Abstract
Regulatory DNAs serve as templates to bring weakly interacting transcription factors into close proximity so they can work synergistically to switch genes on and off in time and space. Most of these regulatory DNAs are enhancers that can work over long distances — a million base pairs or more in mammals — to control gene expression. Critical enhancers are sometimes even found within the introns of neighboring genes. This review summarizes well-defined examples of enhancers controlling key processes in animal development. Potential mechanisms of transcriptional synergy are discussed with regard to enhancer structure and contemporary ChIP-sequencing assays, whereby just a small fraction of the observed binding sites represent bona fide regulatory DNAs. Finally, there is a discussion of how enhancer evolution can produce novelty in animal morphology and of the prospects for reconstructing transitions in animal evolution by introducing derived enhancers in basal ancestors.
Introduction
Shortly after the first glimpse into the molecular organization of eukaryotic genes it became clear that critical regulatory DNAs could be uncoupled from the core promoter, the docking site for RNA Polymerase II (Pol II) [1]. The first eukaryotic enhancers were identified in animal viruses due to their ability to co-opt the transcriptional machinery of host cells upon infection [2–5]. The prototypic enhancer was identified in the SV40 animal virus, an unlikely source for long-range regulatory elements as the SV40 genome is just 5.2 kilobases (kb) in length [1].
The SV40 enhancer contains two 72 base pair (bp) repeats located ~200 bp 5′ of the gene encoding T-antigen, which is essential for viral replication and transcription of late viral genes in infected cells [1]. This 5′-regulatory sequence was shown to work at a distance when attached to a β-globin reporter gene and transfected in cultured monkey kidney cells [1]. Subsequent studies identified several sequence-specific transcription factors that bind to discrete sites within the 72 bp repeats, including the bZIP transcription factor AP1 and the Rel-containing factor NF-κB [6,7]. Both transcription factors are modulated by signaling pathways, receptor tyrosine kinases (RTKs) and Toll, respectively [8,9], and thereby anticipate the importance of enhancers in integrating cell signaling processes — a key insight not appreciated for another decade, e.g., [10]. Indeed, the synergistic activation of the prototypic SV40 enhancer by distinct classes of activators is a common theme of enhancer function and a number of examples will be considered in the course of this review.
Evidence that enhancers might have a more divine purpose than merely augmenting the efficacy of viral infection was obtained by examining genes that exhibit tissue-specific expression. The first cellular enhancers that were identified control the expression of the immunoglobulin (Ig) heavy chain gene in mammalian B lymphocytes [11–13]. Subsequent studies identified a number of sequence-specific transcription factors that bind Ig enhancers, including NF-κB and the basic helix-loop-helix (bHLH) activator E12/E47 [14,15]. Both proteins were subsequently shown to be critical effectors of a variety of processes in animal development and disease, including programmed cell death, inflammation, and lymphocyte differentiation [16,17].
Remote enhancers located tens or even hundreds of kilobases from the target gene are a distinctive property of metazoans that is absent in yeast and rarely seen in plants [18,19]. Such long-range interactions open the door to complex gene control, whereby a given gene can be used in a variety of developmental or physiological processes, as discussed below.
I will first review the general properties of metazoan enhancers, particularly those engaged in developmental processes, and then discuss several well-defined examples.
Principles of Enhancer Function
Activator Synergy
Most developmental enhancers have a number of shared properties, regardless of the overall size of the genomes from which they originate [20–22]. Such enhancers are typically 200 bp to 1 kb in length. They contain multiple binding sites for two or more classes of sequence-specific transcription factors [23]. A recurring theme is the use of at least two different activators to regulate expression, such as NF-κB and AP1 in the case of the SV40 enhancer mentioned above [6,7]. Many enhancers also contain binding sites for sequence-specific repressors, which exclude expression in inappropriate tissues, e.g. [24,25].
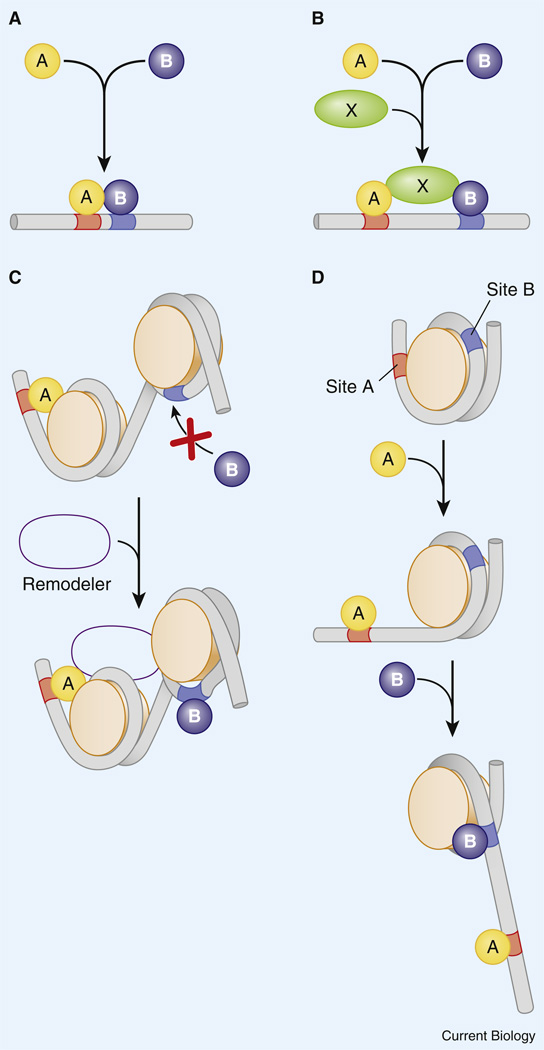
Several different modes of transcriptional synergy are known, including cooperative occupancy of linked sites via protein–protein interactions [26,27] and the coordinate recruitment of co-activators such as CBP to the DNA template [28,29]. It is also possible that different classes of activators recruit distinct co-activators, which in turn function synergistically to activate gene expression (Figure 1). For example, activator A might recruit CBP, which mediates acetylation of core histones, while activator B might recruit Swi/Snf, which remodels chromatin by displacing nucleosomes [30,31]. Finally, indirect modes of cooperative binding have been suggested, whereby activator A binds its target site and helps displace the associated nucleosome to facilitate binding of activator B to a neighboring site [32,33] (Figure 1C,D). Most of these mechanisms of activator synergy are non-exclusive, so it is possible, for example, that two activators bind cooperatively to linked sites and coordinately recruit one or more co-activators to the DNA template. Regardless of the exact mechanism, A–B activator synergy depends on close spatial linkage of the binding sites, typically within a half turn of the nucleosome.
