Background: Deoxycytidylate deaminases are the only allosterically regulated members of the zinc-dependent cytidine deaminase family.
Results: dTTP binding is confirmed and described in the previously defined dCTP allosteric binding site.
Conclusion: Higher affinity of dTTP than dCTP to the allosteric site may confer its inhibitory effect.
Significance: This is the first dTTP-bound crystal structure of deoxycytidylate deaminase.
Keywords: Allosteric Regulation, Cytidine Deaminase, Protein Structure, X-ray Crystallography, Zinc, Deoxycytidylate Deaminase, S-TIM5, dCMP Deaminase, dTTP
Abstract
Deoxycytidylate deaminase is unique within the zinc-dependent cytidine deaminase family as being allosterically regulated, activated by dCTP, and inhibited by dTTP. Here we present the first crystal structure of a dTTP-bound deoxycytidylate deaminase from the bacteriophage S-TIM5, confirming that this inhibitor binds to the same site as the dCTP activator. The molecular details of this structure, complemented by structures apo- and dCMP-bound, provide insights into the allosteric mechanism. Although the positioning of the nucleoside moiety of dTTP is almost identical to that previously described for dCTP, protonation of N3 in deoxythymidine and not deoxycytidine would facilitate hydrogen bonding of dTTP but not dCTP and may result in a higher affinity of dTTP to the allosteric site conferring its inhibitory activity. Further the functional group on C4 (O in dTTP and NH2 in dCTP) makes interactions with nonconserved protein residues preceding the allosteric motif, and the relative strength of binding to these residues appears to correspond to the potency of dTTP inhibition. The active sites of these structures are also uniquely occupied by dTMP and dCMP resolving aspects of substrate specificity. The methyl group of dTMP apparently clashes with a highly conserved tyrosine residue, preventing the formation of a correct base stacking shown to be imperative for deamination activity. The relevance of these findings to the wider zinc-dependent cytidine deaminase family is also discussed.
Introduction
Accurate reproduction of the genomic code, DNA replication, is an essential process in cell division that necessarily requires an available and balanced pool of deoxynucleotide building blocks (1, 2). Within the nucleotide biosynthesis pathway, dCMP may be converted to dUMP, the immediate precursor for thymidine nucleotide synthesis via thymidylate synthase, by the enzyme deoxycytidine-5-monophosphate (dCMP or deoxycytidylate) deaminase (3). Whereas deoxycytidylate deaminases (dCDs)2 have been biochemically characterized for decades (4–7), their catalytic activity has received renewed interest because it can lead to inactivation of the monophosphorylated form of deoxycytidine analogues, which are commonly used in anti-cancer and antiviral therapies (8).
dCD proteins form part of the zinc-dependent deaminase superfamily, which also includes free cytidine deaminases (CDAs) (9) and apolipoprotein B mRNA editing enzyme catalytic polypeptide-like (APOBEC) proteins (which deaminate cytidine within single-stranded DNA/RNA) (10), but not the dCTP deaminases, which possess a different fold and a zinc-independent catalytic mechanism (11). All zinc-dependent deaminases are characterized by an [H][A/V]E and PCX2–4C signature. The histidine and the two cysteines coordinate a zinc atom and form the catalytic core of the enzyme. Despite catalyzing identical chemical reactions, the substrate preferences of zinc-dependent deaminases has been shown to be very specific and must be regulated by structural nuances within an otherwise highly similar protein fold. Presumably, the differences in substrate recognition among these proteins are mainly a result of the length and position of the loops surrounding the catalytic site (9, 10). One interesting exception to this strict substrate selectivity is the report of a bifunctional dCTP-dCMP enzyme from chlorovirus (12).
dCDs are unique within the zinc-dependent deaminase superfamily as being allosterically regulated, activated by dCTP, and inhibited by the end product of this deoxynucleotide synthesis pathway, dTTP. Crystal structures from Streptococcus mutans (apo, PDB code 2HVV; and complexed with dCTP in the allosteric site and DHOMP substrate analogue in the active site, PDB code 2HVW) identified the allosteric binding site as a conserved GYNG motif (13), and although two other structures are available in the PDB from human (apo; PDB code 2W4L) and T4 phage R115E mutant (complexed with inhibitor PdR in the active site, PDB code 1VQ2 (14)), until now only biochemical evidence has driven the presumption that dTTP binds to the same allosteric binding site as its dCTP activator counterpart. A molecular level description of dTTP association with dCD has been lacking, and it is imperative for detailing the allosteric mechanism unique to these enzymes.
