Reannotation of the maize genome using MAKER-P results in many revised and new gene models.
Abstract
The large size and relative complexity of many plant genomes make creation, quality control, and dissemination of high-quality gene structure annotations challenging. In response, we have developed MAKER-P, a fast and easy-to-use genome annotation engine for plants. Here, we report the use of MAKER-P to update and revise the maize (Zea mays) B73 RefGen_v3 annotation build (5b+) in less than 3 h using the iPlant Cyberinfrastructure. MAKER-P identified and annotated 4,466 additional, well-supported protein-coding genes not present in the 5b+ annotation build, added additional untranslated regions to 1,393 5b+ gene models, identified 2,647 5b+ gene models that lack any supporting evidence (despite the use of large and diverse evidence data sets), identified 104,215 pseudogene fragments, and created an additional 2,522 noncoding gene annotations. We also describe a method for de novo training of MAKER-P for the annotation of newly sequenced grass genomes. Collectively, these results lead to the 6a maize genome annotation and demonstrate the utility of MAKER-P for rapid annotation, management, and quality control of grasses and other difficult-to-annotate plant genomes.
Plant genomes, especially grass genomes, are difficult substrates for genome annotation due to regional and whole-genome duplication events and often contain large numbers of pseudogenes. These factors impact every aspect of gene structure annotation, from revision of existing annotations in light of new data to annotation of newly sequenced plant genomes. These aspects of plant genomes also dramatically lengthen compute times, because the many repeated genes and other sequences result in commensurately more sequence alignments and gene predictions. In many ways, annotation of the maize genome epitomizes these problems.
In 2005, the National Science Foundation, U.S. Department of Agriculture, and Department of Energy announced that the approximately 2.3-Gb genome of the maize (Zea mays) inbred line B73, a major contributor to much of the germplasm used for U.S. grain production, would be sequenced using a bacterial artificial chromosome (BAC)-by-BAC approach. The plan was to sequence BACs from a minimal tiling path to approximately 6× coverage and to further improve only the unique genic regions. These sequences would be labeled Phase 1 HTGS_IMPROVED at GenBank, and the GenBank record for each BAC was to include information on the improved regions as well as order and orientation, where available, as comments. The Maize Genome Sequencing Consortium planned to release all data via MaizeSequence.org, a project database, with a plan to transition all data into MaizeGDB (Sen et al., 2009) and Gramene (Monaco et al., 2014), a comparative resource for plant genomics (Youens-Clark et al., 2011), at project close.
Not only did the Maize Genome Sequencing Consortium produce these sequences, they created reference assemblies for each chromosome (the first assembly was named B73 RefGen_v1) as well as structural and functional annotations to genes (Liang et al., 2009; Schnable et al., 2009). The published B73 reference genome (RefGen_v1) available from GenBank consisted of 2,048 Mb in 125,325 sequence contigs (N50 of 40 kb), forming 61,161 scaffolds (N50 of 76 kb) anchored to a high-resolution genetic map (Wei et al., 2009). After predicting transposable elements (TEs), a combination of evidence-based, ab initio approaches and stringent TE filtering resulted in a set of 32,540 high-confidence, predicted protein-encoding genes (the Filtered Gene Set). Due to incomplete sampling of the genome, the B73 reference genome is estimated to be missing approximately 5% to 10% of genes that are physically present in the B73 genome.
Following the release of the first draft, B73 RefGen_v2 improved v1 by the addition of fosmid reads as well as by integrating genetic and optical map information. For B73 RefGen_v2, approximately 80% of the maize genome is ordered and oriented, and optical map and genetic map comparisons suggest that only 2% to 2.5% of the sequences are likely to be misplaced in the assembly (Fusheng Wei, Jeff Glaubitz, and Mike McMullen, personal communication). The set of gene predictions for RefGen_v2 included 110,028 transcript models in the Working Gene Set (5a) with a subset of 39,656 high-confidence structures identified as the Filtered Gene Set (5b). (Note that here we use the naming conventions imposed by the MaizeSequence.org data generators, although alternative naming conventions have been used in some cases for these data sets; e.g. at Phytozome [http://www.phytozome.net/maize.php], the Working Gene Set is called the unfiltered working set.)
In the last year of the project, Roche/454 whole-genome shotgun (WGS) reads were made available to improve the coverage of the gene space not included in the BAC minimal tiling path (and thereby identifying some of the estimated 5%–10% of genes that were missed). Improvements for B73 RefGen_v3 included refinements to contig placement supported by recent improvements to the IBM genetic map and inclusion of 1,844 gene space contigs. These 1,844 contigs were produced from a WGS sequencing library to fill in missing gene space both within and between original BAC sequences. In addition, approximately 65,000 full-length complementary DNAs (cDNAs) were aligned to the RefGen_v2 assembly and the new WGS contigs. The new 5b+ annotation build included 251 new gene models and 213 improved models. The number of protein-coding genes (including all nuclear chromosomes, mitochondrial DNA, chloroplast DNA, and unknown chromosome) actually decreased to 39,475 models due to merging and additional quality control. The annotation consists of 137,208 gene transcripts and 316 short noncoding genes. The maize B73 assemblies and various annotations are represented at Gramene, MaizeGDB, EnsemblPlants, and GenBank.
MaizeGDB, the Maize Genetics and Genomics Database (http://www.maizegdb.org), is the U.S. Department of Agriculture Agricultural Research Service’s long-term model organism database and the maize research community’s data portal. MaizeGDB makes accessible genetic and genomic data and data analysis tools that are used by researchers to investigate basic biological concepts and translate findings into technology that is deployed in farmers’ fields. During the period from 2013 through 2018, the MaizeGDB team is tasked to make accessible high-quality, actively curated, and reliable genetic, genomic, and phenotypic data sets. At the root of a high-quality genome lies a well-supported assembly and annotation. For this reason, the deployment of an automated high-quality genome annotation system is of the utmost importance. As we demonstrate here, MAKER-P will fulfill this need.
Updating a genome’s annotations over time is a complex task, and the rapidly changing data landscape can render annotations obsolete almost as they are created. Continuity is another major issue. Many genome projects have annotations that embody years of manual curation and revision. Simply throwing old annotations away and substituting new ones created by another pipeline is hardly desirable. To be truly effective, any revision process must build upon the foundation of existing annotations and provide incremental means to move forward in light of new data.
Next-generation sequencing data, especially RNA sequencing (RNA-seq) data, also hold great potential for the annotation of newly sequenced plant genomes. But again, making use of them is no easy task. For example, using transcriptome data to train gene finders for use on a newly assembled genome can be a difficult, frustrating task, so much so that many genome projects attempt to leverage gene finders trained for other genomes. As we have demonstrated previously (Holt and Yandell, 2011), both approaches are challenging and fraught with difficulties, and gene model accuracy suffers when gene finders are trained with unmatched species parameters.
Moreover, gene space is not limited to protein-coding genes; increasingly, noncoding RNA (ncRNA) annotations are coming to be considered an essential component of every genome’s annotations. Pseudogenes are also an issue, especially for plant genomes, due to frequent whole-genome duplication and subsequent degeneration of paralogs (Zou et al., 2009). Consider the rice (Oryza sativa) genome, for example, which has approximately 39,000 annotated protein-coding genes and 28,330 pseudogenes (Zou et al., 2009); clearly, means to annotate pseudogenes are needed.
MAKER-P (Campbell et al., 2014) is an easy to use genome annotation pipeline with great software portability, based upon the widely used MAKER genome annotation pipeline (Holt and Yandell, 2011). Designed to address the needs of the plant genomes community, MAKER-P provides means for the annotation of newly sequenced plant genomes and for automated revision, quality control, and management of existing genome annotations. MAKER-P also extends MAKER to include means for pseudogene annotation and noncoding gene finding. MAKER-P provides the plant genomics community early access to new functionalities prior to their later, general release in the MAKER package. Moreover, MAKER-P is dramatically faster than other genome annotation pipelines, allowing it to scale to even the largest plant genomes. MAKER-P is designed to run on Unix-like operating systems, including Linux and Apple OS X. It can run on laptop and desktop machines, but it also has extensions to take advantage of capabilities offered by high-performance computer clusters. Recent work, for example, has shown that the version of MAKER-P available within the iPlant Cyberinfrastructure can reannotate the entire maize genome in less than 3 h (Campbell et al., 2014) and that it can carry out the complete de novo annotation of the 17.83-Gb draft loblolly pine (Pinus taeda) genome in less than 24 h (Neale et al., 2014; Wegrzyn et al., 2014).
Our previous work using the Arabidopsis (Arabidopsis thaliana) genome demonstrated MAKER-P’s effectiveness for the management and quality control of existing annotations and for de novo annotation using this relatively simple plant genome (Campbell et al., 2014). Here, we apply MAKER-P to the much less tractable maize genome, using it for analysis and quality control of the 5b+ annotation build, to systematically compare the 5b and 5b+ annotation builds with one another, for revision of the 5b+ annotations in light of 96 different RNA-seq data sets, and for de novo annotation of the maize genome. Also presented is maize genome annotation build 6a, which is demonstrably superior to the existing 5b+ build, thereby demonstrating MAKER-P’s utility for management and quality control of the maize genome annotations.