Figure 1. Activator synergy.
Several nonexclusive mechanisms can be envisioned by which two transcriptional activators augment each other’s function. (A) Activators A and B cooperatively bind to linked sites. (B) A and B coordinately bind a third protein, X, which stabilizes the binding of A and B. (C) A recruits a histone remodeling protein that facilitates the binding of B. (D) A leads to ‘slippage’ of the nucleosome and thereby uncovers the B binding site. Reproduced with permission from [125].
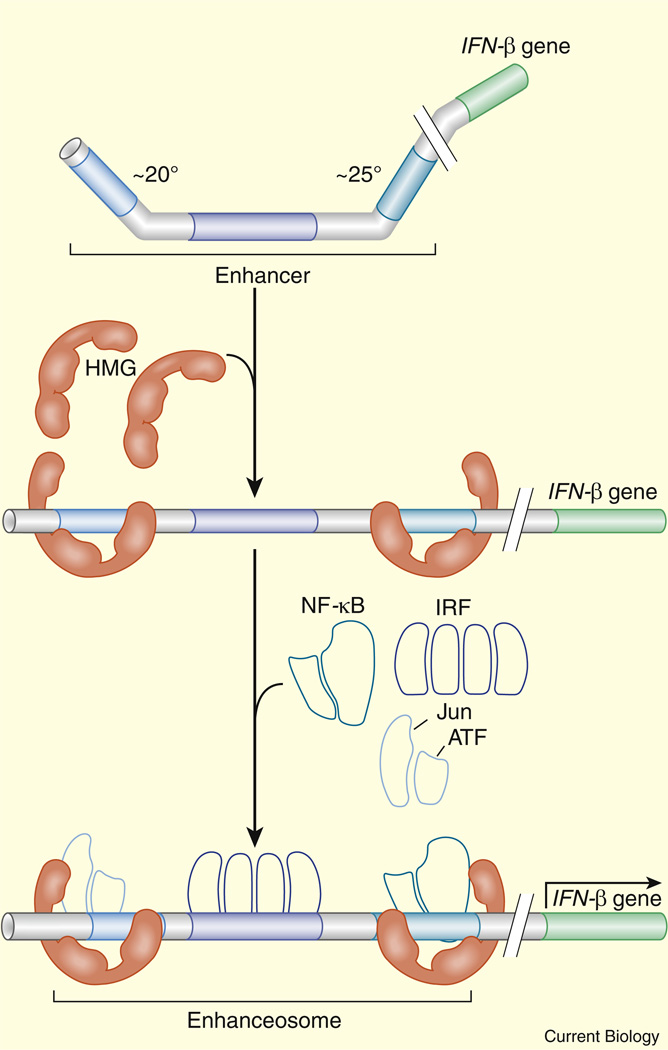
The best-characterized enhancer is the enhanceosome controlling the expression of the mammalian β-interferon gene in response to viral infection [34–36]. The enhanceosome is activated by three heterodimeric protein complexes, c-Jun/ATF-2, IRF3/IRF7 and NF-κB, which bind tightly linked recognition sequences that are highly conserved in evolution. A co-crystal of the enhanceosome along with the DNA binding domains of the three activator complexes reveals an extended composite protein surface spanning the length of the ~200 bp enhanceosome [37]. It has been suggested that this surface provides a template for the effective recruitment of co-activators such as CBP (Figure 2).
Figure 2. Coordinate recruitment of co-activators at the β-interferon enhanceosome.
HMG bends the enhanceosome and facilitates the binding of NF-κB, IRF, and Jun/ATF to linked sites. The three activator complexes form an extended surface for the recruitment of co-activators such as CBP. Reproduced with permission from [125].
DNA Looping
Once an appropriate combination of sequence-specific activators and co-activators has bound to an enhancer how does it control gene expression? There is considerable evidence that active enhancers loop to the promoter regions of target genes [38]. These loops can be visualized using chromosome conformation capture (3C) assays and various modifications thereof [39]. These methods are based on identifying the joining of distant DNA segments via PCR.
The exact relationship between the binding of activators and enhancer looping is unclear [40,41]. The Drosophila melanogaster Pax2 enhancer contains a discrete region that is important for activation at a distance, but not when the enhancer is placed immediately adjacent to a reporter gene [42]. Cohesins have been implicated as mediators of long-range looping [43,44]. Once the enhancer loops to the promoter it might stimulate transcription by the recruitment of general transcription factors such as the Mediator complex (MED) [45]. In cases of genes containing paused Pol II, activation might be achieved by the recruitment of transcriptional elongation factors such as pTEFb [46].
Enhancer–Promoter Interactions
Whole-genome assays suggest that genes are often contained within larger chromosomal domains that are flanked by insulator DNA [47,48]. Insulators — also known as ‘chromosomal barrier elements’ or ‘boundary elements’ — prevent enhancers located within one chromosomal domain from inappropriately activating genes located in neighboring domains [49–55]. In principle, chromosomal inversions or deletions can result in the rearrangement or loss of insulator DNA, and as a result, genes can acquire novel patterns of expression due to activation by enhancers that are normally located within a separate domain.
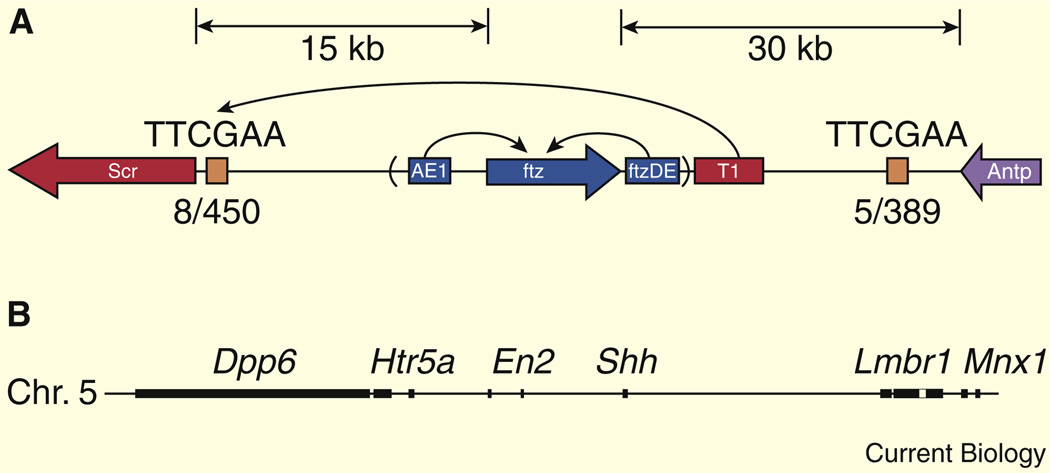
In some cases, enhancers can bypass a nearby gene in order to activate a more distal transcription unit. This is seen in the Drosophila Antennapedia complex, whereby the T1 enhancer is located 3′ of the fushi tarazu (ftz) transcription unit but bypasses ftz in order to activate the more distal Sex combs reduced (Scr) gene [56–58]. Selective T1–Scr interactions depend on a proximal tethering element located immediately 5′ of the Scr core promoter (Figure 3A). T1 can activate gene expression from the ftz promoter upon insertion of the ~300 bp tethering element. This type of enhancer–promoter specificity is also influenced by sequence elements contained within core promoters. TATA-containing promoters might preferentially interact with certain enhancers, while DPE-containing promoters are activated by different enhancers [59–61].
