosinophil levels.
While this work was under review, 2 large-scale GWASs of allergic sensitization56 and self-reported allergy15 were published. Variants in strong LD with rs7009110 (8q21) and rs62026376 (16p13) were found to be associated at the suggestive significance level with self-reported allergy in the Hinds et al15 GWAS, which is consistent with our results. However, the agreement between results from Hinds et al and our asthma with hay fever GWAS was expected given the extensive overlap between the samples included in these 2 studies.
Each of the 11 variants identified in the meta-analysis of asthma with hay fever had a significant but weaker association with asthma and hay fever when both diseases were considered as separate phenotypes. For 3 variants (in/near ZBTB10, SMAD3, and CLEC16A), the association did not reach the genome-wide significance level for risks of asthma or hay fever, despite the larger number of samples (up to 13,000 more) included in the analyses. These results highlight the benefit of considering the combined asthma with hay fever phenotype when searching for variants that influence the risk of both diseases. An additional analysis that we did not consider but that might prove useful to identify shared genetic risk factors is to compare asthma or hay fever cases with control subjects who were both asthma and hay fever free. A similar approach has been successful in identifying risk variants shared between patients with Crohn disease and those with ulcerative colitis.57
Because the association with asthma risk is confounded by hay fever status and vice versa, we also analyzed the 2 mutually exclusive phenotypes of asthma without hay fever and hay fever without asthma to compare the association of each variant with the 2 separate diseases. We found that the TSLP and GSDMA variants have stronger associations with the risk of asthma, whereas the LRRC32 variant has a stronger association with the risk of hay fever.
Our study has identified novel suggestive associations that warrant follow-up, of which we highlight those 20 kb upstream of IL2RA (rs41295115) and in TNS1 (rs76043829). IL2RA is an established risk locus for multiple autoimmune diseases58–63; however, the asthma-associated variant is independent (r2 < 0.1) of all reported autoimmune risk variants. The variant we identified in TNS1 is close to but also independent of (r2 < 0.1) a variant reported to be associated with FEV1.45 Interestingly, the putative asthma risk variant is 307 kb upstream of CXCR2, a gene that encodes the β chain of the IL-8 receptor, which is the major mediator of neutrophil migration to sites of inflammation64 and represents a promising drug target for asthma.65
In conclusion, we have shown that genetic risk factors for allergic disease are located in or near ZBTB10 and CLEC16A. For both associations, there is no functional evidence directly linking any nearby gene with allergic disease; further investigations of the molecular mechanisms underlying both associations might identify previously unrecognized pathways in the pathogeneses of asthma and hay fever. This study has also shown that by focusing GWASs on phenotypes with stronger evidence of a (stronger) genetic cause, greater understanding of the genetic causes of conditions can be discovered.
Supplementary Material
Clinical implications.
Our results implicate ZBTB10, CLEC16A, or other nearby genes in the cause of asthma and hay fever. These might be part of previously unrecognized pathways in the pathogeneses of allergic disease.
Acknowledgments
Supported by the Australian National Health and Medical Research Council (NHMRC; grants 241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498, 613627, 403981, and 003209); the Australian Research Council (grants A7960034, A79906588, A79801419, DP0770096, DP0212016, and DP0343921); the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254); the US National Institutes of Health (grants AA07728, AA07535, AA10248, AA11998, AA13320, AA13321, AA13326, AA14041, AA17688, DA12854, and MH66206); Asthma Foundations in Tasmania, Queensland, and Victoria; the Clifford Craig Trust in Northern Tasmania; the Lew Carty Foundation; the Royal Hobart Research Foundation; the University of Melbourne; the Great Wine Estates of the Margaret River region of Western Australia; the University of Western Australia (UWA); Raine Medical Research Foundation; UWA Faculty of Medicine, Dentistry and Health Sciences; the Telethon Institute for Child Health Research; the Women and Infants Research Foundation; the Canadian Institutes of Health Research (MOP-82893); and the National Heart, Lung, and Blood Institute of the National Institutes of Health under grant no. 1R43HL115873-01.
Queensland Institute of Medical Research: We thank the twins and their families for their participation; Dixie Statham, Ann Eldridge, Marlene Grace, and Kerrie McAloney for sample collection; Lisa Bowdler and Steven Crooks for DNA processing; David Smyth, Harry Beeby, and Daniel Park for IT support; and Professor Peter Visscher for discussions on power improvement through sample selection based on correlated traits. MARF is supported by the National Health and Medical Research Council. Tasmanian Longitudinal Health Study: M.C.M., S.C.D., and J.L.H. are supported by the National Health and Medical Research Council. ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. Raine: We are grateful to the Raine Study participants and their families and to the Raine Study research staff for cohort coordination and data collection. We gratefully acknowledge the National Health and Medical Research Council for their long-term contribution to funding the study over the last 20 years and the assistance of the Western Australian DNA Bank (National Health and Medical Research Council National Enabling Facility). 23andMe: We thank the customers of 23andMe who answered surveys, as well as the employees of 23andMe, who together made this research possible.
