Significance
The envelope subunit gp41 is an attractive target for therapeutic intervention against HIV-1. Interfering with the interaction between the heptad-repeat regions of gp41 is a promising approach to inhibit HIV-1 fusion to the host cell membrane. Here, we present an alternative rational design and protein-engineering approach to produce highly stable single-chain proteins that accurately mimic the trimeric coiled-coil surface of the gp41 N-terminal heptad repeat. This approach has a strong potential for development to HIV-1 drugs, vaccines, or microbicides and could be extendable to the design of proteins interfering with other types of coiled-coil interactions.
Keywords: fusion inhibitor, HIV-1 neutralization, X-ray crystallography, drug design, coiled coil
Abstract
During HIV-1 fusion to the host cell membrane, the N-terminal heptad repeat (NHR) and the C-terminal heptad repeat (CHR) of the envelope subunit gp41 become transiently exposed and accessible to fusion inhibitors or Abs. In this process, the NHR region adopts a trimeric coiled-coil conformation that can be a target for therapeutic intervention. Here, we present an approach to rationally design single-chain protein constructs that mimic the NHR coiled-coil surface. The proteins were built by connecting with short loops two parallel NHR helices and an antiparallel one with the inverse sequence followed by engineering of stabilizing interactions. The constructs were expressed in Escherichia coli, purified with high yield, and folded as highly stable helical coiled coils. The crystal structure of one of the constructs confirmed the predicted fold and its ability to accurately mimic an exposed gp41 NHR surface. These single-chain proteins bound to synthetic CHR peptides with very high affinity, and furthermore, they showed broad inhibitory activity of HIV-1 fusion on various pseudoviruses and primary isolates.
The HIV-1 envelope glycoprotein is a trimer of heterodimers of two noncovalently associated glycoprotein (gp) subunits, gp120 and gp41, that promotes viral infection by mediating the fusion of the viral membrane with the host cell membrane (1). The transmembrane subunit gp41 consists of an extracellular domain (ectodomain), a transmembrane segment, and an intracytoplasmic tail. The gp41 ectodomain contains a hydrophobic N-terminal fusion peptide followed by a polar region, an N-terminal heptad repeat (NHR), a disulfide-bridged loop region, a C-terminal heptad repeat (CHR), and a membrane-proximal external region. The current model of HIV-1 infection suggests that binding of gp120 to the CD4 receptor and a coreceptor triggers the protrusion of the gp41 fusion peptide, which penetrates into the host cell membrane. This event is followed by gp120 shedding and a large conformational change of gp41 from an unstable prefusion state, featuring exposed NHR and CHR regions, to a highly stable postfusion state forming a six-helix bundle, coiled-coil structure (2). This conformational change brings the two membranes into close proximity, facilitating their fusion (3). Because of its involvement in membrane fusion and viral entry as well as its highly preserved sequence among viral strains, gp41 offers potential targets to drugs or Abs.
During gp41-mediated HIV infection, the NHR region forms a parallel trimeric coiled coil that becomes transiently exposed (4). This exposed conformation was shown by the elicitation of Abs that block HIV fusion using immunogens exposing an NHR coiled coil (5). Also, two human mAbs (D5 and Hk20) recognize a highly conserved NHR hydrophobic pocket and neutralize diverse HIV-1 clinical isolates (6, 7). Moreover, potent HIV-1–neutralizing CHR peptides, pioneered by T20, and small molecules bind to the trimeric NHR, interfering with formation of the NHR/CHR six-helix bundle (8–12).
NHR-derived peptides can also inhibit HIV entry by targeting the CHR region, but in monomeric form, they show far less potency because of their low conformational stability and high tendency to aggregate. Potent HIV inhibitors have been designed by sequestering the NHR peptide into a nonaggregating trimeric coiled-coil conformation, which should be able to effectively bind the CHR helix and interfere with six-helix bundle formation. Several design approaches of this type include covalent stabilization of the trimer by disulfide bridges (13–15) and fusion of NHR segments to trimerization domains (16–18). Also targeting the CHR region, a protein denoted 5-Helix was engineered by connecting five of six helices that make up the core of the gp41 postfusion structure using short peptide linkers (19). All of these constructs inhibit HIV-1 entry at nanomolar concentrations.
Despite these advances, only T20 is currently approved for HIV treatment among this type of fusion inhibitors. Unfortunately, the clinical use of T20 has been limited by its short half-life. T20 treatment needs high dosage by two times per day s.c. injection to overcome its proteolysis and rapid renal filtration, producing painful site reactions and other side effects that reduce adherence to treatment. These practical problems result in an expensive drug (∼$30,000 per year) that is only approved for patients experiencing treatment failure caused by multidrug resistance. In addition, continued treatment produces the appearance of T20-resistant viruses. Additional approved fusion inhibitors are, therefore, needed to improve the potency of the therapeutic arsenal and allow for adapting the therapy to the development of resistance. Other gp41-based constructs still have significant problems that make difficult their development as suitable drugs or vaccines. Chemically synthesized peptides, like T20, share a short half-life and are difficult to produce. Some constructs show low solubility at physiological pH, which impairs production and formulation. Aggregation may also alter their conformation and reduce bioavailability. Recombinant expression is an alternative approach, but disulfide-bonded constructs may present considerable difficulties to refolding in the active form. Moreover, the addition of trimerization domains to stabilize the trimer increases the molecular size of the constructs, which can be detrimental to access of sterically obscured targets, and if used as immunogens, the exogenous epitopes might divert the immune responses.
