Abstract
Alterations in RNA splicing are associated with cancer, but it is not clear whether they result from malignant transformation or have a causative role. We show here that hepatocyte-specific deletion of splicing factor SRSF3 impairs hepatocyte maturation and metabolism in early adult life, and mice develop spontaneous hepatocellular carcinoma (HCC) with aging. Tumor development is preceded by chronic liver disease with progressive steatosis and fibrosis. SRSF3 protects mice against carbon tetrachloride-induced fibrosis and carcinogenesis, and suppresses inclusion of the profibrogenic EDA exon in fibronectin 1. Loss of SRSF3 increases expression of insulin like growth factor 2 (Igf2) and the A-isoform of the insulin receptor (Insr-A) allowing aberrant activation of mitogenic signaling, promotes aberrant splicing and expression of epithelial to mesenchymal transition (EMT) genes, and activates Wnt/β-catenin signaling leading to c-Myc induction. Finally, SRSF3 expression is either decreased or the protein miss-localized in human HCC.
Conclusion
Our data suggest a potential role for SRSF3 in preventing hepatic carcinogenesis by regulating splicing to suppress fibrosis, mitogenic splicing and EMT. Thus, these mice may provide an attractive model to discover the pathogenic mechanisms linking aberrant pre-mRNA splicing with liver damage, fibrosis and HCC.
Keywords: RNA, liver, insulin receptor, IGF2, fibronectin
Introduction
Hepatocellular carcinoma (HCC) accounts for 75–80% of primary liver cancer (1) and usually arises after years of liver disease (2). Cirrhosis and hepatitis resulting from chronic fibrosis and inflammation is found in >80% of cases of HCC (3). Although a large number of genes and signaling pathways have been implicated in the development of HCC, the molecular mechanisms underlying both the proliferation, tumorigenesis and metastasis of HCC, and the increased risk conferred by non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) are still poorly understood.
A growing body of evidence suggests that cancer is associated with aberrant RNA splicing (4–7). SRSF3 is the smallest member of the SR protein family of RNA-binding proteins that promote RNA splicing at constitutive as well as alternatively-spliced exons (8, 9). It has been implicated in blastocyst formation (10), B cell development (11) and somatic cell reprogramming (12). SRSF3 auto-regulates its own expression through splicing of an exon containing a premature termination codon (13, 14), but is also regulated by other cues such as TGFβ1 signaling, Wnt signaling and senescence (15, 16). SRSF3 expression is also regulated during G1/S (17) and controls the G2/M transition of immortal rat fibroblasts (18), which has led to the suggestion that SRSF3 is a proto-oncogene.
Recently we demonstrated a role for SRSF3 in controlling hepatocyte differentiation, and glucose and lipid metabolism (19) using a hepatocyte-specific SRSF3 knockout mouse model (SRSF3-HKO). Here we report that genetic deletion of SRSF3 in hepatocytes causes fibrosis, steatohepatitis and the development of metastatic HCC with aging. Our results show that loss of an RNA splicing factor can precede and predispose to carcinogenesis, and in this regard SRSF3 appears to function like a tumor suppressor gene. Dysregulation of splicing may thus be an early event in cancer initiation rather than late event following malignant transformation. We also report that SRSF3 expression is decreased in >50% of human HCC, so the SRSF3-HKO mouse model may be very useful for understanding human liver disease.
Materials and Methods
Full details of experimental procedures and antibodies are provided in the Supporting Material and Methods. Microarray data is available at GEO Accession Number GSE57021.
Results
Spontaneous Development of Hepatocellular Carcinomas in SRSF3-HKO Mice
Given the liver damage and regeneration phenotype in the young SRSF3-HKO (HKO) mice, we were interested in whether this phenotype would worsen with age. Though the HKO mice are lighter at 1 month, the body weights normalize by 6 months and mice are heavier at 12 months. Surprisingly, after 12 months the HKO mice lose weight (Fig. 1A) and show increased mortality (Fig. 1B). HKO mice initially have small livers but liver weight normalizes by 6 months then there is an abrupt increase in liver weight at 18–24 months (Fig. 1C)
Fig. 1. Spontaneous development of HCC in SRSF3HKO liver.
(A) Body weights (BW) of WT and HKO mice at indicated ages. (B) Kaplan-Meier survival plot, n=37/genotype. (C) Liver weight and liver to BW ratio at indicated ages. (D) Representative photographs of livers from WT and HKO mice. Small tumors are indicated by arrows at 12 months. Scale bar represents 0.5 cm. (E) H&E stained liver sections (20×) from 24-month WT and HKO mice. A low power image (5×) of a tumor containing HKO liver is also shown. Small tumors are steatotic with large circular lipid droplets. Late stage metastatic tumors show loss of cell-cell contacts and necrosis. TUNEL stained sections showing apoptotic cell death in HKO liver. Tumor is outlined on the section. Scale bar represents 100 µm. (F) Representative photographs of WT and HKO lungs and spleens. Macro-metastases are marked by arrows.
Macroscopic examination reveals atypical liver nodules at 12 months and HCC at 24 months in 100% of the HKO mice (Fig. 1D). There is no sex-bias for tumor development as both male (8 of 8) and female (15 of 15) mice develop tumors (Table S1). Most tumors are densely cellular with irregular nuclei and morphological pleomorphism (Fig. 1E). Smaller and earlier tumors show a steatotic phenotype, whereas metastatic tumors show evidence for loss of cell-cell contacts and necrosis (Figs. 1E and S1A). Apoptosis is increased in the tumors (Figs. 1F and S1B). Many of tumor-bearing mice (22%) have visible macro-metastases in the lung (Fig. 1F) and most mice (60%) have micro-metastases (Fig. S1C). HKO mice also have enlarged spleens (Fig. 1F) at 24 months.