RESULTS AND DISCUSSION
Overview of the 5b and 5b+ Builds
Our overarching goal in these analyses was to systematically compare the 5b and 5b+ annotation builds with one another using MAKER-P’s management functions, to update and reevaluate the 5b+ annotation build in light of additional RNA-seq evidence, and to determine if MAKER-P was capable of automatically producing an annotation build of comparable quality. Table I summarizes the 5b and 5b+ RefGen builds. The Arabidopsis Information Resource (TAIR) 10 annotations are also included for purposes of comparison. As can be seen, the 5b and 5b+ builds are very similar to one another, differing primarily by 251 new and 213 improved genes in 5b+ (160 new models in chromosomes 1–10). In addition, a higher percentage of 5b+ models have annotated start and stop codons. In what follows, we present a detailed analysis of the relationship of the 5b+ annotation build to its supporting evidence, subjecting it to a series of quality-control analyses. We will also describe three additional annotation builds: a MAKER-P updated version of 5b+; a MAKER-P de novo annotation build; and a new 6a annotation build. The 6a build is a consensus build composed of the MAKER-P updated 5b+ gene models minus a set of 2,647 poorly supported 5b+ gene models. The 6a annotation build also includes 4,466 additional new, but well-supported, gene annotations derived from the MAKER-P de novo build; 102,370 pseudogene fragments; and an additional 2,522 ncRNA gene annotations. Each of these annotation data sets is described in detail below.
Table I. Overview of maize annotation builds.
5b and 5b+ refer to nuclear chromosomes 1 to 10 only in versions 5b and 5b+ of Maize Genome Sequencing Project annotation builds, respectively. Also included is a de novo annotation data set generated by MAKER-P. 5b+ update is a MAKER-P updated version of the 5b+ annotation build. 6a is the final, combined data set consisting of the updated 5b+ gene models with evidence support plus an additional 4,964 new gene models derived from the MAKER-P de novo build. TAIR 10 annotations are included for purposes of comparison.
| Parameter | 5b | 5b+ | MAKER-P | 5b+ Update | 6a | TAIR 10 |
|---|---|---|---|---|---|---|
| Protein-coding genes | 39,024 | 39,155 | 44,200 | 38,783 | 40,602 | 27,206 |
| Average gene length | 4,100 | 4,014 | 3,600 | 4,203 | 4,190 | 1,488 |
| Average protein length per gene | 375 | 366 | 327 | 371 | 366 | 410 |
| Average exons per mRNA | 4.8 | 4.8 | 4.6 | 5 | 5.1 | 5.3 |
| Percentage of genes with UTRs | 81 | 81 | 59 | 85 | 86 | 77 |
| Average UTR length | 397 | 422 | 284 | 515 | 507 | 259 |
| Average 5′ UTR length | 137 | 161 | 107 | 202 | 199 | 94 |
| Average 3′ UTR length | 260 | 261 | 177 | 313 | 308 | 165 |
| Percentage of models with start and stop codons | 84 | 97 | 86 | 98 | 94 | 96 |
| Percentage of genes with a Pfam domain | 64 | 65 | 62 | 65 | 69 | 79 |
Use of RNA-seq Data
RNA-seq data provide means for the independent confirmation and improvement of genome annotations. MAKER-P (Campbell et al., 2014), like its parent pipeline MAKER2 (Holt and Yandell, 2011), provides integrated means for employing RNA-seq data for de novo annotation, for revising existing annotation data sets in light of new RNA-seq data, and for quality-control purposes. MAKER-P uses these data to add additional untranslated region (UTR) and exon sequences to existing gene models and for the creation of new gene models where none existed previously (Holt and Yandell, 2011).
Extensive RNA-seq resources exist for maize, and our goal here was 2-fold: to use these data for purposes of quality control and to determine if MAKER-P could employ them to improve the quality of the 5b+ annotations. For these analyses, we used 96 different RNA-seq data sets downloaded from the Sequence Read Archive repository (Benson et al., 2013). The data sets are derived from various maize genotypes, developmental stages, and plant tissues. The data sets are composed of various read lengths, ranging from single-end 35 bp to 2 × 100 bp (for details, see Supplemental Table S1). Assembly of these data using Trinity (Grabherr et al., 2011; see “Materials and Methods”) produced 5,116,586 different transcripts, all of which were used in the analyses described below.
After assembly with Trinity, we ranked the RNA-seq data sets according to their number of assembled transcripts, our assumption being that data sets with the most transcripts would have the greatest value for annotation and quality control. We also sought to determine if there was a constant or perhaps diminishing benefit of using ever-greater numbers of RNA-seq data sets in the annotation process. Table II documents the power of pooling ever-larger numbers of RNA-seq data sets for discovery and quality-control purposes. Column 2 of Table II tallies the number of all 5b+ annotations on maize chromosome 5 that were overlapped, at least by 1 bp, by one or more transcripts using top one, five, 10, 15, 20, and finally all 96 transcript assemblies. The third column tallies the percentage of 5b+ annotations encoding a protein with a Pfam domain (Finn et al., 2014) but without transcript support, as annotations containing known protein domains are less likely to be false positives. As can be seen, the number of additional confirmed annotations begins to plateau beyond 10 transcript assemblies, with only modest improvements thereafter. These results provide two important facts: first, they place an approximate upper bound on the expected percentage of gene models that can be confirmed using the available RNA-seq data: about 91%; second, they provide some guidance regarding the minimum number of transcript assemblies to employ in quality-control and future reannotation efforts. Properties of RNA-seq data sets such as read depth and heterogeneity make generalizations for other genomes and their RNA-seq data sets problematic, but for these data, it appears that it would be advisable to use at least 10 of the RNA-seq data sets. In the interest of performing as near exhaustive analysis as possible, we employed all available maize RNA-seq transcript assemblies as well as an additional 136,673 maize EST and full-length cDNA sequences from the National Center for Biotechnology Information (NCBI) and 33,635 nonmaize SwissProt plant protein sequences in the analyses that follow.
Table II. Impact of using increasing numbers of RNA-seq data sets for annotation.
Ninety-six different RNA-seq data sets were ranked according to the number of Trinity-assembled transcripts they produced. The number (and percentage) of maize chromosome 5 5b+ genes supported by the top one, five, 10, 15, 20, or all transcript collections was calculated (column 2). Column 3 shows the number (and percentage) of 5b+ genes containing a Pfam domain but not supported by any transcript evidence.
| RNA-seq Data Sets | Transcript-Supported 5b+ Annotations on Chromosome 5 | 5b+ Annotations with Pfam Domains But without Transcript Support |
|---|---|---|
| Best 1 | 2,670 (59.7%) | 886 (19.8%) |
| Best 5 | 3,624 (81.0%) | 314 (7.0%) |
| Best 10 | 3,924 (87.7%) | 159 (3.6%) |
| Best 15 | 4,015 (89.8%) | 130 (2.9%) |
| Best 20 | 4,066 (90.9%) | 115 (2.6%) |
| All assemblies | 4,082 (91.3%) | 121 (2.7%) |
Accuracy of Intron-Exon Structures
MAKER-P provides automated means to assess the accuracy of a genome’s annotations in the context of the evidence used to produce them (Campbell et al., 2014). To do so, it uses a performance measure called annotation edit distance (AED; for review, see Yandell and Ence, 2012). AED measures the goodness of fit of an annotation to the evidence supporting it. AED is a number between 0 and 1, with an AED of 0 denoting perfect concordance with the available evidence and a value of 1 indicating a complete absence of support for the annotated gene model. AED can be calculated relative to any specific sort of evidence: EST and protein alignments, ab initio gene predictions, or RNA-seq data. In each case, the AED score provides a measure of an annotation’s congruency with a particular type or types of evidence. By plotting the cumulative distribution function (CDF) of AED across all annotations, a genome-wide perspective can be obtained of how well the annotations reflect the EST, protein, and RNA-seq evidence. Importantly, this can be done even in the absence of a gold-standard set of reference annotations. AED also makes it possible to compare the annotations of different genomes with one another, making possible many new sorts of cross-genome quality-control analyses (Eilbeck et al., 2009; Holt and Yandell, 2011; Yandell and Ence, 2012). For additional information on AED, see Yandell and Ence (2012).
The top of Figure 1 presents AED CFD curves for the 5b and 5b+ annotation builds. For reference purposes, also included is the TAIR 10 annotation build, presented previously (Campbell et al., 2014). The bottom of Figure 1 summarizes the same AED CFD curves as stack plots, wherein the AED data have been binned into quartiles. In previous work, we advocated that an AED CDF curve wherein more than 90% of genome annotations have an AED score of less than 0.5 is evidence that that genome is well annotated (Yandell and Ence, 2012). The Arabidopsis, human, and mouse genome annotations, for example, all satisfy this criterion (Eilbeck et al., 2009; Holt and Yandell, 2011; Campbell et al., 2014). As can be seen, approximately 90% of maize annotations have AED scores of less than 0.5, indicating that maize is a relatively well-annotated genome, but less so compared with the TAIR 10 reference annotations. Thus, Figure 1 serves to highlight an essential point regarding the maize genome annotations. Despite the complexity of the maize genome, the quality of its existing gene models as measured by their congruency with the available evidence is reasonably high, but nowhere near that of Arabidopsis. Figure 1 also makes it clear that the 5b+ and 5b builds are of very similar quality as judged by AED. This result, taken together with the data presented in Table I, which demonstrate the similarity of the two builds with regard to gene numbers, lengths, exons, and intron content, makes it clear that the two data sets are globally very similar to one another. Also presented in Figure 1 is an AED curve and stack plot for the 160 new gene models present in the 5b+ build. These new genes, on average, are less well supported.
Figure 1.