Coiled-coil motifs are extremely versatile structures sharing a characteristic 7-aa (heptad) repeat, (abcdefg)n, with hydrophobic residues at the first (a) and fourth (d) positions (20). These heptad-repeat sequences can adopt parallel or antiparallel dimeric as well as trimeric and tetrameric coiled-coil conformations. Taking advantage of the plasticity and versatility of the coiled coils, we describe here an approach to design small protein mimetics of a stable NHR coiled-coil surface. We transformed the trimeric parallel-chain topology of gp41 NHR into a single-chain bundle of three antiparallel helices connected by short loops. The engineered protein constructs fold as predicted, interact strongly with CHR peptides, and show a strong antiviral activity against a variety of HIV-1 isolates.
Results
Rational Design of a Single-Chain Mimetic of the gp41 NHR Coiled Coil.
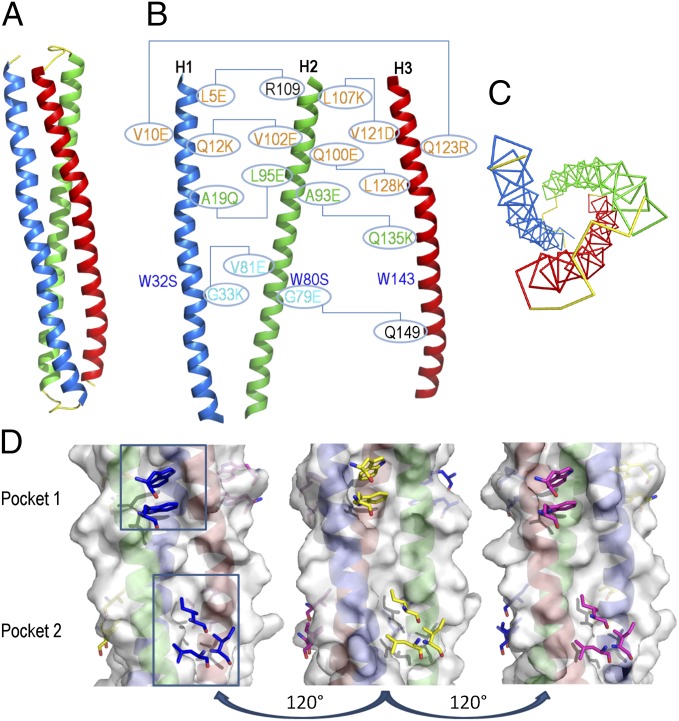
We designed an antiparallel trimeric coiled-coil protein, in which the surface of the two parallel α-helices would mimic that of the parallel NHR coiled coil of gp41. As a template, we used the three NHR helical segments (residues A30–L81) in the postfusion structure model of the gp41 ectodomain [Protein Data Bank (PDB) ID code 1IF3] (21). We modeled by homology one of the α-helices with the inverse NHR polypeptide sequence. Then, we upturned the spatial orientation of the modified helix and docked it onto the other two original helices, preserving the internal core packing of the new antiparallel coiled coil. Steric clashes were removed by optimizing the side-chain rotamers. The three helices were then tethered using short peptide linkers, creating a helix–turn–helix–turn–helix topology (Fig. 1A). Amino acids at the edges of the helices and at the linkers were chosen to create favorable helix-capping stabilizing interactions (22). This first construct (covNHR1) was cloned in Escherichia coli but could not be expressed, possibly because of a low structural stability and/or a strong aggregation propensity. To stabilize the trimeric antiparallel fold and enhance solubility, a second construct (covNHR2) was engineered to reduce surface hydrophobicity by replacing several exposed hydrophobic residues with charged amino acids that could make salt bridges (Fig. 1B). These mutations did not alter the conserved hydrophobic pocket on the NHR surface. Finally, a third construct (covNHR3) was designed with longer loops connecting the helices. The polypeptide sequences are listed in SI Appendix, Table S1. These two first generation covNHR proteins were expressed in E. coli with good yields and purified to homogeneity.
Fig. 1.
Design of single-chain gp41 NHR mimetics. (A) Ribbon representation of the chain topology of the covNHR constructs. The red and blue helices run in parallel to the original NHR gp41 sequence, whereas the green helix is antiparallel. Short linkers colored in yellow connect the helices. (B) Engineered mutations on the surface covNHR constructs are indicated according to the sequence numbering of covNHR3 (SI Appendix, Table S1). Helices have been separated artificially for clarity. Lines connect residues that establish new interhelical stabilizing interactions. Surface mutations for the first generation constructs are colored in orange; groups A, B, and C of the mutations for the second generation constructs are colored in cyan, green, and blue, respectively. (C) Threefold pseudosymmetry of the covNHR molecule topology. Only the backbone atoms have been represented for clarity. Colors are the same as in A. (D) Molecular surface of the three faces of the covNHR3 model. The sticks colored in blue, yellow, and magenta represent the hydrophobic side chains of complementary CHR residues as they would interact on each hydrophobic pocket of the NHR surface.
Biophysical Characterization of the First Generation covNHR Proteins.