Deletion of Hepatic SRSF3 Causes Lipodystrophy and Steatosis
The tumor bearing HKO show reduced liver glycogen storage (Fig. S2A) and mice have lower fasting blood-glucose (Fig. S2B). Despite the lack of glycogen, the mice show normal glucose tolerance at 12 months (Fig. S2C). The mice are insulin sensitive (Fig. S2D) due to an inability to stimulate gluconeogenesis, as demonstrated by reduced PEPCK (Pck1) and glucose-6-phosphatase (G6pc) expression (Fig. S2E) and reduced HOMA-IR (Fig. S2F).
A striking feature of aged HKO mice is the lack of adipose tissue (Fig. 2A). White adipose tissue (WAT) is reduced at 6 months but is severely depleted at 24 months (Fig. 2A). HKO mice have higher hepatic triglyceride at 4 months (19) and micro-steatosis at 6 months (Fig. 2B) which progresses to macro-vesicular steatosis and portal inflammation at 12 months (Figs. 2B,C). Liver histology and serology were similar in male and female HKO mice. Triglyceride secretion is reduced in HKO mice (Fig. 2D) and Pparg, Cidec and Cd36 are increased (Fig 2E).
Fig. 2. Deletion of hepatic SRSF3 causes lipodystrophy and steatosis.
(A) Representative photographs showing smaller body size and decreased intra-abdominal fat in HKO mice. White adipose tissue (WAT) weight and WAT to BW ratio at 6 and 24 months, n = 6–7/group. (B) Liver morphology in WT and HKO mice by H&E staining at 6 and 12 months. Micro-steatosis is indicated by green arrows, macro-steatosis by yellow arrows and portal inflammation by white arrows. (C) Steatosis score at 6 and 12 months and inflammation score at 12 months from evaluation of histopathology of multiple sections. (D) Serum triglycerides after administration of Triton WR-1339 to block lipoprotein lipase, n = 5–10/group. (E) Expression of Pparg, Cidec and Cd36 mRNAs at 1 month by Q-PCR, n = 6–7/group. (F) Stimulation of STAT5, NFkB and JNK phosphorylation following GH injection (5 mg/kg, i.p.) in two representative liver samples at 1 month. Total STAT5 is shown as a loading control. Serum FFA and GH levels at 18 months are shown to the right.
Young HKO mice show loss of Ghr, low IGF-1 and high GH (19). To verify GH resistance, 1-month mice were starved overnight and injected with GH (5 mg/kg). GH strongly activates STAT5 in WT livers but not HKO livers (Fig. 2F). Unexpectedly, GH stimulates JNK and p65-NFkB in HKO liver. GH levels are also significantly higher at 18 months as are serum FFA levels (Fig. 2F). Thus, the increased adipose lipolysis coupled with reduced hepatic TG secretion are consistent with hepatosteatosis in HKO mice.
Deletion of Hepatic SRSF3 Causes Fibrosis in liver
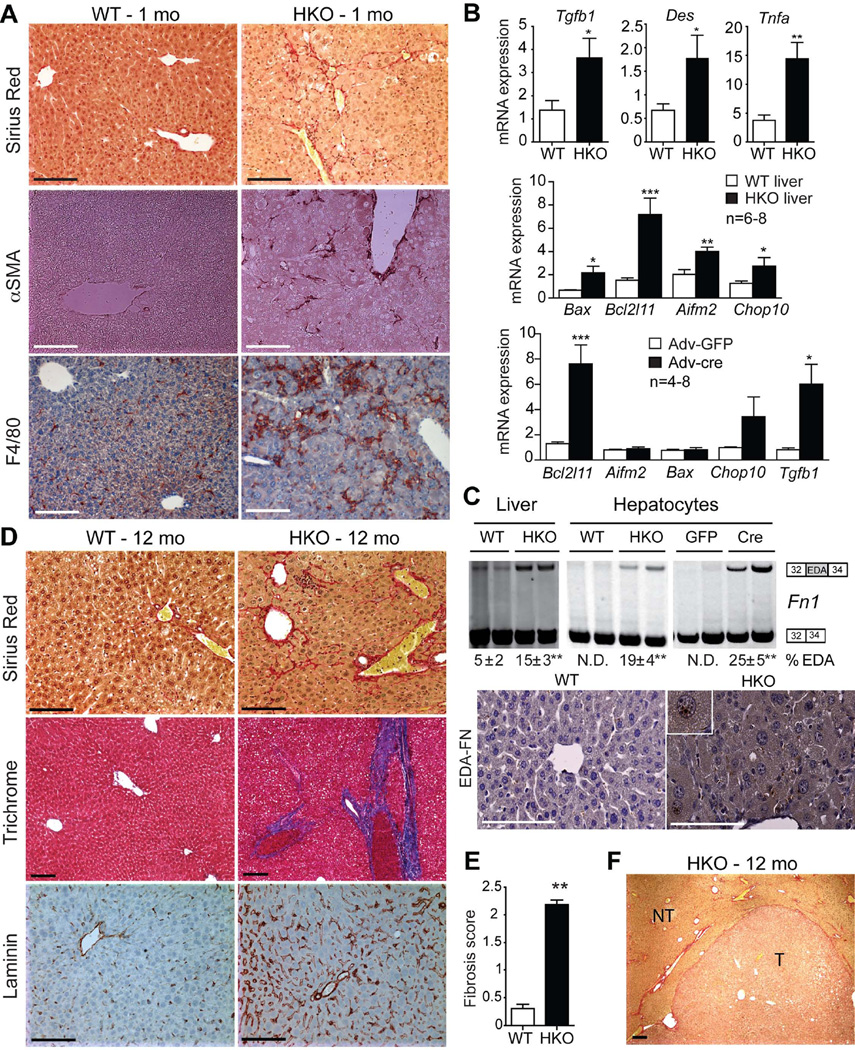
Periportal collagen fiber deposition and activation of myofibroblasts are seen in HKO livers at 1 month (Fig. 3A). Fibrosis is accompanied by increases in transforming growth factor beta (Tgfb1), desmin (Des) and tumor necrosis factor alpha (Tnfa) (Fig. 3B), and other fibrogenic factor mRNAs (Table S2). Livers also showed increased expression of pro-apoptotic genes, Bax, Bcl2l11 (Bim), Aifm2, and Chop10(Gadd153) indicating hepatocyte apoptosis (Fig. 3B). This is a direct effect of loss of SRSF3 since acute deletion of SRSF3 (Fig. S3) in primary Srsf3:flox hepatocytes using adenoviral cre expression causes induction of Bcl2l11 and Tgfb1 (Fig. 3B).
