Significance
Changes in the expression of proteins are often associated with oncogenesis, and are frequently used as cancer biomarkers. Changes in the subcellular location of proteins have been less frequently investigated. In this paper, we describe a robust pipeline for identifying those proteins whose subcellular location undergoes statistically significant changes in cancers of four tissues, and also for identifying biochemical pathways that are enriched for proteins that translocate. Future investigation of these proteins and pathways may provide new insight into oncogenesis. Further, the analysis pipeline is expected to be useful for assessing disease type and severity in a clinical setting.
Keywords: biomarkers, protein subcellular location, image analysis, digital pathology, cancer
Abstract
Molecular biomarkers are changes measured in biological samples that reflect disease states. Such markers can help clinicians identify types of cancer or stages of progression, and they can guide in tailoring specific therapies. Many efforts to identify biomarkers consider genes that mutate between normal and cancerous tissues or changes in protein or RNA expression levels. Here we define location biomarkers, proteins that undergo changes in subcellular location that are indicative of disease. To discover such biomarkers, we have developed an automated pipeline to compare the subcellular location of proteins between two sets of immunohistochemistry images. We used the pipeline to compare images of healthy and tumor tissue from the Human Protein Atlas, ranking hundreds of proteins in breast, liver, prostate, and bladder based on how much their location was estimated to have changed. The performance of the system was evaluated by determining whether proteins previously known to change location in tumors were ranked highly. We present a number of candidate location biomarkers for each tissue, and identify biochemical pathways that are enriched in proteins that change location. The analysis technology is anticipated to be useful not only for discovering new location biomarkers but also for enabling automated analysis of biomarker distributions as an aid to determining diagnosis.
Our understanding of the number and types of changes that occur in various cancers is continuously growing. Previous work to discover proteins that vary significantly between normal and cancer cells has used techniques such as microarray profiling, next-generation sequencing, antibody arrays, and proteomic profiling (1–4). These studies have led to the discovery of proteins (termed expression biomarkers) whose expression levels mark different disease states. However, for some proteins, the extent of localization in the nucleus can be used to predict patient prognosis; β-catenin (5) and NF-κB (6) are examples. The discovery of more proteins that undergo oncogenesis-associated changes in subcellular location (which we term location biomarkers) could potentially improve disease diagnosis in conjunction with traditional protein expression markers. Further, discovering proteins that relocate in the disease state may give new insight into changes driving disease, and such changes would go undetected by measuring only expression.
Immunohistochemistry (IHC) studies are a major source of data on protein expression and location. Most such studies use visual examination to assess changes, a difficult and time-consuming task. With the advent of high-throughput acquisition technologies like tissue microarrays and automated slide scanners, computerized analysis of tissue images is highly desirable, and studies have shown that quantitative software can detect changes in disease states that are missed by visual inspection (7). Methods for analyzing changes in expression and pattern are well established in cultured cells (8), but histological images are typically more difficult to analyze because cellular heterogeneity and the closely packed organization of cells lead to significant cell segmentation challenges. Several projects have been initiated to build workflows that process IHC images (9, 10). Most of this work has been focused on quantifying differences in protein abundance between normal and cancer tissue. However, as discussed earlier, differences in subcellular protein locations could also be critical for understanding and diagnosing disease. Thus, there is a strong need for systems that can analyze protein subcellular location in IHC images.
We have previously described an automated system for recognizing major subcellular patterns in IHC images (11), and presented preliminary results from the use of that system to identify proteins that change location in various cancers (12). These studies used a subset of the extensive collection of IHC images in the Human Protein Atlas (HPA) (13). However, we have found that the performance on a larger collection of proteins with more pattern variation was significantly lower compared with the 16 marker proteins used in our previous study. We therefore sought to develop a system that can identify potential location biomarkers by using new approaches without explicit classification. By using images from the HPA, we show that our system can identify proteins with altered subcellular location directly from tissue images and anticipate that approaches such as this may significantly contribute to diagnosis, treatment, and monitoring of cancers.
Results
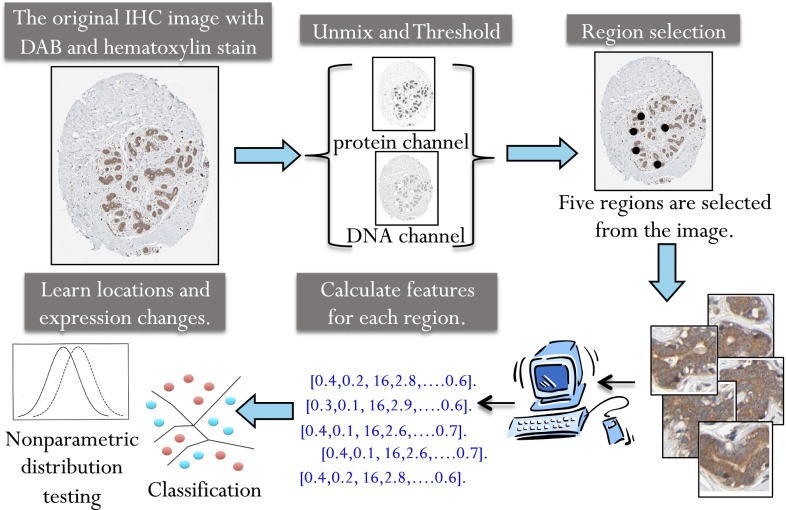
Our analysis pipeline (Fig. 1) consists of five steps.
