Background: Many proteins are modified by lysine methylation.
Results: It is shown that the previously uncharacterized enzyme METTL20 methylates electron transfer flavoprotein β (ETFβ), thereby inhibiting its ability to mediate electron transfer from acyl-CoA dehydrogenases.
Conclusion: METTL20-mediated methylation modulates the function of ETFβ.
Significance: The first mitochondrial lysine-specific protein methyltransferase in animals is reported, and the resulting methylation is shown to have functional consequences.
Keywords: Enzyme Catalysis, Mitochondrial Metabolism, Post-translational Modification (PTM), Protein Methylation, Protein Targeting, Electron Transfer Flavoprotein
Abstract
Proteins are frequently modified by post-translational methylation of lysine residues, catalyzed by S-adenosylmethionine-dependent lysine methyltransferases (KMTs). Lysine methylation of histone proteins has been extensively studied, but it has recently become evident that methylation of non-histone proteins is also abundant and important. The human methyltransferase METTL20 belongs to a group of 10 established and putative human KMTs. We here found METTL20 to be associated with mitochondria and determined that recombinant METTL20 methylated a single protein in extracts from human cells. Using an methyltransferase activity-based purification scheme, we identified the β-subunit of the mitochondrially localized electron transfer flavoprotein (ETFβ) as the substrate of METTL20. Furthermore, METTL20 was found to specifically methylate two adjacent lysine residues, Lys200 and Lys203, in ETFβ both in vitro and in cells. Interestingly, the residues methylated by METTL20 partially overlap with the so-called “recognition loop” in ETFβ, which has been shown to mediate its interaction with various dehydrogenases. Accordingly, we found that METTL20-mediated methylation of ETFβ in vitro reduced its ability to receive electrons from the medium chain acyl-CoA dehydrogenase and the glutaryl-CoA dehydrogenase. In conclusion, the present study establishes METTL20 as the first human KMT localized to mitochondria and suggests that it may regulate cellular metabolism through modulating the interaction between its substrate ETFβ and dehydrogenases. Based on the previous naming of similar enzymes, we suggest the renaming of human METTL20 to ETFβ-KMT.
Introduction
Methylation is a commonly occurring biochemical reaction within the cell, and a number of different S-adenosylmethionine (SAM)-dependent2 methyltransferases (MTases) catalyze the transfer of a methyl group from SAM to a large set of substrate molecules, ranging from small metabolites to the macromolecules, RNA, DNA, and proteins (1). In proteins, lysines and arginines are the primary modification sites, and a lysine residue can receive up to three methyl groups, thereby potentially acquiring several different methylation states. In particular, methylation of histone proteins has been extensively studied, and these methyl modifications play important roles in defining transcriptional activity and chromatin state (2). Numerous lysine methylations have also been reported in non-histone proteins, but the knowledge is so far limited regarding both the functional significance of these modifications and the enzymes introducing them (3, 4).
Among the ∼200 MTases predicted to be encoded by the human genome, most remain yet uncharacterized with respect to the targeted substrate and the functional role (1). The two largest groups of MTases are the so-called seven β-strand (7BS) MTases, which have a characteristic core fold of seven β-strands, and the SET proteins, which contain a defining SET domain (1, 5). Although 7BS MTase have been shown to target a wide range of small molecules and macromolecular substrates, the SET proteins mainly encompass Lys (K) specific MTases (KMTs), many of which target histones. However, recent studies have revealed that also several 7BS MTases methylate Lys residues in proteins. In particular, the group of 7BS MTases denoted MTF16 (methyltransferase family 16), which has 10 human members, was recently shown to consist of KMTs (6). The activity of five of these human enzymes has now been established: CaM-KMT methylates calmodulin (7), VCP-KMT (METTL21D) targets the ATP-dependent chaperone VCP (6), HSPA-KMT (METTL21A) methylates several HSPA (Hsp70) proteins (8, 9), METTL22 methylates Kin17 (9), and eEF2-KMT (FAM86A) methylates eukaryotic elongation factor 2 (eEF2) (10). In addition, DOT1L and METTL10, which are 7BS MTases but not MTF16 members, have been shown to methylate histone H3 and eukaryotic elongation factor 1A (eEF1A), respectively (11, 12).
Electron transfer flavoprotein (ETF) is a FAD-containing, mitochondrial protein composed of two subunits: α and β (ETFα and ETFβ, respectively) (13–16). ETF acts as a mobile electron carrier that shuttles electrons between several FAD-containing dehydrogenases present in the mitochondrial matrix and the membrane-bound ETF:quinone oxidoreductase (17). Electrons are then passed on to the ubiquinone pool of the mitochondrial respiratory chain. Altogether, there are 13 human dehydrogenases that interact with ETF, and it has been proposed that a “recognition loop” within the ETFβ chain, composed of residues 191–200, is responsible for interaction with the dehydrogenases (18). Some of these dehydrogenases, e.g. medium chain acyl-CoA dehydrogenase (MCAD), are involved in β-oxidation of fatty acids (19), whereas others, including glutaryl-CoA dehydrogenase (GCDH) and isovaleryl-CoA dehydrogenase, are involved in the oxidation of amino acids (20, 21). Yet another group is involved in oxidative reactions in the pathway for degradation of choline to glycine, resulting in the transfer of one-carbon moieties. These are dimethylglycine dehydrogenase (DMGDH) and sarcosine dehydrogenase (SARDH), which demethylate dimethylglycine and sarcosine, respectively (22, 23). Both DMGDH and SARDH also contain tetrahydrofolate as a co-factor, which serves to accept formaldehyde released during removal of methyl group from dimethylglycine and sarcosine, thus producing 5,10-methylene-tetrahydrofolate (23). In summary, ETF is involved in oxidation of several types of metabolites, and, in addition to complexes I and II, it is the third major provider of electrons to the ubiquinone pool of the mitochondrial respiratory chain.
Human METTL20 is a MTF16 member, suggesting its involvement in protein methylation, and we set out to investigate the function of this uncharacterized protein. Thus, in the present report, we provide evidence that human METTL20 is a mitochondrial protein containing a functional N-terminal mitochondrial targeting sequence (MTS). Furthermore, by using an activity-based approach, we identified METTL20 as a highly specific MTase responsible for methylation of ETFβ and provide direct biochemical evidence that METTL20 is targeting ETFβ at two residues, namely Lys200 and Lys203. We also show that METTL20 can methylate ETFβ in vivo. Finally, we demonstrate that methylation of ETF impairs its ability to extract electrons from two acyl-CoA dehydrogenases, namely MCAD and GCDH, thus suggesting an important functional role for METTL20-mediated methylation of ETFβ.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit polyclonal anti-ETFβ (ab97936) antibody was from Abcam (Cambridge, UK).
Cloning and Mutagenesis
ORFs for human METTL20, ETFα, ETFβ, MCAD, and GCDH were cloned from cDNA generated from HEK293 or HeLa cells, using Phusion DNA Polymerase HF (Thermo Fisher Scientific, Waltham, MA). PrimerX program was used to design mutagenesis primers. All constructs were sequence-verified, including tags.
Bioinformatics Analysis
The NCBI Basic Local Alignment Search Tool (BLAST) was used to identify sequence homologues of human METTL20 (24). Multiple protein sequence alignments and secondary structure predictions were performed using algorithms embedded in the Jalview (v2.8) interface (25). Prediction of the N-terminal MTS and the location of the putative cleavage site were performed using the MitoProt algorithm (26) available at the ExPASy Bioinformatics Resource Portal.