Fig. 3. Deletion of SRSF3 causes liver fibrosis.
(A) Representative sections from young livers (1 mo) stained for fibrosis (Sirius Red), myofibroblast activation (αSMA) and macrophage infiltration (F4/80), n=8/genotype. (B) Expression of Tgfb1, desmin (Des), Tnfa, and the apoptotic regulators Bax, Bcl2l11, Aifm2 and Chop10 in WT and HKO liver at 1 month by Q-PCR, n=6–8/group. Gene expression after Adv-cre or Adv-GFP infection of Srsf3:floxed hepatocytes, n = 4–8/group. (C). Representative gels showing Fn1 splicing in liver or hepatocytes from 1 month WT and HKO mice, or after Adv-cre or Adv-GFP infection of Srsf3:floxed hepatocytes, with the mean percentage of EDA exon inclusion ± SEM shown below, n=8/group. Photographs show liver sections at 1 month stained for EDA-FN. (D) Sirius Red and Trichrome staining for fibrosis (red and blue, respectively), and laminin staining (brown) in WT and HKO livers at 12 mo. (E) Mean fibrosis score from evaluation of histopathology from multiple liver sections at 12 months. (F) Sirius Red staining (5X) for fibrosis in HKO tumor at 12 months. T and NT indicate tumor and non-tumor areas.
To assess how altered RNA splicing might promote fibrosis we performed RNAseq analysis and aligned exon-junction reads to the genome to identify novel splicing events. HKO livers show aberrant splicing of multiple fibrogenic genes (Fig. S4). In particular, HKO livers and hepatocytes show greater inclusion of exon 33 of the pro-fibrogenic EDA exon in the Fn1 transcript that is confirmed by immunohistochemistry (Fig. 3C). This is a direct target for SRSF3 since acute deletion in primary Srsf3:flox hepatocytes causes a similar change (Fig. 3C).
At 12 months, HKO livers have bridging collagen fibers that extend into the interlobular septum, increased laminin staining around the sinusoids (Fig. 3D), and a significantly higher fibrosis score (Fig. 3E). Mice also have lower white blood cell and platelet counts consistent with fibrosis (Table S3). In tumor-bearing mice, fibrosis is increased in the non-tumorous sections of the liver but is notably absent in the tumors (Fig. 3F).
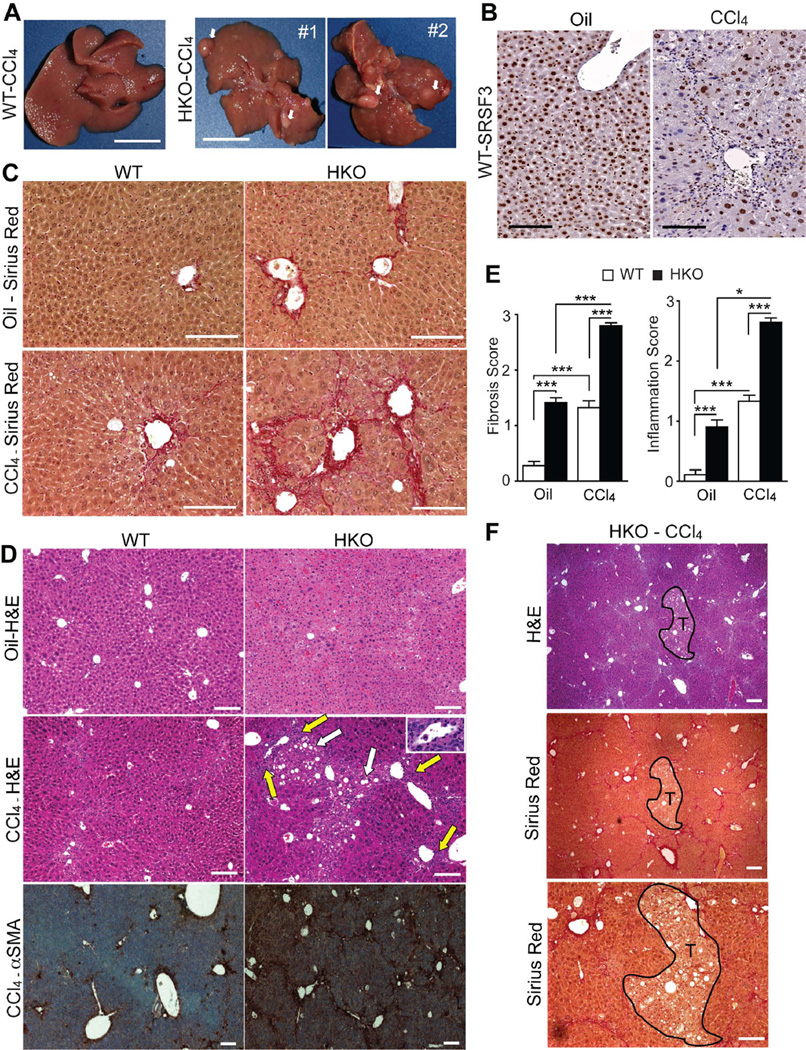
To test whether SRSF3 is protective against a pro-fibrogenic insult, we challenged young mice with CCl4. WT mice show the expected roughened liver morphology after CCl4 treatment (Fig. 4A). HKO livers have the same roughened appearance but in addition 5 out of 6 livers showed visible precancerous nodules (Fig. 4A). Interestingly, SRSF3 expression is greatly reduced in fibrotic areas around the periportal