Abstract
Quantitative Structure-Activity Relationship (QSAR) modeling and toxicogenomics are used independently as predictive tools in toxicology. In this study, we evaluated the power of several statistical models for predicting drug hepatotoxicity in rats using different descriptors of drug molecules, namely their chemical descriptors and toxicogenomic profiles. The records were taken from the Toxicogenomics Project rat liver microarray database containing information on 127 drugs (http://toxico.nibio.go.jp/datalist.html). The model endpoint was hepatotoxicity in the rat following 28 days of exposure, established by liver histopathology and serum chemistry. First, we developed multiple conventional QSAR classification models using a comprehensive set of chemical descriptors and several classification methods (k nearest neighbor, support vector machines, random forests, and distance weighted discrimination). With chemical descriptors alone, external predictivity (Correct Classification Rate, CCR) from 5-fold external cross-validation was 61%. Next, the same classification methods were employed to build models using only toxicogenomic data (24h after a single exposure) treated as biological descriptors. The optimized models used only 85 selected toxicogenomic descriptors and had CCR as high as 76%. Finally, hybrid models combining both chemical descriptors and transcripts were developed; their CCRs were between 68 and 77%. Although the accuracy of hybrid models did not exceed that of the models based on toxicogenomic data alone, the use of both chemical and biological descriptors enriched the interpretation of the models. In addition to finding 85 transcripts that were predictive and highly relevant to the mechanisms of drug-induced liver injury, chemical structural alerts for hepatotoxicity were also identified. These results suggest that concurrent exploration of the chemical features and acute treatment-induced changes in transcript levels will both enrich the mechanistic understanding of sub-chronic liver injury and afford models capable of accurate prediction of hepatotoxicity from chemical structure and short-term assay results.
Keywords: Quantitative Structure Activity Relationship (QSAR) modeling, toxicogenomics, biological descriptors, hepatotoxicity
Introduction
Hepatotoxicity is a major factor contributing to the high attrition rate of drugs. At least a quarter of the drugs are prematurely terminated or withdrawn from the market due to liver-related liabilities (1). As a result, modern drug development has evolved into a complex process relying on the iterative evaluation of multiple data sources to eliminate potentially harmful drug candidates as cheaply and as early as possible. In addition, high throughput, high content, and other data-rich experimental techniques, accompanied by the appropriate informatics tools, are rapidly incorporated into the toxicity testing.
Quantitative structure-activity relationship (QSAR) modeling is widely used as a computational tool that allows one to relate the potential activity (e.g., toxicity) of an agent to its structural features represented by multiple chemical descriptors. As with any multivariate statistical modeling, rigorous validation procedures are necessary to guard against overfitting and overestimating model predictivity (2). QSAR models have demonstrated good predictivity especially for specific endpoints such as solubility or binding affinity to a certain target. However, QSAR predictivity is generally poor in the case of a complex endpoint such as hepatotoxicity where the structure-activity relationship is less straightforward due to multiple mechanisms of action (3).
Toxicogenomics is now routinely used in drug and chemical safety evaluation, providing valuable mechanistic understanding of the molecular changes associated with the disease or treatment (4). In addition, its utility for predicting toxicity has been explored. Blomme et al. (5) developed models using transcriptional changes after short term (5 days) exposure to predict bile duct hyperplasia that otherwise required long-term in vivo experiments. Fielden et al. (6) developed a 37-gene classification model using microarray data following short-term (1–5 days) exposure to predict non-genotoxic hepatocarcinogenicity with over 80% accuracy. Zidek et al. (7) reported high accuracy with a 64-gene classifier for the prediction of acute hepatotoxicity. The Toxicogenomics Project in Japan, set up by the Ministry of Health, Labour and Welfare, National Institute of Health Sciences, and 15 pharmaceutical companies, has also identified several toxicogenomic signatures indicative of the various toxic modes of action such as phospholipidosis (8), glutathione depletion (9), bilirubin elevation (10), non-genotoxic hepatocarcinogenesis (11), and peroxisome proliferation (12).
Most previous studies on statistical modeling of toxicity used either chemical descriptors (conventional QSAR), or toxicogenomic profiles independently for model development. However, in our recent studies, we have demonstrated the benefits of hybrid classification models of in vivo carcinogenicity (13) and toxicity (14), and employing both chemical descriptors and biological assay data (treated as biological descriptors). In the first study of this type (13), we used the results of high-throughput screening assays of environmental chemicals along with their chemical descriptors to arrive at improved models of rat carcinogenicity. This approach was extended to predicting acute toxicity half-maximal lethal dose in rats using dose-response in vitro data as quantitative biological descriptors (14).
Following our hybrid (chemical and biological descriptors) data modeling paradigm, we sought to integrate QSAR and toxicogenomic data to develop classification models of hepatotoxicity using a data set of 127 drugs studied in the Japanese Toxicogenomics Project (15). We built classifiers combining chemical descriptors and toxicogenomic data alongside the conventional QSAR, as well as toxicogenomic models. Our objective was to investigate if chemical descriptors and biological descriptors, such as gene expression, could be complementary. In addition, we sought to enhance the interpretation of the models in terms of elucidating the chemical structural features and biological mechanisms associated with hepatotoxicity. We show that statistically significant and externally predictive models can be developed by combining chemical and biological descriptors and can be used to predict hepatotoxicity and prioritize chemicals for toxicogenomics and other in vivo studies.
Materials and Methods
Data
The chemical name, dosage, administration route and vehicle for the 127 compounds used in this study are summarized in Table I of the Supporting Information. The detailed protocol for the animal study was described previously (15). Briefly, 6-week old male Sprague–Dawley rats (Charles River Japan, Inc., Kanagawa, Japan) with five animals per group were used in the study. Animals were sacrificed 24 hours after a single dose, or 24 hours after repeat daily treatment for 28 days. Blood samples were collected from the abdominal aorta under ether anesthesia. Serum chemical indicators included alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), total bilirubin (TBIL), direct bilirubin (DBIL), and gamma-glutamyl transpeptidase (GGT). The livers were quickly removed following exsanguination and sections of the livers were placed in 10% phosphate-buffered formalin for histopathology. Formalin-fixed liver tissue was embedded in paraffin and sections were stained with hematoxylin and eosin and examined histopathologically under light microscopy. Remaining liver tissues from left lateral lobes were soaked in RNALater (Ambion Inc., Austin, TX) and stored at –80°C until used for microarray analysis. Detailed methods for microarray analysis were previously reported (15). Raw microarray data files with individual animal histopathological data are available (http://toxico.nibio.go.jp/datalist.html). In this study, toxicogenomic data obtained from rats treated with a single dose of a drug or vehicle for 24 hours was used. The experimental protocols were reviewed and approved by the Ethics Review Committee for Animal Experimentation of the National Institute of Health Sciences (Tokyo, Japan).