Figure 3. Long-range enhancer–promoter interactions.
(A) The T1 enhancer bypasses the ftz locus to activate Scr within the Antennapedia gene complex of Drosophila. Reproduced with permission from [58]. (B) The primary enhancer controlling Shh expression in the developing vertebrate limb bud is located within the intron of the Lmbr1 locus (unfilled box), which maps nearly one megabase from the Shh transcription start site. Reproduced with permission from [63].
Tethering elements, and promoter specificity, might enable remote enhancers to ignore nearby genes and activate distal transcription units. These and other mechanisms lead to remarkable examples of uncoupling of enhancers from their target genes. For example, the sonic hedgehog (Shh) gene is essential for the patterning of the neural tube and limbs [62]. These processes are controlled by separate enhancers, and those regulating expression in the notochord and floor plate are located relatively close to the Shh transcription unit. However, the enhancer that regulates Shh expression in the developing limb buds is located nearly one megabase away, within the intron of a neighboring locus (Lmbr) [63] (Figure 3B). Once enhancers are decoupled from their target genes, it is thus easy to imagine that they can be re-routed to produce novel patterns of gene expression during animal evolution.
The Question of Grammar
The enhanceosome represents an extreme example of cis-regulatory ‘grammar’ or ‘syntax’, as it contains a fixed arrangement of binding sites spanning the entire length of the enhancer [37]. ‘Grammar’ here refers to the phenomenon that spacing and arrangement of binding sites matter for the activity of the enhancer, just like the order of words in a sentence can affect its meaning. Just about any change in spacing between adjacent sites disrupts enhanceosome function, except the insertion of 10 bp of DNA, one turn of the helix, which maintains the orientation of adjacent heterodimer complexes [34–36]. Developmental enhancers have a more flexible arrangement of binding sites than the enhanceosome. For example, changing the spacing of adjacent Bicoid and Hunchback activator sites does not alter the even-skipped (eve) stripe 2 expression pattern in Drosophila, although there are diminished levels of expression [64]. Nonetheless, it is conceivable that developmental enhancers contain limited arrangements of fixed binding sites.
Just a handful of well-defined developmental enhancers were identified during the 1980s and 1990s. Back in the day, 5′ flanking regions were ‘blindly’ fused to a reporter gene and the fusion genes were examined in transgenic embryos after incorporation into the germline via microinjection assays [65]. Even so-called ‘short-cuts’, such as transient expression in injected sea urchin and mouse embryos, required the establishment of demanding microinjection methods [66,67].
The post-genome era provided an opportunity to identify putative enhancers using a host of computational methods, such as phylogenetic foot-printing [68,69], the identification of conserved non-coding sequences among distantly related genomes (e.g., pufferfish and mice). Additional methods include the computational identification of clusters of binding sites [70,71] and whole-genome chromatin immuno-precipitation (ChIP) assays [72,73]. Of course, these methods are not sufficient to identify enhancers, but they can be used to pinpoint the locations of putative enhancers within extended genomic intervals. The combination of computer and experiment has greatly augmented the collection of developmental enhancers, providing a foundation for investigating the question of whether developmental enhancers contain fixed arrangements of binding sites.
The analysis of enhancers that direct gene expression in the presumptive neurogenic ectoderm of the early Drosophila embryo suggests a limited grammar in the arrangement of Dorsal and Twist activator binding sites [74]. Dorsal is a Rel-containing transcription factor that is related to mammalian NF-κB, while Twist is a bHLH activator implicated in mesoderm development in a variety of animal embryos [75,76]. Dorsal activates Twist, and low levels of the two proteins work synergistically to activate a number of neurogenic genes (so-called ‘type 2 Dorsal target genes’), which restrict EGF and Dpp (TGFβ) signaling in the early embryo [74].
Although the six known type 2 enhancers are unrelated by simple sequence homology, they all contain closely linked Dorsal and Twist binding sites, and the asymmetric Twist site is oriented towards the adjacent Dorsal site. This arrangement fosters cooperative interactions between Dorsal and Twist, but only a subset of the binding sites displays such linkage. Recent computer simulations have been used to argue that linked binding sites might represent evolutionary ‘mirages’ [77]. However, this example of grammar does not depend on evolutionary constraint, but is seen as a convergent design feature among unrelated enhancers. Moreover, the importance of grammar in the interferon enhanceosome is clear-cut.
Thus, there is no denying that grammar occurs in certain enhancers. The question is whether it is a pervasive feature of developmental enhancers. Later in the review I will describe how changes in the arrangement of binding sites in the Drosophila Pax2 enhancer alter gene expression in the Drosophila eye [42].
The Proposed Importance of Off-Rates
Sequence-specific DNA binding proteins scan the DNA double helix and then dwell at favored recognition sequences [78,79]. The primary determinant of a ‘good’ or high-affinity binding site as opposed to a low-affinity site is the off-rate. DNA binding proteins dwell longer at preferred recognition sequences as compared with sequences that deviate from the optimal consensus sequence.
It is reasonable to suppose that critical recognition sequences must be stably occupied within an enhancer in order to augment transcription. Stable interactions are likely to be required for the recruitment of co-activators, such as CBP, as well as for other aspects of enhancer function such as looping to the core promoter. Stable occupancy depends not only on the intrinsic quality of the binding sites but also on protein–protein interactions, including cooperative binding and interactions with co-activator proteins. It is unlikely that current whole-genome methods, such as ChIP-sequencing, can distinguish between binding sites based on off-rates [80]. Such methods fail to provide kinetic measurements, but represent average states of binding site occupancy. Authentic binding sites responsible for the control of gene expression might depend on regulatory grammar such as protein–protein interactions at linked sites, as discussed earlier.