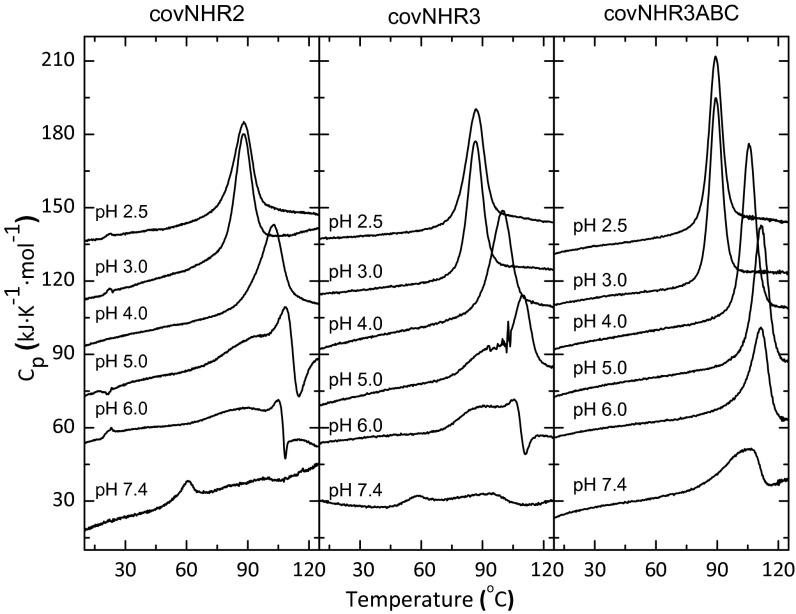
The covNHR2 and covNHR3 constructs were highly soluble at both acid and physiological pH, and they could be lyophilized and redissolved in buffer without significant structural loss. The constructs are monomeric at pH 2.5, whereas at pH 7.4, the proteins tend to self-associate according to dynamic light-scattering (DLS) and static light-scattering measurements (SI Appendix, Fig. S1 and Table S2). The far-UV CD spectra are typical of highly α-helical proteins (SI Appendix, Fig. S2), which is in good agreement with the design models. The near-UV CD spectra showed very low ellipticity because of solvent exposure of the aromatic residues. Differential scanning calorimetry (DSC) with the two proteins showed single sharp thermal unfolding transitions at acid pH with very high Tm (Fig. 2). These transitions are slightly asymmetric and do not accurately follow a two-state unfolding model, but they could be very well-fitted with a model of two sequential unfolding transitions (N ⇄ I ⇄ U) (SI Appendix, Figs. S3 and S4). The unfolding cooperativity was evaluated according to the maximum population of the partially unfolded intermediate state calculated from the thermodynamic parameters of the fits (SI Appendix, Table S3). covNHR3 showed a higher unfolding enthalpy and cooperativity than covNHR2, indicating a more organized tertiary structure. Maximum cooperativity was observed at pH 3.0. As the pH was increased, exothermic effects appeared after thermal unfolding, which becomes irreversible, likely because of thermally induced aggregation. At physiological pH, the DSC thermograms showed two faint peaks. The first one (at about 60 °C) is accompanied by a strong decrease in the α-helical secondary structure, which is shown by CD (SI Appendix, Fig. S5) and aggregation according to DLS measurements. These results indicated that, while at acid pH, the two proteins are highly stable and well-structured monomers, but at physiological pH, they have a lower structural cooperativity and a propensity to self-associate.
Fig. 2.
Thermal stability of covNHR proteins. DSC thermograms were measured at different pH values, which are indicated along each curve. The curves have been arbitrarily shifted along the y axis for the sake of clarity.
Interaction of the covNHR Constructs with Exogenous gp41 CHR Peptides.
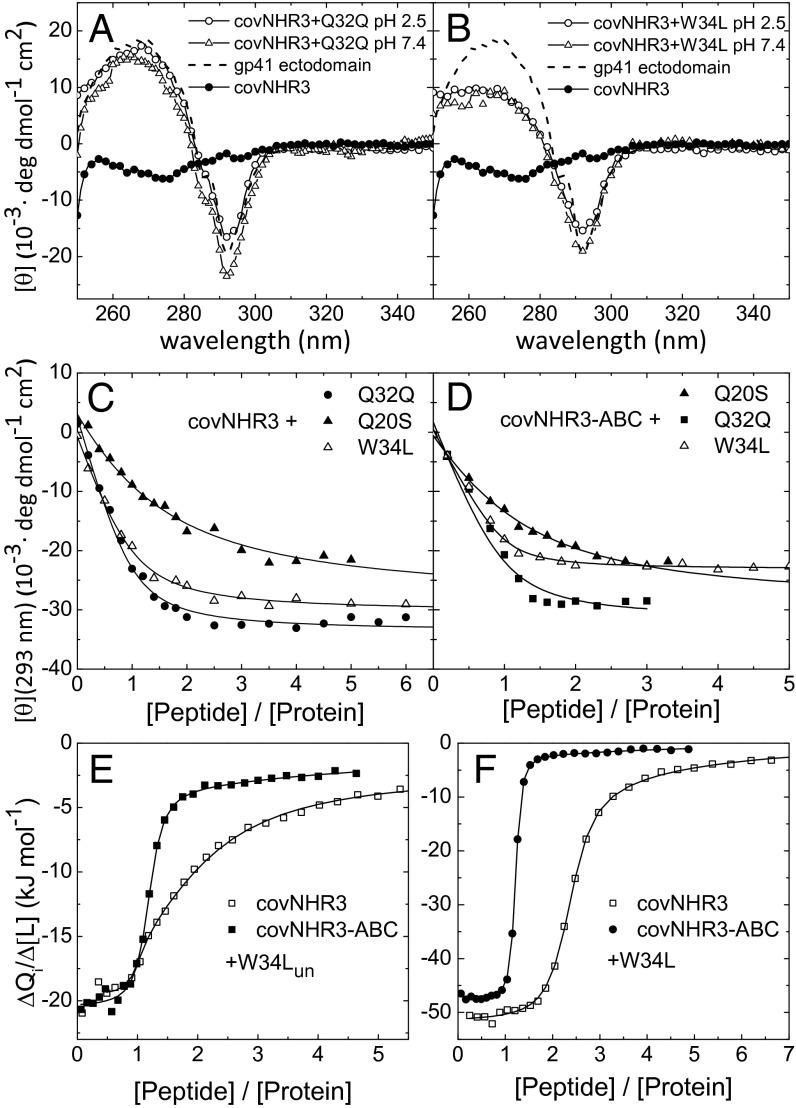
We evaluated the capacity of the covNHR2 and covNHR3 constructs to bind several synthetic peptides encompassing different CHR segments of gp41, namely CHR-127–162 (T20) (23), CHR-117–150 (W34L) (24), CHR-110–141 (Q32Q) (25), and CHR-110–129 (Q20S). All of these peptides bound to covNHR2 and covNHR3 in equimolar mixtures, which were inferred from the changes in CD spectra (SI Appendix, Figs. S6 and S7). The putative NHR–CHR interaction in the postfusion conformation of gp41 involves the staking and insertion of W117 and W120 side chains into the NHR-conserved hydrophobic pocket (2). This interaction results in a characteristic negative band centered at 293 nm in the CD spectrum (26). Because the covNHR proteins lack this signal, binding of CHR peptides containing the two-tryptophan motif restored the negative CD band (Fig. 3 A and B). Titrations of the covNHR proteins with the CHR peptides monitoring the CD signal at 293 nm (Fig. 3C) indicated apparent affinities in the low micromolar range (SI Appendix, Table S4). Furthermore, according to DSC experiments at pH 2.5 and especially, pH 7.4, the two proteins became strongly stabilized by the binding of the CHR peptides (SI Appendix, Fig. S8).