Liver histopathology and serum chemistry in animals treated for 28 days were assessed for the determination of the hepatotoxicity endpoint for prediction. Histopathology was graded by two trained pathologists in a blinded manner as follows: no change, very slight (minimal), slight, moderate, severe. Spontaneously observed lesions (e.g., minimal focal necrosis and microgranuloma) were not used for grading. The results of a histopathology analysis were considered positive if the grade recorded was other than “no change.” Table I of the Supporting Information lists serum chemistry and histopathology classification for each compound. A compound was denoted hepatotoxic if it exhibited histopathology characteristic of hepatotoxicity (e.g., hepatocellular necrosis/degeneration, inflammatory cells infiltration, bile duct proliferation, etc.) regardless of the findings from serum chemistry. Conversely, a compound was deemed non-hepatotoxic if it did not result in adverse histopathological features. When the histopathological observations were inconclusive (e.g., hepatocellular hypertrophy, vacuolization, etc.), serum chemistry data was considered. Under these circumstances, significant changes (Dunnett's test) in at least one enzyme marker would render the compound hepatotoxic. Otherwise, the compounds with inconclusive histopathology and normal serum chemistry were denoted non-hepatotoxic. In total, there were 53 (42%) hepatotoxic and 74 (58%) non-hepatotoxic compounds.
Curation of chemical data
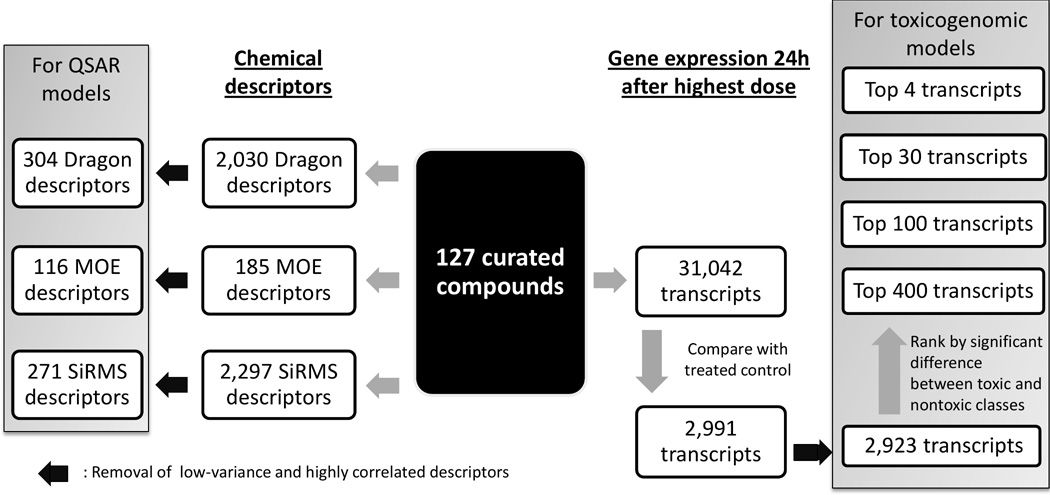
The data set was curated according to the procedures described by Fourches et al. (16). Briefly, counter ions and duplicates were removed, specific chemotypes such as aromatic and nitro groups were normalized using several cheminformatics software ChemAxon Standardizer (v.5.3, ChemAxon, Budapest, Hungary), HiT QSAR (17), and ISIDA (18). Following the automated curation, the data set was inspected manually and two inorganic compounds for which most chemical descriptors were not applicable, cisplatin and carboplatin, were removed. Chemical descriptors were calculated with Dragon (v.5.5, Talete SRL, Milan, Italy), Molecular Operating Environment (MOE, v.2009.10, Chemical Computing Group, Montreal, Canada) software. Simplex representation of molecular structure (SiRMS) descriptors were derived as detailed elsewhere (19). After range scaling (from 0 to 1), low variance (SD < 10−6) and highly correlated descriptors (if pair-wise r2>0.9, one of the pair was randomly removed) were removed. QSAR models were built separately using 304 Dragon, or 116 MOE, or 271 SiRMS descriptors (Figure 1).
Figure 1.
Workflow illustrating data curation and selection for modeling.
Selection of transcripts
Transcripts were selected for modeling using various feature selection methods. Of the 31,042 transcripts measured, we removed those consistently absent across all compounds. Then we extracted 2,991 transcripts with sufficient variation across all the compounds based on the following criteria: the largest change of any transcript over its untreated equivalent must exceed 1.5 fold and the smallest false discovery rate (Welch t-test) must be less than 0.05. Next, transcripts with low variance (all, or all but one value is constant) and high correlation (if pair-wise r2>0.9, one of the pair, chosen randomly, was removed) were excluded leaving 2,923 transcript variables (Figure 1) which were range scaled.
Then, supervised selection methods were used to filter genes differentially expressed between hepatotoxic and non-hepatotoxic compounds. Significance analysis of microarrays (SAM) (20), a permutation variant of the t-test commonly used for transcript selection, was used. Top ranked transcripts were filtered for modeling. Different sets of transcripts were selected for each modeling set and used in 5-fold external cross-validation to avoid selection bias introduced by a supervised selection process.
Modeling and validation
K nearest neighbors (kNN) (21), support vector machines (SVM) (22), random forest (RF) (23) and distance weighted discrimination (DWD) (24) machine learning techniques, designed to effectively handle high dimension-low sample size data, were used for modeling. The modeling workflow (2; 25) used both internal and external validation (Figure 1 of the Supporting Information). In a 5-fold external cross validation, 127 compounds were randomly partitioned into 5 subsets of nearly equal size. Each subset was paired with the remaining 80% of the compounds to form a pair of external and modeling sets respectively. The data within each modeling set were further divided into multiple pairs of training and test sets for internal validation.
Although models were built using the training set, model selection depended on their performance on both the training and test sets (i.e., internal validation) since training set accuracy alone is insufficient to establish robust and externally predictive models (26). The prediction outcome for each model was categorized as “0” for non-toxic compounds, or “1” for toxic ones. Selected models were then pooled into a consensus model by simple averaging and used to predict the hepatotoxicity of compounds in the external sets (i.e., external validation). The toxicity threshold was set at 0.5 unless otherwise mentioned, i.e., a compound is predicted to be non-toxic if a consensus mean is less than 0.5 and toxic otherwise.