Transient Transfection and Generation of Stable Cell Lines
HeLa cells were transiently transfected with pEGFP-N1 plasmid (Clontech), either empty or containing cDNA encoding human METTL20 or its putative MTS (amino acids 1–42). Cells were analyzed by confocal fluorescence microscopy 24 h after transfection. Plasmid pcDNA5/FRT/TO (Invitrogen, Thermo Fisher Scientific) was used for stable incorporation of METTL20-GFP fusion gene into Flp-In T-REx-293 cells (Invitrogen), according to the manufacturer's protocol, and the resulting cells were designated as TRex-METTL20-GFP cells. Overexpression of METTL20-GFP was induced for 48 h with 1 μg/ml doxycycline (Dox), and cells were subjected to confocal fluorescence microscopy or collected for biochemical analysis by scraping.
Confocal Fluorescence Microscopy
Living cells were stained with 50 nm MitoTracker Deep Red FM (Invitrogen) and 0.5 μg/ml Hoechst 33258 (Sigma-Aldrich) to visualize the mitochondria and the nuclei, respectively. Cells were imaged using an Olympus FluoView 1000 (Ix81) confocal fluorescence microscopy system with a PlanApo 60× NA 1.1 oil objective (Olympus, Hamburg, Germany). The different fluorophores were excited at 405 nm (Hoechst), 488 nm (GFP), and 635 nm (MitoTracker), and Kalman averaging (n = 3) was used to record multichannel images. The fluorescent signals emitted from GFP, MitoTracker, and Hoechst were acquired through green, red, and blue channels, respectively, and merged.
Expression and Purification of Recombinant METTL20
Human METTL20 WT and Δ38-METTL20 (with the N-terminal 38 amino acids deleted) were cloned into the plasmid pET28a and expressed as N-terminally His6-tagged proteins in the Escherichia coli strain BL21-CodonPlus(DE3)-RIPL (Agilent Technologies, Santa Clara, CA). Bacteria were lysed in lysis buffer 1 (50 mm NaH2PO4, pH 7.2, 500 mm NaCl, 10% glycerol, 1% Triton X-100, and 30 mm imidazole), supplemented with 1× Complete (EDTA-free) protease inhibitor mixture (Roche) and 10 units/ml benzonase nuclease (Sigma-Aldrich), and His-tagged proteins were purified on nickel-nitrilotriacetic acid-agarose (Qiagen), according to the manufacturer's protocol. Eluted proteins were buffer-exchanged to storage buffer 1 (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, and 10% glycerol), using centrifugal concentrators with a molecular mass cutoff of 10 kDa (Sartorius, Goettingen, Germany). Protein purity was assessed by SDS-PAGE and Coomassie staining, and protein concentrations were measured using Pierce BCA protein assay kit (Thermo Fisher Scientific). Theoretical molecular mass of recombinant proteins was used to calculate the molar concentrations of proteins.
Purification of Recombinant ETFα/β and FAD-containing Dehydrogenases
Human ETFβ WT and mutants, Δ25-MCAD and Δ44-GCDH (mature forms of respective dehydrogenases, with the N-terminal 25 or 44 amino acids deleted) were cloned into pET28a, with the N-terminal His6 tag, whereas Δ19-ETFα (mature form, with the N-terminal 19 amino acids deleted) was cloned into pETDuet-1 without a His tag. To express the ETFα/β heterodimer, E. coli were initially transformed with pET28a containing full-length ETFβ and then co-transformed with pETDuet-1 containing Δ19-ETFα and selected for kanamycin, chloramphenicol, and ampicillin resistance. Cells were lysed in lysis buffer 2 (50 mm NaH2PO4, pH 7.2, 10% glycerol, 30 mm imidazole, and 1 μm FAD), supplemented with 1× Complete (EDTA-free) protease inhibitor mixture and 10 units/ml benzonase nuclease, and cleared lysates were loaded on nickel-nitrilotriacetic acid-agarose, equilibrated in lysis buffer 2. After washing with 15 column volumes of lysis buffer 2, bound proteins were eluted with lysis buffer 2 supplemented with 300 mm imidazole. Eluates containing MCAD or GCDH were rebuffered to Storage Buffer 2 (20 mm Tris-HCl pH 8.0, 10% glycerol and 1 μm FAD), whereas ETFα/β was rebuffered to storage buffer 3 (50 mm NaH2PO4, pH 7.2, 10% glycerol, and 1 μm FAD), using centrifugal concentrators with a molecular mass cutoff of 50 kDa.
Preparation and Fractionation of Cell Extracts
Human cells were lysed for 10 min at 4 °C in lysis buffer 3 (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1% Triton X-100, 5% glycerol, 0.5 mm DTT, and 1× Complete (EDTA-free) protease inhibitor mixture), and the resulting lysates were sonicated and cleared by centrifugation. Cell lysates were fractionated at 4 °C by ion exchange chromatography using either Pierce Strong Anion Exchange Q or Pierce Strong Cation Exchange S Spin Columns (Thermo Fisher Scientific). In short, lysates were diluted with dilution buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, and 5% glycerol), to bring NaCl concentration to 50 mm and then applied onto the Q column equilibrated in dilution buffer. Material bound to the Q column was eluted by a step gradient of increasing NaCl concentrations prepared in dilution buffer, in 100-μl fractions: 0.15Q (eluted between 0.05 and 0.15 M NaCl), 0.3Q (eluted between 0.15 and 0.3 M NaCl), and 0.5Q (eluted between 0.3 and 0.5 M NaCl). Unbound material, which was released in the Q column flow-through (FTQ) was reapplied onto the S column equilibrated in dilution buffer. The unbound material, present in the S column flow-through, was designated FTS, whereas material bound to the S column was eluted in 100-μl fractions by a step gradient of increasing NaCl concentrations prepared in dilution buffer, analogous to the elution from the Q column. To partially purify ETFβ from TRex-METTL20-GFP cells, cell extracts were loaded on the Q column, and ETFβ was recovered in FTQ.
In Vitro Methyltransferase Assays Using [3H]SAM
To test MTase activity of METTL20 on cellular material, 10-μl reactions were set up on ice, in MTase assay buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 50 mm KCl and 10% glycerol) containing 40–80 pmol of Δ38-METTL20, 20–60 μg of protein from cell extracts and/or equivalent fractions obtained from ion exchange chromatography, and 0.5 μCi of [3H]SAM (PerkinElmer Life Sciences) ([SAM]total = 0.64 μm, specific activity = 78.2 Ci/mmol). Reaction mixtures were then incubated at 28 °C for 2 h. Proteins were resolved by SDS-PAGE, transferred to PVDF membrane, and stained with Ponceau S. The membrane was then dried, sprayed with EN3HANCE spray (PerkinElmer Life Sciences), and exposed to Kodak BioMax MS film (Sigma-Aldrich) at −80 °C. For quantitative fluorography analysis, [3H]SAM was diluted with nonradioactive SAM (New England Biolabs) ([SAM]total = 32.6 μm, specific activity = 1.53 Ci/mmol). In titration experiments, reaction mixtures were set up in MTase assay buffer, supplemented with 0.1 mg/ml BSA (as carrier), and contained 0.5 μCi of [3H]SAM ([SAM]total = 32.6 μm), with increasing concentrations of Δ38-METTL20 (0–25 μm) and fixed concentrations of ETFα/β (∼1.5 μm). Reactions were stopped by precipitation with 10% TCA, and TCA-insoluble material was subjected to scintillation counting.
Preparation of Samples for MS Analysis
In vitro methylation of recombinant or cellular (in extract) ETFβ for the purpose of MS analysis was performed as in the above section, except that radiolabeled SAM was replaced by 1 mm nonradioactive SAM. Proteins were resolved by SDS-PAGE and stained with Coomassie, and the portion of gel containing the protein of interest was cut out and subjected to in-gel trypsin (Sigma-Aldrich) or AspN (Roche) digestion, followed by MS analysis of the resulting proteolytic fragments, as previously described (8). MS data were analyzed with an in-house maintained human protein sequence database using SEQUESTTM and Proteome DiscovererTM (Thermo Fisher Scientific). The mass tolerances of a fragment ion and a parent ion were set as 0.5 Da and 10 ppm, respectively. Methionine oxidation and cysteine carbamido-methylation were selected as variable modifications. ETFβ methylated peptide MS/MS spectra were manually searched by Qual Browser (v2.0.7).