veins in CCl4-treated WT mice (Fig. 4B). Cirrhosis is evident in CCl4-treated HKO livers with bridging fibrosis and collagen fibers extending into the interlobular septum (Fig. 4C). Inflammatory infiltrates are apparent in CCl4-treated HKO livers and increased α-SMA staining confirms myofibroblast activation (Fig. 4D). Overall, HKO livers have higher fibrosis and inflammation scores in both oil and CCl4-treated mice (Fig. 4E). As was observed for the spontaneous tumors, the precancerous nodules in the CCl4-treated HKO mice do not exhibit fibrosis but have a steatotic phenotype similar to the early tumors (Fig. 4F).
Fig. 4. SRSF3 protects against CCl4 toxicity.
(A) Gross morphology of 6 mo WT and HKO livers treated with CCl4 for 4 weeks. White arrows indicate pre-cancerous nodules in HKO liver. (B) Liver sections from WT mice treated with oil or CCl4 and stained for SRSF3. (C) Sirius Red staining for fibrosis (red) in oil and CCl4-treated mice. (D) H&E and αSMA stained liver sections from oil- or CCl4-treated mice showing steatosis (white arrows), portal inflammation (yellow arrows and inset) and myofibroblast activation (brown). (E) Mean fibrosis and inflammation scores from evaluation of multiple liver sections. (F) H&E and Sirius Red stained sections through a precancerous nodule in CCl4-treated HKO liver. Nodule is outlined in black and labeled T.
Loss of SRSF3 causes increased insulin-like growth factor signaling
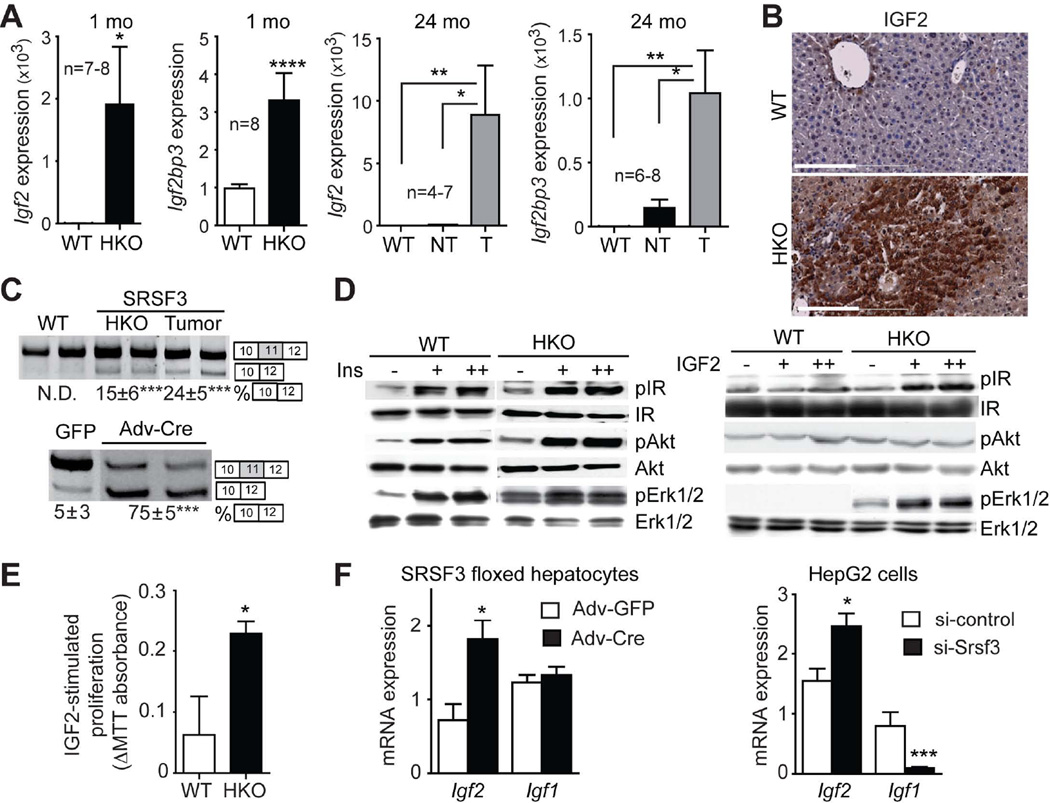
To gain molecular insight in the processes governing the malignant transformation of hepatocytes in HKO animals, we analyzed gene expression and exon usage in tumors compared to matched non-tumor liver. There was no evidence for altered exon usage in the tumors after multiple testing correction, but 265 genes showed evidence for altered expression in the tumors (Table S4). The expression of Igf2 and the insulin-like growth factor 2 mRNA binding protein 3 (Igf2bp3) is strongly up-regulated in the tumor. Both Igf2 and Igf2bp3 are overexpressed at 1 month and expression is greatly increased in tumors at 24 months by Q-PCR (Fig. 5A). At the protein level, expression of IGF2 is significantly higher in HKO liver at 1 month (data not shown) and in the tumors at 24 months (Fig. 5B). We have previously demonstrated that SRSF3 regulates insulin receptor (INSR) alternative splicing in vitro (20). Increased expression of the Insr-A isoform is evident in both non-tumorous HKO livers and in tumors (Fig. 5C). This is a direct effect of SRSF3 as acute deletion with Adeno-cre increases