-
i)
Selecting a set of proteins for analysis guided by staining levels. For a given tissue, we selected antibodies from the HPA whose staining intensity was annotated as moderate or strong, and whose staining quantity was annotated as greater than 75%. As a result of tissue-specific expression and variations in staining, the proteins identified (referred to as the analysis set) were different for each tissue.
-
ii)
Separating the DNA and protein components of each image by unmixing the hematoxylin and diaminobenzidine stains. The HPA images were collected as red, green, and blue (RGB) images in which the two stains appear as purple and brown, respectively. The intensity derived from each stain is therefore a combination of the intensities from the three RGB channels. We unmixed the spectra to give separate images reflecting mainly DNA and protein content (11).
-
iii)
Selecting regions of each image with the highest protein expression, under the assumption that the highest stained regions would be less likely to contain connective tissue, stroma, and other noncellular regions.
-
iv)
Calculating features to describe the location patterns in each region (11).
-
v)
First, estimating the probability that a given protein’s location pattern differs between the two conditions. The nonparametric Friedman–Rafsky (FR) test was used to calculate a P value for the null hypothesis that the sets of regions from normal images and from cancer images show the same pattern. Second, estimating the probability that a given protein’s level of expression differs between the two conditions. Expression P values were calculated by using the Wald–Wolfowitz method to test the null hypothesis that the level of expression in the regions from the normal and cancer images came from the same distribution. Calculations for 35 random samplings of images were averaged, giving very high repeatability of the results (Materials and Methods). Finally, calculating a classification accuracy for separating normal and cancer images by using protein location information.
Fig. 1.
Overview of the location biomarker discovery pipeline. Images with strong or moderate antibody staining were selected. Linear unmixing was used to separate each image into two composite images representing the DNA and protein stains as previously described (11). Regions were selected by convolving the protein image with a low-pass filter and selecting the highest points as region centers. Fifty-seven numerical features were calculated to describe the pattern in each region. The nonparametric FR test was used to calculate a P value and determine whether the null hypothesis, that the features from the normal and cancer image come from the same distribution, should be rejected. The nonparametric Wald–Wolfowitz test was used to calculate a P value to measure how likely the two sets of images are to come from the same expression distribution. A nearest neighbor classifier was also used to determine the ability of each antibody to distinguish normal and cancer images.
We applied this pipeline to images from the HPA for four tissues: breast, liver, prostate, and bladder (the results are contained in Dataset S1). After running the pipeline for each tissue, the proteins were sorted by their location P values to obtain a ranking by extent of subcellular location change. Representative images of the top three hits for each tissue are shown in Fig. S1.
Testing Using Known Location Biomarkers.
We expected that proteins known to change location in cancer would be ranked high on this list. To test this, we constructed validation sets by using pathologists’ annotations of the gross subcellular location provided in HPA: (i) nuclear, (ii) cytoplasm and plasma membrane, (iii) nuclear, cytoplasm, and plasma membrane, and (iv) none. The validation set for a given tissue consisted of those proteins from the analysis set for that tissue that had different location annotations between the normal and cancer images (Materials and Methods). Treating these as true positives, we constructed receiver operating characteristic (ROC) curves in which a threshold on the P value at which a protein was considered positive was varied (Fig. S2). In this case, the area under the curve (AUC) is a measure of how well our test finds the true positives. If the validation markers were the only proteins expected to change location, and if the system performed perfectly, the AUC values should be 1. However, we expect some of the proteins ranked highly by P value may be actual location biomarkers even if they are not in the validation set. For example, proteins may undergo a change in location that was not captured by the gross location annotations used to define true positives. Thus, we do not expect even a very good discovery system to give values near 1. The AUC values for breast, liver, prostate, and bladder were 0.67, 0.59, 0.67, and 0.68, respectively. These are all significantly greater than 0.5, the AUC expected for random performance.
Distinguishing Location and Expression Changes.
The features we used are designed to minimize the effect of differences in protein staining level. Even so, a major change in expression may cause a change in image texture that would be detected by our features even if subcellular location remains the same. This may cause proteins that do not change their location significantly but do change their expression dramatically to rank highly on our lists. We therefore used the expression P values and location P values together to analyze each protein’s change.
