Significance
The Pictet–Spengler (PS) reaction constructs many important phytochemicals such as morphine and camptothecin, but it has not yet been noticed in the fungal kingdom. Here, the startup of the PS reaction-based silent fungal biosynthetic machinery is presented to generate unforeseeably “unnatural” natural products of unprecedented carbon skeletons with antibacterial and acetylcholinesterase inhibitory activities. The gene-implied enzyme inhibition strategy is introduced to facilitate understandings of the key diversification steps. Collectively, the PS reaction-based fungal biosynthetic machinery that used to be silent has been set up to produce unpredictably novel molecules valuable for new biology and biomedicine.
Keywords: Pictet–Spengler reaction, FPS, Chaetomium globosum 1C51, GIEI
Abstract
The Pictet–Spengler (PS) reaction constructs plant alkaloids such as morphine and camptothecin, but it has not yet been noticed in the fungal kingdom. Here, a silent fungal Pictet–Spenglerase (FPS) gene of Chaetomium globosum 1C51 residing in Epinephelus drummondhayi guts is described and ascertained to be activable by 1-methyl-l-tryptophan (1-MT). The activated FPS expression enables the PS reaction between 1-MT and flavipin (fungal aldehyde) to form “unnatural” natural products with unprecedented skeletons, of which chaetoglines B and F are potently antibacterial with the latter inhibiting acetylcholinesterase. A gene-implied enzyme inhibition (GIEI) strategy has been introduced to address the key steps for PS product diversifications. In aggregation, the work designs and validates an innovative approach that can activate the PS reaction-based fungal biosynthetic machinery to produce unpredictable compounds of unusual and novel structure valuable for new biology and biomedicine.
Microbes and plants produce a multitude of unpredictably structured organic molecules known as secondary metabolites (natural products), from which more than half of globally marketed drugs have been developed (1–3). Large-scale genomic mining has indicated that microbial secondary metabolites are substantially underestimated because many biosynthetic genes remain silent or less active in the laboratory cultivation conditions (4, 5). Accordingly, there has long been an urgent need to develop a new strategy that enables microorganisms to produce more unforeseeable bioactive compounds, which are important to the drug discovery efforts to combat life-threatening diseases (6, 7), and to the complexity-based driving force for synthetic and material chemistry (8–10).
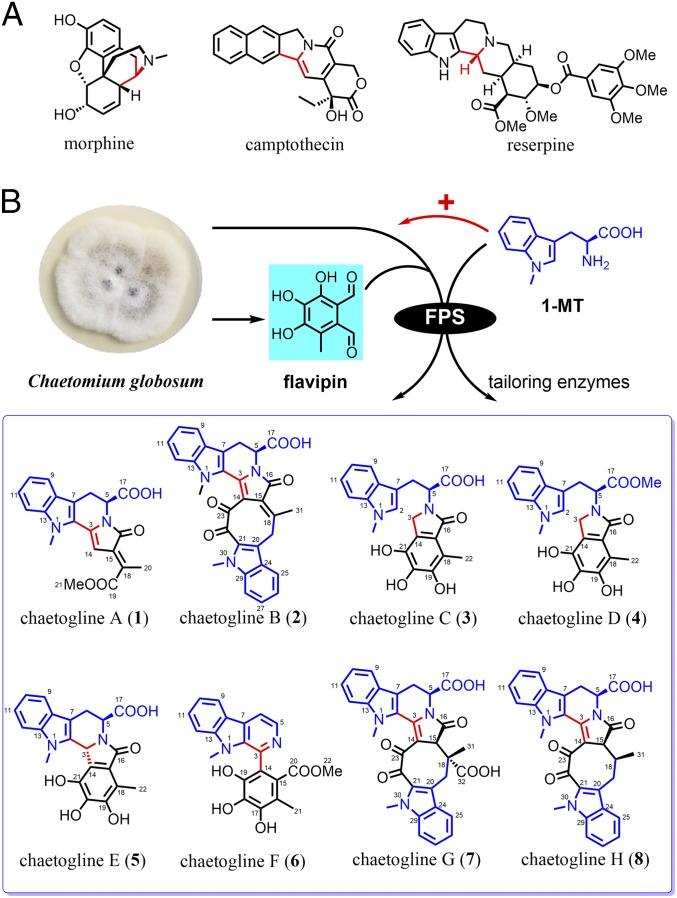
Characterized by forming a piperidine ring through a condensation of β-arylethylamine with an aldehyde, Pictet–Spengler (PS) reaction contributes greatly to the framework diversification of important alkaloidal phytochemicals such as morphine, camptothecin, and reserpine (Fig. 1A), with plant-derived Pictet–Spenglerase (called strictosidine synthetase, STR) mechanistically addressed (11). The PS mechanism has been presumed to involve in the tetrahydroisoquinoline antibiotic biosynthesis in the bacterium Streptomyces lavendulae (12, 13), and likely in the biosynthetic pathway of hyrtioreticulin F in the marine sponge Hyrtios reticulatus (14). However, surprisingly, nothing is known concerning the PS reaction in the fungal kingdom.
Fig. 1.
Alkaloids derived from the PS reaction. (A) Representatives for PS reaction-based phytochemicals. (B) 1-MT has been found to be a potent up-regulator for the FPS gene expression, and a suitable FPS substrate for its PS condensation with flavipin to yield unnatural natural products (1−8) with unprecedented skeletons.