Y-randomization test was employed to ensure that there was no chance correlation between selected descriptors and hepatotoxicity. After random permutation of the hepatotoxicity labels in the modeling sets, models were rebuilt following the same workflow and their CCR values for both training and test sets were collected and compared. This test was repeated at least three times. Models generated from the randomized labels were expected to perform significantly worse than those derived from the original data set.
All reported model predictivity measures, specificity, sensitivity, and correct classification rate, were obtained from 5-fold external cross validation. Specificity denotes the true negative rate, or the rate correctly predicted within the non-hepatotoxic class. Similarly, sensitivity, the true positive rate, measures the rate correctly predicted within the hepatotoxic class. CCR is the average of the rates correctly predicted within each class (CCR = [specificity + sensitivity]/2). Coverage is the percentage of compounds in the external set within the applicability domain (AD) of the model. The AD is a similarity threshold within which compounds can be reliably predicted (27).
Chemical and toxicogenomic descriptors found to be predictive were subsequently analyzed. Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) software was used for the functional analysis of the significant transcripts. The networks were constructed based on pre-defined molecular interactions in the Ingenuity database and the Ingenuity score was used to rank pathways for analysis. Chemicals were clustered by the selected toxicogenomic descriptors using an unsupervised self-organizing map (SOM) in R (Kohonen package). Chemical structural alerts for hepatotoxicity were identified using HiT QSAR (17) and verified with XCHEM (28). Briefly, XCHEM searches for common structural motifs within each class and ranks them by their relative frequencies.
Results
Model development
First, we developed QSAR models of sub-chronic (28 days of treatment) hepatotoxicity using various types of chemical descriptors (Table 1). Prediction performance was generally poor (55–61% CCR) across all descriptor types and classification methods. Three compounds (tannic acid, vancomycin and cyclosporine) with molecular weights exceeding 1,200 (median molecular weight of the data set was 285) were excluded from the data set, corresponding to a coverage of 98% for some of the models. Given the generally unpromising results of the QSAR models described in Table 1, further Combi-QSAR (29) efforts to systematically combine each descriptor type with each classification method were not attempted.
Table 1.
5-fold external cross validation prediction performance of QSAR models.
| Descriptors | Dragon | Dragon | MOE | SiRMS |
|---|---|---|---|---|
| Method | kNN | SVM | kNN | RF |
| Specificity SDa | 0.620.17 | 0.620.16 | 0.600.18 | 0.770.08 |
| Sensitivity SD | 0.560.14 | 0.480.17 | 0.560.16 | 0.450.14 |
| CCR SD | 0.590.11 | 0.550.09 | 0.580.12 | 0.610.10 |
| Coverage (%) | 98 | 98 | 98 | 100 |
SD refers to the standard deviation of the external predictivity measures (e.g. specificity) across the 5 folds.
Second, we developed classification models of sub-chronic (28 days of treatment) hepatotoxicity using liver toxicogenomic data obtained after a single dose treatment as a predictor of future toxicity. To find the optimal number of variables (transcripts), several sets of top ranking transcripts were selected (based on SAM analysis) for modeling by SVM, and the outcomes were compared (Figure 2). CCR ranged from 72% with top 4 significant transcripts per modeling fold to 78% with all 2,923 significant transcripts. An optimal model with a CCR of 76% was achieved when 30 transcripts per fold were used. These 5 sets of 30 transcripts per fold comprised of 85 unique transcripts across all folds, which may serve as predictive biomarkers (Table II of the Supporting Information for a complete list). We used these 85 transcripts to develop additional models employing other classification methods (Table 2). The RF model had the highest performance with a CCR of 76%. DWD was also applied to the full set of 2,923 transcripts and had CCR of 73%. The difference in performance between the QSAR and the toxicogenomic models was significant (p<0.001).
Figure 2.
CCR accuracy of the models with respect to the number of chemical descriptors and transcripts used. All models were generated by SVM classification with 5-fold external cross validation.
Table 2.
5-fold external cross validation prediction performance of toxicogenomic models based on the 85 selected transcripts (see Table II of the Supporting Information for a complete list).
| Method | kNN | SVM | DWD | RF |
|---|---|---|---|---|
| Specificity SD | 0.820.08 | 0.840.10 | 0.770.11 | 0.840.05 |
| Sensitivity SD | 0.570.07 | 0.670.12 | 0.620.17 | 0.660.20 |
| CCR SD | 0.700.06 | 0.760.09 | 0.690.11 | 0.760.10 |
| Coverage (%) | 95 | 99 | 99 | 100 |
Third, we developed hybrid models of sub-chronic (28 days of treatment) hepatotoxicity using both chemical descriptors and single-dose treatment toxicogenomic data as biological descriptors. We studied how SVM model predictivity was affected when both the number of chemical descriptors and the number of transcripts were varied. To that effect, SAM was applied to independently rank chemical descriptors and transcripts, after which, different portions of top ranked variables were used for SVM modeling. Figure 2 shows that the CCR of the hybrid models did not exceed that of the models based on toxicogenomic data alone. However, hybrid models identified both important chemical descriptors and transcripts for the enhanced interpretation of the modeling outcomes. We could not have reliably detected the important chemical features from the poorly fitted QSAR models. Adding transcripts boosted the predictivity of the hybrid models such that important chemical features were identified with greater confidence. Specifically, contributions of SiRMS descriptors used in RF hybrid models were interpreted using the approach of Polishchuk et al. (23) to uncover chemical substructures critical to hepatotoxicity. The substructures obtained through this analysis were compared to the predictions derived using XCHEM (28) and found to be concordant. The largest and most frequent substructures within each toxicity class are listed in Table 3 and provide evidence of structure-activity relationship in the hybrid model. All QSAR, toxicogenomics and hybrid models were significantly better than Y-randomized models (p<0.05 by Z-test), indicating that our models were not the result of chance correlations.
Table 3.
Structural alerts mapped onto example compounds. All compounds are toxic unless denoted with an “*”.
 |
The toxicity threshold of the consensus models was set to 0.5, below which the compounds were classified as non-toxic and above which they were classified as toxic. Because the compounds on the margin are typically predicted with less confidence, we sought to determine the effect of adjusting toxicity threshold on prediction performance. Figure 3A shows the distribution of QSAR-predicted values (using kNN method) for non-toxic and toxic compounds. Overall, the separation was poor due to a large proportion of non-toxic compounds that were predicted as toxic. While alternative thresholds yielding models with very high CCR may be selected (Figure 3C), severely reduced coverage of such models is a considerable drawback (Figure 3E). For example, setting two thresholds (dashed lines in Figure 3A), one at 0.36 (<0.36 are assigned non-toxic) and the second one at 0.56 (>0.56 are assigned as toxic) increased CCR to 68%, as compared to 59% with a single threshold of 0.5. However, the coverage of such model was only 80% because the compounds whose predicted activities were between 0.36 and 0.56 could no longer be classified. Conversely, the toxicogenomic model developed with kNN showed good separation between toxic and non-toxic compounds (Figure 3B). A change in thresholds had a minor effect on the model’s CCR and coverage (Figures 3D and F), showing that a single threshold was sufficient and optimization of the activity thresholds would not be necessary. The optimal thresholds will be useful in the prediction of additional external compounds.