Large Scale Preparation of Methylated Recombinant ETFα/β
Large scale preparation of methylated ETFα/β (ETFα/βmet) was made by overnight incubation at 23 °C of 40 μm ETFα/β and 40 μm Δ38-METTL20, in MTase assay buffer containing 3 mm SAM and 50 μm FAD. For the control reactions, METTL20 and SAM were omitted. Both reactions were subject to ion exchange purification, by loading onto Pierce strong anion exchange Q spin columns, equilibrated in MTase assay buffer, and the flow-through (containing ETFα/β) was collected and rebuffered to storage buffer 3, using centrifugal concentrators with a 50-kDa cutoff. Protein purity and concentration was determined as in previous sections. Both ETFα/β and ETFα/βmet had equal A280/A450 ratios.
DCIP Reduction Assay
The ability of ETFα/β to mediate electron transfer from acyl-CoA dehydrogenases was tested using the previously published DCIP reduction method (19), with some modifications. Typically, 250–350-μl mixtures were prepared, containing ETFα/β (0.2–5 μm), GCDH, or MCAD as acyl-CoA dehydrogenase (0.05–2 μm), 50 μm FAD, and 50 μm DCIP (A600 ≈ 1), all in DCIP assay buffer (10 mm NaH2PO4, pH 7.2, 10% glycerol, and 0.05% Tween 20). When indicated, recombinant ETFα/β (5 μm) was incubated with nonradioactive SAM (1 mm) and recombinant Δ38-METTL20 (5 μm) for 2 h at 28 °C and then diluted 25 times with DCIP assay buffer containing MCAD (0.1–0.2 μm), FAD (50 μm), and DCIP (50 μm). Reaction mixtures were allowed to equilibrate at room temperature for 15 min, and then the reaction was started by adding, through manual mixing, glutaryl-CoA (50 μm) to assay GCDH or octanoyl-CoA (50 μm) to assay MCAD. The kinetics of DCIP reduction (bleaching) was monitored continuously at 600 nm for 3–5 min, using a Shimadzu UV-1601 spectrophotometer. The rate of DCIP reduction was calculated from the slope of the linear part of the kinetic curve, using molecular extinction coefficient of DCIP: ϵ = 21,700 m−1 cm−1 and expressed in μm of DCIP reduced per minute. Student's t test was used to evaluate the probability (p values) that two populations are the same.
RESULTS
Human METTL20 Is an Evolutionarily Conserved Mitochondrial Protein Methyltransferase
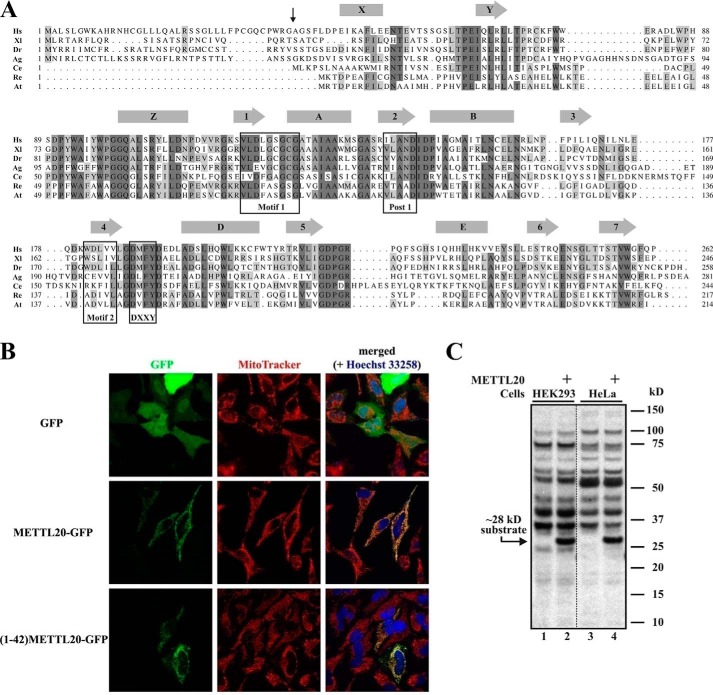
As an initial step in our functional characterization of human METTL20, we performed a BLAST search, which revealed the presence of putative METTL20 orthologues in a wide range of organisms (Fig. 1A). Interestingly, METTL20 shows a rather scattered and unusual distribution across the tree of life. In eukaryotes, METLL20 is found exclusively in multicellular animals and is apparently present in all chordates (animals with a dorsal nerve chord), while showing a more scattered distribution among other animals, such as insects and nematodes. Interestingly, METTL20 orthologues are also found in a small subset of bacteria, primarily those belonging to the Rhizobiales order of α-proteobacteria.
FIGURE 1.
Human METTL20 is an evolutionarily conserved protein methyltransferase localized to mitochondria. A, alignment of METTL20 orthologues from Homo sapiens (Hs; NP_776163.1), Xenopus laevis (Xl; NP_001090037.1), Danio rerio (Dr; NP_001154967.1), Anopheles gambiae (Ag; XP_308218.4), Caenorhabditis elegans (Ce; NP_491943.2), Rhizobium etli (Re; YP_471372.1), and Agrobacterium tumefaciens (At; NP_355584.1). Predicted α-helices (rectangles) and β-strands (arrows) are indicated. Hallmark motifs of 7BS (Motif 1, Post 1, and Motif 2) and MTF16 (DXXY) MTases are boxed. The vertical arrow shows the predicted position of cleavage of putative MTS in human METTL20. B, in vivo confocal fluorescence microscopy images of HeLa cells after 24 h of transient transfection, expressing either GFP, METTL20-GFP, or (1–42)-METTL20-GFP, in the presence of MitoTracker and Hoechst dyes. Data were acquired through green (GFP), red (MitoTracker), and blue (Hoechst) channels and merged. C, recombinant human METTL20 catalyzes methylation of a ∼28-kDa protein in human cell extracts. Extracts from HEK293 or HeLa cells were incubated with [3H]SAM in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of METTL20. Proteins were resolved by SDS-PAGE and transferred onto a PVDF membrane, which was subjected to fluorography. The positions of the ∼28-kDa substrate and molecular mass markers are indicated.
Sequence alignment of putative METTL20 orthologues from various organisms revealed the presence of conserved sequence segments, encompassing the hallmark motifs of the 7BS-MTases, as well as the characteristic (D/E)XX(Y/F) motif shared by the MTF16 members (Fig. 1A). Compared with its prokaryotic counterparts, human METLL20 contains a ∼40-amino acid N-terminal extension, which, by using the MitoProt algorithm (26), was predicted to represent a MTS. Indeed, both full-length METTL20 and the isolated putative MTS were, when expressed as N-terminal fusions with GFP, able to target GFP to mitochondria, indicating that METTL20 is a mitochondrial protein (Fig. 1B).
Previously, we have shown that recombinant MTF16 enzymes with KMT activity can methylate specific proteins in cellular extracts (8, 10), and we investigated METTL20 for such activity. In these assays, we used a METTL20 deletion mutant lacking the MTS, which we reasoned was likely dispensable for enzymatic activity. Methylation reactions with cellular extracts were performed in the presence of [3H]SAM and then subjected to SDS-PAGE and fluorography, enabling the visualization of methylated proteins as distinct bands. Indeed, human METTL20 methylated a protein of ∼28 kDa in extracts from both HeLa and HEK293 cells (Fig. 1C). Several additional labeled proteins were observed, but their labeling was not METTL20-dependent, indicating that they reflect methylations performed by endogenous MTases in the extract.