Fig. 3.
Binding of CHR peptides to the covNHR proteins. (A and B) Near-UV CD spectra of isolated covNHR3 and equimolar mixtures of covNHR3 and the CHR peptides (A) Q32Q and (B) W34L. The spectrum of a recombinant full gp41 ectodomain in the six-helix bundle conformation is also shown as dashed lines. (C and D) Titrations of (C) covNHR3 and (D) covNHR3–ABC with CHR peptides followed by near-UV CD at 293 nm. The lines correspond to the curve fittings using a 1:1 binding model. (E and F) ITC isotherms for the binding of peptides (E) W34Lun and (F) W34L to covNHR3 and covNHR3–ABC. The symbols represent the experimental heats measured for each peptide injection normalized per mole increment in the peptide concentration. The curves correspond to the best fittings using a theoretical binding model of several different and independent binding sites (details are in the text).
We measured the thermodynamic parameters of the binding of the W34L peptide to covNHR3 by isothermal titration calorimetry (ITC) at pH 7.4 and 25 °C. No reproducible ITC data could be measured with the Q32Q and Q20Q peptides because of a propensity of these peptides to self-associate. ITC titrations were carried out with the N-acetylated and C-amidated peptide (W34L) and the same peptide with unprotected termini (W34Lun). Surprisingly, the ITC isotherms did not obey a 1:1 binding stoichiometry (Fig. 3 E and F) but could be fitted using a model of several different and independent binding sites. The thermodynamic magnitudes resulting from this analysis are listed in SI Appendix, Table S5. For the protected W34L peptide, the ITC data complied with two binding sites with similarly high affinity (Kd = 230 ± 30 nM) and a third site with a lower affinity (Kd = 36 μM). In the case of W34Lun, the ITC isotherm could only be described assuming three different and independent sites. The highest affinity site was also in the submicromolar range (Kd = 70 ± 50 nM). The binding enthalpy is much more negative for the protected peptide than for the unprotected one but is compensated for by a more favorable binding entropy for the latter, indicating a less ordered complex, possibly caused by helical fraying at the charged peptide termini. The binding stoichiometry could be explained considering that the covNHR constructs have threefold pseudosymmetry (Fig. 1C), with one native-like face and two nonnative ones having three similar hydrophobic crevices (Fig. 1D), which could hold up to three CHR peptide moieties with different affinities.
In conclusion, the covNHR2 and covNHR3 proteins mimic accurately the gp41 NHR trimeric bundle. However, their decrease in structural cooperativity and their propensity to self-associate at physiological pH may reduce their activity and could be problematic during production development. For these reasons, we designed a second generation of covNHR proteins to improve their conformational stability and focus their ability to bind gp41 CHR peptides only in a native-like arrangement.
Design of Second Generation covNHR Proteins.
Each one of three hydrophobic crevices on the covNHR3 model surface contains several consecutive pockets that may accommodate hydrophobic residues from the CHR region (Fig. 1D). One of these pockets (pocket 1), created by the presence of a Gly residue at position g in the helical coil, holds W117 and W120 side chains of the CHR region in the 6HB postfusion gp41 conformation. An alanine at a similar position creates another shallower pocket (pocket 2), where several CHR hydrophobic residues can interact. Therefore, we substituted the corresponding Gly and Ala residues with bulkier residues to fill the hydrophobic pockets and reduce the interaction capacity of the nonnative-like faces of covNHR3. We also engineered neighboring residues in space to design additional stabilizing interactions (Fig. 1B). Two groups of mutations were designed with this aim: group A removing pocket 1 (G33K, V81E, and G79E) and group B removing pocket 2 (A19Q, L95E, A93E, and Q135K). In addition, the covNHR proteins have three tryptophan residues with outwardly oriented side chains, which could promote self-association. Accordingly, group C of mutations (W32S and W80S) replaced the Trp residues by Ser on the nonnative faces of the trimeric bundle. These three groups of mutations were combined in five different constructs, which were named according to the groups of mutations that they contained (SI Appendix, Table S1).