Function of Individual Enhancers in Animal Development
Below I describe a few specific examples of enhancers mediating localized patterns of gene expression during animal development. There is a particular emphasis on the role of enhancers as DNA templates for integrating complex positional information, including localized signaling molecules.
Xenopus Organizer
The dorsal-ventral patterning of the Xenopus laevis embryo is controlled by the famous Spemann-Mangold organizer, located at the dorsal lip of the blastopore [81]. The cells comprising the dorsal lip secrete a variety of BMP/TGFβ signaling antagonists, including Noggin and Chordin [82]. The homeobox gene, Goosecoid (Gsc), is one of the first genes to be activated in the presumptive organizer, where it is thought to participate in the subsequent regulation of patterning genes such as Noggin [83].
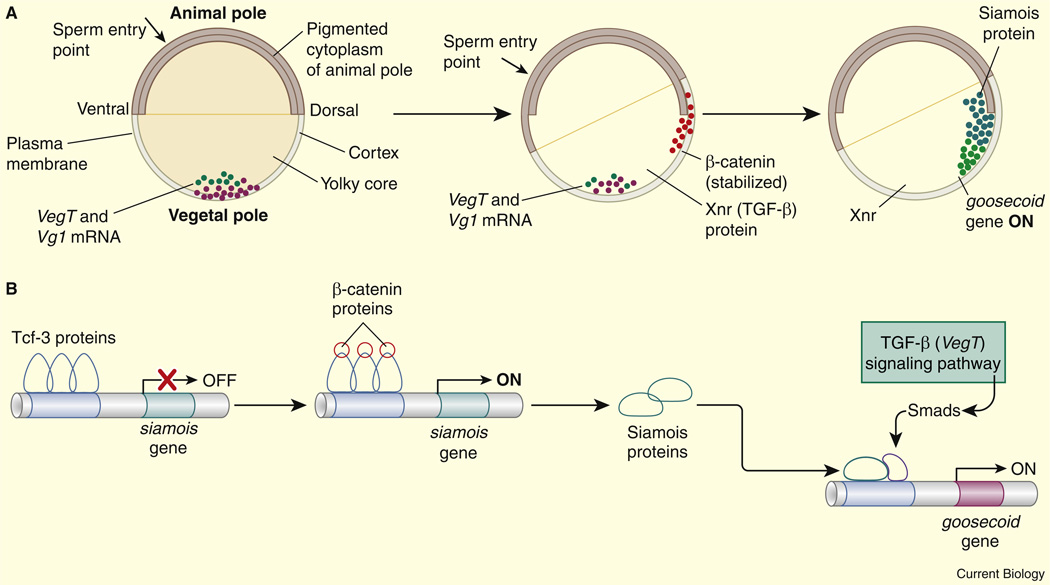
The Xenopus egg contains a number of localized maternal mRNAs, including Vg1 and VegT, the latter of which encodes a T-box transcription factor that activates Nodal-related genes (XNRs) [84,85]. VegT–XNR interactions create localized activation of pSmad transcription factors in vegetal regions of the early embryo. As pSmads become activated in the vegetal embryonic cells (blastomeres) a process of cortical rotation leads to the localized activation of β-catenin and the Lef/Tcf transcription factor along the presumptive dorsal surface of the early embryo [81,82]. Activated Tcf triggers the expression of the Siamois homeobox gene in these dorsal regions [86]. Blastomeres located at the intersection of the vegetal and dorsal regions express both Siamois and pSmads (Figure 4). These two sequence-specific transcription factors interact with a 5′ enhancer in the Gsc locus to activate Gsc expression and thereby help delineate the organizer [87].
Figure 4. Overlapping activators define the organizer.
(A) Xenopus blastula. Xnrs (Xenopus Nodal-related signaling molecules) and pSmads are expressed in vegetal regions of the embryo. The homeobox gene Siamois is selectively expressed along the presumptive dorsal surface of the embryo. The two activators, pSmads and Siamois, are co-expressed in the region of the presumptive organizer where they activate the expression of the Goosecoid (Gsc) gene. (B) Summary of Gsc regulation in the organizer. The Smads and Siamois interact with Gsc regulatory sequences and activate expression. Reproduced with permission from [125].
Ciona Heart Primordium
The beating heart of the adult sea squirt Ciona intestinalis arises from a single pair of blastomeres (B7.5) at the 110-cell stage of embryogenesis [88]. A key step in the specification of the heart is the restricted expression of the bHLH regulatory gene MesP in the B7.5 blastomeres. MesP activates a number of subordinate genes that are required for the directed migration and differentiation of the cardiomyocytes in the developing tadpole [89].
MesP is regulated by a 5′ enhancer that contains linked binding sites for two distinct activators, Lhx3 and Tbx6 [90]. Lhx3 is expressed throughout the presumptive endoderm and extends into the B7.5 blastomere, which is located at the boundary between the endoderm and presumptive tail muscles. Tbx6 is expressed in the presumptive tail muscles as well as B7.5. Thus, only the B7.5 blastomeres express both Lhx3 and Tbx6. An outstanding question, posed earlier, is whether a fixed arrangement of Siamois/pSmad and Lhx3/Tbx6 binding sites are essential for the accurate expression of Gsc in the Xenopus organizer and MesP in Ciona cardiomyocytes.
Drosophila Eye
Pax2 is essential for the specification of cone cells within the adult fly eye. It is regulated by a 362 bp enhancer located ~850 bp upstream of the Pax2 transcription start site [10,42]. Localized expression within presumptive cone cells depends on Notch and EGF signaling, along with cellular determinants, including Lozenge (Lz), a Runx-containing transcription factor. The Pax2 enhancer contains a series of 12 binding sites for all three critical regulators: Su(H) (Notch signaling), Ets (EGF signaling) and Lz. Altering the arrangement of binding sites causes the modified enhancer to be active in the R1 and R6 photoreceptor cells rather than cone cells [42]. These experiments clearly illustrate the importance of enhancer structure, or grammar, in the cell-specific regulation of Pax2 expression. However, there is no evidence for long-range arrangements of binding sites along the length of the enhancer. Rather, there is rapid turnover of binding sites in the Pax2 enhancers of divergent drosophilids, such as D. pseudoobscura [42]. It would appear that grammar may be limited to just a subset of closely linked sites, as seen for the Dorsal/Twist linkage in type 2 neurogenic enhancers.