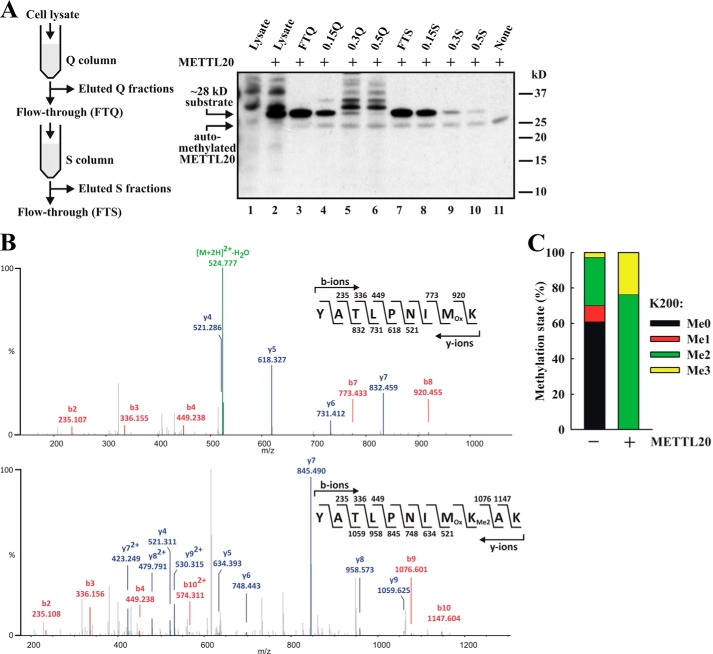
Identification of ETFβ as the Substrate of Recombinant Human METTL20
To reveal the identity of the ∼28-kDa substrate by MS, we applied an activity-based purification strategy to reduce the complexity of the sample. Thus, a HEK293 cell lysate was fractionated using ion exchange chromatography, and the fractions were assayed for METTL20-mediated methylation (Fig. 2A). We found that the ∼28-kDa substrate bound poorly to an anion exchange (Q) column and was found mostly in the flow-through (FTQ) (Fig. 2A). When the FTQ fraction was applied to a cation exchange (S) column, the ∼28-kDa protein was found to partially bind to the S column and was completely eluted with 0.15 m NaCl (0.15 S) (Fig. 2A). To identify the METTL20 substrate, the 0.15 S fraction was resolved by SDS-PAGE, and the region corresponding to the substrate was excised and subjected to trypsin digestion, followed by protein identification through MS. The analysis identified 10 different proteins, and interestingly, and in agreement with the observed mitochondrial localization of METTL20, four of these were mitochondrial proteins, i.e. 3-hydroxyacyl-CoA dehydrogenase type 2, ETFβ, adenylate kinase 2, and malate dehydrogenase (Table 1). To directly identify the METTL20 target, as well as the residue(s) being methylated, a parallel sample had been generated, where the 0.15 S fraction had been treated with METTL20 in the presence of nonradioactive SAM. Intriguingly, only a single peptide was found to display a different degree of methylation between the untreated and the METTL20-treated samples, namely the ETFβ-derived YATLPNIMKAK peptide (residues 192–202), where the methylation level of Lys200 was substantially increased in the METTL20-treated sample (Fig. 2, B and C). In the untreated sample, Lys200 was found to be mostly unmethylated and dimethylated with some monomethylation and trimethylation. In contrast, in the METTL20-treated sample Lys200 was exclusively found in the dimethylated and trimethylated states. Notably, among the four potential METTL20 substrates localized to mitochondria, ETFβ was the one showing the highest sequence coverage and also displayed the second highest score (Table 1). Thus, these results indicate that METTL20 can catalyze the methylation of Lys200 in ETFβ and suggest that this enzyme mediates this reaction also in vivo.
FIGURE 2.
Identification of ETFβ as substrate of recombinant human METTL20. A, partial purification of the ∼28-kDa METTL20 substrate from HEK293 extracts by ion exchange chromatography. Schematic representation of ion exchange-based fractionation procedure (left panel). The various fractions were incubated with [3H]SAM in the presence of METTL20, and methylation was detected by fluorography (right panel). B, ETFβ present in extracts from HEK293 cells is dimethylated at Lys200. The 0.15 S fraction was resolved by SDS-PAGE and Coomassie-stained. The region of gel containing ETFβ was excised and subjected to trypsin digestion, and the resulting tryptic peptides were analyzed by MS. The figure shows fragmentation mass spectra of ETFβ-derived peptide with series of b-ions (red) and y-ions (blue) mapped onto the sequence of the peptide, supporting Lys200 of ETFβ as target of dimethylation. C, METTL20 methylates Lys200 in ETFβ in extracts from HEK293 cells. The 0.15 S fraction was incubated in the absence or presence of METTL20 and nonradioactive SAM (1 mm) and analyzed as in B. The relative intensities of MS signals representing different ETFβ methylation states at Lys200, in samples incubated with or without METTL20, are shown.
TABLE 1.
List of proteins identified in ∼28-kDa region of SDS-PAGE from 0.15 S fraction
| Protein name | Accession number | Score | Coverage | Molecular mass | Subcellular locationa |
|---|---|---|---|---|---|
| % | kDa | ||||
| Triosephosphate isomerase | P60174 | 67.42 | 34.94 | 30.8 | Cyt |
| 3-Hydroxyacyl-CoA dehydrogenase type 2 | Q99714 | 55.02 | 41.76 | 26.9 | Mit |
| Electron transfer flavoprotein subunit β | P38117 | 43.63 | 47.06 | 27.8 | Mit |
| Poly(rC)-binding protein 1 | Q15365 | 31.89 | 9.27 | 37.5 | Nuc, Cyt |
| Adenylate kinase 2 | P54819 | 30.68 | 43.10 | 26.5 | Mit |
| V-type proton ATPase subunit E1 | P36543 | 29.02 | 32.35 | 26.1 | IV |
| Poly(rC)-binding protein 2 | Q15366 | 27.87 | 11.78 | 38.6 | Nuc, Cyt |
| Malate dehydrogenase | P40926 | 18.51 | 22.19 | 35.5 | Mit |
| Endoplasmic reticulum resident protein 29 | P30040 | 18.13 | 19.92 | 29.0 | ER |
| GTP-binding nuclear protein Ran | P62826 | 15.33 | 18.52 | 24.4 | Nuc, Cyt |
a Cyt, cytosol; Mit, mitochondrion; Nuc, nucleus; IV, intracellular vesicles; ER, endoplasmic reticulum.
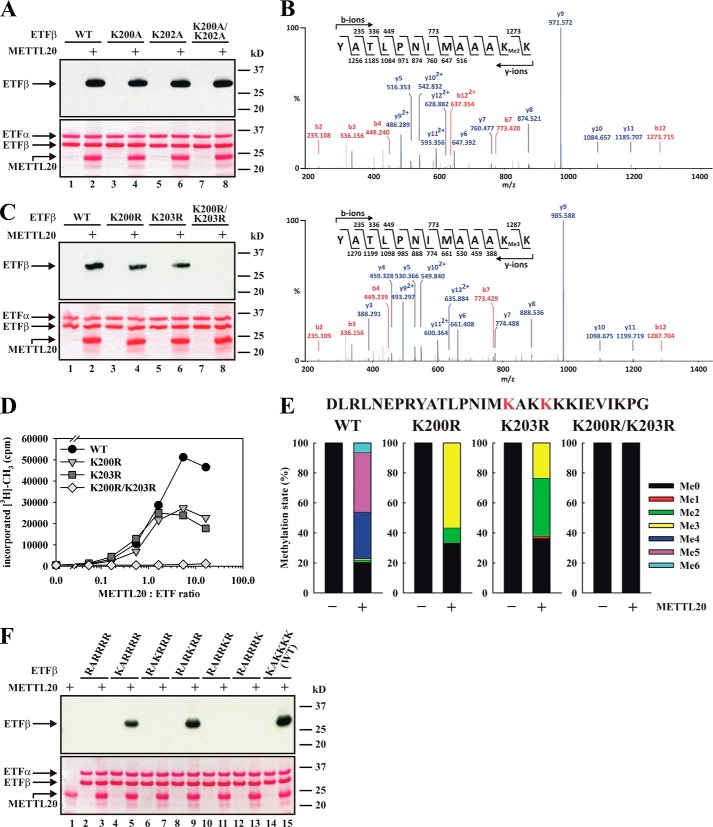
Recombinant METTL20 Catalyzes Methylation of Lys200 and Lys203 in ETFβ in Vitro
To directly demonstrate that METTL20 can methylate ETFβ, we expressed and purified His6-tagged human ETFβ and found recombinant METTL20 to methylate this substrate in vitro (data not shown). However, because the isolated ETFβ subunit displayed stability problems, we generated ETFα/β heterodimers through co-expression and co-purification of untagged ETFα with His6-tagged ETFβ in E. coli. Indeed, the β-subunit of recombinant ETFα/β was subject to methylation by METTL20 in vitro (Fig. 3A).