Figure 3.
External prediction results of the QSAR (A, C, E) and toxicogenomic (B, D, F) models by kNN using different classification criteria. Distribution of the predicted values (A and B) and heat maps illustrating classification accuracy (C and D, CCR) and coverage (E and F, percent chemicals within the applicability domain) results are shown. Dashed (A and B) and diagonal (B–F) lines denote a default single-threshold classification (threshold=0.5). An example of a double-threshold classification (non-toxic if activity < 0.36; toxic if activity > 0.56) is shown by the dashed lines (C and E).
Model interpretation
Toxicogenomic data-based models were most predictive of hepatotoxicity. To explore the biological significance and the mechanistic relevance of the selected 85 transcripts (64 up-regulated and 21 down-regulated), functional pathway analysis was performed. Hepatic nuclear factor 4α (Hnf4a)- and v-myc myelocytomatosis viral oncogene homolog (Myc)-centered interactomes were the two highest ranked networks involving large numbers of the 64 selected up-regulated genes (Figures 5A–B and Table IVa of the Supporting Information). Canonical pathway analysis revealed that the eukaryotic initiation factor (Eif) 2 signaling pathway responsible for protein translation was up-regulated (Table IVb of the Supporting Information). Among the down-regulated genes, the network involving cellular function and maintenance including transporters and inflammatory responses was the highest ranked network (Figure 5C and Table IVc of the Supporting Information). Canonical pathway analysis also revealed that many down-regulated genes were involved in the complement pathway (Table IVd of the Supporting Information).
Figure 5.

Molecular networks representing the toxicogenomic predictors of hepatotoxicity. Hnf4a-centered (A), Myc-centered (B), cellular function and maintenance-related (C) interactomes were selected as the highest ranked networks among the 64 up- or 21 down-regulated genes used in modeling. Red and green represent molecules up-regulated or down-regulated, respectively, by the hepatotoxic compounds. Ellipse, square, triangle, trapezoid, lozenge and circle represent transcription regulator, cytokine, kinase, transporter, enzyme and other molecules, respectively. Arrows indicate molecular interactions while lines indicate binding. Dashed arrows or lines indicate indirect interactions or binding. See Tables IIIa–d in the Supporting Information for a complete list of networks.
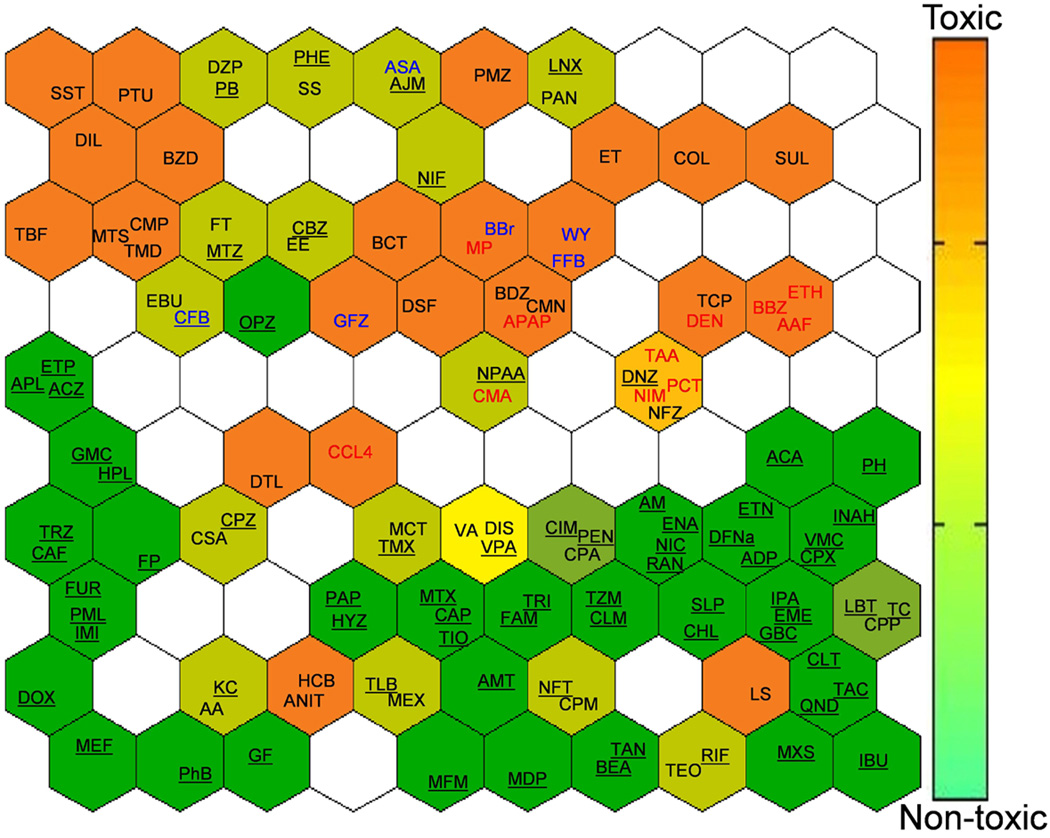
In addition, we used an unsupervised self-organizing map to cluster chemicals based on their gene expression profiles (Figure 6 and Figure 2 of the Supporting Information). The objective was to uncover commonalities within clusters with similar gene expression profiles. As expected, the non-hepatotoxic agents were tightly clustered (green background). Among the hepatotoxic drugs (orange background), there were several clusters of compounds which may act through similar mechanisms of action. For example, oxidative stress-inducing agents (red text) such as acetaminophen, methapyriline, and nimesulide; and peroxisome proliferator-activated alpha (PPARα) agonists (blue text) such as fenofibrate, WY-14643, benzbromarone, clofibrate, and gemfibrozil formed two sub-clusters among the hepatotoxicants. The model-selected 85 transcripts were sufficient to cluster the drugs into toxicologically meaningful groups with similar modes of hepatotoxicity.
Figure 6.
Self-organizing map of the compounds clustered by the expression of the 85 selected transcripts. Non-toxic (underlined) compounds are tightly clustered in the bottom right. PPARα activating and oxidative stress-inducing chemicals are colored in blue and red, respectively.