Here we report the first structure of a dCD in complex with dTTP, proving conclusively that this allosteric inhibitor binds not only to the same site as dCTP but also in a highly similar orientation. The atomic details of this structure provide a valuable description of the allosteric mechanism. The dTTP-bound dCD crystal structure from the bacteriophage S-TIM5 is complemented by structure apo and structures uniquely capturing dTMP and dUMP in the catalytic site, further revealing aspects of substrate specificity.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The gene encoding the full-length (135 amino acids) deoxycytidylate deaminase from S-TIM5 was subcloned into the pET28b vector (Novagen) to give an N-terminal thrombin cleavable His tag. Mutations were created using the QuikChange kit (Agilent Technologies Genomics) and validated by DNA sequencing. Plasmids were transformed into BL21(DE3) and grown in LB medium at 37 °C (to an A600 of 0.6), induced by 1 mm isopropyl β-d-thiogalactopyranoside (supplemented with 50 μm zinc acetate) and harvested 4 h after induction. Protein from lysed cells (lysis buffer containing 500 mm NaCl, 50 mm potassium phosphate, pH 7.5, 5 mm imidazole, and 5 mm β-mercaptoethanol) was purified on HisPur cobalt resin (Pierce). The tag was subsequently cleaved with thrombin (2 units/mg; Novagen), and protein was filtrated through a Superdex 200 size exclusion column (GE Healthcare Life Sciences) in 150 mm NaCl, 20 mm Tris-HCl, pH 7.0, and 1 mm DTT.
Protein Crystallization, Data Collection, and Structure Determination
Protein samples (∼10 mg/ml) were crystallized (hanging drop vapor diffusion at 20 °C) using the following conditions: S-TIM5-A (1.8 m malic acid, pH 7), S-TIM5-C (0.1 m MMT buffer, pH 8.0, 25% (w/v) PEG 1500, 5 mm MgCl2, 5 mm dCTP), and S-TIM5-T (0.2 m potassium formate, 20% (w/v) PEG 3350, 5 mm MgCl2, 5 mm dTTP). Cryoprotectant solutions contained an additional 20% glycerol. Diffraction data were collected at the European Synchrotron Radiation Facility on the beamlines indicated in Table 1. The data were scaled and merged using MOSFLM (15), and molecular replacement was carried out with Phaser (16) using 2HVV (13) as a search model. The structures were refined using Phenix (17) interspersed with manual modifications of the model into calculated electron density using Coot (18). Structural alignments, visualization, and structure figure preparations were performed using the PyMOL Molecular Graphics System (Schrödinger).
TABLE 1.
Data collection and refinement statistics
ASU, asymmetric unit.
| S-TIM5-A | S-TIM5-C | S-TIM5-T | |
|---|---|---|---|
| Data collection | |||
| Space group | P6322 | C2 | P212121 |
| Mol/ASU | 1 | 12 | 6 |
| Beamline | ESRF ID23-1 | ESRF ID14-4 | ESRF ID14-4 |
| Cell dimensions | |||
| a, b, c (Å) | 78.1, 78.1, 80.6 | 134.5, 146.6, 100.0 | 75.9, 79.0, 126.5 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 96, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-2.6 | 53.4-2.6 | 67.0-2.9 |
| Rsym or Rmerge | 0.07 (0.283)a | 0.088 (0.325) | 0.089 (0.349) |
| I/σI | 23.0 (8.3) | 11.1 (3.8) | 11.7 (4.0) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) | 99.8 (99.9) |
| Redundancy | 10.7 (11.4) | 4.0 (4.1) | 4.1 (4.3) |
| Refinement | |||
| Resolution (Å) | 2.6 | 2.6 | 2.9 |
| No. unique reflections | 4840 (672) | 60,000 (8747) | 17,452 (2511) |
| Reflections (work/test) | 4591/215 | 56,969/2905 | 17,399/880 |
| Rwork/Rfree (5%) | 0.215/0.272 | 0.233/0.268 | 0.212/0.268 |
| B-factors (mean) | 51.3 | 36.5 | 48.9 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.01 | 0.011 | 0.13 |
| Bond angles (°) | 1.3 | 1.1 | 1.4 |
| Ramachandran (%) | |||
| Favored | 100 | 98 | 97 |
| Outliers | 0 | 0 | 1 |
| PDB code | 4P9E | 4P9C | 4P9D |
| Ligands/Mol | 1Zn, 1Cl | 1Zn, 1dCMP, 1dUMP | 1Zn, 1Cl, 1Mg, 1dTTP, 1dTMP |
a The numbers in parentheses corresponds to the highest resolution shell.