FIGURE 3.
Recombinant human METTL20 methylates ETFβ on Lys200 and Lys203in vitro. A, METTL20 methylates the β-subunit of the ETFα/β heterodimer. ETFα/β containing ETFβ WT or mutant as indicated was incubated with [3H]SAM in the absence or presence of METTL20. Proteins were resolved by SDS-PAGE and then transferred to a PVDF membrane that was stained with Ponceau S (bottom panel). The membrane was subsequently subjected to fluorography (top panel). B, ETFβ mutant (K200A/K202A) is methylated on Lys203 by METTL20. ETFα/β with mutant (K200A/K202A) β-subunit was treated with METTL20 in the presence of nonradioactive SAM. Proteins were analyzed as in Fig. 2B. The figure shows fragmentation mass spectra of ETFβ-derived peptides with series of b-ions (red) and y-ions (blue) mapped onto the sequence of the peptide, supporting Lys203 as target of di- and trimethylation. C, METTL20 targets Lys200 and Lys203 in ETFβ. ETFα/β with WT or mutant (K200R, K203R, and K200R/K203R) β-subunit was incubated with [3H]SAM in the absence or presence of METTL20, and methylation was analyzed as in A. D, Lys200 and Lys203 of ETFβ are independently methylated by METTL20. ETFα/β heterodimer (∼1.5 μm) containing either WT or mutant β-subunit was incubated with various concentrations (0–25 μm) of METTL20, in the presence of [3H]SAM ([SAM]total = 32.6 μm). Proteins were precipitated with 10% TCA and subjected to scintillation counting. E, METTL20-catalyzed methylation introduces up to three methyl groups at each of the sites Lys200 and Lys203 in ETFβ. Mutant or WT ETFα/β was incubated with nonradioactive SAM in the presence or absence of METTL20. Proteins were then resolved by SDS-PAGE and subjected to Coomassie staining. The ETFβ-containing region of the gel was excised and subjected to AspN digestion, and the resulting proteolytic peptides were analyzed by MS. The figure shows the relative intensities of signals corresponding to the different methylation states of the AspN-digested peptide encompassing residues 184–212 (red color indicates the location of the two methylation sites Lys200 and Lys203 in the peptide sequence). F, METTL20-mediated lysine methylation of ETFβ occurs specifically at residues Lys200 and Lys203. ETFα/β with β-subunit WT or mutant (with indicated sequence at residues 200–205) was incubated with [3H]SAM in the absence or presence of METTL20, and methylation was assessed by fluorography, as in A.
Our previous results (Fig. 2, B and C) indicated that Lys200 is target of METTL20-mediated methylation, and importantly, this residue has already been annotated as methylated in vivo, according to the Phosphosite database (27). Interestingly, Phosphosite also reports another methylation site in ETFβ, namely the neighboring Lys202. To further investigate the potential methylation at these two sites, we generated K200A, K202A, and K200A/K202A mutants, which were tested (in the context of ETFα/β) for METTL20-mediated methylation. Somewhat surprisingly, mutating both these residues did not abolish methylation, suggesting the presence of other methylation sites (Fig. 3A). Indeed, when the K200A/K202A mutated ETFα/β was methylated in vitro and subjected to MS analysis, di- and trimethylation at a neighboring site, Lys203, was observed (Fig. 3B). Thus, we next tested a K200R/K203R double mutant and found that METTL20 mediated methylation was completely abolished (Fig. 3C). In contrast, the K200R and K203R single mutants were methylated, albeit at reduced levels. Furthermore, when the WT and mutant ETFα/β substrates were treated with different amounts of METTL20, the titration curves for K200R and K203R were similar and plateaued off at a level corresponding to ∼50% of that observed with the WT protein (Fig. 3D). This suggested that Lys200 and Lys203 are methylated at similar efficiencies and that their methylation together account for all the observed METTL20-mediated methylation of ETFβ.
To specifically investigate the methylation state of Lys200 and Lys203 after METTL20-mediated methylation, mutant or WT ETFα/β was incubated with recombinant METTL20, and an AspN generated peptide encompassing both methylation sites was analyzed by MS. We found the peptide derived from WT ETFβ to be primarily tetra-, penta-, or hexamethylated after METTL20 treatment, whereas corresponding peptides from the K200R and K203R mutants were mostly di- or trimethylated (Fig. 3E). As expected, no methylation was observed for the K200R/K203R double mutant. These results show that METTL20 is a nonprocessive enzyme capable of generating several different methylation states.
Clearly, the above results show that Lys200 and Lys203 are targets of METTL20, but, because we were unable to obtain sequence information for the AspN-generated peptide by MS, we set out to obtain independent evidence that these are the only methylation sites. Lys200 and Lys203 are found as part of the sequence KAKKKK (residues 200–205), i.e. in the context of several other lysines, and to individually investigate the potential methylation at these residues, the KAKKKK segment was mutated into RARRRR, and the individual Lys residues were reintroduced. When these mutants were tested in MTase assays with METTL20, only the ones with Lys in position 200 or 203, i.e. the sequences KARRRR or RARKRR, respectively, were methylated (Fig. 3F). In summary, the data presented above show that METTL20 is a highly specific methyltransferase, capable of independently methylating the two sites Lys200 and Lys203 in ETFβ in vitro.
METTL20-mediated Methylation of ETFβ in Cells
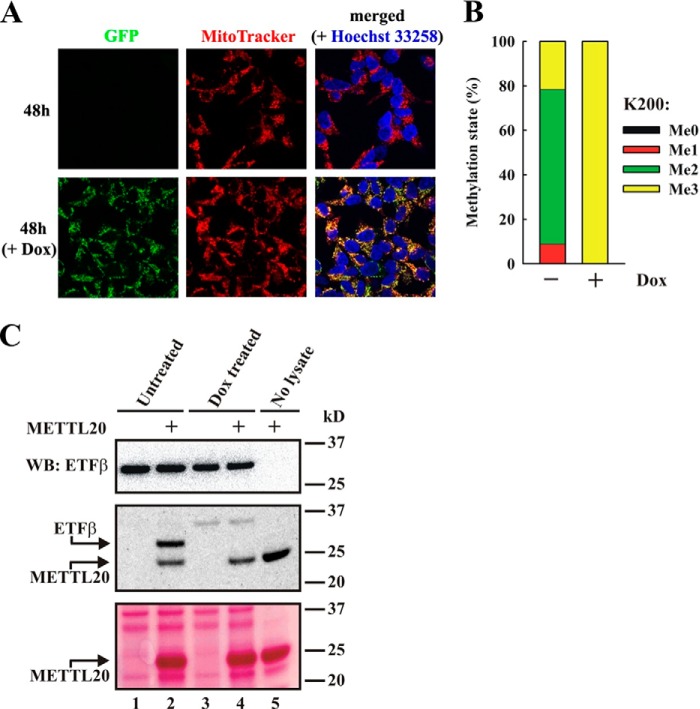
Having demonstrated that cellular ETFβ is methylated and that recombinant METTL20 efficiently methylates ETFβ in vitro, we set out to show that METTL20-mediated methylation of ETFβ also can occur in a cellular context. To study this, we generated a human cell line expressing a fusion protein between GFP and METTL20 (METTL20-GFP) under a tetracycline inducible promoter. To verify the inducible expression of METTL20-GFP, cells were treated with the tetracycline analogue Dox, and, indeed, a strong GFP signal was observed in mitochondria (Fig. 4A).