Understanding this difference in performance between the QSAR and the toxicogenomic models warrants an in-depth examination of the spatial distribution of compounds in their chemical and toxicogenomic space. Principal component analysis of the chemical features (Dragon descriptors, Figure 4A) and toxicogenomic data (85 selected transcripts, Figure 4B) demonstrated that the separation between non-toxic and toxic classes was poor in the chemical space. Table IIIa of the Supporting Information lists 40 most chemically similar pairs of compounds. Half of them had opposite toxicities. Conversely, among pairs of compounds with the most similar gene expression profiles, only 23% exhibited opposite toxicities (Table IIIb of the Supporting Information). In other words, pairs of compounds with similar gene expression profiles were more likely to have the same hepatotoxicity than pairs of chemically similar compounds.
Figure 4.
Principal component analysis of the chemical (A) and toxicogenomic (B) descriptors. Toxic and non-toxic compounds are colored red and black, respectively. Compounds mispredicted by the toxicogenomic model but correctly predicted by the QSAR model are marked as crosses (×) or circles (●) otherwise. An example of a non-toxic compound (danazol, DNZ) which has distant toxic toxicogenomic neighbors but close non-toxic chemical neighbors is shown.
The best hybrid model had similar performance to the best toxicogenomic model (76–77% CCR), differing only in the predictions of 3 compounds (ajmaline, griseofulvin, propylthiouracil). Examining the compounds in their chemical and toxicogenomic spaces separately (by QSAR and toxicogenomic models respectively) revealed instances for which the models were complementary and thus boosted the overall predictivity. When both QSAR and toxicogenomics models were in agreement, it implied greater reliability of the prediction (Table 4). When they were discordant, deferring to the toxicogenomics model (superior to the QSAR model) would more likely return correct predictions. However, of note were 19 compounds (shaded in Table 4) mispredicted by the toxicogenomics model but correctly predicted by the QSAR model. PCA showed that many of these compounds (denoted by crosses in Figures 4A and B) were surrounded by toxicogenomic neighbors of opposite toxicities (Figure 4B) but have chemical neighbors of similar toxicities (Figure 4A). For example, non-toxic danazol has toxic toxicogenomic neighbors (Figure 4B) but non-toxic chemical neighbors (Figure 4A). Some of these mispredicted compounds, e.g. gemfibrozil (PPARα activator) and lomustine (genotoxic hepatocarcinogen), exhibit late-onset toxicity which could explain the failure of 24h expression profiles to capture relevant changes and consequently, to predict their 28-day hepatotoxicity.
Table 4.
Confusion table showing predictions by QSAR model and toxicogenomics model. Compounds mis-predicted by the toxicogenomics model but correctly predicted by the QSAR model are identified in italicized font. Compounds mis-predicted by both the QSAR model, and by the toxicogenomics model are underlined.
| Toxicogenomic model (85 transcripts, kNN) | Predicted as toxic |
Actually non-toxic
|
Actually toxic
|
Actually non-toxic
|
Actually toxic
|
| Predicted as non-toxic |
Actually non-toxic
|
Actually toxic
|
Actually non-toxic
|
Actually toxic
|
|
| Predicted as non-toxic | Predicted as toxic | ||||
| QSAR model (Dragon descriptors, kNN) | |||||
While toxicogenomics was in general better at discriminating between toxic and non-toxic drugs, there remained a few exceptions whose chemical neighbors were more indicative of their hepatotoxicity. In such cases, overall predictivity would be augmented by complementing toxicogenomic models with QSAR models.
Discussion
Our study showed that chemical features and toxicogenomic data were useful and relevant for the development of classification models for understanding and predicting hepatotoxicity. High classification accuracy of toxicogenomic models supports the use of early transcriptional response as an indicator for long term toxicity and for understanding a potential mode of action. Even though chemical-based QSAR models were less predictive, they will continue to be used for initial virtual screening in cases where no experimental data (e.g., toxicogenomics) are available. By developing hybrid models using both chemical descriptors and toxicogenomic data, we identified both chemical features and transcripts, which provided additional insights into understanding drug-induced liver injury.
Biological pathways involved in liver injury
Toxicogenomic data from single exposure were not only useful for the classification of 28-day liver injury phenotype, but they also provided important mechanistic insights into pathways that may lead to long-term toxicity. Pathway analysis showed that the 85 most predictive transcripts were in Hnf4α-, Myc-, and Eif2-centered networks, all of which have been implicated in hepatotoxicity. Hnf4α, a transcriptional factor of the nuclear hormone receptor family, is known to play an important role in the liver function, morphological and functional differentiation of hepatocytes, cell proliferation and detoxification (30). Although the Hnf4α gene itself was not among the selected transcripts, Hnf4α-regulated genes were up-regulated in the early stage of hepatocellular injury.
In addition, Hnf4α is essential for controlling the acute phase response of the liver induced by endoplasmic reticulum (ER) stress (31). ER stress is a common response to many toxicants and under conditions of severe or prolonged ER stress, apoptosis is triggered by accumulation of incompletely assembled or misfolded proteins (32). Activation of Eif2 signaling pathway is widely recognized as a key contributor to ER stress. In the present study, we found the characteristic up-regulation of several genes involved in Eif2 signaling pathway after treatment with several hepatotoxicants, such as Eif2 subunit 1 alpha (Eif2s1), Eif3 subunits G (Eif3G) and J (Eif3J), and Eif4a1. Thus, our analysis provided additional supporting evidence that Eif2 signaling pathway may be a common mechanism involved in early liver damage through ER stress.
Myc is a transcription factor which regulates cell proliferation, differentiation and apoptosis (33). In the present study, we found up-regulations of several genes in the Myc-centered network including transcription factors nucleophosmin 1 (Npm1), TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor (Taf9), Eif4a1, and general transcription factor IIIC polypeptide 3 (Gtf3c3). While further studies are needed to link the effects of individual chemicals to transcriptional changes in the Myc-centered network, our analysis shows that these transcripts may be important early predictive biomarkers for sub-chronic hepatocellular injury.
Biological pathway analysis revealed down-regulation of genes involved in cellular function and maintenance, consisting of transporters and inflammatory response, such as the complement system pathway. Abnormal homeostasis and cellular function are often associated with hepatotoxicity. In particular, coagulopathy is often involved because many factors in coagulation system are synthesized in the liver. Recently, toxicogenomic biomarkers for diagnosis and prognosis of hepatotoxicity-related coagulation abnormalities have been reported (34). Our results further support that malfunction of the coagulation system is a common feature in liver injury and the down-regulation of complement 8, beta polypeptide (C8b), complement 9 (C9), and complement factor B (Cfb) may be an early indicator of impaired liver function by different types of drugs.