Most if not all Chaetomium fungi in the Chaetomiaceae family produce l-tryptophan–derived alkaloids, but “refuse” to generate any PS reaction-based secondary metabolite (15–18). However, a comparative genomic analysis has clarified that C. globosum 1C51 does have an FPS gene (CHGG_06703, STR-like) (SI Appendix, Fig. S25), but remains silent or poorly activated in the laboratory cultivations because no PS-derived secondary metabolite has been detected in the fungal culture. Therefore, this C. globosum 1C51 strain was adopted here to test for the activation of its “unworking” PS reaction-based biosynthetic machinery. As a result, 1-methyl-l-tryptophan (1-MT) was demonstrated to be able to up-regulate the FPS expression and condense with the fungal aldehyde flavipin (3,4,5-trihydroxy-6-methyl phthalaldehyde) to form unexpectedly a family of skeletally unprecedented alkaloids, trivially named chaetoglines A−H (1−8) (Fig. 1B). A gene-implied enzyme inhibition (GIEI) strategy, derived from the hypothesis-based enzyme modulation described elsewhere (19, 20), was introduced to identify the key diversification steps for the PS reaction-derived compounds (Figs. 2–4). Chaetoglines B (2) and F (6) have been found to be more antibacterial than tinidazole (a coassayed drug prescribed in clinic for bacterial infections) against pathogenic anaerobes Veillonella parvula, Bacteroides vulgatus, Streptococcus sp., and Peptostreptococcus sp. Moreover, alkaloid 6 is potently inhibitory on acetylcholinesterase (AChE), an effective target enzyme exploited for the treatment of Alzheimer’s disease (21, 22).
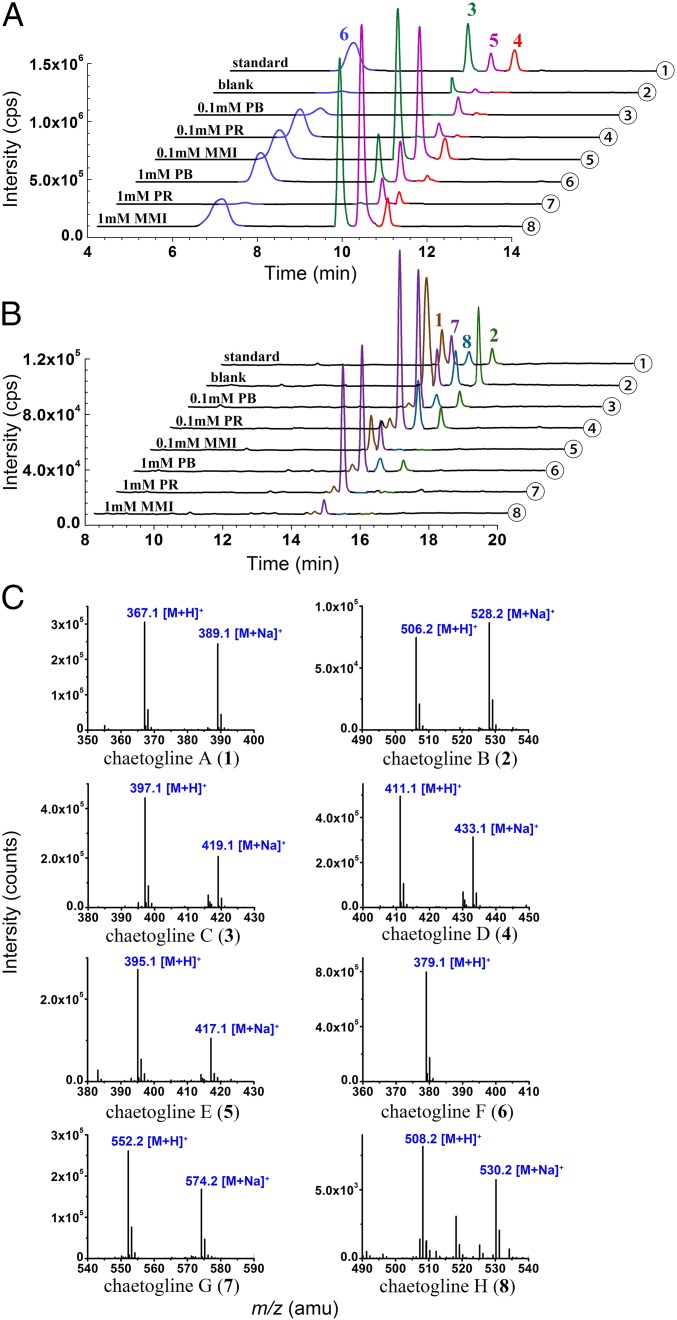
Fig. 2.
LC-MS profile-based comparisons for the chaetogline production in monooxygenase inhibitor exposed fungal cultures: (A) for 3–6; (B) for 1–2 and 7–8. The ESI-MS spectra (C) of 1−8 (①) displayed the corresponding protonated and Na+-liganded molecular ions. Samples were ethyl acetate extracts derived from 1-MT supplemented C. globosum 1C51 cultures without (②) and with the separate exposure to PB, PR, and MMI at 0.1 (③∼⑤) and 1.0 mM (⑥∼⑧), respectively.
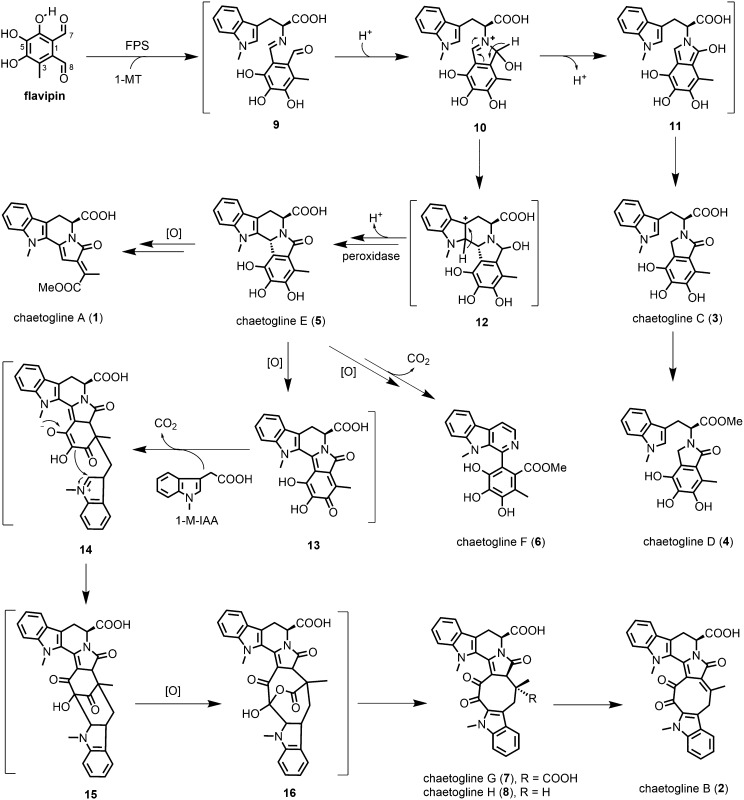
Fig. 4.