Insr-A expression in floxed hepatocytes (Fig. 5C). Although the change in splicing is relatively modest, it leads to a dramatic difference in response to IGF2. Insulin treatment causes comparable activation of AKT and ERK1/2 in WT (Fig. 5D) but, in contrast, IGF2 treatment strongly activates INSR and ERK1/2 only in the INSR-A expressing HKO hepatocytes (Fig. 5E). IGF2 also leads to significant increase of proliferation in HKO hepatocytes (Fig. 5E). Acute loss of SRSF3 in both primary hepatocytes and HepG2 cells leads to increased Igf2 expression (Fig. 5F) suggesting that SRSF3 suppresses autocrine activation of the INSR by IGF2.
Fig. 5. Loss of SRSF3 increases insulin/insulin-like growth factor signaling.
(A) Igf2 and Igf2bp3 mRNA expression by Q-PCR. T indicates tumor and NT indicates non-tumor part of HKO liver.. (B) IGF2 immunostaining of WT and HKO liver at 24 mo (brown). (C) Analysis of exon 11 skipping in Insr transcripts from livers at 24 mo and from Srsf3:floxed hepatocytes after Adv-cre infection. Representative gels are shown with the % exon11 skipping underneath. (D) Activation of insulin and IGF2 signaling in primary hepatocytes. Cells were stimulated with 30 (+) or 100 nM (++) insulin or 50 (+) or 100 ng/ml (++) IGF2, then extracts were immunoblotted for phospho and total IR, Akt and Erk1/2. (E) IGF2-stimualted proliferation of WT and HKO hepatocytes by MTT assay. (F) Igf2 and Igf1 mRNA expression by Q-PCR following Adv-cre infection of Srsf3:floxed hepatocytes or siRNA knockdown in HepG2 cells.
Loss of SRSF3 leads to aberrant splicing of EMT genes, activation of β-catenin signaling and c-MYC expression, and induction of liver progenitor cell markers
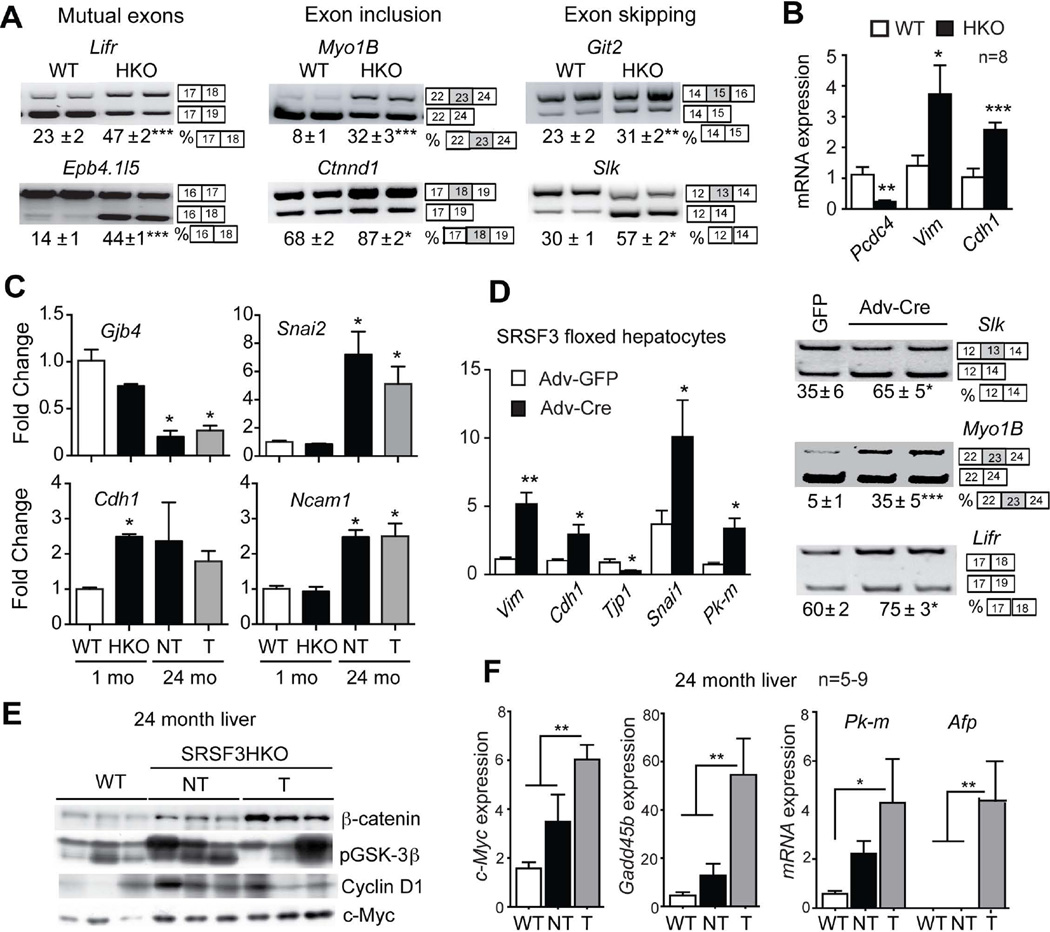
EMT is important for tumor cell dissemination and has been implicated in HCC. A number of EMT-associated genes show altered splicing or altered expression in livers of young HKO mice (Fig. 6A and B). Additional genes are altered in older mice and in tumors (Fig. 6C and S5). Again, acute loss of SRSF3 causes similar changes in EMT markers (Fig. 6D) suggesting that SRSF3 is a direct regulator of EMT. EMT is often associated with activation of the Wnt/β-catenin signaling. HKO hepatocytes show stabilization of β-catenin, elevation of glycogen synthase kinase 3β phosphorylation, and increased cyclin D1 expression (Fig. 6E). β–catenin levels are further elevated in the tumors, as are c-Myc and Gadd45b, two β–catenin targets (Fig. 6F). Consistent with elevated Wnt signaling, Wnt9, Wnt11 and Frzd5 are also increased (Fig. S5).