Many of the 85 selected transcripts have also been previously implicated with liver diseases by the same chemicals on the Comparative Toxicogenomics Database (http://ctd.mdibl.org/). For instance, ubiquitin specific peptidase 10 (Usp10) has been associated with the Myc-centered network in acetaminophen-induced liver toxicity (35). It is also closely related to ubiquitin specific peptidase 2 (Usp2) which is among the 37 genes used to derive a toxicogenomic model for hepatotumorigenesis by Fielden et al. (6). The agreement with previous findings lends credence to our selected list of transcripts as biomarkers for hepatotoxicity.
Hybrid models afford exploration of chemical structural alerts
Development of QSAR models of hepatotoxicity for structurally diverse chemicals is a challenge (36) and the results of this study show that a correct classification rate of such models ranged between 55 and 61%. Thus, interpretation of such models with regards to the potential chemical “structural alerts” for hepatotoxicity may be futile. However, when chemical descriptors and toxicogenomic data were used together to develop hybrid models, significantly higher predictive accuracy (as high as 77%) of the models provided additional confidence for considering the chemical fragments selected by the models as potentially predictive of an increased risk of liver toxicity. By examining the chemical substructures suggested by the predictive models (see Table 3), we observe that an unbiased feature selection identified several well-known toxicophores. This finding provides a strong indication of the value of the hybrid modeling for identification of the toxicophores as compared to the traditional QSAR which is plagued by a much weaker predictive power.
Substructure A (Acetanilide). Toxic species formed: N-hydroxylamines and nitroso compounds
The acetanilide substructure was present in several hepatotoxic drugs, as well as the non-toxic phenylbutazone. The acetanilide substructure is especially susceptible to N-oxidation (37). The N-hydroxylamine and nitroso products are highly reactive. However, some compounds may be toxic due to activation at sites outside of the acetanilide substructure. For example, acetaminophen owes much of its toxicity to the quinone imine metabolite despite its chemical similarity with phenacetin. Its only difference from phenacetin is its 4-hydroxyl group which is preferentially oxidized by CYP2E1 to the reactive quinone imine. In phenacetin and bucetin, the 4-hydroxyl group is replaced by an alkoxyl substituent which renders them less susceptible to quinone formation and more likely to be activated by N-hydroxylation (38). Phenylbutazone also undergoes another transformation (aromatic hydroxylation) instead of N-hydroxylation (39). This probably explains its lack of rat hepatotoxicity in this study despite containing the acetanilide substructure.
Substructure B (Thioamide). Toxic species formed: sulfur species of various oxidation states
Our models showed that the presence of thioamide (Table 3, substructure B) is associated with hepatotoxicity. Thiocarbonyls are often oxidized or desulfurated by to produce toxic sulfur-containing species. Thioacetamide S-oxide is highly polar and forms adducts with proteins (40). Disulfiram, despite being a dithiocarbamate instead of a thioamide, also forms a sulfoxide that binds to proteins and inhibits their activity. Such protein binding is also responsible for disulfiram’s therapeutic inhibition of aldehyde dehydrogenase (41). The only non-toxic drug that has this substructure was methimazole. Although methimazole was defined as non-hepatotoxic in this study, it has been reported to yield atomic sulfur species that bind and inhibit P450 activity, possibly leading to liver necrosis (42).
Substructure C (Alkyl chloride). Toxic species formed: alkyl radicals
Hepatotoxicity of alkyl chloride compounds has been attributed to the homolytic cleavage of the C-Cl bond which produces damaging free radicals. This is a well-studied phenomenon best exemplified by carbon tetrachloride and its alkyl halide analogues such as chloroform and bromotrichloromethane (43). However, other chlorinated alkanes studied here, cyclophosphamide, lomustine and chloramphenicol, do not share the same toxic mechanism as carbon tetrachloride and cannot be attributed to the C-Cl bond. For instance, the ultimate toxicant responsible for cyclophosphamide hepatotoxicity is acrolein which is formed independently of the alkyl chloride group.
Substructure D (Styrene). Toxic species formed: epoxides
The non-aryl double bond in substructure D when it is part of a benzofuran or benzopyran is especially prone to epoxide formation (44). Such epoxides often form DNA and protein adducts (45). Coumarin’s toxicity requires the formation of an epoxide which is followed by subsequent rearrangement of the epoxide to o-hydroxyphenylacetaldehyde which is considered to be the hepatotoxic intermediate (46). Hence, it is comparatively more toxic in rat than in humans because of the rat’s metabolism via the 3,4-epoxide (47) while in humans, coumarin primarily undergoes aromatic hydroxylation instead of forming the above mentioned epoxide (46; 48). The three benzofurans in our study, benziodarone, benzbromarone and amiodarone, are known hepatotoxic agents whose toxicity has been attributed to the 2-substituted benzofuran (44). Although amiodarone was not found to be hepatotoxic based on its 28-day histopathology and serum chemistry results, hepatocellular vacuolization indicative of phospholipidosis was noted (Table I of the Supporting Information).
Limitations
The performance of QSAR models generally suffers when predicting complex toxicity endpoints such as hepatotoxicity, a phenotype with several complex mechanisms. There are numerous examples of chemically similar compounds with widely divergent liver effects. While ibuprofen is safe in humans, ibufenac, lacking a methyl group, is toxic (36). In our data set, non-toxic caffeine and toxic theophylline differ by a methyl group. This phenomenon is known as an “activity cliff” where very similar molecules possess disparate activities, such that the profile of activity plotted against compound is akin to a rugged landscape with many cliffs (49). QSAR can be realistically applied if there are enough compounds to adequately represent the complex activity landscape. Unfortunately, this was not the case for our data set. The high proportion (50%) of opposite activities among chemically similar pairs compounded by the lack of congeners in our chemically diverse set posed further challenges to QSAR modeling. Hence, it was not surprising that the CCR of the QSAR models could barely exceed 60% in predicting the biologically complex hepatotoxicity endpoint.