Caenorhabditis elegans Nervous System
The gustatory (taste) neurons of the nematode C. elegans, ASER and ASEL, are specified by a zinc finger transcription factor, CHE-1 [91]. A combination of conventional genetics and microarray assays identified a number of putative CHE-1 target genes that are specifically expressed in the taste neurons. The 5′ regulatory regions of a number of these genes contain a critical cis-regulatory element that is essential for their expression in taste neurons [91]. The CHE-1 target gene cog-1 contains two copies of this element [92], which might work in a partially redundant fashion to ensure robust expression of the gene in response to environmental fluctuations, as seen for ‘shadow enhancers’ in Drosophila (see below). It is unlikely that the CHE-1 recognition sequence is sufficient for the activation of gene expression in taste neurons. Many target genes exhibit asymmetric expression in the left-right pair of taste neurons, ASELand ASER, respectively. For example, cog-1 is specifically expressed in the ASER neuron, and the 5′ regulatory region contains conserved binding motifs that are likely to work in concert with CHE-1 to generate a restricted expression pattern.
Examples of Modular Enhancers in Development
While the preceding examples focused on the regulation of gene expression by individual enhancers, I now consider examples of genes that are regulated by multiple enhancers.
Drosophila segmentation
The expression of pair-rule genes in two-segment wide stripes in the Drosophila blastoderm embryo provides one of the most dramatic examples of combinatorial gene control in animal development [93,94]. The pair-rule stripes arise after broadly expressed maternal activators (Bicoid and Caudal) and localized gap repressors (Hunchback, Krüppel, Knirps and Giant). The first hint regarding how these broadly distributed regulatory factors produce sharp stripes of gene expression came from the analysis of the 500 bp eve stripe 2 enhancer [95,96]. This enhancer contains twelve binding sites; six activator sites and six repressor sites. Bicoid and Hunchback bind the activator sites to augment eve expression throughout the anterior half of the embryo (where the two proteins are present at highest levels). The Giant and Krüppel gap proteins bind to the 6 repressor sites to define the anterior and posterior borders of the stripe 2 pattern, respectively [64,97].
eve is regulated by 5 separate enhancers located upstream and downstream of the transcription unit [98]. Each of these enhancers employs the same basic logic described for the stripe 2 enhancer: broadly distributed activators working together with localized gap repressors to define the stripe borders. Three of the gap repressors, Krüppel, Knirps, and Giant, interact with a common co-repressor protein, CtBP, which functions as a short-range repressor [99]. That is, CtBP must bind within 50–100 bp of upstream activators or the core promoter in order to inhibit gene expression. Such short-range repression ensures that the five eve enhancers work independently of one another to produce the complete seven-stripe pattern of gene expression.
Mouse Hindbrain
Hox genes control the anterior-posterior patterning of most or all metazoans [100]. They are usually located within tightly regulated complexes or clusters. Both local and long-range cis-regulatory DNA elements control complex patterns of Hox expression during development. The best-characterized vertebrate Hox enhancers are those controlling the expression of Hoxb1 in the mouse hindbrain [101,102]. A 3′ enhancer located downstream of the Hoxb1 transcription unit activates expression in the neurogenic ectoderm, in a broad domain spanning rhombomeres 3 through 5 (R3–R5). A 5′ silencer element containing retinoic acid response elements and Krox20 binding sites represses Hoxb1 expression in R3 and R5 and restricts expression to the R4 rhombomere.
Hoxb1 expression is subsequently maintained in R4 via a 335 bp 5′ enhancer that mediates auto-regulation. The 5′ auto-regulatory enhancer (R4 ARE) contains a series of linked binding sites for the Hoxb1 protein and a ‘ubiquitous’ homeobox activator, Pbx. Hox–Pbx protein–protein interactions are thought to result in cooperative occupancy of the linked binding sites, and also foster synergistic recruitment of co-activators [103]. Hoxb1 regulation is reminiscent of eve stripe 2 regulation, in that both systems employ spatially localized repressors for delineating the limits of gene expression. However, the retinoic acid repressor functions over long distances, nearly 2 kb, to silence the Hoxb1 transcription unit in the R3 and R5 rhombomeres.
In addition to local enhancers that regulate individual Hox genes, such as Hoxb1, it appears that at least some vertebrate Hox clusters are coordinately regulated by remote sequences that have the properties of the locus control region (LCR) controlling the temporal order of globin gene expression during hematopoiesis [104,105]. The global control region (GCR) of the Hoxd complex ensures the sequential expression of the individual Hoxd genes along the anterior-posterior axis of developing limbs. There is no evidence that such long-range elements regulate Hox genes in invertebrates such as Drosophila. LCR/GCR regulatory DNAs might thus be a distinctive innovation of vertebrates.
Evolution of Gene Expression Patterns
There is growing evidence that changes in gene expression underlie the evolution of novelty and divergence of animal morphology, although there are warring schools of thought regarding the molecular basis for animal diversity [106]. There are those who ascribe most such change to sequential modifications in protein coding sequences, while others insist that changes in cis-regulatory DNA are the key agent of animal diversity. There is little doubt that both mechanisms contribute, with DNA duplication as a critical driving force for both types of sequence evolution.
A classical example that highlights the contributions of both protein and cis-regulatory evolution in animal physiology is seen at the β-globin locus of vertebrates [107]. The β-globin gene, and its flanking regulatory sequences, underwent a series of duplication events. γ-globin genes are selectively expressed during embryogenesis and the encoded proteins possess a high-affinity for oxygen. This permits the mammalian fetus to compete for the oxygen supply. Upon birth, the embryonic globin genes are shut off and the β-globin genes are activated. The adult globin protein has a lower affinity for oxygen, which is important for gas exchange in deep tissues. In this example, DNA duplication and divergence generated both regulatory DNAs and protein coding sequences with related but distinct activities.
Insect Patterning Genes
Developmental control genes often exhibit distinctive expression patterns in divergent insects. For example, the dorsal-ventral patterning gene sim is critical for the specification of the ventral midline of the insect central nervous system [108]. In Drosophila, the sim expression pattern encompasses just a single row of cells spanning the length of the nerve cord. However, in other insects, particularly the honeybee (Apis mellifera), the sim expression pattern is broader and encompasses several cells in width, resulting in an expanded midline [109].
In both Drosophila and Apis, the sim expression pattern is controlled by a proximal enhancer located immediately upstream of the core promoter. The Drosophila sim enhancer is regulated by a combination of Dorsal, Twist, and Notch signaling. The latter regulatory input restricts sim expression to a single line of cells on either side of the presumptive mesoderm (which is the source of the localized Notch signaling protein, Delta). However, the Apis sim enhancer contains high-affinity Twist binding sites in place of Notch response elements [109], resulting in an expanded sim expression pattern.