Proposed generation of the fungal PS-derived products. Catalyzed by FPS, 1-MT and flavipin (a fungal aldehyde) undergo PS reaction to form chaetoglines A−H (1−8) in concert with tailing reactions including oxidation, decarboxylation, and Aldol reaction. The Schiff base intermediate 9 tends to tautomerize via 10 and 11 to give chaetogline C (3) that can be methyl-esterified into chaetogline D (4). Intramolecular cyclization of 10 gives 12, which is oxidizable into chaetoglines A (1) and E (5), the latter yielding chaetogline F (6) after the oxidative and decarboxylative aromatization. Chaetogline E (5) can also be oxidized to intermediate 13, which gives 14 after condensing presumably via the decarboxylative Aldol reaction (34, 35) with 1-M-IAA derived from 1-MT by the fungi (SI Appendix, Fig. S24). Intermediate 14 undergoes intramolecular cyclization, monooxygenation, and isomerization to form chaetogline G (7), which after decarboxylation gives chaetogline H (8), a precursor of chaetogline B (2).
Results and Discussion
Design for the Startup of Fungal PS Reaction.
Some fungi biosynthesize aryl aldehydes through the nonreducing polyketide synthase (23, 24), which may be hypothetically condensable with β-arylethylamine derivatives under the FPS catalysis according to the logic of PS reaction (13). Accordingly, our endeavor began with the screening for the activator of the cryptic FPS gene and a suitable β-arylethylamine substrate that may undergo the PS reaction with the fungal aldehyde upon the microbial cultivation. To minimize the “chemical disturbance” to the fungal growth, we screened for the FPS activator from a small collection of tryptophan derivatives with the desired β-arylethylamine motif (SI Appendix, Table S1). Luckily, 1-MT was justified to be appropriate for the purpose by (i) its activation of the FPS gene (SI Appendix, Fig. S1), and (ii) its inhibition of tryptophan-catabolizing indoleamine 2,3-dioxygenase (25), which may facilitate the PS reaction by preventing the presumable decomposition of tryptophan derivatives.
Validation of PS Reaction in Fungal Culture.
In view of the distinct resonance of 1-MT originated protons, the ethyl acetate extract derived from the 1-MT supplemented fungal culture was fractionated by tracing the unique 1H-NMR signal due to the 1-MT motif in presumed PS reaction products (SI Appendix, Fig. S2). The first compound thus isolated was chaetogline A (1), with its molecular formula of C20H18N2O5 corresponding to the protonated molecular ion at m/z 367.1284 in its high-resolution electrospray mass spectrometry (HR-ESI-MS). The 1H-NMR and 1H-1H chemical shift correlation spectroscopy (COSY) spectra of 1 exhibited a coupling sequence ascribable to a 2-substituted 1-methyl-tryptophan motif, indicating that it was a PS reaction-condensed alkaloid (SI Appendix, Table S2). The 13C-NMR, heteronuclear singular quantum correlation (HSQC), heteronuclear multiple-bond coherence (HMBC), and rotating frame Overhauser effect spectroscopy (ROESY) spectra of 1 collectively highlighted that its skeleton was unexpectedly modified after the PS condensation (SI Appendix, Fig. S3). The postulation, along with its 5S configuration, was confirmed by a low-temperature single-crystal X-ray diffraction of 1 with monochromatic Cu Kα radiation (SI Appendix, Fig. S4).
The second PS reaction-derived alkaloid was chaetogline B (2), which was evidenced to have a molecular formula of C30H23N3O5 from the protonated molecular ion at m/z 506.1715 in its HR-ESI-MS. The 1H- and 13C-NMR spectra of 2 disclosed the presence of a 2-substituted 1-MT scaffold and a 2,3-disubstituted N-methyl indole moiety (SI Appendix, Table S3). The 1H-1H COSY, ROESY, HSQC, and HMBC spectra of 2 corroborated the presence of substructures A and B (SI Appendix, Fig. S5), being edited into the molecule according to key HMBC correlations. However, no HMBC correlations could be discerned for the “isolated” quaternary carbons resonating at δC 187.0 (C-22) and 194.5 (C-23). The isotope-labeling experiments had to be carried out to clarify the uncertainty. Feeding sodium [1-13C]-acetate enriched resonances of C-3, C-15 and C-23, demonstrating that they originated from C-1 of supplemented acetates. Administration of sodium [2-13C]-acetate enhanced the 13C signal intensity of C-14, C-16, and C-22, being thereby derived from C-2 of incorporated acetates. As anticipated, the supplementation of sodium [1,2-13C2]-acetate intensified three pairs of carbon signals C-3/C-14, C-23/C-22, and C-15/C-16. The deduction was confirmed by the long-range couplings of the carbon pairs in the INADEQUATE spectrum (SI Appendix, Fig. S6). The observation, along with the unsaturation index of 2, could only be explained by assuming a macrocycle formed by inserting the 1,2-diketone motif between C-14 and C-21. The absolute configuration of 2 was disclosed by theoretical simulation (SI Appendix, Fig. S7), and supported by its CD spectral resemblance to 1, indicating that both of the alkaloids possess the same chirality-dependent chromophore despite the distinct structural difference (SI Appendix, Fig. S8).
Searching for the Fungal-Derived FPS Substrate.