FIGURE 4.
METTL20 methylates ETFβ in vivo. A, confocal fluorescence microscopy images of Flp-In T-REx-293 cells with stably inserted METTL20-GFP fusion gene, in the absence or presence of 1 μg/ml Dox to induce expression of the fusion protein for 48 h, in the presence of MitoTracker and Hoechst dyes. The data were acquired as in Fig. 1B. B, overexpression of a METTL20-GFP fusion protein results in complete trimethylation of Lys200 in ETFβ. Cells as in A were either untreated or treated with Dox for 48 h. ETFβ was partially purified from cell extracts by anion exchange chromatography on a Q column. The ETFβ-containing flow-through was resolved by SDS-PAGE, and the ETFβ-containing region was excised from the Coomassie-stained gel and then subjected to trypsin digestion and MS analysis. The relative intensities of MS signals representing different ETFβ methylation states at Lys200, as in Fig. 2B, are shown. C, overexpression of a METTL20-GFP fusion protein abolishes hypomethylation of ETFβ. Cells, as in A, were incubated for 48 h in the absence or presence of Dox. ETFβ was partially purified as in B and incubated with [3H]SAM in the absence or presence of recombinant METTL20, resolved by SDS-PAGE, transferred to PVDF membrane, and stained with Ponceau S (bottom panel). The membrane was probed with anti-ETFβ antibody as loading control (top panel). The membrane was then stripped, dried, and subjected to fluorography (middle panel). Arrows show the positions of methylated ETFβ and automethylated METTL20 (middle panel), as well as the position of METTL20 on Ponceau S stained membrane (bottom panel).
To address whether ectopically expressed METTL20 methylates cellular ETFβ, we analyzed the methylation status of the tryptic peptide encompassing Lys200 by MS (unfortunately, we were not able to detect the AspN-generated peptide that encompassed both methylation sites, shown in Fig. 3E, in these cellular samples). In the absence of Dox treatment, Lys200 was found predominantly in the dimethylated state but also showed some mono- or trimethylation. However, after induction of METTL20-GFP expression through Dox treatment, Lys200 was shifted to nearly complete (>99.5%) trimethylation (Fig. 4B). Because ETFβ from cells with ectopically expressed METTL20 is completely trimethylated, one would predict that ETFβ from such cells is not substrate for further methylation by METTL20. To test this, thereby providing independent evidence for METTL20-mediated methylation of ETFβ in cells, we subjected an extract from the Dox-treated cells to in vitro methylation by METTL20 in the presence of [3H]SAM. Indeed, extracts from the Dox-induced cells showed no radiolabeled band corresponding to ETFβ in fluorography, whereas the noninduced cells showed a strong band (Fig. 4C). Taken together, the results described above show that METTL20 is capable of methylating ETFβ also in vivo.
ETFα/β Methylation Reduces Electron Transfer from Acyl-CoA Dehydrogenases
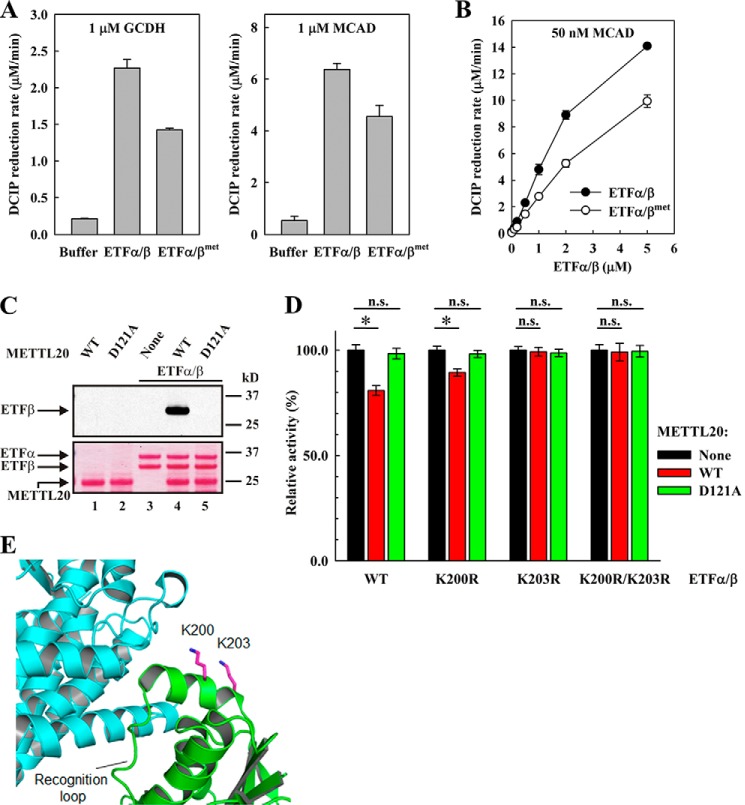
ETFα/β acts as a mobile electron carrier that shuttles electrons from 13 different mitochondrial dehydrogenases to ETF:quinone oxidoreductase (16). Apparently, the interaction between ETFα/β and these dehydrogenases is mediated by a recognition loop in ETFβ encompassing residues 191–200 (18), i.e. a region that partially overlaps with the segment methylated by METTL20. Thus, to investigate the interesting possibility that METTL20-induced methylation has a modulatory/regulatory function, we assessed the effect of methylation on the ability of ETFα/β to receive electrons from dehydrogenases in vitro. We found that some dehydrogenases were difficult to express and purify or failed to show ETF-dependent activity in vitro (data not shown), However, we were able to express and purify recombinant MCAD and GCDH, and both these enzymes displayed ETFα/β-dependent reduction of their respective substrates, octanoyl-CoA and glutaryl-CoA (Fig. 5A). In these experiments, DCIP was used as the final electron acceptor, and ETF-dependent dehydrogenase activity was monitored as bleaching of DCIP, resulting from its reduction. Interestingly, when we compared methylated (ETFα/βmet) and unmethylated ETFα/β for their ability to transfer electrons from the dehydrogenases to DCIP, we found that methylation substantially reduced the extent of electron transfer from both MCAD and GCDH, i.e. by ∼30 and 40%, respectively (Fig. 5A). To exclude the possibility that the observed differences between ETFα/β and ETFα/βmet originated from small errors in protein concentration determination, we also performed an experiment where the concentration of ETFα/β or ETFα/βmet was varied, whereas that of MCAD was kept constant. The resulting titration curves show that the observed difference between ETFα/β and ETFα/βmet would approximately correspond to a 2–3-fold difference in ETFα/β concentration, i.e. far more than what would expectedly be caused by inaccurate protein concentration measurements (Fig. 5B).
FIGURE 5.