AED analyses of the 5b, 5b+, and TAIR 10 annotation builds. Top, AED CDF curves; bottom, stack plots with the same data broken down into quartiles. 5b+ new models are those models that are not present in 5b.
AED and Gene Category
Closer inspection of Figure 1 reveals that the maize 5b and 5b+ annotation builds, as well as the TAIR 10 build, contain a significant fraction of gene models with very little or no evidence supporting them. These models, with an AED score of 1 or nearly so, produce the sudden ramp present at the far right end of their AED curves. These models are shown in purple in the stack plots.
The TAIR 10 annotation for Arabidopsis can be used to better understand this ramp. TAIR employs a five-star ranking system for quality control of its genome annotations (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR_gene_confidence_ranking/DOCUMENTATION_TAIR_Gene_Confidence.pdf). In the TAIR schema, the best-supported transcripts are afforded five stars or four stars, with less supported annotations assigned three-, two-, and one-star status. Annotations with no support are assigned to the no-star category. In previous work (Campbell et al., 2014), we cross-validated MAKER-P’s AED and TAIR 10’s star ratings. For five-star TAIR 10 transcripts, 94% have AED scores of less than 0.5, whereas only 33% of one-star transcripts have an AED less than 0.5. All of the 604 TAIR 10 no-star annotations have AED’s of one, indicating that they have no evidence support.
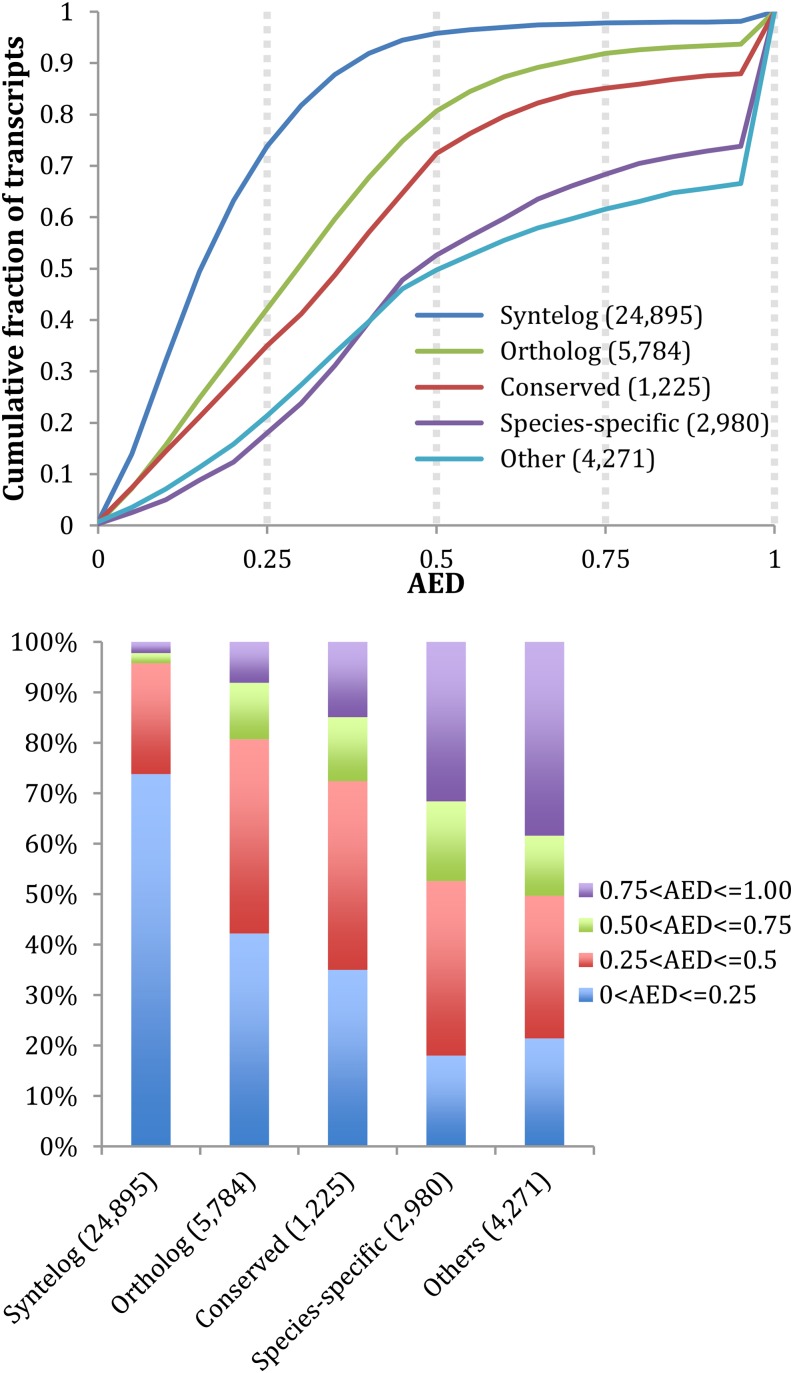
In order to better understand the characteristics of the poorly supported gene models in the maize v3 build, we divided the 5b+ maize annotations into five categories based upon the following categories of homologous relationships: Syntelogs, Orthologs, Conserved, Species-specific, and Other. We term Syntelogs as those gene annotations with syntenic orthologs in rice and/or sorghum (Sorghum bicolor). We classified as Orthologs those models with an ortholog in rice and/or sorghum that is not syntenic. Conserved are those gene models that are identified in a multispecies tree but where no orthologous relationships were found. Species-specific are those annotations encoding proteins with one or more paralogs in maize but not found elsewhere. And by Other, we mean gene models not meeting any of the above criteria. The results of this process are shown in Figure 2. As can be seen, the overall level of support and the congruency of the 5b+ gene models’ intron-exon structures with their supporting evidence differ in a consistent fashion across the categories. Syntelogs, for example, are characterized by much lower (better) AED scores than the other categories. The 160 new genes in the current 5b+ build are distributed across these five categories as follows: 68 in the Syntelog category, 23 in the Ortholog category, 11 in the Conserved category, three in the Species-specific category, and 55 in the Other category.
Figure 2.
5b+ annotations with stronger evidence of conservation have correspondingly better AED values. 5b+ maize annotations are broken into five categories: Syntelog, Ortholog, Conserved, Species-specific, and Other. For details of the classification system, see text. Note the extreme AED ramp of the Other category due to a lack of supporting evidence for these gene models. Top, AED curves; bottom, stack plots for the same data broken down into quartiles.
Poorly Supported Annotations in 5b+
Of the five categories, presented in Figure 2, Other is clearly the most problematic. Over 30% of these annotations have AED scores of greater than 0.75. By comparison, less than 1% of Syntelogs fall into this AED quartile. Given that the Other category comprises almost 4% of the 5b+ annotation build, the question naturally arises whether these are real maize genes, but inaccurately annotated, or false positives (i.e. not actually protein-coding genes). Our analyses call into question a considerable portion of genes in the Other category as well as unsupported annotations present in the rest of the categories. Using our evidence data sets (see “Materials and Methods”), a total of 3,141 (8%) of the 5b+ annotations have no supporting experimental evidence (e.g. RNA-seq, protein, and EST or encode Pfam domains). The results from Table II suggest that we should expect around 3% of the 5b+ annotations with protein support or containing a domain to lack transcript support. Although there may have been support for these annotations in prior annotation builds, 3,141 5b+ models have no support (transcript, protein, or domain) in our analysis. These facts suggest that these 3,141 5b+ annotations should be considered questionable and, in turn, that the 5b+ gene build contains 36,014 supported gene models.
MAKER-P Updates to the 5b+ Build
MAKER-P has the capacity to automatically revise an annotation build using new evidence (Campbell et al., 2014). This functionality is especially useful for updating annotations in light of new RNA-seq data. When run in update mode, MAKER-P revises the intron-exon structures of a reference annotation data set, adding additional 5′ and 3′ exons and UTRs to the reference annotations as suggested by the new evidence; reference annotations are split and merged in order to improve their fit to the supporting evidence; and new gene models are created in regions of the genome where experimental evidence supports the existence of a gene but where the reference build has no annotation. Importantly, when run in update mode, MAKER-P will not delete a reference gene model, even when MAKER-P fails to find evidence to support it.