The unexpected substitution pattern of 1 and 2 suggested that the PS condensation might be accompanied or followed by other tailing reactions such as oxidation. Indeed, this blocked our identification of the fungal-derived FPS substrate. To bypass the obstacle, we have introduced the GIEI approach that was expected to facilitate the identification of precursors or intermediates suggestive of the fungal-derived substrate. The C. globosum genome was scrutinized to carry as many as 90 cytochrome P450 (CYP450) and 2 flavin-containing monooxygenases genes (SI Appendix, Table S6), which might have expressed enzymes capable of diversifying the PS reaction products (23, 26). The inhibition of monooxygenases was hypothesized to stabilize the intact or less-oxidized PS reaction product(s). Accordingly, the 1-MT supplemented fungal cultures were exposed separately to three monooxygenase inhibitors: methimazole (MMI), phenylbutazone (PB), and proadifen (PR). To our assumption, the abundance of 1 and 2 was substantially attenuated, but those of others were increased in the inhibitor added cultures (Fig. 2). Subsequently, the less-oxidized products named chaetoglines C−H (3−8) were isolated from the extract derived from the scale-up 1-MT supplemented fermentations with exposure to MMI (for 3−6) and PR (for 7 and 8), respectively.
Chaetoglines C (3) and D (4) were shown to have molecular formulae of C21H20N2O6 and C22H22N2O6 by their HR-ESI-MS spectra, respectively. The observation, along with their 1H- and 13C-NMR spectra, suggested that the latter was a methyl ester of the former. Scrutiny of the 1H-1H COSY, ROESY, HSQC, and HMBC spectra of 3 indicated the presence of a 4,5,6-trihydroxy-7-methylisoindolin-1-one motif (27, 28), which was shown to share the nitrogen atom with the 1-MT moiety (SI Appendix, Fig. S9). The low-temperature single-crystal X-ray crystallographic analysis of 4 confirmed the above elucidation and clarified the absolute stereochemistry in concert with the CD spectra of the two alkaloids (SI Appendix, Figs. S10 and S11).
Chaetogline E (5) was evidenced to have a molecular formula of C23H17N3O3 by its HR-ESI-MS. The spectral data of 5 were comparable to those of 3. However, the singlet at δH 7.09 and N-methylene doublets (J = 13.4 Hz) at δH 4.27 and 4.19 in the 1H-NMR spectrum of 3 were collectively replaced by a one-proton singlet of H-3 at δH 6.22 in that of 5. The observation could be explained by forming the piperidine ring, which was confirmed by the HMBC correlations (SI Appendix, Fig. S12). In the ROESY spectrum of 5, no correlation could be found between H-3 and H-5, suggesting that the two protons likely positioned at the opposite side of the piperidine cycle. Provided that the 5S configuration “inherited” from 1-MT, the absolute configuration of 5 was most likely deduced to be 3R,5S, which was reinforced by theoretical simulation using the time-dependent density functional theory (TD-DFT) (SI Appendix, Fig. S13). Chaetogline F (6) was determined to have a molecular formula of C21H18N2O5 by its HR-ESI-MS. Its 1H-NMR spectra acquired in CDCl3, acetone-d6, DMSO-d6, and methanol-d4 had poor resolution owing to its basicity. Thus, it was transformed into its TFA salt crystal from methanol, and subsequent single-crystal X-ray diffraction clarified its structure (SI Appendix, Fig. S14).
Chaetoglines G (7) and H (8) were analyzed to have molecular formulae of C31H25N3O7 and C30H25N3O5 by their HR-ESI-MS spectra, respectively, suggesting that the latter could be the decarboxylative product of the former. The assumption was reinforced by the 1H and 13C NMR spectra of 7 and 8, which were comparable to those of 2 (SI Appendix, Table S3). The structure determination of 7 and 8 was accomplished by the 1H-1H COSY, HSQC, HMBC, and ROESY spectra (SI Appendix, Figs. S15 and S16). With relative configurations in hand, the absolute stereochemistry of 7 and 8 was established subsequently by comparing their CD spectra with the electronic circular dichroism curves calculated by TD-DFT for all optional stereoisomers (SI Appendix, Figs. S17 and S18).
The characterization of 3–8 suggested that the aldehyde substrate of the PS reaction could be flavipin, which has been reported from some fungal cultures (28–33). To confirm the postulation, C. globosum 1C51 was fed with sodium [1-13C]-, [2-13C]-, and [1,2-13C2]-acetates, respectively, to lead to the expected 13C enrichment of the 4,5,6-trihydroxyisoindolin-1-one motif of chaetoglines E (5) and F (6), but not the aromatic methyl carbons (C-22 in 5 and C-21 in 6), whose intensity could only be enhanced by supplementing the fungal culture with l-[methyl-13C]-methionine (SI Appendix, Tables S9 and S10). The structural feature of 1–8 suggested that the two aldehyde groups of flavipin are not equally reactive to 1-MT for the fungal PS reaction. This agreed with the flavipin 1H-NMR spectrum (SI Appendix, Fig. S19) and the difference in the positive charge distributions on the two aldehyde carbons [SI Appendix, Table S12, calculated by quantum-mechanical (QM) method at Becke’s three-parameter hybrid functional applying Lee–Yang–Parr correlation (B3LYP) with the basis set of 6–31G(d,p) level], highlighting collectively that the electrophilic effect of 2-fomyl carbon was increased by its hydrogen bonding with the 6-hydroxyl group.
Diversification of the PS Reaction Products.