METTL20-mediated methylation of ETFβ impairs its ability to mediate electron transfer from acyl-CoA dehydrogenases. A, methylation of ETFβ inhibits the rate of ETFα/β-dependent oxidation of glutaryl-CoA by GCDH and oxidation of octanoyl-CoA by MCAD. Reactions containing GCDH or MCAD (as indicated), in the absence or presence of ETF (0.2 μm), either nonmethylated (ETFα/β) or methylated (ETFα/βmet), were started by the addition of glutaryl-CoA or octanoyl-CoA, respectively, and their oxidation was followed by measuring the rate of DCIP reduction. B, concentration dependence of the effect of ETFα/β or ETFα/βmet on the oxidation of octanoyl-CoA by MCAD. Reactions containing MCAD in the presence of increasing ETF, either nonmethylated (ETFα/β) or methylated (ETFα/βmet), were started by the addition of octanoyl-CoA, and its oxidation was followed as in A. C, D121A mutated METTL20 is unable to methylate ETFβ. Recombinant ETFα/β was incubated with [3H]SAM in the absence or presence of recombinant METTL20, either WT or mutant (D121A). Material was analyzed as in Fig. 3A. The positions of protein bands are indicated by arrows. D, enzymatically active METTL20 inhibits the ability of ETFβ to support oxidation of octanoyl-CoA by MCAD. ETFα/β containing either WT or mutant (K200R, K203R, or K200R/K203R) β-subunit was incubated with nonradioactive SAM, in the absence or presence of recombinant human METTL20, either WT or mutant (D121A). Reactions were then diluted with DCIP assay buffer containing MCAD, octanoyl-CoA was added, and its oxidation was followed as in A. Results from individual experiments were normalized for the activity of either ETFα/β WT or ETFα/β mutants, measured in the absence of METTL20. Sample variation is expressed as standard deviation (n = 10). *, p value < 0.01; n.s., not significant. E, structure of the region of interaction between ETFβ (green) and MCAD (cyan) (Protein Data Bank code 1T9G) (18). The recognition loop followed by Lys200 and Lys203 (magenta) of ETFβ is indicated. The ϵ-nitrogen (site of methylation) in Lys residues is indicated in dark blue.
To further establish that the observed effects of METTL20 on the electron transferring ability of ETF reflect METTL20-mediated methylation, we performed various control experiments. First, we generated a catalytically inactive METTL20 enzyme by mutating a key conserved residue in motif I (D121A) (Fig. 5C), and we found that the mutant METTL20, unlike its wild-type counterpart, had no significant effect on ETFβ-mediated electron transfer (Fig. 5D). Second, we also tested the effect of METTL20 on ETFβ mutants where the methylated Lys residues had been individually or collectively replaced by Arg, i.e. K200R, K203R, and K200R/K203R. Notably, the mutants K203R and K200R/K203R ETFβ were capable of mediating electron transfer, but this was not influenced by neither wild-type nor mutant METTL20 (Fig. 5D). Electron transfer by the K200R mutant was significantly reduced by enzymatically active METTL20, but the effect was substantially weaker than for WT ETFβ. In summary, the above experiments strongly indicate that METTL20-mediated methylation of ETFβ has an inhibitory effect on the transfer of electrons from MCAD and GCDH and indicate that methylation at both Lys200 and Lys203 mediates this effect.
DISCUSSION
We have here identified the biochemical function of the human lysine-specific MTase METTL20 using both in vitro and in vivo approaches. We found METTL20 to be associated with mitochondria and to specifically methylate ETFβ on two adjacent lysine residues, thereby establishing METTL20 as the first mitochondrial KMT in multicellular organisms. Moreover, methylation reduced the ability of ETFβ to receive electrons from the dehydrogenases MCAD and GCDH, demonstrating its functional significance and suggesting a regulatory role.
Previously, we reported, based on bioinformatics analysis, that METTL20 belonged to a family of 10 putative and established human KMTs (6). Other characterized members are, similarly to METTL20, highly specific KMTs that methylate unique cellular substrates, i.e. the chaperones VCP (METTL21D/VCP-KMT) and Hsp70/HSPA (METTL21A/HSPA-KMT), calmodulin (C2orf34/CaM-KMT), the DNA repair protein KIN17 (METTL22), and the eukaryotic translation elongation factor 2 (FAM86A/eEF2-KMT) (6–10). Based on the previous nomenclature for naming these enzymes, we suggest that METTL20 is redubbed ETFβ-KMT. The substrates of these enzymes have been mostly identified in screens for interacting proteins, but we have here used an activity-based purification scheme for identifying ETFβ as the substrate of METTL20, thus demonstrating this as an alternative and viable approach for identifying the substrates of uncharacterized KMTs.
Very recently, while the present manuscript was in preparation, Rhein et al. (28) actually implicated METTL20 in methylation of ETFβ. Tandem affinity purification tagging experiments revealed ETFβ as a binding partner of METTL20, and ETFβ was found to be methylated at Lys200 and Lys203. METTL20 was suggested to be responsible for ETFβ methylation, because modulation of cellular METTL20 levels through ectopic overexpression or siRNA mediated knockdown caused increased or decreased ETFβ methylation, respectively. The study by Rhein et al. (28) nicely complements the present work, in that it demonstrated methylation of ETFβ on both Lys200 and Lys203 in cellular samples, whereas we, for technical reasons, were unable to show methylation at Lys203 in vivo. However, unlike the present study, which includes extensive in vitro characterization of the activity of recombinant METTL20 on various ETFβ-derived substrates, the paper by Rhein et al. (28) did not directly demonstrate the ability of METTL20 to methylate ETFβ, thus leaving the possibility that the effect of METTL20 on ETFβ methylation levels could be indirect. The relevance of such a concern is illustrated by a recent study that concluded, from similar overexpression and knockdown experiments, that the MTase SETD1A was responsible for methylation of Lys561 in Hsp70 (HSPA1) (29). However, two subsequent, independent studies have established that this methylation reaction is catalyzed by METTL21A (8, 9). Furthermore, the study by Rhein et al. (28) exclusively used peptide sequencing by MS/MS to identify Lys200 and Lys203 as methylation sites, and other similar analyses had actually identified Lys200 and Lys202 as methylation sites (27). Thus, an additional contribution of the present study is the firm establishment of Lys200 and Lys203 as the sites of METTL20-mediated methylation.
Lysine methylation of nuclear proteins, in particular histones, has been subject to extensive study, both with respect to the responsible enzymes and functional consequences. In recent years, it has become clear that Lys methylation of cytosolic proteins is also important, because several highly specific KMTs with cytosolic targets have been identified. The knowledge on Lys methylation of mitochondrial proteins is, however, very limited. Until the discovery of METTL20, only a single eukaryotic mitochondrial KMT had been identified, namely the Saccharomyces cerevisiae MTase Ctm1p, which methylates Lys72 in cytochrome c (30). However, this methylation is not conserved in mammals, which lack a Ctm1p sequence homologue. Notably, several additional mitochondrial proteins have been identified as targets of lysine methylation, e.g. ATP synthase, citrate synthase, and large ribosomal protein 39 (MRPL39), as well as several members of solute carrier family 25 (SLC25) (27, 31). This predicts the existence of additional, yet undiscovered, mitochondria-associated KMTs in humans.
Sequence homologues of METTL20 show a rather unusual distribution on the tree of life, because these proteins are limited to animals in the eukaryotes but also are found in a small subset of bacteria. Interestingly, most bacteria that have METTL20-like proteins belong to the alpha class of the phylum proteobacteria, and this group of bacteria has been indicated as the likely evolutionary precursor of mitochondria (32). This may suggest that methylation of ETF is a primordial function, acquired before the appearance of mitochondria. Indeed, ETFβ from METTL20-containing bacteria has strong sequence homology to mammalian ETFβ, but future studies are required to address the possible role of ETFβ methylation in bacteria.