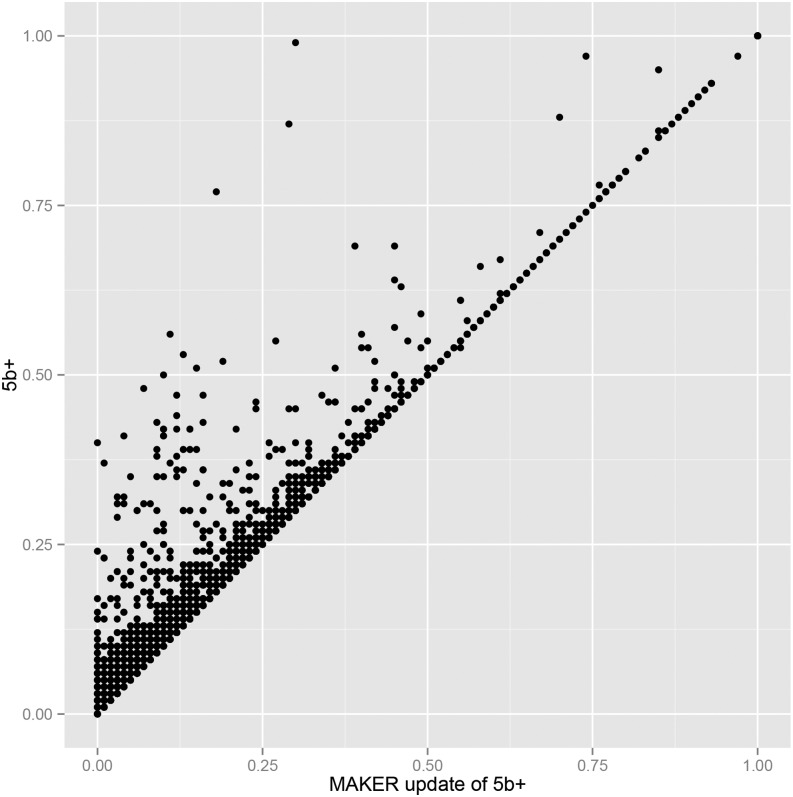
The MAKER-P revision process for 5b+ merged 31 annotations, slightly decreasing the 5b+ gene set from 39,155 (nuclear chromosomes 1–10 only) to 38,783 annotations (for additional details, see Table I). Figure 3 illustrates the impact of revision upon the maize chromosome 10 5b+ gene models. Points along the diagonal line denote models unchanged by the revision process. Note that with MAKER-P revision, AED only improves; it never worsens. This is because MAKER-P defaults to the original reference annotation whenever it is unable to improve upon it. Note too that most changes are to those models having the lowest (best) AED scores in the reference set. This is because it is often the best-annotated models that have the richest supporting evidence: with 96 different RNA-seq data sets and 5,116,586 different assembled transcripts, highly expressed genes are often overlapped by such a superabundance of evidence, some supportive, some not, that human annotators are simply stymied. MAKER-P, in contrast, is able to effectively revise the gene models regardless of the complexity or quantity of evidence. For more on this point, see Campbell et al. (2014).
Figure 3.
AED-based comparison of the 5b+ and 5b+ updated gene models for maize chromosome 10. Circles represent annotations with physical overlap between a 5b+ and its corresponding updated MAKER-P gene model. x axis, AED of the corresponding MAKER-P updated 5b+ gene model; y axis, AED of 5b+ models.
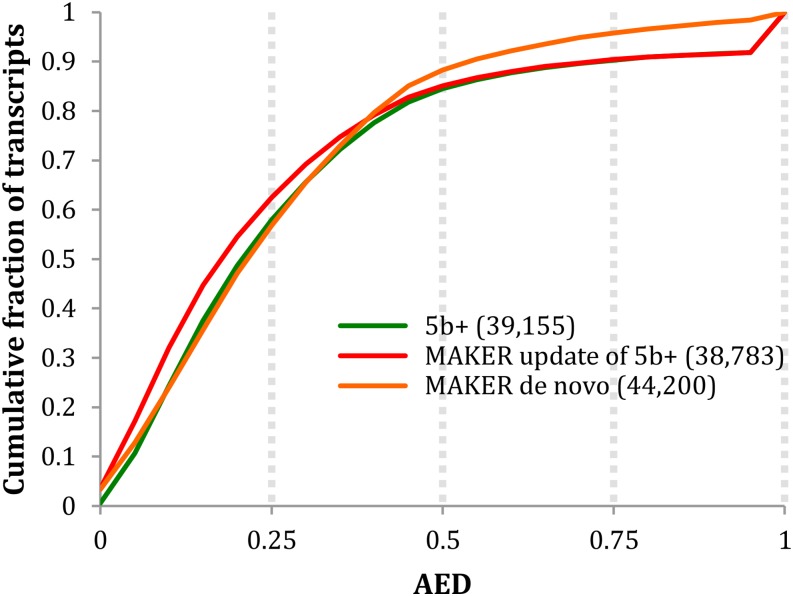
Figure 4 presents the AED CDF curves for the MAKER-P update in the context of both the 5b+ annotations and a MAKER-P de novo annotation build (discussed below). As can be seen, revision of the 5b+ build by MAKER-P shifts its AED CDF curve toward lower AEDs, indicating that the revision process has brought the 5b+ build into still better congruence with the available evidence. Note, however, that the AED ramp at the right side of the curve is unaffected; this is because the MAKER-P revision process has retained every gene model in the 5b+ build for which there was no supporting evidence. As shown, overall, the MAKER-P revised gene models have the highest proportion of genes with AEDs of less than 0.2. Table I summarizes the global differences between the 5b+ build and the MAKER-P 5b+ updated build. As can be seen, the MAKER-P revised models on average have more exons (five versus 4.8), contain additional UTR sequence (515 versus 422 bases of UTR), and the percentage of genes having any UTR at all increases from 81% to 85%. Collectively, these facts demonstrate the power of MAKER-P’s update functionality to revise and improve even high-quality maize 5b+ gene models.
Figure 4.
AED analyses of the MAKER-P updated 5b+ gene models. For ease of reference, also included are the MAKER-P de novo annotations and the original 5b+ annotations.
The MAKER-P de Novo Annotations
We also generated a MAKER-P de novo annotation build for the maize genome, using the same evidence data sets as the analyses presented in Table I and Figures 1 to 4 (for details, see “Materials and Methods”). Our goal here was to 2-fold: (1) to measure the performance of MAKER-P on the maize genome by comparing its annotations with the 5b+ annotation build in order to gain an indication of what to expect when using MAKER-P on other difficult-to-annotate plant genomes; and (2) to determine if MAKER-P might identify additional maize genes absent from the 5b+ annotation build.
Training MAKER-P
Given sufficient training data (i.e. gold-standard gene models), ab initio gene predictors can deliver very accurate gene models (Guigó et al., 2006; Yandell and Ence, 2012). However, for newly sequenced genomes, no training data are usually available. In previous work (Holt and Yandell, 2011; Campbell et al., 2014), we described a procedure whereby MAKER-P can be used to train Augustus (Stanke and Waack, 2003; Stanke et al., 2008) and SNAP (Korf, 2004), two widely used ab initio gene finders. This training process uses RNA-seq data and ESTs in lieu of a preexisting gold-standard set of gene models. These data are aligned to the genome using the splice-aware aligner Exonerate (http://www.ebi.ac.uk/~guy/exonerate/), and an automatically identified postprocessed subset of high-quality alignments is used for gene-finder training.
Grass genomes are generally repeat rich and harbor the results of multiple polyploidization events, making them difficult substrates for annotation. It seemed likely that these same features of grass genomes might negatively impact the effectiveness of MAKER-P’s gene-finder training procedures. Maize thus provides an opportunity to examine this problem. The genome is typical of grass genomes: there is a preexisting gold standard of reference annotations (e.g. the conserved Syntelogs of the 5b+ build), and there exist a plethora of maize RNA-seq and EST data. Equally important, the popular and very accurate gene finder Augustus (Stanke and Waack, 2003; Stanke et al., 2008) comes pretrained for maize, providing an opportunity to benchmark the performance of a version of Augustus trained by MAKER-P using maize RNA-seq and EST data to one trained by the authors of Augustus using the maize reference annotations. Supplemental Figure S1 shows the AED CDF curves for these two versions of Augustus. As expected, the version trained by the Augustus group using the 5b gene models is more accurate than the MAKER-P version trained using the noisy RNA-seq and EST data, but not greatly so. The MAKER-P-trained version of Augustus, for example, calls about 5% more genes, and 87%, as opposed to 91%, of its models have an AED of less than 0.5, indicating that the intron-exon structures of the MAKER-P-trained version of Augustus are nearly as accurate. These results demonstrate that MAKER-P’s training procedure is effective even for difficult-to-annotate grass genomes. We used the MAKER-P-trained version of Augustus for the de novo annotation run described below.
MAKER-P de Novo Results
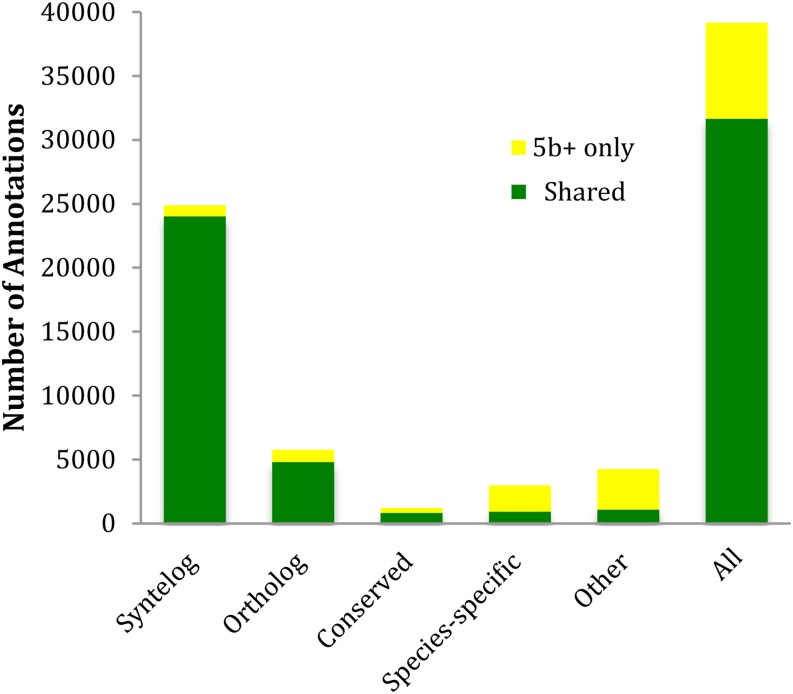
AED curves and stack plots comparing the MAKER-P de novo build with the 5b+ and updated 5b+ builds are presented in Figure 4. As can be seen, overall, its models are nearly as congruent with the evidence as the updated 5b+ build. Figure 5 summarizes the intersections between the 5b+ build and the MAKER-P gene set, broken down by gene category. As shown, there is almost perfect agreement among the Syntelog gene set, with less, but still considerable, congruence for the Ortholog and Conserved categories. However, of the 5,401 models comprising the 5b+ Other category, only 1,347 have supporting evidence and are also called by MAKER-P, again suggesting that many of 5b+ genes belonging to the Other category should be considered provisional.