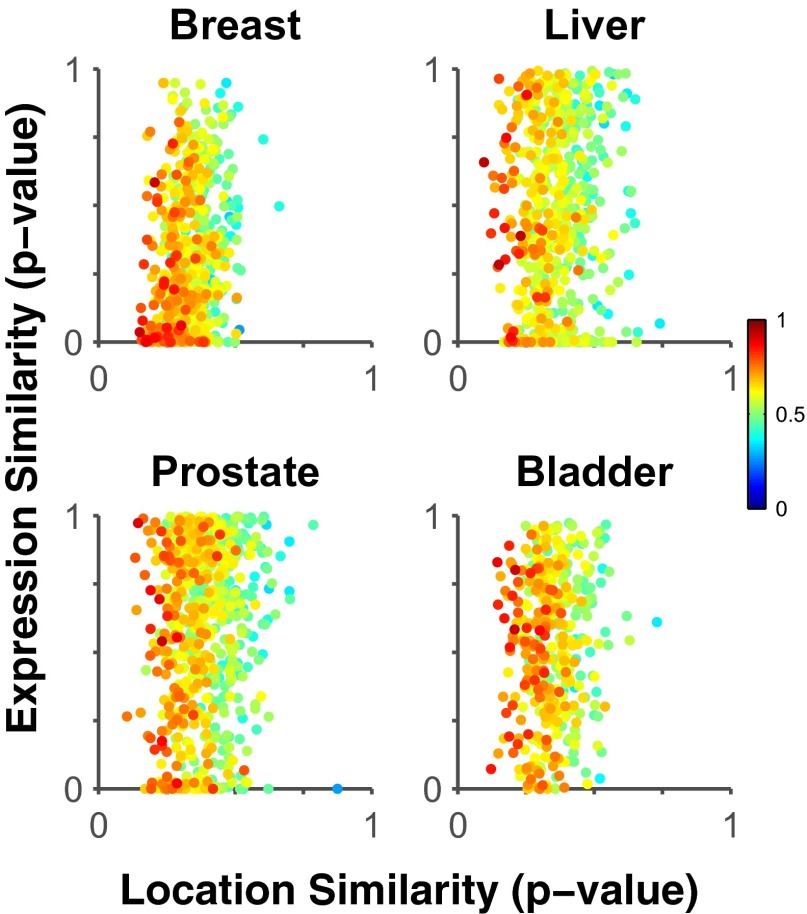
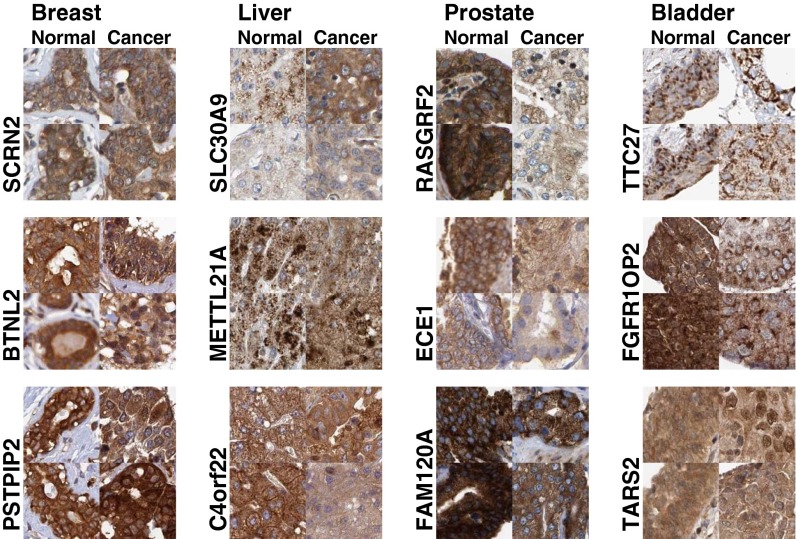
Fig. 2 shows the relationship between the expression change and location change for proteins in various tissues. The first conclusion we can draw is that the two values are not correlated, suggesting that proteins that change location do not always change expression, and vice versa. Second, the points in the upper left corner of each scatter plot in Fig. 2 represent proteins that have significantly changed location (low P values) but have not changed expression (high P values). The color of each point indicates how well that protein can be used to train a classifier to distinguish images from normal and cancerous tissue (Materials and Methods; the accuracy values are listed in Dataset S1). Thus, we expect proteins whose symbols are dark red and in the upper left corner to be potential biomarkers useful in a clinical setting for recognizing cancerous tissue by measuring differences in subcellular location. These proteins would not have been identified as potential markers by measuring expression changes alone. Dataset S1 is ranked for each tissue using the Euclidian distance from the upper left corner, that is, proteins that change location and do not change expression. The five top-ranked proteins for each tissue using this criterion are shown in Table 1, and images of the top three from each tissue are shown in Fig. 3.
Fig. 2.
Distinguishing between intensity and location changes. Each dot shows the P values for the hypotheses that location or expression are different between normal and tumor tissue for a given protein. The correlation between location and expression P values is weak, suggesting that proteins that change location in the cancer state do not necessarily change expression, as seen in the top left corner. The color indicates the classification accuracy for separating normal and cancer images of that protein by using subcellular location information alone. Proteins with high classification accuracy for distinguishing normal and cancer images are represented by a red dot. Red proteins closest to the top left corner are potential location biomarkers, and their discovery would have been missed by traditional experiments that measure changes in protein expression.
Table 1.