The structure pattern of 1–8 suggested that the PS reaction products might have been structurally diversified in the fungal culture. The proposal was confirmed by a combination of the GIEI approach and QM calculation. As anticipated theoretically (SI Appendix, Fig. S20 and Tables S13–S16), the PS reaction intermediate 9 tautomerizes via 10 and 11 with a lower reaction barrier around 5.17 kcal/mol to give an energetically much more stable product of chaetogline C (3) (−31.97 kcal/mol), whose methyl esterification yields 4. Driven by the C-2 nucleophilicity of indole motif, 10 undergoes an intramolecular cyclization to form 12, which gives 1 and 5 after successive oxidations. Chaetogline E (5) seems subject to two types of oxidations with its piperidine ring aromatized to afford chaetogline F (6). These were ascertained to agree with a set of enzymatic transformation tests. Coexposure of flavipin and 1-MT to the fungal enzyme formed 3 and 4, with the latter derived from the former (Fig. 3 A and C). The conversion from 5 into 1 and 6 was also realized by the enzymatic catalysis experimentation (Fig. 3B). Meanwhile, chaetogline E (5) could be further oxidized to form intermediate 13, which might give 14 by condensing with 1-methylindole-3-acetic acid (1-M-IAA, a fungal metabolite from 1-MT; SI Appendix, Fig. S24) most likely through the decarboxylative Michael addition noticed earlier (34, 35). This process would be followed by an intramolecular cyclization of 14 to form 15. Monooxygenation of 15 gave 16 that might be isomerized to 7. Decarboxylation of 7 yielded 8 as a precursor of 2 (Fig. 4). The 13C-labeled pattern of 2 supported the aforementioned monooxygenation, isomerization, and decarboxylation (SI Appendix, Table S8). To test this hypothesis, another set of enzyme inhibition experiments was carried out. Compounds 2, 7, and 8 could not be detected in the fungal culture supplemented with methimazole, a flavin-containing monooxygenase inhibitor (36) (Fig. 2). The observation highlighted that the fungal monooxygenation may involve in the macrocycle construction of the three alkaloids. Furthermore, the production of 1, 4, and 6 was catalyzed by carboxylesterase because their abundances in the fungal culture substantially decreased upon the exposure to 3-(trifluoromethyl)-phenylacetone, a carboxylesterase inhibitor (37) (SI Appendix, Fig. S21). By analogy, the biosynthesis of 1, 2, and 5–8 was abolished by supplementing NaN3, a peroxidase inhibitor (38), suggesting that the transformation from 12 to 5 was catalyzed by a fungal peroxidase (SI Appendix, Fig. S22). It is noteworthy that the fungal genome contains too many (92) monooxygenase genes to identify specific enzymes responsible for the oxidations leading to the structural feature of chaetoglines, and that the genes around FPS may not involve in the chaetogline formation according to our bioinformatic analysis (SI Appendix, Table S18).
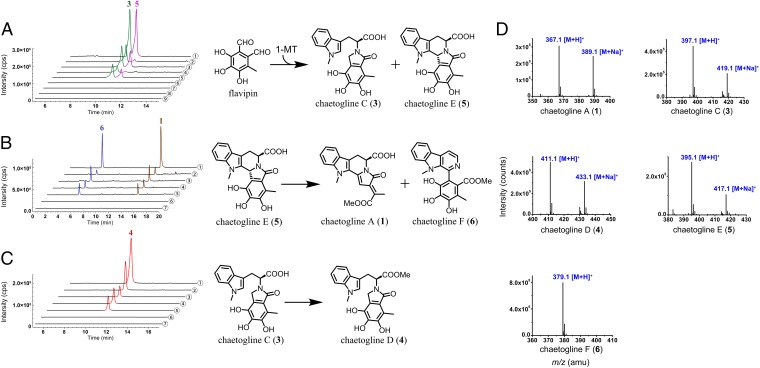
Fig. 3.
Enzymatic transformation in vitro. (A) Compounds 3 and 5 generated upon the fungal protein coexposure to 1-MT and flavipin. (B) Enzymatic treatment of 5 gave 1 and 6; (C) Fungal protein exposure to 3 formed 4. Controlled by LC-MS profiles of authentic chaetoglines (①), the enzymatic catalysis tests were performed by using intracellular (IP) and extracellular protein (EP) from C. globosum 1C51 precultured with (IP, ④; EP, ⑤) or without 1-MT (IP, ②; EP, ③). Pure IP (⑥) and EP (⑦) were equally processed but gave no products to exclude the possibility that the detected products could be endogenous. The individual exposure of 1-MT or flavipin to IP (⑧) and EP (⑨) produced none of chaetoglines 1−8. (D) The ESI-MS spectra of authentic chaetoglines.
FPS Substrate Promiscuity.
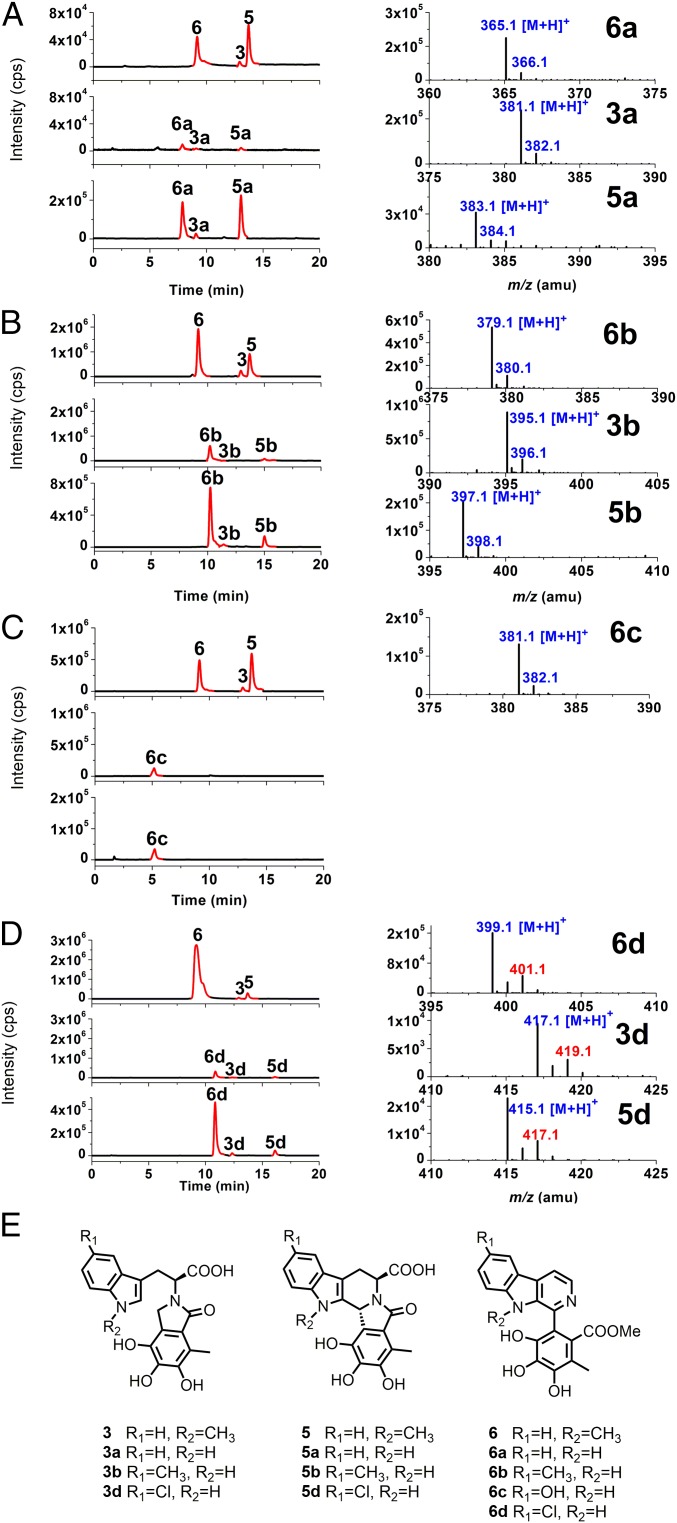
To understand the FPS substrate promiscuity, the 1-MT supplemented fungal culture was allowed to expose separately to tryptophan, 5-methyl-tryptophan (5-MT), 5-hydroxyl-l-tryptophan (l-5-HTP), and 5-chloro-tryptophan (5-Cl-Trp). The extracts derived from these cultivations were analyzed by liquid chromatography-mass spectrometry (LC-MS). With the FPS gene activated by 1-MT, the main 1-MT–derived PS products chaetoglines C (3), E (5), and F (6) were detected and accompanied by chaetogline-like derivatives 3a, 3b and 3d; 5a, 5b and 5d; and 6a–6d (Fig. 5, structures by analogy with those of 3, 5, and 6 as well as 3d, 5d, and 6d with the chlorine-derived isotopic peaks in their MS spectra). Surprisingly, most of those chaetogline-like derivatives remained detectable in the 1-MT free fungal cultures with separate supplementation of tryptophan, 5-MT, l-5-HTP, and 5-Cl-Trp [Fig. 5 A–D (③)]. This agreed with the slight FPS activating activity of the tryptophan derivatives (SI Appendix, Fig. S1). Thus, the fungal FPS is presumably substrate-promiscuous, and 1-MT seems to be the best FPS activator and FPS substrate among the tested tryptophan derivatives (SI Appendix, Table S1).