Figure 5.
Shared and unique gene models in the 5b+ and the MAKER-P gene de novo gene sets. To facilitate comparison, both builds were broken down into the same five gene categories described for Figure 2. Intersecting genes are shown in green, and gene models unique to the MAKER-P de novo build are shown in yellow.
Table I summarizes the relevant statistics of the MAKER-P de novo gene models. Globally, the MAKER-P de novo build is quite similar to the 5b+ build, but it differs in three regards: (1) fewer of its gene models contain UTRs; (2) its gene models are shorter; and (3) it contains 5,045 additional annotations that do not overlap 5b+ gene models. Point 2 is largely a consequence of the additional gene models not present in the 5b+ build. The 5,045 additional gene models tend to be short and are predominantly single-exon genes. In these respects, they are quite similar to the majority of 5b+ genes in the Other category. But they differ in one vital regard: every MAKER-P gene is supported by transcript, protein, and/or domain evidence, whereas the majority of the 5b+ Other genes are supported only by ab initio gene predictions, a point we return to in “Conclusion.” Collectively, analyses presented in Figures 4 and 5 and Table I indicate that, globally, the MAKER-P de novo build is slightly inferior to the curated 5b+ build with regard to protein-coding genes, but not dramatically so, demonstrating that MAKER-P is capable of producing a high-quality de novo gene build for a grass genome, one that is a suitable starting point for further manual and automated curation. Moreover, as we document below, the MAKER-P de novo build has no unsupported models and contains additional pseudogene, ncRNA, and well-supported protein-coding gene models not present in the curated 5b+ build.
Nonprotein-Coding Genes
MAKER-P’s annotations are not limited to protein-coding genes alone. The MAKER-P toolkit provides a process for the annotation of pseudogenes. The ability to annotate and identify pseudogenes is particularly important for grass genomes, given their abundance. MAKER-P also provides means for the identification of known and new classes of ncRNAs.
Pseudogenes
In total, 102,370 putative partial or complete pseudogenes were identified in maize with MAKER-P. These pseudogenes have a mean length of 191 bp, similar to what was found in Arabidopsis and rice (Zou et al., 2009b; Campbell et al., 2014), with a significant positive skew, indicating that the majority of pseudogenes were on the shorter end of the spectrum. This can be a consequence of the inability to connect pseudoexons of a pseudogene together. Nonetheless, the same MAKER-P pipeline identified only 4,204 pseudogenes in Arabidopsis, far less than what we have recovered in maize. One explanation is that the gene deletion rate was higher in the Arabidopsis lineage, consistent with the finding that genome size differences between Arabidopsis (150 Mb) and Arabidopsis lyrata (207 Mb) is due to extensive DNA loss (Hu et al., 2011). Another possibility is that pseudogenes were generated or retained at a greater rate in the maize lineage. This is consistent with a much more recent whole-genome duplication in the maize lineage (approximately 11 million years ago; Gaut and Doebley, 1997) compared with that in Arabidopsis (α-genome duplication, approximately 50 million years ago; Bowers et al., 2003). In addition, in maize, there is an overabundance of Helitrons carrying gene fragments (Du et al., 2009; Yang and Bennetzen, 2009). Among 272 manually annotated Helitrons, 94% of them carry captured sequences from 376 genes (Du et al., 2009). There is also evidence suggesting that more than 20,000 gene fragments in the B73 genome are transduplicated and reshuffled due to Helitron activities (Yang and Bennetzen, 2009). Together with the suggestion that Helitrons are involved in exon shuffling (Feschotte and Wessler, 2001), these findings are consistent with the possibility that Helitrons have contributed significantly to the high pseudogene fragment number observed.
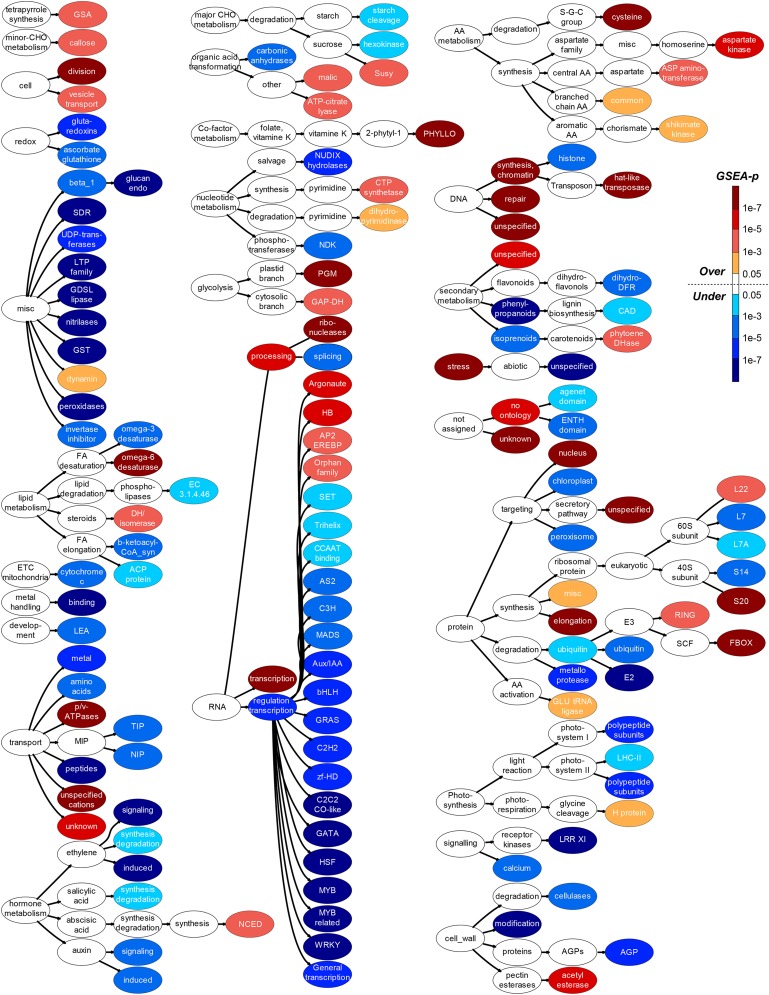
To better understand what kinds of duplicates tend to become pseudogenes, MapMan (Thimm et al., 2004) annotations were assigned to pseudogenes based on the maize protein sequences used to identify them. As a result, 54.6% of pseudogenes have one or more MapMan annotations. Fisher’s exact test was used to identify MapMan annotations associated with overrepresented and underrepresented numbers of pseudogenes (Figure 6). Overrepresented terms include stress, protein degradation (via ubiquitin), and secondary metabolism (unspecified), which are also known to be overrepresented in Arabidopsis (Zou et al., 2009). Similarly, the Argonaute gene family involved in small RNA biogenesis has 43 annotated, presumably functional, members and 127 pseudogenes (Figure 6). Argonaute genes are important for viral defense in plants (Qu et al., 2008). In addition, genes involved in external stimulus responses tend not only to experience lineage-specific duplication (Hanada et al., 2008) but also to pseudogenize at a higher rate (Zou et al., 2009). Taken together, the significant overrepresentation of Argonaute pseudogenes may be the product of viral defense genes that were no longer useful. We also found that most transcriptional regulators are among the underrepresented class of pseudogenes, except the Homeobox and APETALA2/ethylene response element binding protein families (Figure 6). The underrepresentation of transcription factor pseudogenes is consistent with higher retention rates among plant transcription factor duplicates (Schnable et al., 2009), particularly those derived from whole-genome duplications (Blanc and Wolfe, 2004; Seoighe and Gehring, 2004; Shiu et al., 2005). Therefore, in spite of differences in the number of pseudogenes identified, the pseudogenization of duplicates in Arabidopsis and maize follows similar trends.
Figure 6.