Potential location biomarkers
| Gene name | HPA Ab | P value | Accuracy | |
| Location | Expression | |||
| Breast | ||||
| SCRN2 | HPA023434 | 0.24 | 0.95 | 0.69 |
| BTNL2 | HPA039844 | 0.24 | 0.95 | 0.59 |
| PSTPIP2 | HPA040944 | 0.28 | 0.95 | 0.58 |
| USP10 | HPA006749 | 0.27 | 0.90 | 0.58 |
| NT5DC3 | HPA041634 | 0.27 | 0.89 | 0.62 |
| Liver | ||||
| SLC30A9 | HPA004014 | 0.15 | 0.96 | 0.81 |
| METTL21A | HPA034712 | 0.15 | 0.87 | 0.65 |
| C4orf22 | HPA043383 | 0.19 | 0.92 | 0.65 |
| WDR24 | HPA039506 | 0.16 | 0.85 | 0.67 |
| PARP12 | HPA003584 | 0.22 | 0.94 | 0.78 |
| Prostate | ||||
| RASGRF2 | HPA018679 | 0.14 | 0.97 | 0.90 |
| ECE1 | HPA001490 | 0.17 | 0.99 | 0.77 |
| FAM120A | HPA019734 | 0.18 | 0.94 | 0.73 |
| PLA2G4C | HPA043083 | 0.19 | 0.95 | 0.69 |
| TMEM194A | HPA014394 | 0.13 | 0.85 | 0.79 |
| Bladder | ||||
| TTC27 | HPA031246 | 0.19 | 0.89 | 0.84 |
| FGFR1OP2 | HPA038696 | 0.14 | 0.83 | 0.89 |
| TARS2 | HPA028626 | 0.25 | 0.96 | 0.54 |
| STAC | HPA035143 | 0.19 | 0.83 | 0.71 |
| — | CAB009119 | 0.20 | 0.82 | 0.72 |
The five proteins with the greatest location change and the smallest expression change are shown (the full ranked list is in Dataset S1). Classification accuracies for distinguishing normal and cancer are also shown.
Fig. 3.
Example regions from top location biomarker predictions with very small mean intensity changes. For every protein, the features from each disease state were clustered by using k-means (k = 2), and the region closest to each centroid is displayed.
Of course, we expected that classic biomarkers that are known to translocate in cancer, such as E-cadherin, β-catenin, and NF-κB, would be ranked highly in this list. These proteins were not part of our analysis sets because the HPA did not contain a sufficient number of images to meet the threshold of our pipeline. We therefore separately calculated location P values for those proteins by using the images that were available for breast and prostate cancers. The P values for two E-cadherin antibodies with high reliability, CAB000087 and HPA004812, were higher than 0.20. The P values for three β-catenin antibodies, CAB000108, HPA029159, and HPA029160, were higher than 0.32. The two antibodies against NF-κB in prostate cancer are CAB004031 and HPA027305, with P values greater than 0.22. Thus, our tests indicate that none of these are strong location biomarkers in these tissues, contrary to expectation based on previous literature reports. In addition, on visual inspection of the HPA images, we did not observe a pattern change; the pathologist annotations also did not indicate a location change. All the antibodies for these three proteins had identical location annotations in the two disease states with the exception of HPA004812, which moved from nuclear and cytoplasmic and membranous to mostly cytoplasmic membranous in the cancer state. The basis for this difference in this dataset from previous results is unclear.
Given that we were evaluating a large number of antibodies, we were also interested in estimating the generalizability of the relative performance of the antibodies. In other words, how likely it is that proteins with low P values or high classification accuracies would show similar values in future experiments? To do this, we calculated P values and accuracies for a smaller number of images and compared them with the values with the larger number (SI Text; note that this is different from the repeatability of the rankings using the same number of images). The results shown in Fig. S3 indicate a high correlation in the two estimates, indicating that the generalizability of our results to future experiments should be high.
Features Reveal Visually Distinguishable Changes Useful for Distinguishing Tumor From Healthy Tissue.
To determine whether the differences in location being identified by our pipeline were visually distinguishable changes, we performed hierarchical clustering and optimal leaf ordering to order the regions for a given antibody using our features (Materials and Methods). Fig. 4 shows 10 representative regions ordered for two example proteins from the top-ranked proteins. In breast tissue stained for SLC36A4, the normal regions clustered near each other on the left. Further, the regions appear ordered by increasing nuclear localization, suggesting our features can detect incremental and possibly continuous changes in this location pattern. In liver, GSTZ1 showed a decrease in nuclear localization from left to right, and also an increase in cytoplasmic graininess. The clustering grouped the normal and cancer regions separately.
Fig. 4.
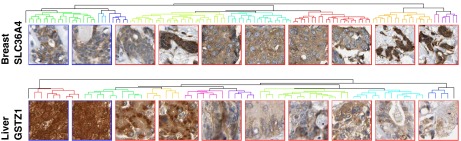
Ordering regions by location change progression. We selected one top-ranking protein from breast and liver: antibodies HPA017887 and HPA004701, respectively. The Euclidean distances between every pair of regions were calculated by using the features and clustered into a binary hierarchical tree. The leaves were ordered to maximize the sum of similarities between adjacent leaves across the tree. The tree was cut at 10 clusters, and leaves contained in each cluster are indicated by color. The region closest to the mean of each cluster is displayed below the tree from left to right. Normal tiles are outlined in blue; cancer tiles are outlined in red.