Biophysical Characterization of Second Generation covNHR Proteins.
The five modified constructs were produced by E. coli expression with high yields and are highly soluble at both acid and physiological pH levels. At pH 2.5, all constructs are monomeric according to DLS measurements, similar to the parental molecule covNHR3, whereas at pH 7.4, the hydrodynamic radii were more variable (SI Appendix, Table S2). The constructs containing group A of mutations showed radii closer to those of monomers. This size was confirmed for covNHR3–ABC by static light-scattering analysis (SI Appendix, Fig. S1). All of the mutant proteins are also highly α-helical at both pH 2.5 and 7.4 (SI Appendix, Fig. S2). The thermal stability and cooperativity of all mutants are higher than that of covNHR3 to different extents, which was observed by DSC (Fig. 2 and SI Appendix, Fig. S9 and Table S3). The unfolding cooperativity of covNHR3–ABC turned out to be very high at acid pH, and in fact, the DSC transition followed well the two-state unfolding model. However, at pH 7.4, the thermal transitions were irreversible for all of the mutants, although they showed higher thermal stability and sharper denaturation profiles than covNHR3, indicating more stable and cooperative structures. Overall, the covNHR3–ABC construct showed the best biophysical properties among five mutants. Moreover, this construct exhibited considerably higher resistance than T20 against proteolysis by thermolysin at physiological pH (SI Appendix, Fig. S10).
Binding of Second Generation covNHR Proteins to CHR Peptides.
All of the second generation constructs were able to bind exogenous synthetic CHR peptides to different extents (SI Appendix, Fig. S11). Titrations of covNHR3–ABC with CHR peptides followed by CD indicated apparent binding affinities in the low micromolar range (Fig. 3D and SI Appendix, Table S4). The ITC titrations of covNHR3–ABC with each of W34L and W34Lun peptides (Fig. 3 E and F) indicated a main 1:1 high-affinity binding event accompanied by small residual heats. The binding isotherms could be fitted with a model of one site with very high affinity (Kd = 40 and 180 nM for W34L and W34Lun, respectively) plus two sites with very low affinity attributable to weak unspecific binding (SI Appendix, Table S5). These results showed that the mutations engineered in covNHR3–ABC selected specifically the native-like binding mode.
Crystal Structure of the covNHR3–ABC Construct.
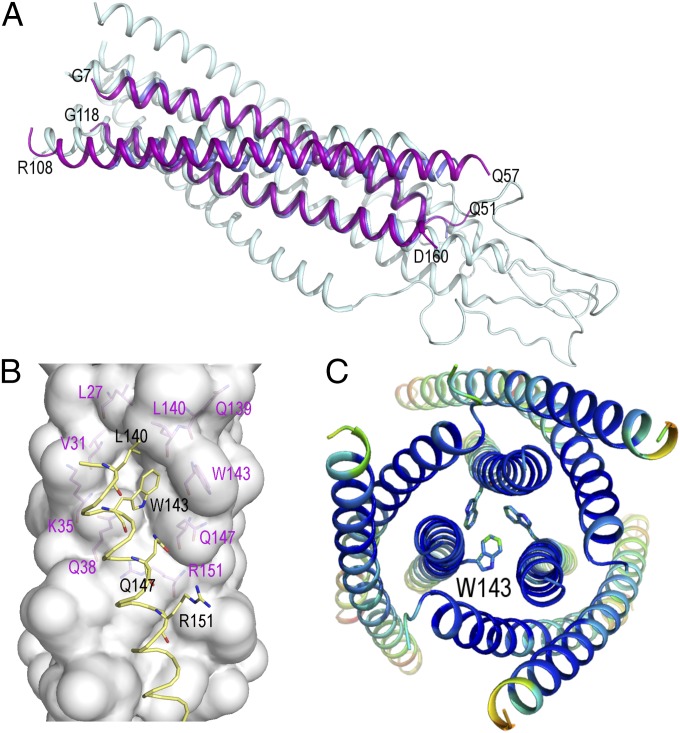
The crystal structure of the covNHR3–ABC protein was determined at a resolution of 3.1 Å, confirming the characteristic antiparallel trimeric coiled-coil structure. Several regions of the polypeptide chains were not visible in the electron density map (SI Appendix, Fig. S12) and were assumed to be disordered, namely residues 1–7 and 162–175 (including the 11-residue C-terminal His tag) at the N and C termini and residues 53–57 and 110–118 corresponding to the loops connecting the α-helices. Fig. 4A shows a superposition of the covNHR3–ABC structure to the theoretical model of the gp41 ectodomain generated from the coordinates of the simian immunodeficiency virus gp41 ectodomain (1IF3) (21) and the crystallographic structure of the five-helix construct (2XRA) (7). The backbones of the two parallel NHR helices are well-superimposed, with an rmsd of 1.4 Å. Moreover, the hydrophobic pocket on the surface of the two parallel helices is very well-preserved (Fig. 4B), which is in good agreement with the design models. Interestingly, a Trp cluster involving the conserved hydrophobic pocket mediates intermolecular association between three protein monomers within the crystal (Fig. 4C).
Fig. 4.
Crystallographic structure of the covNHR3–ABC construct. (A) Cartoon representation of the superposition of covNHR3–ABC (purple) onto the theoretical model of the gp41 ectodomain in the postfusion conformation (PDB ID code 1IF3; gray) (21) and the three NHR helices of 5-Helix (PDB ID code 2XRA; blue) (7). (B) Upper view of the packing of three covNHR3–ABC molecules in the crystal. The side chains of W143 form an aromatic cluster that brings together three covNHR3–ABC molecules. The helices are colored by B-factor spectrum (from blue to red). (C) Conserved hydrophobic pocket on the surface of the two parallel helices of covNHR3–ABC. The side chains of residues lining the pocket are labeled in magenta. The helix of another covNHR3–ABC molecule (in yellow) packs in the crystal against this hydrophobic cavity with the insertion the side chain of W143. All of the figures were made using Pymol.