‘Shadow’ enhancers represent another potential source for the evolution of novel patterns of gene expression. Whole-genome ChIP-chip assays suggest that many developmental patterning genes active in the early Drosophila embryo contain secondary enhancers, which produce patterns of gene expression that are the same or similar to those generated by more proximal primary enhancers [110,111]. For example, sog encodes a secreted BMP inhibitor that is related to Xenopus Chordin [112]. The computational identification of clustered Dorsal binding sites identified an enhancer within the first intron of the sog transcription unit that recapitulates the endogenous expression pattern in early embryos [70]. ChIP-chip assays identified a cluster of Dorsal, Twist, and Snail binding sites that co-incides with this intronic enhancer [110]. These assays also identified a second binding cluster located nearly 25 kb upstream of the sog transcription start site, on the other side of a neighboring gene. Despite this remote location, the 5′ binding cluster functions as a sog enhancer when tested in transgenic embryos [111]. The shadow enhancer produces a pattern of gene expression that is similar to that seen for the primary, intronic enhancer. Recent studies suggest that shadow enhancers might buffer the expression of developmental patterning genes in response to environmental and genetic fluctuations, such as changes in temperature [113–115]. Although fixed in populations by conferring developmental fitness, shadow enhancers might be able to deviate from primary enhancers to produce novel patterns of gene expression at later stages of development.
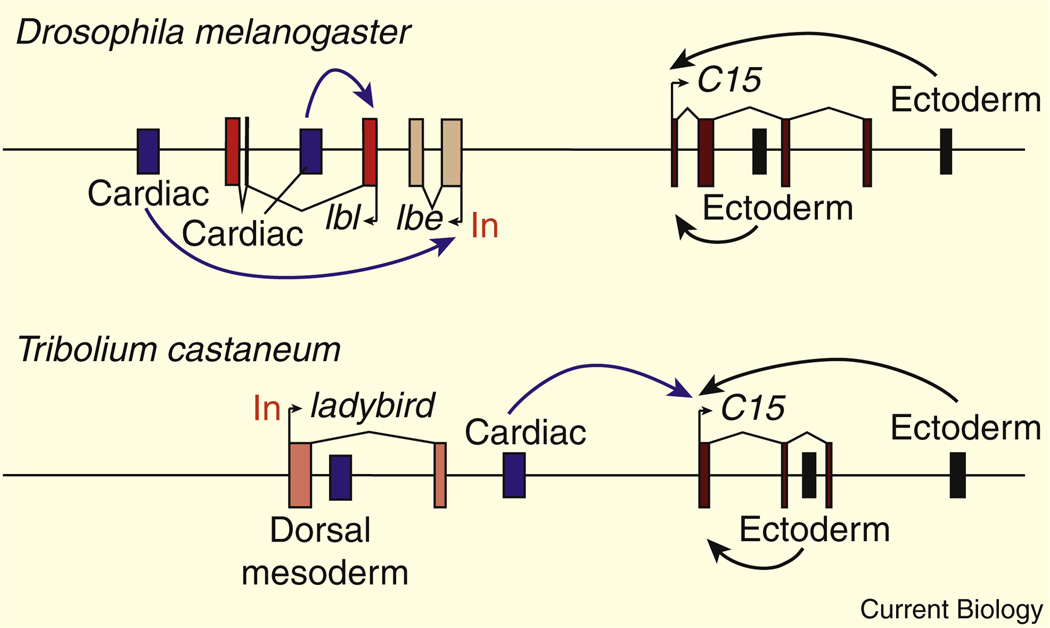
The preceding examples focus on changes in enhancer sequences that can produce novel patterns of gene expression in evolution. An example of re-directing enhancer–promoter interactions is seen in the Tinman gene complex (Tin-C), which contains a series of homeobox genes that control the patterning of cardial and pericardial cells in the Drosophila heart [116]. The ladybird gene within the Tin-C is regulated by a 3′ enhancer, which mediates expression in pericardial cells within the developing heart [117]. The ladybird promoter region contains paused Pol II and works as an insulator [118]. Consequently, the 3′ enhancer does not activate the C15 gene, which is located upstream of ladybird (summarized in Figure 5). However, in the flour beetle Tribolium castaneum a chromosomal inversion positions the ladybird 3′ enhancer upstream of the C15 gene [118], such that it is now able to activate C15 expression (Figure 5).
Figure 5. Redirecting a conserved enhancer.
In Drosophila melanogaster, 3′ enhancers (blue) activate ladybird expression (both the lbl and lbe genes) in the developing cardiac mesoderm. The lbe promoter contains paused Pol II, and has an enhancer blocking activity, preventing the activation of the neighboring C15 gene. In the flour beetle (Tribolium castaneum) the single ladybird gene is inverted relative to the orientation of the C15 locus. As a result the 3′ cardiac enhancer is able to activate C15 expression. Reproduced with permission from [118].
Pelvic Fin Reduction in Sticklebacks
While the preceding examples produce only modest changes in morphology, the reduction of pelvic fins in stickleback fish provides a more dramatic example. Sticklebacks are found worldwide in both ocean and fresh water habitats. These different populations have been isolated for over 10,000 years due to the recession of glaciers from the ice age [119]. The ancestral fish contained a pair of prominent pelvic fins, and this is the prevailing phenotype among open ocean populations where vigorous and extended swimming is a mandatory aspect of life. However, a number of isolated fresh water populations have independently loss their pelvic fins. This loss might help sticklebacks escape the grasping jaws of predatory fish.
Crosses between sticklebacks from different populations allowed the genetic mapping of a number of loci responsible for various morphological variations, and a single locus was found to be a major determinant of reduced pelvic fins [119]. This region contains the Pitx1 homeobox gene, which has been implicated in a variety of developmental processes, including the specification of the mandible, anterior pituitary, and hindlimbs in mammalian embryos. The Pitx1 regulatory region appears to contain a series of separate enhancers for these various regulatory activities [120].
Pitx1 is selectively expressed in the developing hindlimbs, but not forelimbs. Ectopic expression of Pitx1 in the forelimbs is sufficient to transform forelimbs into hindlimbs [121]. The pelvic fins of sticklebacks are thought to be homologous to the hindlimbs of higher vertebrates. Causal DNA sequence polymorphisms associated with the loss of pelvic fins do not map within the Pitx1 coding region. Instead, they map within the 5′ regulatory region. Different stickleback populations lacking pelvic fins exhibit deletions of a 5′ ~500 bp enhancer that activates Pitx1 expression in the developing pelvic fins [122]. This enhancer is located in a ‘fragile’ region of the genome, near the telomere of linkage group 7. It is therefore possible that the adaptive loss of pelvic fins is facilitated by the fortuitous location of Pitx1 in the stickleback genome, a location that might foster a high rate of chromosomal deletions.