Location Biomarkers Can Distinguish Between Cancer Grades.
Each cancer in the HPA has a specified grade or subtype. We partitioned the images by grade and ran the pipeline to compare the two grades to each other for the prostate and bladder cancer set (Dataset S2). We also asked how well each protein could be used as a potential biomarker in a classifier trained to distinguish three disease states: normal tissue and low-grade and high-grade tumors. Fig. S4 shows the location P value and the expression P value for each protein when comparing the two subtypes versus each other. Points that fall in the upper left corner (Fig. S4) have different subcellular locations between the two grades but similar expression levels. The color of each point represents that protein’s three-class classification accuracy.
Example images for the proteins with the greatest three-class classification accuracies are shown in Fig. S5. The best classification accuracy was obtained for S100A6 in bladder: it has a classification accuracy of 83% (compared with 33% expected at random), a location P value of 0.081, and an expression P value of 0.34. This protein is the best example of a potential location biomarker (one that changes location but not expression) in bladder. These results provide further support for the utility of our system for identifying important location changes between disease states.
Identification of Biochemical Pathways Enriched in Translocated Proteins.
Finally, we were interested to find out whether our analysis could suggest entire pathways, or major portions thereof, that might undergo translocation together in cancer (the simplest example would be proteins that are part of a translocating complex). To answer this, for each Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, we calculated the probability that all of the proteins in it changed location or expression compared with a randomly sampled background distribution (SI Text and Fig. S6). (Note that this represents an underestimate of the change in a pathway if it contains subcomponents that do not translocate.) We calculated pathway changes by using our image processing pipeline or pathologist annotations. Pathways with the largest change in either location or expression are listed in Table 2. As discussed later, some of these pathways have been previously implicated in cancer and some are novel predictions.
Table 2.
Pathways with the largest location or expression changes
| Tissue | L:P | E:P | L:A | E:A | Pathway |
| Breast | + | — | — | — | HTLV I infection |
| + | — | — | — | One carbon pool by folate | |
| — | + | — | — | Proteasome | |
| — | + | — | — | ErbB signaling | |
| Liver | + | — | — | — | Axon guidance |
| — | + | — | — | GPI anchor biosynthesis | |
| — | + | — | — | Hypertrophic cardiomyopathy | |
| — | + | — | — | Dilated cardiomyopathy | |
| Prostate | + | — | — | — | Fatty acid elongation |
| + | — | — | — | HIF-1 signaling | |
| — | + | — | — | Oxidative phosphorylation | |
| — | + | — | — | PPAR signaling | |
| — | + | — | — | Viral myocarditis | |
| Bladder | + | — | + | — | Hippo signaling |
| — | + | — | — | NF-κB signaling | |
| — | + | — | — | p53 signaling | |
| — | + | — | — | Transcription misregulation in cancer | |
| — | + | — | — | Apoptosis | |
| — | + | — | — | Cell cycle | |
| — | + | — | — | mRNA surveillance | |
| — | + | — | — | Ribosome biogenesis |
Pathway P values were calculated by using individual protein location (L) or expression (E) P values from the pipeline (L:P, E:P) and using P values from pathologist annotations (L:A, E:A). Plus symbol indicates pathway P < 0.01. Values for all pathways are in Dataset S3.
Discussion
We have described a workflow to identify proteins that change their subcellular location between normal and cancerous tissue without requiring classification. We confirmed our ability to detect changes in location classes by using annotations provided by the HPA database.
Upon visual inspection of top hits (Fig. 3), we noted that our system was able to find texture changes between the two disease states; however, these did not always represent changes between distinct subcellular location classes. In some cases, our texture features detected changes in tissue structures and morphology.
In Fig. 3, BTNL2 in breast showed a decrease in nuclear localization. SLC30A9 in liver decreased in cytoplasmic graininess, and C4orf22 decreased in plasma membrane and vesicle localization in the cancer state. In prostate cancer, ECE1 decreased in plasma membrane accumulation. In bladder cancer, FGFR1OP2 changed from cytoplasmic and nuclear localization to mostly a grainy cytoplasmic localization in cancer. TARS2 in bladder increased in nuclear membrane localization in cancer. In some cases, our texture features picked up changes in tissue structure but not necessarily subcellular location, as was seen in SCRN2 in breast and TTC27 in bladder.
It was of interest to consider whether our top predictions had previously been implicated as being altered in cancers, and which ones were new discoveries. In breast cancer, very few studies have reported on BTNL2; however, there is strong evidence that variants of this gene play a role in susceptibility to sporadic and familial prostate cancer (14). PSTPIP2 has not been reported in breast cancer; however, it is implicated in the expansion of macrophage progenitors leading to autoinflammatory disease (15). USP10 is translocated to the nucleus upon DNA damage and regulates p53 (16).