HIV Inhibitory Activity of covNHR Constructs.
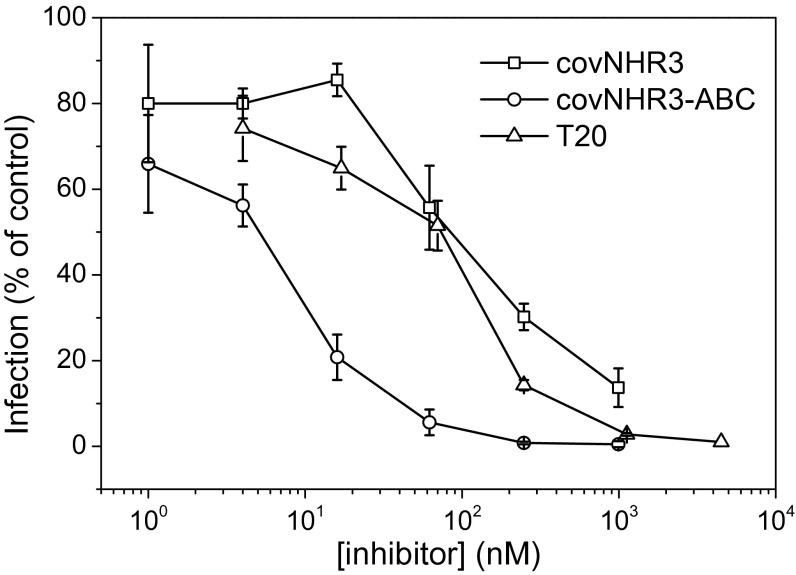
Because the covNHR constructs showed a strong capability to interact with the gp41 CHR sequences, the HIV-1 inhibitory power of covNHR3 and covNHR3–ABC was tested compared with T20 (used as a positive control). Strong inhibition of HIV-1 infection was observed in both TZM-bL and peripheral blood mononuclear cells (PBMC) neutralization assays and for all of the viruses tested. In the neutralization assay with TZM-bL cells infected with HIV-1 SF162 pseudovirus, covNHR3 and T20 induced a similar dose–response inhibitory activity, with an IC50 of about 100 nM (Fig. 5). Interestingly, covNHR3–ABC showed a strongly increased inhibitory activity compared with T20 and covNHR3, with an IC50 of about 7 nM. Also, covNHR3 and covNHR3–ABC displayed a broad inhibitory profile on the infection of PBMCs with various primary isolates (Table 1). covNHR3 inhibited these primary isolates at submicromolar concentration, and again, about a 10-fold increased inhibitory activity was observed with covNHR3–ABC (IC80 in the nanomolar range). These results show that the improved conformational and binding properties obtained with covNHR3–ABC result in an increased HIV-1 inhibitory profile. Interestingly, T20 was a less efficient inhibitor than covNHR3–ABC for most of the HIV-1 strains tested.
Fig. 5.
Inhibition of HIV-1 by covNHR proteins. In vitro inhibition of infection of TZM-bL cells with SF-162 pseudoviruses by different concentrations of covNHR3 and covNHR3–ABC. Inhibition by T20 was also measured as a control. Data are the means of three independent measurements, and error bars correspond to the SDs.
Table 1.
In vitro HIV-1 inhibition by covNHR constructs and T20
| Virus | Subtype (coreceptor use) | T20 (nM) | covNHR3 (nM) | covNHR3–ABC (nM) |
| SF162 | B (R5) | 254 ± 364 | 147 ± 4 | 63 ± 43 |
| QH0692 | B (R5) | 1,124 ± 22 | 232 ± 29 | 53 ± 6 |
| KON | CRF02-AG (X4) | 124 ± 16 | 215 ± 57 | 36 ± 11 |
| 92UG024 | C (R5) | 20 ± 3 | 199 ± 86 | 22 ± 3 |
| TV1 | C (R5) | 157 ± 95 | 132 ± 103 | 17 ± 13 |
| 89.6 | B (X4R5) | 45 ± 32 | 247 ± 3 | 87 ± 18 |
| DU174 | C (R5) | 337 ± 159 | 331 ± 143 | 51 ± 3 |
| 92BR025 | C (R5) | 101 ± 48 | 124 ± 35 | 66 ± 29 |
PBMCs were infected using various HIV-1 primary isolates in the presence of the constructs. Values (means ± SDs of three independent experiments) are IC80.