Fig. 6. Loss of SRSF3 leads to aberrant splicing of EMT genes, activation of β-catenin signaling and c-MYC expression, and induction of liver progenitor cell markers.
(A) Analysis of splicing of EMT-associated transcripts (Lifr, Epb4.1l5, Myo1b, Ctnnd1, Git2 and Slk) at 1 mo. Representative gels are shown with the percentage of exon use given below the gels. The exons involved in the alternative splicing event are shown to the right of the gel images. (B) Quantification of mRNA expression of the EMT-associated genes programmed cell death 4 (Pcdc4), vimentin (Vim) and E-cadherin (Cdh1) at 1 mo by Q-PCR. (C) Quantification of gap-junction protein 4 (Gjb4), slug/snail2 (Snai2), Cdh1 and neuronal cell adhesion molecule 1 (Ncam1) by microarray. Results are presented as mean fold change vs. WT ± SEM. (D) Change of EMT gene expression and alternative splicing following Adv-cre infection of primary Srsf3:floxed hepatocytes. (E) Immunoblotting for non-phospho β-catenin, phospho-GSK3β, Cyclin D1 and c-Myc in livers at 24 months. Three representative samples per group are shown. (F) Quantification of c-Myc, Gadd45, Pkm and Afp mRNA expression at 24 mo by Q-PCR. T and NT indicate tumor and non-tumorous areas of HKO liver.
The expression of liver progenitor cell markers Afp and Pkm is increased in the tumors (Fig. 6F) as is expression of the stem cell markers CD133/Prom1, Ly6d, SCA1/Thy1.1, Ggt1, Flt3 and Epcam and the cell differentiation markers Cd36, Cd44, Cd59a/b, and Cd63 (Fig. S6). The presence of these markers suggests that tumors in the HKO mice have a precursor/stem-cell phenotype.
SRSF3 expression is altered in human HCC
To establish the relevance of our model to human HCC we assessed SRSF3 protein expression on a liver cancer tissue microarray (TMA) containing 120 samples of HCC (grade 2 and 3), 15 bileduct adenocarcinoma (BDAC), 2 combined HCC and BDCa, 2 adenocarcinoma (HCA), 6 cirrhosis and 4 normal liver. SRSF3 protein was detected in all of the normal liver samples, 46% of the HCC, 50% of the BDCa, and 33% of the cirrhosis samples (Fig. 7A). To confirm the TMA data, we obtained 4 HCC, 5 HCA and 4 normal liver samples from the Moores Cancer Center (MCC) Biorepository. The majority of nuclei in normal samples stain strongly for nuclear SRSF3, whereas the majority of nuclei in the adenoma and HCC samples stain weakly (Fig. 7B,C). Interestingly, some of the HCC and adenoma samples show cytoplasmic staining. Human hepatoma cell lines, Hep3B, HepG2 and Huh7 also show aberrant splicing of INSR, FN1 and SLK similar to the HKO livers (Fig. 7D) and si-RNA mediated knock-down of SRSF3 (Fig. S3) leads to greater inclusion of EDA-Fn1 exon in HepG2 and Huh7 cell lines (Fig. 7D). Human adenoma and HCC samples also show expression of EDA-FN like HKO tumor (Fig. 7E).
Fig. 7. SRSF3 expression is altered in human HCC.