The modularity of the Pitx1 regulatory DNA is crucial for this loss of pelvic fins without compromising its other activities in development. In mammals, 3′ enhancers are important for different aspects of Pitx1 function, such as the development of the anterior pituitary. These enhancers are unaffected by the chromosomal deletions that remove the 5′ enhancer in sticklebacks.
Reconstructing the Past
The study of pelvic fin reduction in sticklebacks represents a harbinger of the future of ‘evo-devo’ research, namely, the ability to reconstruct past evolutionary processes in the laboratory. The ancestral pelvic fin enhancer was attached to the Pitx1 transcription unit and the fusion gene was introduced into a stickleback population containing rudimentary pelvic fins [122]. Remarkably, the transgene was sufficient to restore the pelvic fins in this population, even though they lost their fins over 10,000 years ago (Figure 6). Despite this loss, the entire genetic machinery is competent to respond to the critical expression of Pitx1 and form fully normal pelvic fins. Thus, it would appear that there has not been significant genetic drift in the regulatory DNAs of the Pitx1 downstream target genes required for fin morphogenesis.
Figure 6. Reconstructing the past.
Ancestral stickleback populations contain pelvic fins (A,C). Certain freshwater populations have reduced fins (B; arrowhead). The fins are restored in these populations upon expression of a transgene containing 2.5 kb of the 5′ flanking region of the Pitx1 locus from a population with pelvic fins. This regulatory sequence was attached to the coding region of Pitx1 derived from a population lacking pelvic fins. Expression of this transgene restores the pelvic fins in populations normally lacking them — compare (C) with transgene to (D) lacking the transgene. Reproduced with permission from [122].
The loss of stickleback pelvic fins is the premier example of retrograde evolution, the loss of complexity. What is the molecular basis for the acquisition of complexity during evolution? In future years, it might be possible to reconstruct key morphological transitions in animal evolutionary diversity. For example, the gene network underlying the emigration of neural crest cells from the dorsal neural tube of vertebrate embryos is nearly elucidated [123]. It might be possible to promote neural crest formation in an ancestral chordate lacking neural crest, such as amphioxus, by expressing key regulatory genes in the dorsal neural tube (e.g., FoxD and Msx) [124]. Such genes are present in the amphioxus genome, but they are not normally expressed in the dorsal neural tube as seen in vertebrates. The forced expression of these genes using appropriate FoxD and Msx enhancers from vertebrates might be sufficient to produce aspects of neural crest formation in amphioxus. Similarly, forced expression of Shh in the fleshy fins of ‘transitional’ fish might be sufficient to trigger the development of rudimentary tetrapod limbs.
Acknowledgements
I have always relied on the kindness of strangers and in this regard I thank the anonymous reviewers of the manuscript for correcting many errors and greatly improving the text. I also thank the NIH and NSF for providing research support.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers J, Olson L, Tyndall C, Schaffner W. Transcriptional ‘enhancers’ from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982;10:7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schirm S, Weber F, Schaffner W, Fleckenstein B. A transcription enhancer in the Herpesvirus saimiri genome. EMBO J. 1985;10:2669–2674. doi: 10.1002/j.1460-2075.1985.tb03986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spandidos DA, Wilkie NM. Host-specificities of papilloma-virus, Moloney murine sarcoma virus and simian virus 40 enhancer sequences. EMBO J. 1983;2:1193–1199. doi: 10.1002/j.1460-2075.1983.tb01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen U, Sharp PA. Sequences controlling in vitro transcription of SV40 promoters. EMBO J. 1983;2:2293–2303. doi: 10.1002/j.1460-2075.1983.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- 7.Phares W, Herr W. Functional similarities between human immunodeficiency virus type 1 and simian virus 40 kappa B proto-enhancers. J. Virol. 1991;65:2200–2210. doi: 10.1128/jvi.65.5.2200-2210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochem. Biophys. Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 9.Vallabhapurapu S, Karin M. Regulation and function of NF-kB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 10.Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 11.Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 12.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 13.Mercola M, Wang XF, Olsen J, Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983;221:663–665. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- 14.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 15.Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 16.Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–239. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 17.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 18.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 19.Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 20.Kirchhamer CV, Yuh CH, Davidson EH. Modular cis-regulatory organization of developmentally expressed genes: two genes transcribed territorially in the sea urchin embryo, and additional examples. Proc. Natl. Acad. Sci. USA. 1996;93:9322–9328. doi: 10.1073/pnas.93.18.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veitia RA. One thousand and one ways of making functionally similar transcriptional enhancers. Bioessays. 2008;30:1052–1057. doi: 10.1002/bies.20849. [DOI] [PubMed] [Google Scholar]
- 22.Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 23.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1991;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 25.Mellerick DM, Nirenberg M. Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev. Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- 26.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. lambda Repressor and cro–components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 27.Oehler S, Alberti S, Müller-Hill B. Induction of the lac promoter in the absence of DNA loops and the stoichiometry of induction. Nucleic. Acids Res. 2006;34:606–612. doi: 10.1093/nar/gkj453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and co-activators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 29.Struhl K. Transcriptional activation: mediator can act after preinitiation complex formation. Mol. Cell. 2005;17:752–754. doi: 10.1016/j.molcel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Ptashne M, Gann A. Transcription initiation: imposing specificity by localization. Essays Biochem. 2001;37:1–15. doi: 10.1042/bse0370001. [DOI] [PubMed] [Google Scholar]
- 32.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuey DJ, Parker CS. Bending of promoter DNA on binding of heat shock transcription factor. Nature. 1986;323:459–461. doi: 10.1038/323459a0. [DOI] [PubMed] [Google Scholar]
- 34.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 35.Merika M, Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 36.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 37.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 39.van Berkum NL, Dekker J. Determining spatial chromatin organization of large genomic regions using 5C technology. Methods Mol. Biol. 2009;567:189–213. doi: 10.1007/978-1-60327-414-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeller RW, Griffith JD, Moore JG, Kirchhamer CV, Britten RJ, Davidson EH. A multimerizing transcription factor of sea urchin embryos capable of looping DNA. Proc. Natl. Acad. Sci. U S A. 1995;92:2989–2993. doi: 10.1073/pnas.92.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostyuchenko M, Savitskaya E, Koryagina E, Melnikova L, Karakozova M, Georgiev P. Zeste can facilitate long-range enhancer-promoter communication and insulator bypass in Drosophila melanogaster. Chromosoma. 2009;118:665–674. doi: 10.1007/s00412-009-0226-4. [DOI] [PubMed] [Google Scholar]