Discussion
We have designed several single-chain protein mimetics of a fully exposed gp41 NHR trimer. The success of these designs relies on the extraordinary versatility of coiled-coil structures and the combination of several elements to stabilize specifically the antiparallel trimeric arrangement. Building up an antiparallel trimer was challenging, because the altered chain topology could have resulted in considerable perturbations in the knobs-into-holes packing of the original parallel NHR trimer, reduction of the coiled-coil stability, and change in the oligomerization state and/or chain topology, which was observed, for example, in the general control protein (GCN4) leucine zipper, in which a single N16A mutation changes its fold into an antiparallel trimer from the parallel dimer of the WT form (27). However, the gp41 NHR sequence lacks a strong preference for hydrophobic residues that favor each specific oligomerization arrangement, allowing a higher permissibility in chain orientation within a trimeric bundle as long as the hydrophobic/hydrophilic complementarity pattern is maintained. In addition, the presence of three Gln and one Thr side chains at core positions would also favor the trimeric packing (20). Moreover, tethering the consecutive helices with short loops stabilized by helix-capping motifs and addition of other stabilizing interactions also helped to stabilize the antiparallel orientation of the coil. As a result, the covNHR constructs show very high thermodynamic stability and structural cooperativity. At acid pH, the specific unfolding enthalpies measured by DSC are about 3.2–3.6 kJ per residue, which are slightly lower than those typically observed for compact globular proteins (4–4.5 kJ per residue) at similar temperatures (28) but close to those reported for a similarly sized three-stranded coiled-coil of the E. coli outer membrane lipoprotein Lpp-56 (about 3.9 kJ res−1) (29); this finding indicates a good degree of packing within the core of the coil, especially for the improved second generation constructs, which was confirmed by the crystal structure of covNHR3–ABC.
An interesting result is the capability of the covNHR2 and covNHR3 constructs to bind more than one CHR peptide moiety, despite the fact that two of three faces of the trimeric helical bundle are formed by antiparallel helices as opposed to the parallel arrangement in the gp41 native NHR trimer. Our structural model for the constructs suggests a remarkable surface resemblance between the native and the nonnative faces, allowing similar coiled-coil interactions with CHR peptides. This result was further supported by the removal of hydrophobic pockets on the nonnative faces of covNHR3–ABC, practically abolishing the 3:1 binding stoichiometry.
The binding thermodynamics measured by ITC between the covNHR constructs and the W34L peptide, also known as C34 in the literature, indicate nanomolar affinity. For covNHR3–ABC, Kd is 40 ± 8 nM, which is among the highest affinities found for this type of NHR inhibitors. W34L binding is driven by a favorable negative enthalpy and partially opposed by a small negative entropy. For the unprotected peptide W34Lun, the enthalpy is smaller, but binding is also entropically favored. He et al. (30) reported a binding affinity of N36 for the W34L peptide in the submicromolar range (Kd = 300 nM). However, both the binding enthalpy and entropy values were much more negative than those reported here, likely because of the contribution of the folding and association of the NHR trimer. Similarly, higher enthalpy and entropy magnitudes than those measured by us were observed for the binding of nonnatural small cyclic disulfide D peptides to the trimeric coiled-coil IQN17 (31) or W34L or two recombinant peptide inhibitors to 5-Helix (32). The relatively low entropy penalty accompanying the complex formation for the covNHR proteins may arise from a favorable balance between the entropy gain of the hydrophobic interactions and a moderate reduction of configurational freedom caused by the highly stable binding surface offered by the covNHR proteins.
Interestingly, the exceptional binding capacities of the covNHR constructs are associated with an efficient and broad inhibitory activity on various HIV-1 pseudoviruses and primary isolates. This result shows that, like T20, the covNHR constructs can interfere with the HIV-1 fusion process in physiologically relevant HIV-1 infection models. The inhibitory activity of covNHR3–ABC is higher and broader than that of T20. This higher inhibitory potency is likely because of the fact that it interferes with highly preserved interactions participating in HIV fusion, such as those mediated by the conserved NHR hydrophobic pocket (9). Noteworthy, the inhibitory activity of the covNHR proteins is observed despite their higher molecular weight, indicating that any possible structural hindrances engendered by the highly structured proteins may be compensated for by its unusually high affinity for gp41.
In conclusion, these protein mimetics present a well-ordered and stable gp41 NHR binding surface for CHR peptides. The high structural stability of the constructs contributes positively to the high affinity observed as well as their potent and broad HIV-1 inhibitory activity. These constructs may have very good potential as entry inhibitors as well as vaccine candidates. Their main advantages are that they are constituted of a single polypeptide chain, can be readily expressed in E. coli with high yield, and do not need any additional chemical modification. Their highly stable structure makes them less sensitive to proteolysis than unstructured peptides, such as T20, and should improve their pharmacokinetic properties. In addition, our design approach may have broader applicability to produce other stable mimetics targeting different coiled-coil interactions, such as those present in other retroviruses.
Methods
Molecular models of the proteins were built and analyzed using Swiss-Pdb Viewer (33) based on a homology model structure of the HIV gp41 ectodomain (PDB ID code 1IF3) (21). The different protein constructs were produced by overexpression in E. coli cells and biophysically characterized as described in SI Appendix. The interaction between the covNHR constructs and the gp41 CHR region was investigated by mixing each covNHR protein with exogenous synthetic peptides, encompassing different segments of the gp41 CHR region using titration experiments monitored by near-UV CD and ITC. Structure determination of the covNHR3–ABC construct was made by X-ray crystallography. The coordinates were deposited with the Research Collaboratory for Structural Bioinformatics (RCSB) PDB with ID code 4R61. The HIV-1 inhibitory activity of the covNHR constructs was studied using neutralizing assays on TZM-bL cells infected with HIV-1 SF-162 pseudoviruses and PBMCs using HIV-1 primary isolates from various subtypes sensitive to neutralization (34, 35). Comprehensive experimental details are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Jordi Juanhuix at the beamline XALOC from the ALBA synchrotron for assistance in the measurement of the crystals. This research was funded by European Union Seventh Framework Programme Grant HEALTH-F3-2007-201038 and cofinanced by Andalusia Regional Government Grant Incentivos-BOJA-5-1-2008, Spanish Ministry of Economy and Competitiveness Grants BIO2012-39922-C02-01/02 and BIO2008-00750-E, Andalusia Regional Government Grant P09-CVI-5063, and the European Fund for Regional Development (FEDER) (EU). X-ray diffraction data collection was supported by European Synchrotron Radiation Facility block allocation group (BAG) Proposals MX-1406 and MX1541 and Spanish Synchrotron Radiation Facility ALBA Proposals 2012010072 and 2012100378.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4R61).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413592112/-/DCSupplemental.