MapMan terms with overrepresented or underrepresented numbers of maize pseudogenes. The ovals indicate overrepresented (shades of red) and underrepresented (shades of blue) terms and their parent terms (white). Some terms are truncated or abbreviated. For full terms and associated statistics, see Supplemental Table S4.
ncRNA Genes
The MAKER-P toolkit identified 2,192 total tRNA genes. Of these annotated tRNA genes, 1,398 decode the standard amino acids, four decode seleno-Cys, seven are possible suppressor tRNAs, 12 are undetermined, and 771 appear to have been pseudogenized (Table III). Ultimately, these data contain slight differences from tRNA analyses of previous maize genome assemblies in maize secondary to changes in the v3 assembly (http://lowelab.ucsc.edu/GtRNAdb/Zmays/Zmays-stats.html). Using 12 small RNA-seq experiments, the MAKER-P toolkit also identified 183 microRNAs (miRNAs). As mentioned previously (Campbell et al., 2014), the number of miRNAs predicted by the MAKER-P toolkit is dependent on the small RNA evidence; thus, this number represents a lower bound of miRNAs in the v3 assembly. Most of the predicted mature miRNAs are of length 21, which is the typical plant miRNA length. Of the 183 predictions, 87 of them overlap with the existing 5b+ annotation of miRNAs and others are new predictions. The discrepancy mainly stems from the different methods used for miRNA annotation by MAKER-P and the existing maize miRNA identification method (Zhang et al., 2009). While the miRNA prediction pipeline miR-PREFeR of MAKER-P follows the criteria for plant miRNA annotation (Meyers et al., 2008), 5b+ miRNA annotations were created by aligning genomic sequences against miRBase (Griffiths-Jones et al., 2008) sequences using BLASTN (Altschul et al., 1990). Thus, the reliability of 5b+ miRNA annotation relies heavily on the quality of miRBase collections. Although the underlying annotations in miRBase are generally experimentally determined or experimentally verified, errors have been detected in miRBase annotations (Kozomara and Griffiths-Jones, 2014). In addition, many 5b+ miRNA annotations lack expression evidence in our 12 small RNA-seq samples. Finally, the homology search-based annotation method we adopted may miss miRNAs that are specific to maize. Using the same small RNA-seq data sets, the MAKER-P toolkit identified 727 small nucleolar RNAs (snoRNAs) with AEDs less than 0.5. (See Supplemental Text S1 for the link to the GFF file containing the tRNA, miRNA, and snoRNA predictions.)
Table III. Summary of ncRNA annotations.
Numbers of ncRNAs are broken down by type for 5b, 5b+, and 6a annotation builds. The last column gives corresponding numbers in the TAIR 10 annotation of Arabidopsis for reference. NA denotes classes of annotations not present in the non-MAKER-P-derived builds.
The 6a Gene Annotation Build
Table I also provides a summary of an annotation build termed 6a. Our goal in creating the 6a build was to provide the maize community with a single annotation build comprising the best-possible annotated gene models drawn from the 5b+, 5b+ updated, and MAKER-P de novo annotation builds. Thus, the 6a build is a synthetic data set composed of the MAKER-P updated 5b+ gene models, which contain additional 5′ and 3′ exons and UTR sequences, together with additional new, but well-supported, genes derived from the MAKER-P de novo build. We also excluded from 5b+ 2,647 5b+ gene models for which we could find no supporting evidence and 249 models that overlapped with our predicted ncRNA models. These gene models are included in a separate file (Supplemental Table S2) under the title Provisional v3 Gene Models.
The 44,200 MAKER-P de novo protein-coding genes (Table I; Fig. 5) comprised the starting point for our attempt to identify a core set of additional high-quality gene models for inclusion in the 6a build. To identify these models, we first removed any unique MAKER-P de novo gene models that resided within transposons, as these might represent gene fragments carried by transposons; this reduced the number by about 10%. We then broke the remaining MAKER-P unique protein-coding gene models into two classes: (1) multiexon models with at least one splice site perfectly confirmed by RNA-seq or EST alignments; and (2) single-exon models that encode a domain and have annotated start and stop codons. Our reasoning was that models supported by spliced transcript data and having canonical splice sites were reasonable candidates for additional genes. We also enforced an additional criterion on these genes: they must have at least one coding exon predicted by a gene finder. With regard to the unique MAKER-P single-exon gene models, because single-exon genes are often spuriously overlapped by transcript data, we did not consider transcript support as proof of a single-exon gene’s existence. Thus, enforcing the additional criteria that these single-exon genes encode a known domain, their single exon be predicted by a gene finder, and they have annotated start and stop codons should diminish the proportion of the models that constitute a common form of false-positive annotation: random open reading frames fortuitously overlapped by RNA-seq data from noisy transcription data. Likewise, the requirement for start and stop codons should avoid false positives where the supposed single-exon gene consists of portions of a pseudogene with a partial open reading frame encoding a remnant portion of a protein domain. Of course, none of these criteria can guarantee that every one of the additional new genes is truly a new maize protein-coding gene, but what is true is that each of the new gene models identified in the analysis meets a stringent set of criteria for inclusion in the 6a build. Certainly, they are better candidates than the 2,647 provisional gene models we identified in our analyses of the 5b+ build, none of which meet any of these criteria; hence, replacing those provisional models with these additional MAKER-P-derived new models seems reasonable.
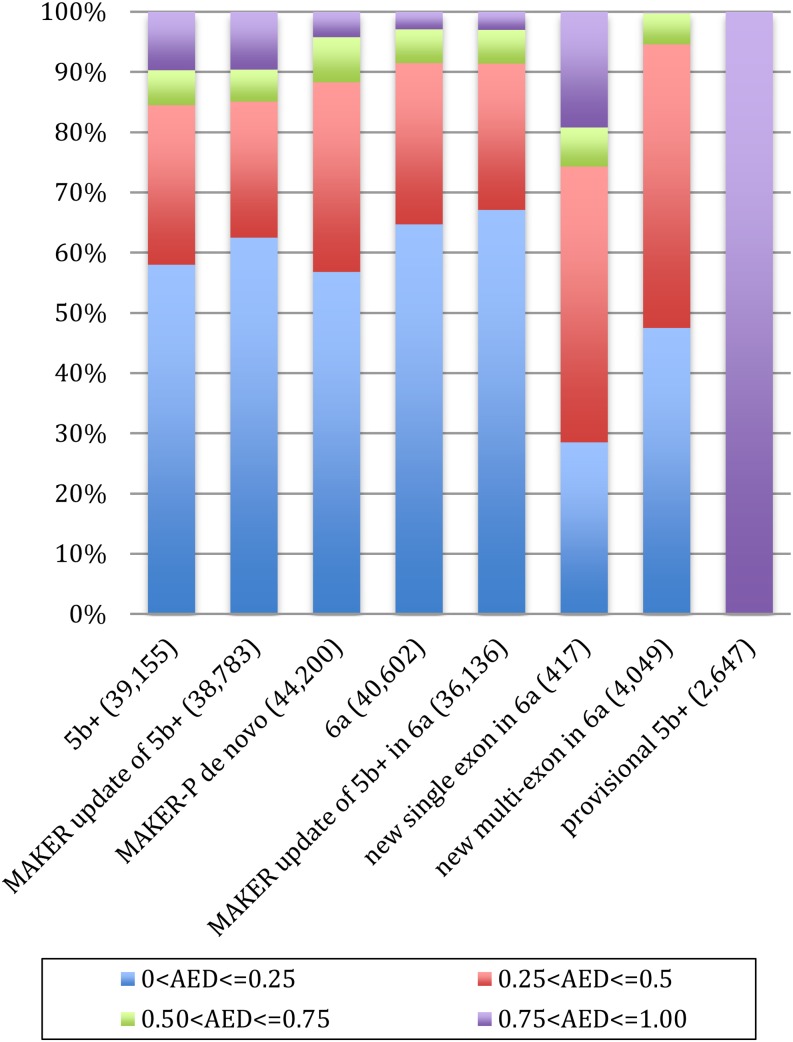
Table IV summarizes the results of this analysis. In total, 4,049 of the new MAKER-P gene models encode multiexon transcripts with at least one confirmed splice site. Note that the average number of exons is 4.9, and 45% of these putative genes encode a Pfam domain. Thus, although they are shorter than the average 5b+ annotation (2,836 versus 4,014), many are sizable, multiexon gene models that contain domains. All 417 of the single-exon models encode a domain, have transcript support, and have annotated start and stop codons. In addition, all of the new models have gene-finder support. Figure 7 presents AED stack plots for the 6a build and various portions thereof. Also included for reference purposes are the 5b+ reference build and the subset of models that we identified as provisional and, thus, that are not included in the 6a build. As can be seen from an inspection of Tables I, III, and IV, the 6a build contains more supported gene models and more models with 5′ and 3′ UTRs, and its gene models have longer UTRs compared with the original 5b+ build, contain more exons, and encode longer proteins. The 6a models are also more congruent with the available evidence as judged by AED. Also included are an additional 3,006 ncRNA genes and 102,370 pseudogene annotations not present in the 5b+ build.
Table IV. Summary of new gene models included in the 6a build.
| Parameter | Multiexon MAKER-P de Novo | Single-Exon MAKER-P de Novo | 6a |
|---|---|---|---|
| Protein-coding genes | 4,049 | 417 | 40,602 |
| Average gene length | 2,836 | 676 | 4,190 |
| Average exons per mRNA | 4.9 | 1 | 5.1 |
| Average exon length | 195 | 648 | 315 |
| Average protein length | 216 | 221 | 366 |
| Percentage of genes with a Pfam domain | 45 | 100 | 68 |
Figure 7.
AED analyses of the 6a build. AED stack plots are broken down into quartiles: 5b+ build, MAKER update of 5b+, MAKER-P de novo, 6a build, 5b+ models in 6a, new MAKER de novo multiexons and single exon in 6a, and provisional 5b+ models. Numbers in parentheses indicate the number of annotations in each gene set.
CONCLUSION
We have carried out systematic analyses of the maize 5b+ annotation build using MAKER-P’s management and quality-control functions. This work has allowed us to reevaluate the 5b+ annotation build in light of additional RNA-seq evidence and to update the 5b+ build using these same data. We have also compared MAKER-P de novo annotations with those of the 5b+ reference build in order to gain an indication of what to expect when using MAKER-P on other difficult-to-annotate plant genomes. These same analyses have identified additional maize genes absent from the 5b+ annotation build.