Abbreviations used
- A+H+
Subjects with asthma and hay fever
- A+H−
Subjects with asthma but not hay fever
- A−H+
Subjects with hay fever but not asthma
- A−H−
Subjects without asthma and without hay fever
- A+
Subjects with asthma, with or without hay fever
- H+
Subjects with hay fever, with or without asthma
- A−
Subjects without asthma, with or without hay fever
- H−
Subjects without hay fever, with or without asthma
- AAGC
Australian Asthma Genetics Consortium
- AD
Atopic dermatitis
- ALSPAC
Avon Longitudinal Study of Parents and Their Children
- GWAS
Genome-wide association study
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- QC
Quality control
- SNP
Single nucleotide polymorphism
Footnotes
Disclosure of potential conflict of interest: M. A. R. Ferreira has received research support from the Australian National Health and Medical Research Council (NHMRC). A. K. Kiefer has received research support from the National Institutes of Health (NIH) and is employed by and has stock/stock options in 23andMe. D. L Duffy has received research support from the NHMRC. N. Eriksson has received research support from the NIH and is employed by and has stock/stock options in 23andMe. P. A. Madden has received research support from the NIH (R01DA012854 [NAG] and R25DA027995 [R25]) and has received lecture fees. M. J. Abramson has received research support from Pfizer and has received travel support from Boehringer Ingelheim. A. C. Heath has received research support from the National Institutes of Health. C. F. Robertson has received research support from the NHMRC (grants 436959, 490321, 491246, 1006215, 044816, and 1044829), the MCRI, the US Cystic Fibrosis Foundation, the Australian Cystic Fibrosis Research Trust, and the NHMRC Centre for Research Excellence. G. W. Montgomery has received research support from the NHMRC. A. J. Henderson has received research support from the Medical Research Council UK and the Wellcome Trust. J. Y. Tung has received research support from the NIH (1R43HL115873-01) and is employed by and has stock/stock options in 23andMe. C. Pennell has received research support from the NHMRC (APP572613, 2009–2012), the NIH, CIHR, and Channel 7 Telethon; is a board member for the Raine Executive Committee; is employed by the University of Western Australia; has patents from the United States; and has received travel expenses from the March of Dimes Preventing Prematurity meeting, the GAPPS meeting, and PreHOT meetings from 2009–2012. D. M. Evans has received research support from the MRC and the Wellcome trust (supply core support to Avon Longitudinal Study of Parents and Their Children). D. A. Hinds has received research support from the NIH (1R43HL115873-01) and is employed by and has stock/stock options in 23andMe. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Guerra S, Sherrill DL, Martinez FD, Barbee RA. Rhinitis as an independent risk factor for adult-onset asthma. J Allergy Clin Immunol. 2002;109:419–25. doi: 10.1067/mai.2002.121701. [DOI] [PubMed] [Google Scholar]
- 2.Leynaert B, Bousquet J, Neukirch C, Liard R, Neukirch F. Perennial rhinitis: An independent risk factor for asthma in nonatopic subjects: results from the European Community Respiratory Health Survey. J Allergy Clin Immunol. 1999;104:301–4. doi: 10.1016/s0091-6749(99)70370-2. [DOI] [PubMed] [Google Scholar]
- 3.Shaaban R, Zureik M, Soussan D, Neukirch C, Heinrich J, Sunyer J, et al. Rhinitis and onset of asthma: a longitudinal population-based study. Lancet. 2008;372:1049–57. doi: 10.1016/S0140-6736(08)61446-4. [DOI] [PubMed] [Google Scholar]
- 4.Burgess JA, Walters EH, Byrnes GB, Matheson MC, Jenkins MA, Wharton CL, et al. Childhood allergic rhinitis predicts asthma incidence and persistence to middle age: a longitudinal study. J Allergy Clin Immunol. 2007;120:863–9. doi: 10.1016/j.jaci.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Togias A. Rhinitis and asthma: evidence for respiratory system integration. J Allergy Clin Immunol. 2003;111:1171–84. doi: 10.1067/mai.2003.1592. [DOI] [PubMed] [Google Scholar]
- 6.Rimmer J, Ruhno JW. 6: Rhinitis and asthma: united airway disease. Med J Aust. 2006;185:565–71. doi: 10.5694/j.1326-5377.2006.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 7.Duffy DL, Martin NG, Battistutta D, Hopper JL, Mathews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–8. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 8.Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy. 2006;36:1382–90. doi: 10.1111/j.1365-2222.2006.02512.x. [DOI] [PubMed] [Google Scholar]
- 9.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, et al. [Accessed March 2013.];A catalog of published genome-wide association studies. Available at: www.genome.gov/gwastudies.