References
- 1.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 2.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 3.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93(5):681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 4.Eckert DM, et al. Characterization of the steric defense of the HIV-1 gp41 N-trimer region. Protein Sci. 2008;17(12):2091–2100. doi: 10.1110/ps.038273.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis JM, Bewley CA, Gustchina E, Aniana A, Clore GM. Characterization and HIV-1 fusion inhibitory properties of monoclonal Fabs obtained from a human non-immune phage library selected against diverse epitopes of the ectodomain of HIV-1 gp41. J Mol Biol. 2005;353(5):945–951. doi: 10.1016/j.jmb.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 6.Luftig MA, et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat Struct Mol Biol. 2006;13(8):740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- 7.Sabin C, et al. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog. 2010;6(11):e1001195. doi: 10.1371/journal.ppat.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilby JM, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4(11):1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 9.Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95(26):15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrer M, et al. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat Struct Biol. 1999;6(10):953–960. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 11.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: Discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 12.Eron JJ, et al. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J Infect Dis. 2004;189(6):1075–1083. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 13.Louis JM, Nesheiwat I, Chang L, Clore GM, Bewley CA. Covalent trimers of the internal N-terminal trimeric coiled-coil of gp41 and antibodies directed against them are potent inhibitors of HIV envelope-mediated cell fusion. J Biol Chem. 2003;278(22):20278–20285. doi: 10.1074/jbc.M301627200. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi E, et al. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc Natl Acad Sci USA. 2005;102(36):12903–12908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong P, et al. An engineered HIV-1 gp41 trimeric coiled coil with increased stability and anti-HIV-1 activity: Implication for developing anti-HIV microbicides. J Antimicrob Chemother. 2013;68(11):2533–2544. doi: 10.1093/jac/dkt230. [DOI] [PubMed] [Google Scholar]
- 16.Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci USA. 2001;98(20):11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis JM, Bewley CA, Clore GM. Design and properties of N(CCG)-gp41, a chimeric gp41 molecule with nanomolar HIV fusion inhibitory activity. J Biol Chem. 2001;276(31):29485–29489. doi: 10.1074/jbc.C100317200. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, et al. Novel recombinant engineered gp41 N-terminal heptad repeat trimers and their potential as anti-HIV-1 therapeutics or microbicides. J Biol Chem. 2010;285(33):25506–25515. doi: 10.1074/jbc.M110.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291(5505):884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 20.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 21.Caffrey M. Model for the structure of the HIV gp41 ectodomain: Insight into the intermolecular interactions of the gp41 loop. Biochim Biophys Acta. 2001;1536(2-3):116–122. doi: 10.1016/s0925-4439(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 22.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7(1):21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: Correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89(21):10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu M, Kim PS. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J Biomol Struct Dyn. 1997;15(3):465–471. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 25.He Y, et al. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: Implications for designing novel anti-HIV fusion inhibitors. J Virol. 2008;82(13):6349–6358. doi: 10.1128/JVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peisajovich SG, Blank L, Epand RF, Epand RM, Shai Y. On the interaction between gp41 and membranes: The immunodominant loop stabilizes gp41 helical hairpin conformation. J Mol Biol. 2003;326(5):1489–1501. doi: 10.1016/s0022-2836(03)00040-8. [DOI] [PubMed] [Google Scholar]
- 27.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA. 2004;101(6):1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makhatadze GI, Privalov PL. Contribution of hydration to protein folding thermodynamics. I. The enthalpy of hydration. J Mol Biol. 1993;232(2):639–659. doi: 10.1006/jmbi.1993.1416. [DOI] [PubMed] [Google Scholar]
- 29.Dragan AI, Potekhin SA, Sivolob A, Lu M, Privalov PL. Kinetics and thermodynamics of the unfolding and refolding of the three-stranded alpha-helical coiled coil, Lpp-56. Biochemistry. 2004;43(47):14891–14900. doi: 10.1021/bi048365+. [DOI] [PubMed] [Google Scholar]
- 30.He Y, et al. Conserved residue Lys574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J Biol Chem. 2007;282(35):25631–25639. doi: 10.1074/jbc.M703781200. [DOI] [PubMed] [Google Scholar]
- 31.Cole JL, Garsky VM. Thermodynamics of peptide inhibitor binding to HIV-1 gp41. Biochemistry. 2001;40(19):5633–5641. doi: 10.1021/bi010085w. [DOI] [PubMed] [Google Scholar]
- 32.Deng Y, Zheng Q, Ketas TJ, Moore JP, Lu M. Protein design of a bacterially expressed HIV-1 gp41 fusion inhibitor. Biochemistry. 2007;46(14):4360–4369. doi: 10.1021/bi7001289. [DOI] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 34.Holl V, et al. Involvement of Fc gamma RI (CD64) in the mechanism of HIV-1 inhibition by polyclonal IgG purified from infected patients in cultured monocyte-derived macrophages. J Immunol. 2004;173(10):6274–6283. doi: 10.4049/jimmunol.173.10.6274. [DOI] [PubMed] [Google Scholar]
- 35.Fenyö EM, et al. International network for comparison of HIV neutralization assays: The NeutNet report. PLoS ONE. 2009;4(2):e4505. doi: 10.1371/journal.pone.0004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.