In liver cancer, PARP12 has been reported to play a role in genome surveillance and DNA repair pathways, and it is being recognized as a new potential therapeutic target (17).
In prostate cancer, three of our top findings have been linked to prostate cancer development. Methylation of the RASGRF2 gene was found to be associated with prostate cancer (18). ECE1 has been implicated in prostate cancer cell invasion, in which different isoforms of the protein were found to play different roles (19). PLA2G4C is regulated by EGR, a gene that is rearranged in approximately 50% of prostate cancer (20). In bladder cancer, very few of the top findings have been published in association with disease.
Pathway Changes.
Some of the top-ranking pathways had location P values that were approximately two orders of magnitude smaller than the expression P values based on the pipeline results (Table 2 and Fig. S6). In the HTLV-1 infection pathway, the HTLV-I Tax oncoprotein initiates malignancy development in leukemia by creating an environment to facilitate DNA damage (21). To our awareness, the molecular mechanism of this pathway has not been studied in the contexts of breast or urothelial cancer. Our results, together with other literature reports, indicate that subcellular location changes in components of HTLV-I infection pathway could play a role in driving cancer. Further, the high rank of this pathway across the four tissues indicates that these changes may be important in identifying and understanding other cancers as well.
The “one carbon pool by folate” pathway was also found to change location in breast cancer. It is known to play an important role in DNA global hypomethylation, which can lead to DNA strand breaks (22). As expected, when changes in expression are used to rank pathways, the ErbB pathway ranks near the top for breast cancer (23).
In liver cancer, the axon guidance pathway was found to change location more than it did expression. One of the genes contributing to the axon guidance pathway, ROBO1, was found to be overexpressed in hepatocellular carcinoma (24). The axon guidance pathway has not been implicated as a whole in liver cancers, but it is known to be altered in pancreatic cancers (25). Our results, with previous reports of ROBO1, suggest this pathway may play a role in liver cancer. The importance of the top pathways seen to change expression in liver is unclear.
In prostate cancer, HIF-1 signaling is known to play an important role in hypoxia adaption of tumors, and HIF-1α is known to be overexpressed in early tumors (26). Our findings suggest that the proteins in this pathway undergo location changes, possibly contributing to the pathway’s dysregulation in cancer. PPAR is a known prostate cancer marker (27), and its pathway is identified as changing expression.
Finally, in urothelial cancer, the hippo signaling pathway was the top-ranking pathway to change location. Hippo signaling is responsible for tissue size and is known to lead to uncontrolled cellular proliferation and blocking of apoptosis when misregulated (28). When we ranked the pathways in urothelial cancer by expression changes, a number of signaling pathways known to be involved in cancers ranked at the top.
We also calculated the product of the P values for each pathway across all four tissues to find those pathways changing in all four cancers (Dataset S3). Three of the top-ranking pathways for location changes were already identified in individual tissues. In addition, the p53 signaling pathway (which is known to involve location changes) was also identified. For expression changes, five pathways previously associated with cancers were highly ranked (which is encouraging with respect to the accuracy of our automated methods).
Our analyses suggest that location changes of these pathways may be important for understanding their role in disease. In addition, our results link previously implicated pathways to new cancers for further investigation.
Conclusion
By using staining patterns of proteins in four tissues, we have identified proteins that show altered subcellular location in cancer and/or whose patterns can be used to distinguish normal and cancerous tissue or different cancer subtypes. Many of these proteins do not have significant expression level changes and would not have been found as biomarkers if we had considered expression level changes alone. Further, some proteins have high classification accuracies but visually similar location patterns. The subtle changes that are being detected may nonetheless be useful for distinguishing disease states. Extended analysis with more images of the potential markers we have identified will be necessary to assess their utility or significance. We note that the analysis pipeline we have described is not only useful for identifying cancer biomarkers, but should also be valuable for automating the process of analyzing IHC images to assess disease state. We are currently carrying out collaborative translational studies to determine whether our technology combined with any of the potential biomarkers is useful for distinguishing lesions with various diagnoses or prognoses.
Materials and Methods
Data.
We used images from the HPA (www.proteinatlas.org) that appeared online on September 24, 2013 (see Fig. S7 for evaluation of the effects of image compression). Proteins were placed in the analysis set for each tissue if they met three criteria: (i) the staining annotation was strong or moderate, (ii) if the quantity field was annotated as greater than 75%, and (iii) at least three images of that protein were available for the normal tissue. Approximately 500 proteins per tissue passed this filtering procedure (Dataset S1 provides a list of proteins in the full analysis set for each tissue).
Identification of Validation Sets.
A validation set of proteins whose location was known to change (which we define as true positives) was created for each tissue. These were found by using HPA annotations. We identified the set of true positives for a given tissue by finding those proteins for which the set of location annotations for all normal images did not intersect the set of location annotations for all cancer images. In the data set for breast, liver, prostate, and bladder there were 5, 3, 7, and 13 true positives, respectively.
Selecting Regions.
For each image, we selected regions that showed significant staining. A low-pass filter was applied to each protein image and we selected regions centered on the peaks of the filtered image. This was done under the assumption that the cellular regions of the tissue would have the highest staining levels, as opposed to the connective tissue, stroma, and other noncellular regions, which would have much lower levels of staining primarily as a result of nonspecific antibody binding.