- 10.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 11.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–8. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13. 5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–11. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN, et al. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS One. 2012;7:e44008. doi: 10.1371/journal.pone.0044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasky-Su J, Himes BE, Raby BA, Klanderman BJ, Sylvia JS, Lange C, et al. HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy. 2012;42:1724–33. doi: 10.1111/cea.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Andiappan AK, de Wang Y, Anantharaman R, Parate PN, Suri BK, Low HQ, et al. Genome-wide association study for atopy and allergic rhinitis in a Singapore Chinese population. PLoS One. 2011;6:e19719. doi: 10.1371/journal.pone.0019719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopper JL, Bui QM, Erbas B, Matheson MC, Gurrin LC, Burgess JA, et al. Does eczema in infancy cause hay fever, asthma, or both in childhood? Insights from a novel regression model of sibling data. J Allergy Clin Immunol. 2012;130:1117–22. e1. doi: 10.1016/j.jaci.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort profile: the ‘children of the 90s’–the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–27. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newnham JP, Evans SF, Michael CA, Stanley FJ, Landau LI. Effects of frequent ultrasound during pregnancy: a randomised controlled trial. Lancet. 1993;342:887–91. doi: 10.1016/0140-6736(93)91944-h. [DOI] [PubMed] [Google Scholar]
- 27.Williams LA, Evans SF, Newnham JP. Prospective cohort study of factors influencing the relative weights of the placenta and the newborn infant. BMJ. 1997;314:1864–8. doi: 10.1136/bmj.314.7098.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans S, Newnham J, MacDonald W, Hall C. Characterisation of the possible effect on birthweight following frequent prenatal ultrasound examinations. Early Hum Dev. 1996;45:203–14. doi: 10.1016/0378-3782(96)01728-8. [DOI] [PubMed] [Google Scholar]
- 29.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 31.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–56. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–93. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 34.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2012;44:187–92. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 38.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–89. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 41.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira MA, Hottenga JJ, Warrington NM, Medland SE, Willemsen G, Lawrence RW, et al. Sequence variants in three loci influence monocyte counts and erythrocyte volume. Am J Hum Genet. 2009;85:745–9. doi: 10.1016/j.ajhg.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira MA, Mangino M, Brumme CJ, Zhao ZZ, Medland SE, Wright MJ, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tillotson LG. RIN ZF, a novel zinc finger gene, encodes proteins that bind to the CACC element of the gastrin promoter. J Biol Chem. 1999;274:8123–8. doi: 10.1074/jbc.274.12.8123. [DOI] [PubMed] [Google Scholar]
- 47.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–91. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 48.Tone M, Tone Y, Babik JM, Lin CY, Waldmann H. The role of Sp1 and NF-kappa B in regulating CD40 gene expression. J Biol Chem. 2002;277:8890–7. doi: 10.1074/jbc.M109889200. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K, Hayashi N, Shimokawa T, Umehara N, Kaminogawa S, Ra C. Cooperative regulation of Fc receptor gamma-chain gene expression by multiple transcription factors, including Sp1, GABP, and Elf-1. J Biol Chem. 2008;283:15134–41. doi: 10.1074/jbc.M800498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosmarin AG, Luo M, Caprio DG, Shang J, Simkevich CP. Sp1 cooperates with the ets transcription factor, GABP, to activate the CD18 (beta2 leukocyte integrin) promoter. J Biol Chem. 1998;273:13097–103. doi: 10.1074/jbc.273.21.13097. [DOI] [PubMed] [Google Scholar]
- 51.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davison LJ, Wallace C, Cooper JD, Cope NF, Wilson NK, Smyth DJ, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21:322–33. doi: 10.1093/hmg/ddr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leikfoss IS, Mero IL, Dahle MK, Lie BA, Harbo HF, Spurkland A, et al. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013;14:62–6. doi: 10.1038/gene.2012.52. [DOI] [PubMed] [Google Scholar]
- 56.Bonnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–6. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 59.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–7. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med. 2010;362:1686–97. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, et al. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265:682–4. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 65.Nair P, Gaga M, Zervas E, Alagha K, Hargreave FE, O’Byrne PM, et al. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebo-controlled clinical trial. Clin Exp Allergy. 2012;42:1097–103. doi: 10.1111/j.1365-2222.2012.04014.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.