Fig. 5.
FPS substrate promiscuity of C. globosum 1C51. LC-MS analyses were performed to compare the presence of (E) chaetoglines 3, 5, and 6, and their derivatives (3a−3d, 5a−5d, and 6a−6d) in the 1-MT (1 mM) exposed (① and ②) and free (③) cultures supplemented separately with (A) tryptophan, (B) 5-MT, (C) l-5-HTP, and (D) 5-Cl-Trp at 1.0 (① and ②) and 2.0 mM (③), respectively. (Right) MS spectra of tryptophan analogs derived chaetogline (3, 5, and 6)-like compounds with 3c and 5c (l-5-HTP derived) undetectable.
Bioactivity Assays.
In vitro antibacterial assay showed that 2 is inhibitory against V. parvula, B. vulgatus, and Peptostreptococcus sp. [minimum inhibitory concentration (MIC): 0.24 μM], with 6 against V. parvula, Streptococcus sp., and Peptostreptococcus sp. (MICs: 0.32, 0.32, 0.66 μM, respectively). Such activities are well comparable to that of tinidazole, a prescribed antibacterial drug (SI Appendix, Table S19). Moreover, the AChE inhibitory activity of 6 was detected as well (IC50: 4.13 μM).
Conclusion
To cope with the changing or growing selection pressure in nature, organisms have to evolve more efficiently to acquire sufficiently advanced biosynthetic machinery for the construction of small-molecule compounds with unpredictable architectures and functions. This is why natural products remain a common stimulating force both for synthetic and material chemistry (8–10), and for pharmaceutical and agrochemical industries (6, 7, 39). Concerning the microbial generation of natural products, the long-time evolution has enabled microbes to organize the biosynthetic machineries in an ingenious way so that they can produce the organic molecular diversity as case-dependent responses to various outside events such as symbiosis (40), gene transfer (41), and host invasion (42). To get more structurally unprecedented small molecules, it is urgently necessary to activate the silent or less-active biosynthetic pathways in the producing organisms. By chemical modulation of cryptic genes, the work presents the startup of the PS reaction-based fungal biosynthetic machinery that usually remains silent in ordinary laboratory cultivations. In particular, the external chemical (1-MT) functions both as an efficient activator for the silent FPS gene, and as a suitable substrate processable by the activated fungal assembly line to produce unnatural natural products with unpredictable skeletons. Understanding generations of these unforeseeable molecules also requires the biochemical approaches to address the key transformational steps. Such a necessity has led to the present introduction of the GIEI protocol, which works generally by taking advantage of microbial genomic information. In this sense, the investigation strengthens the access to novel biocatalysts organized cryptically in the microbial biosynthetic machineries. As a result of complicated microbial interactions with external chemicals such as 1-methyltryptophan used here, more potentially bioactive unnatural natural products can be generated as exemplified by the characterization of chaetoglines A−H (1–8), of which 2 and 6 are potently antibacterial to pathogens as health risks (43), with the latter being additionally inhibitory on AChE as a proven target for treating Alzheimer’s disease (21, 22).
In summary, the present startup of the PS reaction-based fungal biosynthetic machinery provides a conceptually innovative and generally profitable access to unforeseeably structured unnatural natural products. The new chemical space of the “highly chimeric” compounds lies in both the unexpected combination of natural and unnatural building blocks, and from the unpredictable diversification of the hybridized intermediates in microbial cultures. With the microbial genomic information continuing to increase, the GIEI strategy introduced here can substantially rationalize the exploration of the biochemical process for the unnatural compound generation. Finally, some unpredictably chimerized molecules thus produced may play uniquely driving or inspiring roles in the synthetic and materials chemistry, and in the development of new pharmaceuticals and agrochemicals.