Removing Outlier Images.
Next, we removed outlier regions and images based on DNA and protein intensity. For each tissue, we calculated the mean and SD of the protein and DNA stains for all images. This same process was repeated for all regions from each tissue. We removed images and regions from the dataset that were more than 4 SDs from the mean.
Pipeline for Testing Changes in Location or Expression.
Our pipeline calculates P values for the hypotheses that the location or expression of each protein are the same between normal and cancer images. The pipeline requires inputs for the number of images to use, the number of regions to select per image, the region size, and the number of estimates to average when reporting P values and accuracies (choice of these parameters is discussed later).
For location testing, a set of features that do not require segmentation of the image into individual cell regions was extracted from each region as described previously (11), with the modification that horizontal and vertical features were combined to produce a set of 592 rotation invariant features. These include texture features at many different levels of resolution and nuclear overlap features. We used the 57-feature subset that was previously selected to be able to distinguish eight subcellular location classes with high accuracy (ref. 11; see also Fig. S8). The equivalence of the distributions of these features for normal and cancer regions was evaluated by the FR test. Because the test is nonparametric and does not make assumptions about the distributions from which the samples are drawn, it is suitable for small numbers of regions and large numbers of features.
Expression P values were calculated by normalizing the mean protein intensity level across each region used in the location analysis by the respective mean nuclear intensity level. This results in a one-dimensional set of points corresponding to the regions for normal protein expression and cancer protein expression. We calculated a P value that the points in the two sets were drawn from the same distribution by using the Wald–Wolfowitz test, the one-dimensional version of the FR test.
The reported P values and accuracies for each protein were calculated by taking the average of 35 estimates that we found produced consistent ranked lists (SI Text and Fig. S9).
Selecting Image Sets, Number of Regions, and Region Size.
The database has as many as 3 images for each normal tissue and as many as 30 images for each cancer tissue. To have the same null distribution for the nonparametric P values, we needed to use the same number of normal and cancer images for each antibody. To identify the optimal number of each, we randomly selected 200 antibodies for each tissue, selected 2 regions from each image, and assessed the extent to which the validation markers were ranked highly by location P values (using the AUC). The number of normal images was varied from 1 to 3 and the number of cancer images from 3 to 24. We found that the best AUC average over all 200 antibodies resulted from using 2 normal images and 17 cancer images.
Next, the optimal number of regions was found by using the same 200-antibody training set and 2 normal and 17 cancer images. We varied the region count from 2 to 5 from each of the images. The best performance as measured by AUC resulted from using 5, 3, 3, and 4 regions per image for breast, liver, prostate, and bladder tissues, respectively, and we therefore used these values for the full analysis sets. Differences in the optimal number of regions for different tissues presumably reflect tissue-specific variations across the normal and cancer states. Limiting the number of regions per image prevents the sampling of noncellular regions in each tissue.
The optimal region size was chosen by assessing the performance of 100 randomly chosen proteins in ROC curves for each tissue. Ideally the region should be small enough so as to only capture cellular areas from a tissue image, as capturing noncellular regions introduces new textures to the analysis that would affect the subcellular location features. The optimal size was selected to be 75 pixels, with an average AUC of 0.67 for the four tissues.
Classification.
We used nearest neighbor classifiers with the 57 z-scored features and used cross-validation to estimate of the ability of a given protein to distinguish normal from cancer images, or to distinguish low-grade from high-grade cancer images. Images were assigned the majority class of their regions.
Distinguishing Cancer Grades.
The prostate and bladders cancers are identified as high- or low-grade in the HPA, with approximately equal numbers of each. We partitioned the cancer images by grade and ran the pipeline to compare them. We also randomly selected three images from each set and calculated the three-class classification accuracy for a nearest neighbor classifier (using leave-one-out cross-validation). This was repeated 35 times (producing a consistent ranking, as explained earlier) for different sets of randomly selected images, and the average of the 35 accuracies is reported. Proteins that did not have at least three images from each disease state were excluded. The results are contained in Dataset S2.
Supplementary Material
Acknowledgments
We thank the HPA project team for making the valuable collection of IHC images publicly available; E. Lundgren and M. Uhlén for helpful discussions; members of the laboratory of R.F.M. for valuable suggestions; and the anonymous reviewers of the manuscript for comments resulting in significant improvements in the analysis. This work was supported by NIH Grant GM075205, NIH Training Grant EB009403 (to A.K.), Commonwealth of Pennsylvania Commonwealth Universal Research Enhancement Program Grant 4100059192, and a postdoctoral fellowship from the Lane Fellows Program (to A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.W.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415120112/-/DCSupplemental.