In conclusion, this study shows that while QSAR and toxicogenomics are both important predictive tools on their own, concomitant exploration in chemical and transcriptomics spaces, through hybrid models, will elicit deeper insight. Consistent with results from other toxicogenomics studies, we showed that toxicogenomics is predictive and provides valuable mechanistic information. The pathways suggested several mechanisms such as ER stress and coagulopathy that could be related to hepatotoxicity. As QSAR is entirely computational and obviates the need for experiments, it will remain an important virtual screening tool. In agreement with previous reports of improved predictivity after adding in vitro assays as biological descriptors into QSAR modeling. The performance of QSAR can be augmented by the addition of toxicogenomic data which helps to predict complex biological endpoints unexplained by chemical descriptors alone. Consequently, structural alerts can be identified with greater confidence from the better fitted hybrid models. Although hybrid models were not more predictive than toxicogenomics models, examining the compounds in multiple spaces afforded additional neighbors for inferring hepatotoxicity. The development of novel consensus models may be needed to further exploit the complementarities between QSAR and toxicogenomic models.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Ministry of Health, Labor and Welfare of Japan (H14-Toxico-001, H19-Toxico-001), NIH (GM66940, ES015241) and EPA (RD 83382501, RD83272001). The research described in this article has not been subjected to each agency’s policy review and therefore does not necessarily reflect their views, and no official endorsement should be inferred.
Footnotes
Supporting Information Available: Entire data set of compounds (identified by their CAS numbers) with the dosage information, histopathology and serum chemistry results; list of predictive gene biomarkers; list of pathways involving the predictive gene biomarkers; list of compounds paired by chemical and transcriptional similarities. This material is available via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schuster D, Laggner C, Langer T. Why drugs fail--a study on side effects in new chemical entities. Curr. Pharm Des. 2005;11:3545–3559. doi: 10.2174/138161205774414510. [DOI] [PubMed] [Google Scholar]
- 2.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inf. 2010;29:1868–1751. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 3.Hou T, Wang J. Structure-ADME relationship: still a long way to go? Expert Opin. Drug Metab Toxicol. 2008;4:759–770. doi: 10.1517/17425255.4.6.759. [DOI] [PubMed] [Google Scholar]
- 4.Cui Y, Paules RS. Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics. 2010;11:573–585. doi: 10.2217/pgs.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomme EA, Yang Y, Waring JF. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol Lett. 2009;186:22–31. doi: 10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- 7.Zidek N, Hellmann J, Kramer PJ, Hewitt PG. Acute hepatotoxicity: a predictive model based on focused illumina microarrays. Toxicol Sci. 2007;99:289–302. doi: 10.1093/toxsci/kfm131. [DOI] [PubMed] [Google Scholar]
- 8.Hirode M, Ono A, Miyagishima T, Nagao T, Ohno Y, Urushidani T. Gene expression profiling in rat liver treated with compounds inducing phospholipidosis. Toxicol Appl Pharmacol. 2008;229:290–299. doi: 10.1016/j.taap.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Kiyosawa N, Uehara T, Gao W, Omura K, Hirode M, Shimizu T, Mizukawa Y, Ono A, Miyagishima T, Nagao T, Urushidani T. Identification of glutathione depletion-responsive genes using phorone-treated rat liver. J Toxicol Sci. 2007;32:469–486. doi: 10.2131/jts.32.469. [DOI] [PubMed] [Google Scholar]
- 10.Hirode M, Horinouchi A, Uehara T, Ono A, Miyagishima T, Yamada H, Nagao T, Ohno Y, Urushidani T. Gene expression profiling in rat liver treated with compounds inducing elevation of bilirubin. Hum. Exp Toxicol. 2009;28:231–244. doi: 10.1177/0960327109104528. [DOI] [PubMed] [Google Scholar]
- 11.Uehara T, Hirode M, Ono A, Kiyosawa N, Omura K, Shimizu T, Mizukawa Y, Miyagishima T, Nagao T, Urushidani T. A toxicogenomics approach for early assessment of potential non-genotoxic hepatocarcinogenicity of chemicals in rats. Toxicology. 2008;250:15–26. doi: 10.1016/j.tox.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Tamura K, Ono A, Miyagishima T, Nagao T, Urushidani T. Profiling of gene expression in rat liver and rat primary cultured hepatocytes treated with peroxisome proliferators. J Toxicol Sci. 2006;31:471–490. doi: 10.2131/jts.31.471. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Rusyn I, Richard A, Tropsha A. Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity. Environ Health Perspect. 2008;116:506–513. doi: 10.1289/ehp.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedykh A, Zhu H, Tang H, Zhang L, Richard A, Rusyn I, Tropsha A. Use of in Vitro HTS-Derived Concentration-Response Data as Biological Descriptors Improves the Accuracy of QSAR Models of in Vivo Toxicity. Environ Health Perspect. 2011;119:364–370. doi: 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uehara T, Ono A, Maruyama T, Kato I, Yamada H, Ohno Y, Urushidani T. The Japanese toxicogenomics project: application of toxicogenomics. Mol Nutr. Food Res. 2010;54:218–227. doi: 10.1002/mnfr.200900169. [DOI] [PubMed] [Google Scholar]
- 16.Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuz'min VE, Artemenko AG, Muratov EN. Hierarchical QSAR technology based on the Simplex representation of molecular structure. J Comput. Aided Mol Des. 2008;22:403–421. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 18.Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov'yev V, Hoonakker F, Tetko IV, Marcou G. ISIDA - Platform for Virtual Screening Based on Fragment and Pharmacophoric Descriptors. Curr. Comput. Aided Drug Des. 2008;4:191–198. [Google Scholar]
- 19.Muratov EN, Artemenko AG, Varlamova EV, Polischuk PG, Lozitsky VP, Fedchuk AS, Lozitska RL, Gridina TL, Koroleva LS, Sil'nikov VN, Galabov AS, Makarov VA, Riabova OB, Wutzler P, Schmidtke M, Kuz'min VE. Per aspera ad astra: application of Simplex QSAR approach in antiviral research. Future. Med Chem. 2010;2:1205–1226. doi: 10.4155/fmc.10.194. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng W, Tropsha A. Novel variable selection quantitative structure--property relationship approach based on the k-nearest-neighbor principle. J. Chem. Inf. Comput. Sci. 2000;40:185–194. doi: 10.1021/ci980033m. [DOI] [PubMed] [Google Scholar]