Structural and biochemical studies indicated residues 191–200 in ETFβ to form a recognition loop involved in the interaction between ETF and some of the mitochondrial dehydrogenases (16, 18). Because Lys200 and Lys203 are located adjacent to this ETFβ recognition loop, one may speculate that ETFβ methylation may differentially affect its interaction with the various mitochondrial dehydrogenases, thus providing a regulatory mechanism for modulating the metabolism of fatty acids, certain amino acids, and choline. The three-dimensional structure of a complex between human ETFα/β and one of its cognate dehydrogenases is available only in the case of MCAD (18), and in this structure, the side chains of Lys200 and Lys203, which are part of the same α-helix, are localized in the vicinity of MCAD and point in the direction of the MCAD surface (Fig. 5E). In support of the notion that methylation of Lys200 and Lys203 can modulate the interaction of ETFα/β with dehydrogenases, we showed that methylated ETFα/β has reduced capability in mediating electron transfer from MCAD and GCDH to the artificial electron acceptor DCIP. We also found that mutation of Lys200 or Lys203 in ETFβ partially or completely, respectively, abolished the observed effect of METTL20 on ETFβ-mediated electron transfer, suggesting that methylation of both these sites is functionally important. In the recent study by Rhein et al. (28), siRNA-mediated knockdown of METTL20 was shown to cause a moderate decrease in the rate of oxygen consumption in permeabilized cells supplied with palmitoyl-l-carnitine, further supporting the notion that ETFβ methylation influences metabolism. The ETF-dependent dehydrogenases involved in one-carbon metabolism, DMGDH and SARDH, are structurally unrelated to those acting on acyl-CoAs, and their interaction with ETF has not been characterized. Thus, the interesting possibility exists that the interaction of these enzymes with ETF is more sensitive to ETFβ methylation status than what is the case for the enzymes tested here. Recent studies have indicated that cellular SAM levels are subject to tight regulation, and the one-carbon units generated by DMGDH and SARDH can actually be used for replenishment of the cellular SAM pool (33). Thus, the interesting, though speculative, possibility exists that METTL20 acts as SAM sensor, which in a SAM-dependent manner regulates the flow of electrons from DMGDH and/or SARDH to ETF, thereby regulating the level of one-carbon units available for SAM synthesis. Further studies addressing the metabolic consequences of METTL20 inactivation will likely shed light on the very interesting potential role of METTL20 in regulating metabolism.
Acknowledgments
We thank Dr. Michael McDonough for assistance in protein structure visualization. We thank Oslo NorMIC Imaging Platform (located at the Department of Biosciences, University of Oslo) for performing the cell imaging.
This work was supported by the University of Oslo and grants from the Norwegian Cancer Society and the Research Council of Norway.
- SAM
- S-adenosylmethionine
- 7BS
- seven β-strand
- Dox
- doxycycline
- DMGDH
- dimethylglycine dehydrogenase
- ETF
- electron transfer flavoprotein
- GCDH
- glutaryl-CoA dehydrogenase
- MTase
- methyltransferase
- KMT
- lysine methyltransferase
- MCAD
- medium-chain acyl-CoA dehydrogenase
- MTS
- mitochondrial targeting sequence
- MTF16
- Methyltransferase Family 16
- MS
- mass spectrometry
- SARDH
- sarcosine dehydrogenase
- DCIP
- 2,6-dichloroindophenol.
REFERENCES
- 1. Petrossian T. C., Clarke S. G. (2011) Uncovering the human methyltransferasome. Mol. Cell. Proteomics 10.1074/mcp.M110.000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer E. L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang J., Berger S. L. (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 4. Clarke S. G. (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Del Rizzo P. A., Trievel R. C. (2011) Substrate and product specificities of SET domain methyltransferases. Epigenetics 6, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Maelandsmo G. M., Egge-Jacobsen W., Falnes P. O. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038. [DOI] [PubMed] [Google Scholar]
- 7. Magnani R., Dirk L. M., Trievel R. C., Houtz R. L. (2010) Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1, 43. [DOI] [PubMed] [Google Scholar]
- 8. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., Falnes P. Ø. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9, e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davydova E., Ho A. Y., Malecki J., Moen A., Enserink J. M., Jakobsson M. E., Loenarz C., Falnes P. O. (2014) Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2). J. Biol. Chem. 289, 30499–30510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 12. Shimazu T., Barjau J., Sohtome Y., Sodeoka M., Shinkai Y. (2014) Selenium-based S-adenosylmethionine analog reveals the mammalian seven-β-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS One 9, e105394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Husain M., Steenkamp D. J. (1983) Electron transfer flavoprotein from pig liver mitochondria. Biochem. J. 209, 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKean M. C., Beckmann J. D., Frerman F. E. (1983) Subunit structure of electron transfer flavoprotein. J. Biol. Chem. 258, 1866–1870 [PubMed] [Google Scholar]
- 15. Roberts D. L., Frerman F. E., Kim J. J. (1996) Three-dimentional structure of human electron transfer flavoprotein to 2.1-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 93, 14355–14360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toogood H. S., Leys D., Scrutton N. S. (2007) Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes. FEBS J. 274, 5481–5504 [DOI] [PubMed] [Google Scholar]
- 17. Ramsay R. R., Steenkamp D. J., Husain M. (1987) Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein:ubiquinone oxidoreductase. Biochem. J. 241, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toogood H. S., van Thiel A., Basran J., Sutcliffe M. J., Scrutton N. S., Leys D. (2004) Extensive domain motion and electron transfer in the human electron transferring flavoprotein:medium chain acyl-CoA dehydrogenase complex. J. Biol. Chem. 279, 32904–32912 [DOI] [PubMed] [Google Scholar]
- 19. Ikeda Y., Okamura-Ikeda K., Tanaka K. (1985) Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. J. Biol. Chem. 260, 1311–1325 [PubMed] [Google Scholar]
- 20. Goodman S. I., Kratz L. E., DiGiulio K. A., Biery B. J., Goodman K. E., Isaya G., Frerman F. E. (1995) Cloning of glutaryl-CoA dehydrogenase cDNA, and expression of wild type and mutant enzymes in Escherichia coli. Hum. Mol. Genet. 4, 1493–1498 [DOI] [PubMed] [Google Scholar]
- 21. Tiffany K. A., Roberts D. L., Wang M., Paschke R., Mohsen A.-W., Vockley J., Kim J.-J. (1997) Structure of human isovaleryl-CoA dehydrogenase at 2.6 Å resolution: structural basis for substrate specificity. Biochemistry 36, 8455–8464 [DOI] [PubMed] [Google Scholar]
- 22. Wittwer A. J., Wagner C. (1981) Identification of folate-binding proteins of rat liver mitochondria as dimethylglycine dehydrogenase and sarcosine dehydrogenase. J. Biol. Chem. 256, 4109–4115 [PubMed] [Google Scholar]
- 23. Steenkamp D. J., Husain M. (1982) The effect of tetrahydrofolate on the reduction of electron transfer flavoprotein by sarcosine and dimethylglycine dehydrogenases. Biochem. J. 203, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGinnis S., Madden T. L. (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 27. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rhein V. F., Carroll J., He J., Ding S., Fearnley I. M., Walker J. E. (2014) Human METTL20 methylates lysine residues adjacent to the recognition loop of the electron transfer flavoprotein in mitochondria. J. Biol. Chem. 289, 24640–24651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho H. S., Shimazu T., Toyokawa G., Daigo Y., Maehara Y., Hayami S., Ito A., Masuda K., Ikawa N., Field H. I., Tsuchiya E., Ohnuma S., Ponder B. A., Yoshida M., Nakamura Y., Hamamoto R. (2012) Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 3, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Polevoda B., Martzen M. R., Das B., Phizicky E. M., Sherman F. (2000) cytochrome c methyltransferase, Ctm1p, of yeast. J. Biol. Chem. 275, 20508–20513 [DOI] [PubMed] [Google Scholar]
- 31. Chen R., Fearnley I. M., Palmer D. N., Walker J. E. (2004) Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease. J. Biol. Chem. 279, 21883–21887 [DOI] [PubMed] [Google Scholar]
- 32. Esser C., Ahmadinejad N., Wiegand C., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Bryant D., Steel M. A., Lockhart P. J., Penny D., Martin W. (2004) A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 [DOI] [PubMed] [Google Scholar]
- 33. Lu S. C., Mato J. M. (2012) S-Adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 92, 1515–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]