References
- 1.Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 2.Khan J, et al. Expression profiling in cancer using cDNA microarrays. Electrophoresis. 1999;20(2):223–229. doi: 10.1002/(SICI)1522-2683(19990201)20:2<223::AID-ELPS223>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER, Wilson RK. Cancer genome sequencing: A review. Hum Mol Genet. 2009;18(R2):R163–R168. doi: 10.1093/hmg/ddp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung F, Diamandis EP, Kulasingam V. From bench to bedside: Discovery of ovarian cancer biomarkers using high-throughput technologies in the past decade. Biomarkers Med. 2012;6(5):613–625. doi: 10.2217/bmm.12.70. [DOI] [PubMed] [Google Scholar]
- 5.Chung GG, et al. Tissue microarray analysis of beta-catenin in colorectal cancer shows nuclear phospho-beta-catenin is associated with a better prognosis. Clin Cancer Res. 2001;7(12):4013–4020. [PubMed] [Google Scholar]
- 6.Lessard L, et al. Nuclear localization of nuclear factor-kappaB p65 in primary prostate tumors is highly predictive of pelvic lymph node metastases. Clin Cancer Res. 2006;12(19):5741–5745. doi: 10.1158/1078-0432.CCR-06-0330. [DOI] [PubMed] [Google Scholar]
- 7.Guillaud M, et al. Subvisual chromatin changes in cervical epithelium measured by texture image analysis and correlated with HPV. Gynecol Oncol. 2005;99(3, suppl 1):S16–S23. doi: 10.1016/j.ygyno.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 8.Shariff A, Kangas J, Coelho LP, Quinn S, Murphy RF. Automated image analysis for high-content screening and analysis. J Biomol Screen. 2010;15(7):726–734. doi: 10.1177/1087057110370894. [DOI] [PubMed] [Google Scholar]
- 9.Lejeune M, et al. Quantification of diverse subcellular immunohistochemical markers with clinicobiological relevancies: Validation of a new computer-assisted image analysis procedure. J Anat. 2008;212(6):868–878. doi: 10.1111/j.1469-7580.2008.00910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matos LL, Trufelli DC, de Matos MG, da Silva Pinhal MA. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:9–20. doi: 10.4137/bmi.s2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newberg J, Murphy RF. A framework for the automated analysis of subcellular patterns in Human Protein Atlas images. J Proteome Res. 2008;7(6):2300–2308. doi: 10.1021/pr7007626. [DOI] [PubMed] [Google Scholar]
- 12.Glory E, Newberg J, Murphy RF. Automated comparison of protein subcellular location patterns between images of normal and cancerous tissues. Proc IEEE Int Symp Biomed Imaging. 2008;2008:304–307. doi: 10.1109/ISBI.2008.4540993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlén M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald LM, et al. Germline missense variants in the BTNL2 gene are associated with prostate cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2013;22(9):1520–1528. doi: 10.1158/1055-9965.EPI-13-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chitu V, et al. Primed innate immunity leads to autoinflammatory disease in PSTPIP2-deficient cmo mice. Blood. 2009;114(12):2497–2505. doi: 10.1182/blood-2009-02-204925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140(3):384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1(3):328–346. [PMC free article] [PubMed] [Google Scholar]
- 18.Mahapatra S, et al. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18(10):2882–2895. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- 19.Lambert LA, Whyteside AR, Turner AJ, Usmani BA. Isoforms of endothelin-converting enzyme-1 (ECE-1) have opposing effects on prostate cancer cell invasion. Br J Cancer. 2008;99(7):1114–1120. doi: 10.1038/sj.bjc.6604631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massoner P, et al. Characterization of transcriptional changes in ERG rearrangement-positive prostate cancer identifies the regulation of metabolic sensors such as neuropeptide Y. PLoS ONE. 2013;8(2):e55207. doi: 10.1371/journal.pone.0055207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7(4):270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Chen J. One-carbon metabolism and breast cancer: An epidemiological perspective. J Genet Genomics. 2009;36(4):203–214. doi: 10.1016/S1673-8527(08)60108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe LR, Brown PH. Targeting the HER/EGFR/ErbB family to prevent breast cancer. Cancer Prev Res (Phila) 2011;4(8):1149–1157. doi: 10.1158/1940-6207.CAPR-11-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito H, et al. Identification of ROBO1 as a novel hepatocellular carcinoma antigen and a potential therapeutic and diagnostic target. Clin Cancer Res. 2006;12(11 pt 1):3257–3264. doi: 10.1158/1078-0432.CCR-05-2787. [DOI] [PubMed] [Google Scholar]
- 25.Biankin AV, et al. Australian Pancreatic Cancer Genome Initiative Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimbro KS, Simons JW. Hypoxia-inducible factor-1 in human breast and prostate cancer. Endocr Relat Cancer. 2006;13(3):739–749. doi: 10.1677/erc.1.00728. [DOI] [PubMed] [Google Scholar]
- 27.Collett GP, et al. Peroxisome proliferator-activated receptor alpha is an androgen-responsive gene in human prostate and is highly expressed in prostatic adenocarcinoma. Clin Cancer Res. 2000;6(8):3241–3248. [PubMed] [Google Scholar]
- 28.Barron DA, Kagey JD. The role of the Hippo pathway in human disease and tumorigenesis. Clin Ttransl Med. 2014;3:25. doi: 10.1186/2001-1326-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.