(A) Immunohistochemical staining for SRSF3 on human liver cancer tissue microarray. Normal liver shows strong nuclear staining, whereas cirrhotic liver and most HCC samples stain weakly. Representative HCC sections showing positive and negative staining for SRSF3 are shown. Insets show higher magnification of nuclear staining. Melanoma shows very strong SRSF3 staining and is used as positive control. (B) Pathological samples of normal human livers show strong nuclear staining for SRSF3. Carcinoma and adenoma samples show either weak staining or cytoplasmic staining for SRSF3 (arrows). (C) Quantification of SRSF3 nuclear staining from normal liver (Nrml), adenoma (Ade) and HCC tissue (4 fields per section, n=4). Black bars indicate strongly staining nuclei, and gray bars weakly staining nuclei. (D) Analysis of splicing of INSR, FN1 and SLK in human liver cancer cell lines Hep3B, HepG2 and Huh7 compared to WT and HKO livers, and splicing of Fn1 in HepG2 and Huh7 cells following SRSF3 siRNA knockdown. The mean percentage of exon skipping or EDA exon inclusion is given below the gel image. (E) Immunohistochemical staining for EDA-FN on human liver samples compared to HKO tumor. (F) Schematic of how loss of SRSF3 activates multiple pathways causing mitogenesis, steatosis, fibrosis and EMT that eventually cause HCC. Solid lines show activation or increased expression, dotted line indicates loss of expression
Discussion
Hepatocellular carcinoma usually arises after years of liver disease that is caused most often by chronic hepatitis B or C virus infection, alcoholism, chemical toxicants, or metabolic dysregulation. The ongoing liver damage triggers a regenerative response with proliferation of dedifferentiated hepatocytes, but also triggers activation of myofibroblasts, leading to fibrosis, inflammation, oxidative stress and eventually cirrhosis. This toxic microenvironment promotes genetic mutations that cause transformation and ultimately tumorigenesis. With the increase in obesity, NAFLD and NASH are becoming more prevalent, and both are known risk factors for HCC (21, 22). Tumor development in SRSF3-HKO liver occurs in the context of progressive steatosis, fibrosis and hepatitis resembling the changes in human liver disease. Tumors take 12–15 months to arise, which suggests that secondary genetic events are needed. Hence SRSF3 has the characteristics of a tumor suppressor gene in the liver, in that its loss predisposes to but does not cause cancer. This idea is supported by the finding that SRSF3 protects mice against CCl4-induced hepatotoxicity and carcinogenesis.
The role of SRSF3 in human HCC has not been established, but a number of observations point to a potential role. SRSF3 mRNA expression is uninformative as the gene auto-regulates its expression and protein levels do not correlate with RNA (23). Indeed, the most recent release of Oncomine reports overexpression of SRSF3 in 489 studies and underexpression in 435 studies. The RIKEN database also reports that 6.7% of HCC samples have mutations in SRSF3. In other cancers, the Cancer Genome Atlas database (TCGA) reports that SRSF3 is mutated and deleted in a fraction of bladder, cervix, lung, melanoma, colorectal and prostate cancer, and 8.7% of tumors in the COSMIC database have LOH at the SRSF3 locus. If SRSF3 is lost in human HCC, we would expect to find a SRSF3-dependent expression signature in HCC. Comparison of our mouse data to 69 HCC profiling studies using Gene Set Enrichment Analysis shows enrichment for 11 genesets in the SRSF3-HKO data (Table S5). These included HBV and HCV positive HCC, hepatoblastoma, and proliferative and progenitor/stem cell subclasses of HCC (24–28). In contrast, the WT data was enriched for 23 genesets down-regulated in liver cancer or associated with better prognosis and survival. Similarly we would expect genes altered in the mouse to be altered in human HCC. According to TCGA, 97% of HCC cases show alteration of one or more of the top 100 up-regulated mouse genes (Fig. S7), and 93% of HCC cases show alteration in one or more of the top 100 down-regulated genes. From all these comparisons we conclude that the changes seen in our SRSF3-HKO mouse are similar to those seen in both viral and non-viral-associated human HCC.
Though previous reports (18, 29) had suggested that SRSF3 is a protooncogene, our findings are supported by a recent report showing that knockdown of the splicing factor Slu7 phenocopies many of the changes seen in our SRSF3 knockout, most likely as Slu7 regulates SRSF3 expression (30). These opposing phenotypes are likely related to ability of the liver to regenerate in response to chronic damage. Loss of Mdr2, Mcl1, Tak1, Atg5 or a number of other genes causes a similar phenotype of apoptosis, liver damage and proliferation leading to fibrosis and inflammation and, ultimately, hepatocellular carcinoma. Although SRSF3 is not directly involved in these processes, acute loss of SRSF3 causes over-expression of Chop-10 and Bcl211l that might trigger apoptosis and liver damage, and EDA-Fn that can cause fibrosis (31–33).. Loss of SRSF3 also impairs hepatocyte differentiation and maturation, causing cells to express a hepatic progenitor phenotype, but these cells are not transformed and do not form tumors. Subsequent genetic alterations acquired during the ongoing liver damage likely transforms these progenitor cells allowing the acquisition of additional stem cell markers. Global disruption of splicing is not the origin of these phenotypes since loss of a related protein SRSF1 does not cause fibrosis or HCC despite global changes in RNA splicing (data not shown).
Disruption of Insr splicing may also contribute to the proliferative phenotype, as the increased expression of INSR-A allows inappropriate mitogenic stimulation due to the dramatically increased IGF2 expression, which is also a common feature of human HCC (34). Our data is supported by a recent study reporting that the INSR is over-expressed in human HCC samples (35) and the ratio of INSR-A/INSR-B is significantly higher. Tumor cells must also disrupt normal cell-cell contacts to proliferate and metastasize and SRSF3 loss activates Wnt/β-catenin signaling and promotes EMT. The EMT-associated splicing changes are mediated by the epithelial specific splicing factors (Esrp1/2) but our results suggest that these changes may be due to SRSF3 independent of the ESRPs.