As we have shown, MAKER-P can further improve an existing genome annotation build. The MAKER-P 5b+ update, for example, contains every model present in the 5b+ build but adds additional exons and UTR sequences. It also contains a number of gene splits and merges suggested by the RNA-seq data. The result is an updated 5b+ build that is demonstrably in better agreement with the available evidence. Importantly, these results also show how using MAKER-P for the management of a genome’s annotations does not necessitate a switch from one pipeline’s annotations to another. MAKER-P can improve an existing community annotation resource without introducing any break in continuity (i.e. the existing models are kept but brought forward incrementally to reflect additional evidence).
Our de novo training results demonstrate that MAKER-P also can be used to train a widely used gene finder such as Augustus for employment on newly sequenced plant genomes and that the resulting performance is a close match to that obtained using a gold-standard training set. This is important because previous work by our group and others has made it clear that attempts to leverage gene finders trained from other genomes rarely produce accurate gene predictions. Our analysis of the MAKER-P de novo annotations demonstrates that, although the MAKER-P de novo models are slightly inferior with regard to the accuracy of its intron-exon structures, it is demonstrably superior in its relationship to the available evidence (i.e. the average model is more congruent with its overlapping evidence, and importantly, every one of its annotations has supporting evidence). Collectively these results make clear that MAKER-P provides an effective means for de novo annotation of even difficult-to-annotate grass genomes.
The 6a annotation build provides the maize community a genome annotation data set that is notably superior to both the 5b+ and MAKER-P de novo builds. Informed by new expression evidence assembled from an extensive collection of RNA-seq studies, the 6a build contains the MAKER-P updated 5b+ gene models together with an additional 4,466 new genes not contained in the 5b+ annotation build.
The 6a build also lacks 2,647 5b+ genes for which we could find no support, despite the number and diversity of evidence data sets used. Thus, the improvements offered by the 6a build are not limited solely to new contents. Considering these 2,647 5b+ genes as provisional has important consequences for future work: first, these poorly supported gene models, for example, will no longer introduce biases into comparative studies with regard to statistics such as domain content, UTR lengths, and exon number sets; second, knowledge that these 5b+ genes are provisional will provide a starting point for focused experimental follow-up studies aimed at confirming or denying their existence.
Collectively, the 6a build is a demonstrable improvement upon the 5b+ build. Its genes have more exons, have longer UTRs, and are more congruent with the evidence. Furthermore, the 6a build also supplements the 5b+ build with 102,370 pseudogene and 3,006 ncRNA annotations.
Recent work has shown that the version of MAKER-P available within the iPlant Cyberinfrastructure can reannotate the entire maize genome using the same evidence data sets described here in less than 3 h (Campbell et al., 2014) and that it can carry out a complete de novo annotation of the 20-Gb draft loblolly pine genome in less than 24 h (Neale et al., 2014; Wegrzyn et al., 2014).
These facts have important implications for the future of plant genome annotation. First, they show that MAKER-P provides effective means for the annotation of plant genomes; second, its update mode provides a means to refresh the annotations of established plant genomes to reflect new data; and third, these updates can be carried out much more rapidly and frequently than has heretofore been possible. Perhaps even more important is that MAKER-P’s speed and flexibility will enable individual iPlant users to generate their own custom genome annotation data sets using public annotation builds as starting points but embodying their own data. The 6a annotations and related documents are available for download at http://documents.maizegdb.org/makerp/. The latest version of MAKER-P is available as part of the MAKER package download at http://www.yandell-lab.org/software/maker-p.html
MATERIALS AND METHODS
Transcripts and Protein Evidence
Transcripts and transcript assemblies were used as evidence for gene predictions and MAKER updates. Maize (Zea mays) ESTs and full-length cDNAs were downloaded from the NCBI GenBank. Ninety-five RNA-seq data sets were downloaded from NCBI’s Sequence Read Archive (Supplemental Table S1). One additional RNA-seq data set was described by Takacs et al. (2012) and can be obtained from the authors (Supplemental Table S1). The RNA-seq reads from these data sets were cleaned using tools from the FASTX toolkit (version 0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/). The fastx-clipper program removed adapter sequences from all reads, and the fastx-artifacts-filter was used to remove aberrant reads. These steps were followed by running the fastx-trimmer program, which removed bases with quality scores less than 20 and discarded reads that were less than 30 bases in length. Cleaned RNA-seq reads from individual studies (Supplemental Table S1) were assembled using the Trinity transcript assembly package (Grabherr et al., 2011) and used for annotation. SwissProt plant protein sequences were downloaded from UniProt. Maize protein sequences were removed, and the remaining plant protein sequences were used as annotation evidence. The maize genome (Zea_mays.AGPv3.21.dna.genome.fa.gz) was downloaded from ftp://ftp.ensemblgenomes.org/pub/release-21/plants/fasta/zea_mays/dna/. MAKER-P analyses focused on all nuclear chromosomes 1 to 10 unless specified otherwise.
Classification of the 5b+ Annotation Set Using Comparative Genomics Criteria
We utilized the output of Ensembl Compara Gene Trees and associated synteny builds available from Gramene release 39 (October 2013), currently archived at http://archive.gramene.org/. The Ensembl method identifies ortholog and paralog relationships between genes using phylogenetic inference (Vilella, et al., 2009; see also http://useast.ensembl.org/info/genome/compara/homology_method.html). The Gramene project subsequently maps collinear and near-collinear orthologous genes between related species (Youens-Clark et al., 2011), adapting a protocol originally developed for the analysis of synteny in maize (Schnable et al., 2009; for details, see supporting online materials: http://www.sciencemag.org/content/suppl/2009/11/18/326.5956.1112.DC1/Schnable.SOM.pdf), which uses DAGChainer (Haas et al., 2004). The Compara Gene Trees in Gramene release 39 incorporated gene sets for 25 plant and five nonplant species. This release also included synteny maps for maize-sorghum (Sorghum bicolor) and maize-rice (Oryza sativa). From these data, we classified the maize 5b+ annotation set as follows: Syntelog, having orthologs in rice and/or sorghum that are arranged in a collinear or near-collinear fashion; Ortholog, having a called ortholog in rice and/or sorghum that is not a Syntelog; Conserved, found in a multispecies tree but lacking an identified ortholog; Species-specific, found in a maize-specific gene tree (i.e. having paralogs in maize but without homology to other species); and Other, not found in a tree (thus having no detectable homology with other species in the set).
Repeat Library and Examination of New Genes for Transposons
The repeat library used in this study was derived from the following two sources. First, 1,526 transposon exemplar sequences were downloaded from the maize TE database (http://maizetedb.org/~maize/). Second, 10,619 maize Sirevirus sequences were downloaded from MASiVEdb (Bousios et al., 2012) and masked by the 1,526 transposon sequences from the maize TE database. For a Sirevirus sequence, if 90% of the length was masked with a similarity of 80% or higher, it was excluded, since it was considered to be already present in the 1,526 sequences. Exemplar sequences were chosen from the remainder of the Sirevirus sequences to reduce the redundancy as follows: all sequences were compared using BLASTN. The element with the most matches (cutoff at 80% identity in 90% of the element length) was considered as the first exemplar. Thereafter, this element and its matches were excluded from the group and a second-round BLASTN search was conducted with the remainder of the elements, leading to the generation of the second exemplar. This process was repeated until all elements were excluded. These exemplar sequences were combined with the 1,526 transposon sequences from the maize TE database, and the combined library was used in this study.
Since the combined library only contains true transposon sequences, gene fragments that are carried with transposons such as those in Pack-Mutator-like transposable elements (MULEs) were not included in the library. To test whether the new MAKER-P genes identified in this study were actually gene fragments inside transposons, the relevant gene coordinates were first compared with previously identified Pack-MULEs in maize (Jiang et al., 2011). If over 50% of the mRNA sequence of a gene was located inside a Pack-MULE, this gene was considered a transposon and excluded from the 6a build. For the remainder of the genes, the gene and the 5-kb flanking sequence on both sides of the gene were retrieved and the transposons in the entire fragment were annotated using RepeatMasker with the library mentioned above. If the gene was flanked by two transposons from the same superfamily of transposon and both transposons were truncated by 30 bp or more on the side facing the gene, this gene was considered to reside inside a transposon and excluded from 6a. If only part of the gene was inside the transposon, a 50% cutoff of the transcribed sequences was taken for consideration. In summary, if 50% or more of the mRNA of a gene is inside a transposon, the gene is considered a transposon.
MAKER-P de Novo Annotation and Update of 5b+
RNA-seq data sets from public repositories (Supplemental Table S1) were assembled and used as evidence in MAKER-P 2.31 r1081, along with Uniprot/SwissProt protein evidence and a set of traditional full-length cDNAs. A custom repeat library (see above) was used to mask the repetitive regions (for details, see preceding paragraph). Genes were predicted using Augustus (Stanke and Waack, 2003; Stanke et al., 2008) trained in an iterative fashion in MAKER-P as described before (Campbell et al., 2014). The MAKER de novo annotation set represents those predictions that are supported by evidence or contained a Pfam domain. To obtain a set of MAKER-P revised annotations, maize 5b+ models are passed to MAKER-P as gene predictions, together with the same evidence set and RepeatMasker as above.