Deamination Activity Assays
dCMP/dCTP/CMP deamination was monitored by measuring absorption at 290 nm every 15 s for 15 min in 100 μl of reaction solution containing 2 mm MgCl2 and 20 mm Tris-HCl, pH 7.0. Substrates were used at a concentration of 3.6 mm, activators/inhibitors were used at a concentration of 1 mm, and protein was used at 40 μm (for dCMP deamination) or 80 μm (for CMP or dCTP deamination reactions). The reactions were initiated by the addition of protein and were carried out in UV transparent plates incubated at 37 °C before reading. The reaction rates were determined from the linear portion of the graph obtained from plotting absorption as a function of time, and all reactions were carried out in triplicate on two biological repeats. The reaction rates relative to the wild type were calculated as average fractions with the standard deviation indicated in the error bars.
dCTP/dTTP Affinity Measurements
S-TIM5-dCD was labeled with Monolith NT protein labeling kit BLUE-NHS (Nano Temper Technologies) and eluted in microscale thermophoresis (MST) buffer (50 mm Tris, pH 7.0, 150 mm NaCl, 10 mm MgCl2, 0.05% Tween 20). S-TIM5-dCD (250 nm) was incubated with serial dilutions of either dCTP (Thermo Scientific and Invitrogen) or dTTP (Invitrogen), and MST measurements were carried out using the Monolith NT.115 in standard capillaries with 20% LED and 20% MST power.
RESULTS
Deoxycytidylate Deaminase Isolated from S-TIM5 Functions as a Typical dCMP Deaminase
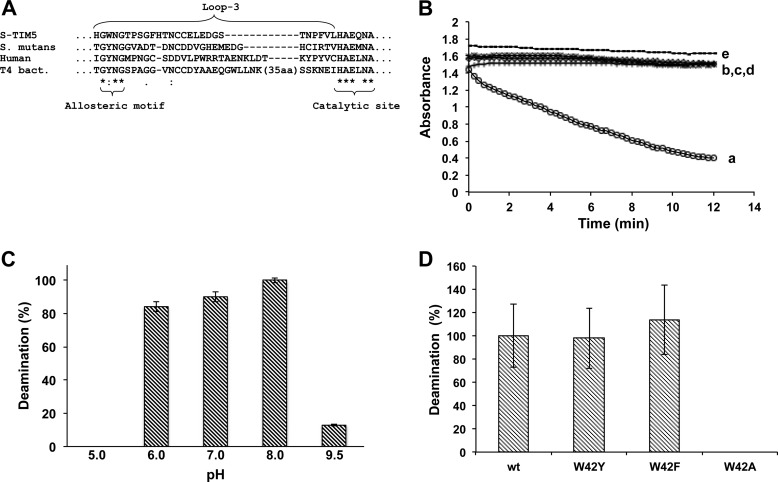
S-TIM5 is a recently isolated cyanophage that infects Synechococcus and was characterized as uniquely belonging to the Myoviridae family (19). Sequence analysis of the S-TIM5 dCD highlights a distinct larger tryptophan corresponding to the conserved aromatic tyrosine within the previously identified and conserved allosteric binding motif, GYNG (13) (Fig. 1A).
FIGURE 1.
Functional characterization of S-TIM5-dCD. A, sequence alignment of S-TIM5-dCD with structurally characterized dCDs. T4 bact., T4 bacteriophage. Displayed are only sequences of loop 3, including the catalytic motif and the allosteric motif highlighting the distinct tryptophan of S-TIM5-dCD allosteric motif. B, deamination activity of S-TIM5-dCD shown in absorbance (290 nm) as a function of time. Protein incubated with dCMP, dCTP, and Mg2+ (line a); dCMP and Mg2+ (line b); dCTP and Mg2+ (line c); CMP, dCTP, and Mg2+ (line d); and dCMP, dCTP, dTTP, and Mg2+ (line e). The ratio of dCTP to dTTP is 1:1. C, relative dCMP deamination activity of S-TIM5-dCD as a function of pH. The error bars represent triplicates. D, relative dCMP deamination activity of S-TIM5-dCD WT and Trp42 mutants in the presence of dCTP and Mg2+. The error bars represent three samples each of three biological replicates.