Together, our data indicate that SRSF3 is a crucial regulator of steatosis, fibrosis, mitogenesis, and EMT in hepatocytes that eventually leads to HCC (Fig. 7F). Given the increasing incidence of chronic liver disease due to obesity (36), the SRSF3HKO mouse represents an attractive model that recapitulates most of the changes seen in human disease and may be valuable tool to further explore the mechanisms of carcinogenesis and response to therapies. Our results also underscore the importance of RNA splicing in disease, and further studies are needed to establish the role of such splicing alterations in normal cellular functions and pathological states.
Supplementary Material
Acknowledgements
We would like to thank Dr. David Tarin (UCSD) for his help in assessing the tumor pathology, the GeneChip Microarray Core at the VASDHS/VMRF for their help exon array analyses, the UCSD Murine Hematology and Coagulation Core Laboratory for blood analysis, the MCC Microscope Facility and Histopathology Core, and Dr. Scott Vandenberg and the MCC Biorepository for provision of human liver samples. N.J.G.W. is faculty member of the UCSD Biomedical Sciences Graduate Program.
Financial Support: The study was funded by a Merit Award BX000130 from the Department of Veterans Affairs, and NIH grants P30DK063491, P30CA023100 and U54CA155435.
Abbreviations
- HCC
hepatocellular carcinoma
- EMT
epithelial to mesenchymal transition
- NAFLD
non alchoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- HOMA-IR
homeostatic model assessment of insulin resistance
Footnotes
The authors declare no competing financial interests.
Supporting Information
The Supplemental Data include Supporting Materials and Methods, seven supporting figures and five supporting tables.
Contributor Information
Supriya Sen, Email: ssen@ucla.edu.
Magda Langiewicz, Email: mtlangiewicz@gmail.com.
Hassan Jumaa, Email: jumaa@immunbio.mpg.de.
Nicholas J.G. Webster, Email: nwebster@ucsd.edu.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. The New England journal of medicine. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Kensler TW, Qian GS, Chen JG, Groopman JD. Translational strategies for cancer prevention in liver. Nature reviews. Cancer. 2003;3:321–329. doi: 10.1038/nrc1076. [DOI] [PubMed] [Google Scholar]
- 4.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature structural & molecular biology. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO reports. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Current genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & development. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Molecular and cellular biology. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Current biology : CB. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 11.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell reports. 2013;5:357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. The EMBO journal. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jumaa H, Nielsen PJ. Regulation of SRp20 exon 4 splicing. Biochimica et biophysica acta. 2000;1494:137–143. doi: 10.1016/s0167-4781(00)00233-5. [DOI] [PubMed] [Google Scholar]
- 15.Hallgren O, Malmstrom J, Malmstrom L, Andersson-Sjoland A, Wildt M, Tufvesson E, Juhasz P, et al. Splicosomal and serine and arginine-rich splicing factors as targets for TGF-beta. Fibrogenesis & tissue repair. 2012;5:6. doi: 10.1186/1755-1536-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM, Stauffer JK, et al. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene. 2013;32:2792–2798. doi: 10.1038/onc.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Molecular and cellular biology. 1997;17:3116–3124. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. International journal of biological sciences. 2010;6:806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nature communications. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Molecular and cellular biology. 2009;29:871–880. doi: 10.1128/MCB.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angulo P. Nonalcoholic fatty liver disease. The New England journal of medicine. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World journal of gastroenterology : WJG. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki K, Yasugi E, Okuma E, Breit SN, Nakamura M, Toda T, Kaburagi Y, et al. Proteomic analysis on insulin signaling in human hematopoietic cells: identification of CLIC1 and SRp20 as novel downstream effectors of insulin. American journal of physiology. Endocrinology and metabolism. 2005;289:E419–E428. doi: 10.1152/ajpendo.00512.2004. [DOI] [PubMed] [Google Scholar]
- 24.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, Herault A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 25.Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, Goga A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer research. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, et al. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer research. 2002;62:3939–3944. [PubMed] [Google Scholar]
- 29.He X, Arslan AD, Pool MD, Ho TT, Darcy KM, Coon JS, Beck WT. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2011;30:356–365. doi: 10.1038/onc.2010.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elizalde M, Urtasun R, Azkona M, Latasa MU, Goni S, Garcia-Irigoyen O, Uriarte I, et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124:2909–2920. doi: 10.1172/JCI74382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarnagin WR, Rockey DC, Koteliansky VE, Wang SS, Bissell DM. Expression of variant fibronectins in wound healing: cellular source and biological activity of the EIIIA segment in rat hepatic fibrogenesis. The Journal of cell biology. 1994;127:2037–2048. doi: 10.1083/jcb.127.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muro AF, Moretti FA, Moore BB, Yan M, Atrasz RG, Wilke CA, Flaherty KR, et al. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. American journal of respiratory and critical care medicine. 2008;177:638–645. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohan M, Muro AF, White ES, Berkman N. EDA-containing cellular fibronectin induces fibroblast differentiation through binding to alpha4beta7 integrin receptor and MAPK/Erk 1/2-dependent signaling. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:4503–4512. doi: 10.1096/fj.10-154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirier K, Chalas C, Tissier F, Couvert P, Mallet V, Carrie A, Marchio A, et al. Loss of parental-specific methylation at the IGF2 locus in human hepatocellular carcinoma. The Journal of pathology. 2003;201:473–479. doi: 10.1002/path.1477. [DOI] [PubMed] [Google Scholar]
- 35.Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Claperon A, Chretien Y, Rey C, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer research. 2013;73:3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- 36.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.