Utility of Transcript Assembly Evidence for Gene Predictions
Our Trinity-derived transcript assemblies from 96 different RNA-seq data sets were ranked by the number of sequences in each assembly. While this approach may not recover the best RNA-seq data sets in all cases (e.g. a data set might contain genomic contamination, resulting in large numbers of spurious transcripts), we found that this simple procedure provided a practical means to select subsets of RNA-seq data when many different data sets are available. Collections of the top one, five, 10, 15, 20, or all transcript assemblies were used as evidence in MAKER-P runs. MAKER-P was run in pass-through mode using the 5b+ gene predictions and the different collections of transcript assemblies as evidence. The 5b+ gene models were unmodified but were assigned AED scores based on the transcript support for each model. Genes with AED scores less than 1 were scored as being supported by the given transcript evidence set.
6a Annotations
MAKER de novo annotations that were not overlapped by MAKER updated 5b+ gene models were retained when (1) single-exon models encoded a domain and contained annotated start and stop codons and (2) multiexon models with at least one splice site was confirmed by EST alignment. Maize 5b+ updated models with domain support or RNA-seq evidence support were combined, along with MAKER-P ncRNA annotations with these two classes of MAKER de novo annotations, to generate the final 6a build. 5b+ models without evidence support (AED = 1.00) and/or encoded Pfam domains were classified as provisional. MAKER de novo annotations residing within transposons were also excluded.
ncRNA Annotation
tRNAs were identified using tRNAscan-SE (Lowe and Eddy, 1997) within the parallelized MAKER-P framework. The snoRNAs were predicted using snoscan (Lowe and Eddy, 1999) also within the parallelized MAKER-P framework. To limit the inevitable false positives resulting from the genome-scale use of stochastic context-free grammars in snoscan, we limited our results to snoscan predictions that matched a ribosomal RNA (rRNA) O-methylation site and had an AED of less than 0.5. rRNA O-methylation sites for maize 26S (Refseq accession no. NR_028022 version NR_028022.2) and 17S (Refseq accession no. NR_036655 version NR_036655.1) rRNAs were inferred based on homology to known rRNA methylation sites (http://lowelab.ucsc.edu/snoscan/default-files/Hu-meth.sites) in human 28S (GenBank accession no. M11167 version M11167.1) and 18S (GenBank accession no. NR_003286 version NR003286.2) rRNA, respectively.
The miRNAs were identified using miR-PREFeR pipeline (Lei and Sun, 2014), which is an improved version of the miRNA annotation pipeline described previously (Campbell et al., 2014). Expression of these miRNAs was confirmed within the miR-PREFeR pipeline using 12 small RNA sequencing experiments from seven tissues (Supplemental Table S3). miR-PREFeR utilizes expression patterns of miRNAs and follows the criteria for plant miRNA annotation (Meyers et al., 2008) to accurately predict plant miRNAs from one or more small RNA-seq samples. The primary criterion is that the small RNA-seq data should provide evidence of precise miRNA/passenger miRNA (miRNA*) excision. Specifically, there should exist abundant reads corresponding to the mature miRNA sequence, and there should be at least one read that can be precisely mapped back to the miRNA* sequence. The miRNA and miRNA* sequences should form a duplex with two-nucleotide 3′ overhangs. In addition, the miRNA/miRNA* duplex needs to present the following structural characteristics: there are typically four or fewer unpaired bases in the miRNA/miRNA* duplex, and asymmetric bulges are rare and small in size.
As the expression of miRNAs can be tissue or condition specific, we aimed to provide a comprehensive miRNA annotation by using multiple RNA-seq samples from different tissues/conditions/developmental stages. There are two advantages of predicting miRNAs from multiple RNA-seq samples. First, some miRNAs are poorly expressed and cannot be identified in a single RNA-seq sample. miR-PREFeR can predict poorly expressed miRNAs by combining all reads from multiple samples. Second, due to fast degradation, some miRNAs lack reads mapping to their miRNA* region and will not satisfy the strict plant miRNA annotation criteria. In our method, if the corresponding miRNA loci from multiple samples demonstrate other typical miRNA characteristics, including high expression, the existence of a well-formed stem loop, and precise miRNA/miRNA* excision in the predicted stem loop, we conclude that this locus contains a true miRNA gene by dropping the requirement for the presence of the star sequence. In this implementation, when there is no read corresponding to the star sequence, we require that there should be at least 1,000 reads in all samples and at least 100 reads in each sample.
Pseudogene Identification
Pseudogenes were identified by MAKER-P according to the method described previously (Campbell et al., 2014). Annotated protein sequences were searched against a version of the genome masked for 6a annotations and filtered using four criteria: e value (<1e-5), identity (greater than 40%), length (more than 30 amino acids), and coverage of the query sequence (5%). Using a maximum interval of 2,032 bp (95th percentile intron length), 510,259 pseudoexons were combined into putative pseudogenes, which were subsequently filtered if they overlapped with annotated gene regions and/or known Viridiplantae repeats. Note that some of these putative pseudogenes are substantially shorter than their annotated, presumably functional, paralogs but do not have disabling mutations (stop or frame shift). In addition, some pseudogenes may be functional genes that are split between contigs or scaffolds. Thus, we only examined putative pseudogenes with one or more disabling mutations or those located distantly from the ends of contigs based on a threshold distance. This threshold distance is defined as the sum of the 95th percentile intron length and a consideration of functional paralog length. Suppose a functional paralog to a pseudogene has length L and the pseudogene match is from M1 and M2, functional paralog length is defined as the larger of M1 or L − M2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparing two versions of trained Augustus within MAKER-P on Chromosome 10.
Supplemental Table S1. RNA-seq data sources used for transcript assemblies.
Supplemental Table S2. Provisional 5b+ gene models.
Supplemental Table S3. Small RNA-seq experiments used in miRNA identification.
Supplemental Table S4. MapMan terms and statistics.
Supplemental Text S1. GFF file containing the tRNA, miRNA, and snoRNA predictions.
Supplementary Material
Acknowledgments
We thank iPlant, Texas Advanced Computing Center, and MaizeGDB support personnel for their efforts.
Glossary
- BAC
bacterial artificial chromosome
- TE
transposable element
- WGS
whole-genome shotgun
- cDNA
complementary DNA
- RNA-seq
RNA sequencing
- ncRNA
noncoding RNA
- TAIR
The Arabidopsis Information Resource
- UTR
untranslated region
- NCBI
National Center for Biotechnology Information
- AED
annotation edit distance
- CDF
cumulative distribution function
- miRNA
microRNA
- snoRNA
small nucleolar RNA
- MULE
Mutator-like transposable element
- rRNA
ribosomal RNA
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1126998 to S.-H.S., Y.S., N.J., K.L.C., and M.Y. and grant no. MCB–1119778 to S.-H.S.), the U.S. Department of Agriculture-Agricultural Research Service, and Iowa State University (MaizeGDB and contributions by C.M.A. and C.J.L.).
Articles can be viewed without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41: D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousios A, Minga E, Kalitsou N, Pantermali M, Tsaballa A, Darzentas N (2012) MASiVEdb: the Sirevirus Plant Retrotransposon Database. BMC Genomics 13: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. (2014) MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol 164: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Fefelova N, Caronna J, He L, Dooner HK (2009) The polychromatic Helitron landscape of the maize genome. Proc Natl Acad Sci USA 106: 19916–19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M (2009) Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Wessler SR (2001) Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes. Proc Natl Acad Sci USA 98: 8923–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E, et al. (2006) EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol (Suppl 1) 7: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646 [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Ferguson AA, Slotkin RK, Lisch D (2011) Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci USA 108: 1537–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. (2004) Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Sun Y (2014) miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 30: 2837–2839 [DOI] [PubMed] [Google Scholar]
- Liang C, Mao L, Ware D, Stein L (2009) Evidence-based gene predictions in plant genomes. Genome Res 19: 1912–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1999) A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. (2008) Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MK, Stein J, Naithani S, Wei S, Dharmawardhana P, Kumari S, Amarasinghe V, Youens-Clark K, Thomason J, Preece J, et al. (2014) Gramene 2013: comparative plant genomics resources. Nucleic Acids Res 42: D1193–D1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sen TZ, Andorf CM, Schaeffer ML, Harper LC, Sparks ME, Duvick J, Brendel VP, Cannon E, Campbell DA, Lawrence CJ (2009) MaizeGDB becomes “sequence-centric.” Database (Oxford) 2009: bap020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Shih MC, Li WH (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644 [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S (2003) Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics (Suppl 2) 19: ii215–ii225 [DOI] [PubMed] [Google Scholar]
- Takacs EM, Li J, Du C, Ponnala L, Janick-Buckner D, Yu J, Muehlbauer GJ, Schnable PS, Timmermans MCP, Sun Q, et al. (2012) Ontogeny of the maize shoot apical meristem. Plant Cell 24: 3219–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E (2009) EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Liechty JD, Stevens KA, Wu LS, Loopstra CA, Vasquez-Gross HA, Dougherty WM, Lin BY, Zieve JJ, Martínez-García PJ, et al. (2014) Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196: 891–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K, Kudrna D, Faga BP, Wissotski M, Golser W, et al. (2009) The physical and genetic framework of the maize B73 genome. PLoS Genet 5: e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Ence D (2012) A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 13: 329–342 [DOI] [PubMed] [Google Scholar]
- Yang L, Bennetzen JL (2009) Distribution, diversity, evolution, and survival of Helitrons in the maize genome. Proc Natl Acad Sci USA 106: 19922–19927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan AS, et al. (2011) Gramene database in 2010: updates and extensions. Nucleic Acids Res 39: D1085–D1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thibaud-Nissen F, Prakash T, Buell CR, Shiu SH (2009) Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol 151: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.