Methods
Fungus.
C. globosum 1C51 was isolated from guts of the marine fish Epinephelus drummondhayi collected in the Yellow Sea, China, and preserved on potato dextrose agar slants containing 10% NaCl and stored at 4 °C, before experimentation.
The Real-time Quantitative PCR Analysis for FPS Gene Expression.
After a 5-d culture with each tryptophan derivative (SI Appendix, Table S1), the total RNA of the fungus was isolated using the RNAiso Plus (Takara). cDNA was generated from the RNAs of assayed strain using an PrimeScript II first Strand cDNA Synthesis Kit (Takara). The qPCR analysis was performed with One Step SYBR PrimeScript RT-PCR Kit (Takara) using the C1000 Thermal Cycler (Bio-Rad) and quantified by CFX96 Real-Time System (Bio-Rad). Primers used for qPCR are listed in SI Appendix, Table S11. The gene expression was assessed in terms of the variation in RNA quantity and quality with the endogenous actin gene as a reference.
Feeding Experiments for 1-MT.
The fermentation was accomplished on a rotary shaker at 160 rpm and 28 °C in 1-L Erlenmeyer flasks containing 400 mL Czapek’s medium (30 g sucrose, 1 g yeast extract, 3 g NaNO3, 0.5 g MgSO4⋅7H2O, 10 mg FeSO4⋅7H2O, 1 g K2HPO4, and 0.5 g KCl per 1 L water), after inoculation at 5% with the starter culture precultivated for 3 d. About 48, 60, 72, and 84 h later, 1-MT in an aqueous stock solution was added to the fungal culture (with the 1-MT concentration adjusted to 0.5, 1.0, 1.5, and 2.0 mM, respectively), followed by another 10-d cultivation.
Detailed experimental procedures and spectral interpretations are included in SI Appendix.
Supplementary Material
Acknowledgments
The work was cofinanced by the Ministry of Science and Technology 2013AA092901 and 2013AA092903 and National Natural Science Foundation of China Grants 81421091, 21132004, 21072092, 81330079, 91313303, 81172948, and UM.C/625/1/HIR/247.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallographic data, atomic coordinates, and structure factors in CIF format have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre (CCDC), Cambridge CB2 1EZ, United Kingdom, www.ccdc.cam.ac.uk/deposit (accession codes: CCDC 1018681, CCDC 1018683, and CCDC 1018682).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417304111/-/DCSupplemental.
References
- 1.Kingston DGI. Modern natural products drug discovery and its relevance to biodiversity conservation. J Nat Prod. 2011;74(3):496–511. doi: 10.1021/np100550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lachance H, Wetzel S, Kumar K, Waldmann H. Charting, navigating, and populating natural product chemical space for drug discovery. J Med Chem. 2012;55(13):5989–6001. doi: 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- 3.Cragg GM, Grothaus PG, Newman DJ. New horizons for old drugs and drug leads. J Nat Prod. 2014;77(3):703–723. doi: 10.1021/np5000796. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11(1):21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 5.Sheridan C. Recasting natural product research. Nat Biotechnol. 2012;30(5):385–387. doi: 10.1038/nbt.2208. [DOI] [PubMed] [Google Scholar]
- 6.Appendino G, Minassi A, Taglialatela-Scafati O. Recreational drug discovery: Natural products as lead structures for the synthesis of smart drugs. Nat Prod Rep. 2014;31(7):880–904. doi: 10.1039/c4np00010b. [DOI] [PubMed] [Google Scholar]
- 7.Molinski TF. All natural: The renaissance of natural products chemistry. Org Lett. 2014;16(15):3849–3855. doi: 10.1021/ol501917g. [DOI] [PubMed] [Google Scholar]
- 8.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater. 2014;13(5):515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunz H, Müllen K. Natural product and material chemistries—separated forever? J Am Chem Soc. 2013;135(24):8764–8769. doi: 10.1021/ja309186q. [DOI] [PubMed] [Google Scholar]
- 10.Moulay S. Dopa/Catechol-tethered polymers: Bioadhesives and biomimetic adhesive materials. Pol Rev. 2014;54(3):436–513. [Google Scholar]
- 11.Stöckigt J, Antonchick AP, Wu F, Waldmann H. The Pictet-Spengler reaction in nature and in organic chemistry. Angew Chem Int Ed Engl. 2011;50(37):8538–8564. doi: 10.1002/anie.201008071. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, et al. Discovery of McbB, an enzyme catalyzing the β-carboline skeleton construction in the marinacarboline biosynthetic pathway. Angew Chem Int Ed Engl. 2013;52(38):9980–9984. doi: 10.1002/anie.201303449. [DOI] [PubMed] [Google Scholar]
- 13.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat Chem Biol. 2010;6(6):408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 14.Imada K, et al. Reticulatins A and B and hyrtioreticulin F from the marine sponge Hyrtios reticulatus. Tetrahedron. 2013;69(34):7051–7055. [Google Scholar]
- 15.Jiang MY, Feng T, Liu JK. N-containing compounds of macromycetes. Nat Prod Rep. 2011;28(4):783–808. doi: 10.1039/c0np00006j. [DOI] [PubMed] [Google Scholar]
- 16.Takayama H, et al. New procedure to mask the 2,3-π bond of the indole nucleus and its application to the preparation of potent opioid receptor agonists with a Corynanthe skeleton. Org Lett. 2006;8(25):5705–5708. doi: 10.1021/ol062173k. [DOI] [PubMed] [Google Scholar]
- 17.Ge HM, et al. Precursor-directed fungal generation of novel halogenated chaetoglobosins with more preferable immunosuppressive action. Chem Commun (Camb) 2011;47(8):2321–2323. doi: 10.1039/c0cc04183a. [DOI] [PubMed] [Google Scholar]