- 42.Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Négre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 49.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 50.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief. Funct. Genomic Proteomic. 2009;8:276–282. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 2009;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 53.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 55.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 56.Gindhart JG, Jr, King AN, Kaufman TC. Characterization of the cis-regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995;139:781–795. doi: 10.1093/genetics/139.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 64.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 65.Hiromi Y, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 66.Hough-Evans BR, Franks RR, Cameron RA, Britten RJ, Davidson EH. Correct cell-type-specific expression of a fusion gene injected into sea urchin eggs. Dev. Biol. 1987;121:576–579. doi: 10.1016/0012-1606(87)90193-x. [DOI] [PubMed] [Google Scholar]
- 67.Krumlauf R, Holland PW, McVey JH, Hogan BL. Developmental and spatial patterns of expression of the mouse homeobox gene, Hox 2.1. Development. 1987;99:603–617. doi: 10.1242/dev.99.4.603. [DOI] [PubMed] [Google Scholar]
- 68.Muller F, Blader P, Straehle U. Search for enhancers: teleost models in comparative genomic and transgenic analysis of cis regulatory elements. Bioessays. 2002;24:564–572. doi: 10.1002/bies.10096. [DOI] [PubMed] [Google Scholar]
- 69.Satija R, Pachter L, Hein J. Combining statistical alignment and phylogenetic footprinting to detect regulatory elements. Bioinformatics. 2008;24:1236–1242. doi: 10.1093/bioinformatics/btn104. [DOI] [PubMed] [Google Scholar]
- 70.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papatsenko D, Levine M. A rationale for the enhanceosome and other evolutionarily constrained enhancers. Curr. Biol. 2007;17:R955–R957. doi: 10.1016/j.cub.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 75.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo–shaping and transducing a morphogen gradient. Curr. Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 76.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev. Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 77.Lusk RW, Eisen MB. Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers. PLoS Genet. 2010;6:e1000829. doi: 10.1371/journal.pgen.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. Trends Biochem. Sci. 1988;13:207–211. doi: 10.1016/0968-0004(88)90085-0. [DOI] [PubMed] [Google Scholar]
- 79.Barker A, Fickert R, Oehler S, Müller-Hill B. Operator search by mutant Lac repressors. J. Mol. Biol. 1998;278:549–558. doi: 10.1006/jmbi.1998.1729. [DOI] [PubMed] [Google Scholar]
- 80.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 81.Moon RT, Kimelman D. From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus. Bioessays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 82.Harland RM. Neural induction in Xenopus. Curr. Opin. Genet. Dev. 1994;45:543–549. doi: 10.1016/0959-437x(94)90070-j. [DOI] [PubMed] [Google Scholar]
- 83.Cho KW, Blumberg B, Steinbeisser H, De Robertis EM. Molecular nature of Spemann’s organizer: the role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51:861–867. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- 85.Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- 86.Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 87.Koide T, Hayata T, Cho KW. Xenopus as a model system to study transcriptional regulatory networks. Proc. Natl. Acad. Sci. USA. 2005;102:4943–4948. doi: 10.1073/pnas.0408125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev. Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 90.Christiaen L, Stolfi A, Davidson B, Levine M. Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev. Biol. 2009;328:552–560. doi: 10.1016/j.ydbio.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 91.Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Meara MM, Bigelow H, Flibotte S, Etchberger JF, Moerman DG, Hobert O. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics. 2009;181:1679–1686. doi: 10.1534/genetics.108.097832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaeger J, Reinitz J. On the dynamic nature of positional information. Bioessays. 2006;28:1102–1111. doi: 10.1002/bies.20494. [DOI] [PubMed] [Google Scholar]
- 94.Levine M. A systems view of Drosophila segmentation. Genome Biol. 2008;9:207. doi: 10.1186/gb-2008-9-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 96.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 98.Jaynes JB, Fujioka M. Drawing lines in the sand: even skipped et al. and parasegment boundaries. Dev. Biol. 2004;269:609–622. doi: 10.1016/j.ydbio.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 101.Ferretti E, Cambronero F, Tümpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 2005;25:8541–8552. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferretti E, Marshall H, Pöpperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 103.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahajan MC, Karmakar S, Weissman SM. Control of beta globin genes. J. Cell Biochem. 2007;102:801–810. doi: 10.1002/jcb.21507. [DOI] [PubMed] [Google Scholar]
- 105.Spitz F, Gonzalez F, Duboule DA. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 106.Pennisi E. Deciphering the genetics of evolution. Science. 2008;321:760–763. doi: 10.1126/science.321.5890.760. [DOI] [PubMed] [Google Scholar]
- 107.Noordermeer D, de Laat W. Joining the loops: beta-globin gene regulation. IUBMB Life. 2008;60:824–833. doi: 10.1002/iub.129. [DOI] [PubMed] [Google Scholar]
- 108.Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 109.Zinzen RP, Cande J, Ronshaugen M, Papatsenko D, Levine M. Evolution of the ventral midline in insect embryos. Dev. Cell. 2006;11:895–902. doi: 10.1016/j.devcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 110.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Francois V, Solloway M, O’Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 1994;8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 113.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perry MW, Boettiger AN, Bothma J, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hobert O. Gene regulation: enhancers stepping out of the shadow. Curr. Biol. 2010;20:R697–R699. doi: 10.1016/j.cub.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 116.Zaffran S, Xu X, Lo PC, Lee HH, Frasch M. Cardiogenesis in the Drosophila model: control mechanisms during early induction and diversification of cardiac progenitors. Cold Spring Harb. Symp. Quant. Biol. 2002;67:1–12. doi: 10.1101/sqb.2002.67.1. [DOI] [PubMed] [Google Scholar]
- 117.Liu J, Qian L, Han Z, Wu X, Bodmer R. Spatial specificity of mesodermal even-skipped expression relies on multiple repressor sites. Dev. Biol. 2008;313:876–886. doi: 10.1016/j.ydbio.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cande JD, Chopra VS, Levine M. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development. 2009;136:3153–3160. doi: 10.1242/dev.038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 120.Lanctôt C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 121.DeLaurier A, Schweitzer R, Logan M. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev. Biol. 2006;299:22–34. doi: 10.1016/j.ydbio.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 122.Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell. Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 124.Yu JK. The evolutionary origin of the vertebrate neural crest and its developmental gene regulatory network–insights from amphioxus. Zoology (Jena) 2010;113:1–9. doi: 10.1016/j.zool.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 125.Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 6th Edition. Benjamin Cummings; 2007. [Google Scholar]