- 22.Fan RE, Chen PH, Lin CJ. Working set selection using the second order information for training SVM. J Mach Leaning Res. 2005;6:1889–1918. [Google Scholar]
- 23.Polishchuk PG, Muratov EN, Artemenko AG, Kolumbin OG, Muratov NN, Kuz'min VE. Application of random forest approach to QSAR prediction of aquatic toxicity. J Chem Inf. Model. 2009;49:2481–2488. doi: 10.1021/ci900203n. [DOI] [PubMed] [Google Scholar]
- 24.Marron JS, Todd MJ, Ahn J. Distance Weighted Discrimination. J. Am. Stat. Assoc. 2007;102:1267–1271. doi: 10.1198/jasa.2010.tm08487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tropsha A, Golbraikh A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharm Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- 26.Golbraikh A, Tropsha A. Beware of q2! J Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 27.Tropsha A, Gramatica P, Gombar VK. The Importance of Being Earnest: Validation is the Absolute Essential for Successful Application and Interpretation of QSPR Models. Quant Struct Act Relat Comb Sci. 2003;22:69–77. [Google Scholar]
- 28.Sedykh AY, Klopman G. A structural analogue approach to the prediction of the octanol-water partition coefficient. J Chem Inf. Model. 2006;46:1598–1603. doi: 10.1021/ci0505269. [DOI] [PubMed] [Google Scholar]
- 29.Kovatcheva A, Golbraikh A, Oloff S, Xiao YD, Zheng W, Wolschann P, Buchbauer G, Tropsha A. Combinatorial QSAR of ambergris fragrance compounds. J Chem. Inf. Comput. Sci. 2004;44:582–595. doi: 10.1021/ci034203t. [DOI] [PubMed] [Google Scholar]
- 30.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 31.Luebke-Wheeler J, Zhang K, Battle M, Si-Tayeb K, Garrison W, Chhinder S, Li J, Kaufman RJ, Duncan SA. Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology. 2008;48:1242–1250. doi: 10.1002/hep.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji C, Kaplowitz N. ER stress: can the liver cope? J Hepatol. 2006;45:321–333. doi: 10.1016/j.jhep.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F constitute a feedforward loop that regulates cell growth: implications for anticancer therapy. Cancer Res. 2009;69:7491–7494. doi: 10.1158/0008-5472.CAN-09-0813. [DOI] [PubMed] [Google Scholar]
- 34.Hirode M, Omura K, Kiyosawa N, Uehara T, Shimuzu T, Ono A, Miyagishima T, Nagao T, Ohno Y, Urushidani T. Gene expression profiling in rat liver treated with various hepatotoxic-compounds inducing coagulopathy. J Toxicol Sci. 2009;34:281–293. doi: 10.2131/jts.34.281. [DOI] [PubMed] [Google Scholar]
- 35.Beyer RP, Fry RC, Lasarev MR, McConnachie LA, Meira LB, Palmer VS, Powell CL, Ross PK, Bammler TK, Bradford BU, Cranson AB, Cunningham ML, Fannin RD, Higgins GM, Hurban P, Kayton RJ, Kerr KF, Kosyk O, Lobenhofer EK, Sieber SO, Vliet PA, Weis BK, Wolfinger R, Woods CG, Freedman JH, Linney E, Kaufmann WK, Kavanagh TJ, Paules RS, Rusyn I, Samson LD, Spencer PS, Suk W, Tennant RJ, Zarbl H. Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol Sci. 2007;99:326–337. doi: 10.1093/toxsci/kfm150. [DOI] [PubMed] [Google Scholar]
- 36.Rodgers AD, Zhu H, Fourches D, Rusyn I, Tropsha A. Modeling liver-related adverse effects of drugs using knearest neighbor quantitative structure-activity relationship method. Chem Res Toxicol. 2010;23:724–732. doi: 10.1021/tx900451r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loew GH, Goldblum A. Metabolic activation and toxicity of acetaminophen and related analogs. A theoretical study. Mol Pharmacol. 1985;27:375–386. [PubMed] [Google Scholar]
- 38.Peters JM, Morishima H, Ward JM, Coakley CJ, Kimura S, Gonzalez FJ. Role of CYP1A2 in the toxicity of long-term phenacetin feeding in mice. Toxicol Sci. 1999;50:82–89. doi: 10.1093/toxsci/50.1.82. [DOI] [PubMed] [Google Scholar]
- 39.Aarbakke J, Bakke OM, Milde EJ, Davies DS. Disposition and oxidative metabolism of phenylbutazone in man. Eur. J Clin Pharmacol. 1977;11:359–366. doi: 10.1007/BF00566533. [DOI] [PubMed] [Google Scholar]
- 40.Porter WR, Neal RA. Metabolism of thioacetamide and thioacetamide S-oxide by rat liver microsomes. Drug Metab Dispos. 1978;6:379–388. [PubMed] [Google Scholar]
- 41.Shen ML, Johnson KL, Mays DC, Lipsky JJ, Naylor S. Determination of in vivo adducts of disulfiram with mitochondrial aldehyde dehydrogenase. Biochem Pharmacol. 2001;61:537–545. doi: 10.1016/s0006-2952(00)00586-4. [DOI] [PubMed] [Google Scholar]
- 42.Lee PW, Neal RA. Metabolism of methimazole by rat liver cytochrome P-450-containing monoxygenases. Drug Metab Dispos. 1978;6:591–600. [PubMed] [Google Scholar]
- 43.Rechnagel RO, Glende EA., Jr Carbon tetrachloride hepatotoxicity: an example of lethal cleavage. CRC Crit Rev Toxicol. 1973;2:263–297. doi: 10.3109/10408447309082019. [DOI] [PubMed] [Google Scholar]
- 44.Kaufmann P, Torok M, Hanni A, Roberts P, Gasser R, Krahenbuhl S. Mechanisms of benzarone and benzbromarone-induced hepatic toxicity. Hepatology. 2005;41:925–935. doi: 10.1002/hep.20634. [DOI] [PubMed] [Google Scholar]
- 45.Adam W, Ahrweiler M, Saha-Moller CR, Sauter M, Schonberger A, Epe B, Muller E, Schiffmann D, Stopper H, Wild D. Genotoxicity studies of benzofuran dioxetanes and epoxides with isolated DNA, bacteria and mammalian cells. Toxicol Lett. 1993;67:41–55. doi: 10.1016/0378-4274(93)90045-y. [DOI] [PubMed] [Google Scholar]
- 46.Vassallo JD, Hicks SM, Daston GP, Lehman-McKeeman LD. Metabolic detoxification determines species differences in coumarin-induced hepatotoxicity. Toxicol Sci. 2004;80:249–257. doi: 10.1093/toxsci/kfh162. [DOI] [PubMed] [Google Scholar]
- 47.Lake BG, Gray TJ, Evans JG, Lewis DF, Beamand JA, Hue KL. Studies on the mechanism of coumarin-induced toxicity in rat hepatocytes: comparison with dihydrocoumarin and other coumarin metabolites. Toxicol Appl Pharmacol. 1989;97:311–323. doi: 10.1016/0041-008x(89)90336-0. [DOI] [PubMed] [Google Scholar]
- 48.Felter SP, Vassallo JD, Carlton BD, Daston GP. A safety assessment of coumarin taking into account species-specificity of toxicokinetics. Food Chem Toxicol. 2006;44:462–475. doi: 10.1016/j.fct.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Maggiora GM. On outliers and activity cliffs--why QSAR often disappoints. J Chem Inf. Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.