- 18.Ding G, et al. Chaetoglobosin U, a cytochalasan alkaloid from endophytic Chaetomium globosum IFB-E019. J Nat Prod. 2006;69(2):302–304. doi: 10.1021/np050515+. [DOI] [PubMed] [Google Scholar]
- 19.Oikawa H, Murakami Y, Ichihara A. Useful approach to find the plausible biosynthetic precursors of secondary metabolites using P-450 inhibitors: Postulated intermediates of chaetoglobosin A. J Chem Soc, Perk Trans. 1992;1(21):2949–2953. [Google Scholar]
- 20.Bradley EL, et al. The biosynthesis of the Streptomyces antibiotic bicyclomycin. Tetrahedron Lett. 1996;37(38):6935–6938. [Google Scholar]
- 21.Ge HM, et al. Hopeahainol A: An acetylcholinesterase inhibitor from Hopea hainanensis. Chem Eur J. 2008;14(1):376–381. doi: 10.1002/chem.200700960. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, et al. Hopeahainol A attenuates memory deficits by targeting β-amyloid in APP/PS1 transgenic mice. Aging Cell. 2013;12(1):85–92. doi: 10.1111/acel.12022. [DOI] [PubMed] [Google Scholar]
- 23.Davison J, et al. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc Natl Acad Sci USA. 2012;109(20):7642–7647. doi: 10.1073/pnas.1201469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Beissner M, Zhao H. Aryl-aldehyde formation in fungal polyketides: Discovery and characterization of a distinct biosynthetic mechanism. Chem Biol. 2014;21(2):257–263. doi: 10.1016/j.chembiol.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuasa HJ, Ball HJ. Molecular evolution and characterization of fungal indoleamine 2,3-dioxygenases. J Mol Evol. 2011;72(2):160–168. doi: 10.1007/s00239-010-9412-5. [DOI] [PubMed] [Google Scholar]
- 26.Gibson M, Nur-e-alam M, Lipata F, Oliveira MA, Rohr J. Characterization of kinetics and products of the Baeyer-Villiger oxygenase MtmOIV, the key enzyme of the biosynthetic pathway toward the natural product anticancer drug mithramycin from Streptomyces argillaceus. J Am Chem Soc. 2005;127(50):17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 27.Talontsi FM, Dittrich B, Schüffler A, Sun H, Laatsch H. Epicoccolides: Antimicrobial and antifungal polyketides from an endophytic fungus Epicoccum sp. associated with Theobroma cacao. Eur J Org Chem. 2013;(15):3174–3180. [Google Scholar]
- 28.El Amrani M, et al. Protein kinase and HDAC inhibitors from the endophytic fungus Epicoccum nigrum. J Nat Prod. 2014;77(1):49–56. doi: 10.1021/np4005745. [DOI] [PubMed] [Google Scholar]
- 29.Yuan D, et al. Alkaloids from the leaves of Uncaria rhynchophylla and their inhibitory activity on NO production in lipopolysaccharide-activated microglia. J Nat Prod. 2008;71(7):1271–1274. doi: 10.1021/np8000305. [DOI] [PubMed] [Google Scholar]
- 30.Ye YH, et al. Flavipin in Chaetomium globosum CDW7, an endophytic fungus from Ginkgo biloba, contributes to antioxidant activity. Appl Microbiol Biotechnol. 2013;97(16):7131–7139. doi: 10.1007/s00253-013-5013-8. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Y, et al. Antifungal screening of endophytic fungi from Ginkgo biloba for discovery of potent anti-phytopathogenic fungicides. FEMS Microbiol Lett. 2013;339(2):130–136. doi: 10.1111/1574-6968.12065. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson G. The biosynthesis of flavipin. I. Incorporation of acetate and methionine. Acta Chem Scand. 1965;19:35–40. doi: 10.3891/acta.chem.scand.19-0035. [DOI] [PubMed] [Google Scholar]
- 33.Pettersson G. The biosynthesis of flavipin. II. Incorporation of aromatic precursors. Acta Chem Scand. 1965;19(7):1724–1732. doi: 10.3891/acta.chem.scand.19-1724. [DOI] [PubMed] [Google Scholar]
- 34.Bae HY, Sim JH, Lee JW, List B, Song CE. Organocatalytic enantioselective decarboxylative aldol reaction of malonic acid half thioesters with aldehydes. Angew Chem Int Ed Engl. 2013;52(46):12143–12147. doi: 10.1002/anie.201306297. [DOI] [PubMed] [Google Scholar]
- 35.Kang YK, Lee HJ, Moon HW, Kim DY. Organocatalytic enantioselective decarboxylative Michael addition of β-ketoacids to α,β-unsaturated ketones. RSC Adv. 2013;3(5):1332–1335. [Google Scholar]
- 36.Lai WG, Farah N, Moniz GA, Wong YN. A Baeyer-Villiger oxidation specifically catalyzed by human flavin-containing monooxygenase 5. Drug Metab Dispos. 2011;39(1):61–70. doi: 10.1124/dmd.110.035360. [DOI] [PubMed] [Google Scholar]
- 37.Costa SP, et al. Automated evaluation of pharmaceutically active ionic liquids’ (eco)toxicity through the inhibition of human carboxylesterase and Vibrio fischeri. J Hazard Mater. 2014;265(0):133–141. doi: 10.1016/j.jhazmat.2013.11.052. [DOI] [PubMed] [Google Scholar]
- 38.Kumar R, Singh KA, Singh VK, Jagannadham MV. Biochemical characterization of a peroxidase isolated from Caribbean plant: Euphorbia cotinifolia. Process Biochem. 2011;46(6):1350–1357. [Google Scholar]
- 39.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA. 2014;111(25):9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwan JC, et al. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci USA. 2012;109(50):20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziemert N, et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci USA. 2014;111(12):E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiemann P, et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9(6):e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer Sauteur PM, van Rossum AM, Vink C. Mycoplasma pneumoniae in children: Carriage, pathogenesis, and antibiotic resistance. Curr Opin Infect Dis. 2014;27(3):220–227. doi: 10.1097/QCO.0000000000000063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.