To probe the functional implications of this tryptophan distinct in S-TIM5-dCD, the wild type protein together with mutants harboring substitutions to the aromatic residue of the allosteric binding motif (W42Y, W42F, and W42A) were assayed for deamination activity (based on difference in absorption coefficient of the cytidine substrate and uridine product at 290 nm (13, 20)). Recombinant S-TIM5-dCD was found to function in a manner typical to dCMP deaminases that are strictly controlled by dCTP activation: in solution, it exists in the hexameric form (size exclusion chromatography results not shown) and catalyzes deoxycytidine deamination only on the dCMP substrate and only in the presence of dCTP allosteric activator (Fig. 1B, lines a and b). No deamination activity was detected when using dCTP (Fig. 1B, line c) or CMP (Fig. 1B, line d) as substrates. dTTP completely abolished deamination activity at a 1:1 ratio with dCTP (Fig. 1B, line e). The dependence of activity on pH was also investigated, and S-TIM5-dCD was shown to be active from pH 6.0 to 8.0 with the highest activity at pH 8.0 (Fig. 1C). The identity of the aromatic residue, Trp, Tyr, or Phe, in the allosteric binding motif has no significant effect on activity, although aromaticity at this position is imperative with an Ala substitution at this position, rendering the protein completely inactive (Fig. 1D).
Crystallization of S-TIM5-dCD Yields the First dTTP-bound Structure
The first ever structure of a deoxycytidylate deaminase in complex with the allosteric inhibitor dTTP was obtained by crystallization of purified S-TIM5-dCD in the presence of dTTP and MgCl2. This structure, in which dTMP was also identified at the active site, is complemented with two other S-TIM5-dCD crystal structures apo and cocrystallized with dCTP and MgCl2. Unexpectedly, cocrystallization with dCTP yielded only monophosphate nucleotides both at the catalytic and allosteric binding sites. Although purified protein was mixed with triphosphate nucleotides, dTTP or dCTP, apparently these solutions were partially degraded at the onset and/or degraded over the course of crystallization (20 °C incubation). Although presumably in minority, these monophosphate entities apparently favored crystallization over other complexes possibly present in solution, probably because of stabilizing the protein units needed to build the repeating crystal lattice. Data collection and refinement statistics for each of the structures, apo (S-TIM5-A) and cocrystallized with dTTP/dTMP (S-TIM5-T) or dCTP/dCMP (S-TIM5-C), are summarized in Table 1. Together, these structures detail for the first time interactions of dTTP and dCMP in the allosteric site, as well as dTMP and dUMP in the active site. Table 2 lists all the dCD structures available today and the identity of the entities bound at the active and allosteric sites.
TABLE 2.
Active and allosteric site occupancy of dCD crystal structures
| PDB code | Species | Active site | Allosteric site | Reference |
|---|---|---|---|---|
| IVQ2 | T4 bacteriophage R115E mutant | DHOMP | Ref. 14 | |
| 2HVV | S. mutans | Ref. 13 | ||
| 2HVW | S. mutans | DHOMP | dCTP allosteric activator | Ref. 13 |
| 2W4L | Human | |||
| 4P9E | S-TIM5 | dTMP | dTTP allosteric inhibitor | This study |
| 4P9D | S-TIM5 | dUMP | dCMP | This study |
| 4P9C | S-TIM5 | This study |
Although the composition of the asymmetric unit cell differed in each case (S-TIM5-A: monomeric, S-TIM5-T: hexameric, and S-TIM5-C: dodecameric), equivalent hexamers were formed in crystallo. The association of monomers into hexamers is analogous to that described in the previously reported structures with the same AB and AC interfaces (13, 14) (Fig. 2A). The most obvious global difference between the structures is a slightly tighter packing of the unbound S-TIM5-A pulling opposite subunits ∼2.5 Å closer as compared with the ligand-bound structures of S-TIM5-C and -T (Fig. 2A, inset). Superimposition of the monomeric units from each of the different structures reveals high similarity with less than 0.6 Å overall root mean square deviation (Fig. 2B). The most variable regions of the backbone are in the position of loops 3 and 7 reminiscent of the variability of these loops in the apo (PDB code 2HVV) and dCTP-bound (PDB code 2HVW) structures from S. mutans (13). In all three S-TIM5-dCD structures, these loops have the highest B-factors, apparently being so flexible in the S-TIM5-A structure that amino acids 55–60 of loop 3 and amino acids 116–118 of loop 7 were unstructured and lack detectable electron densities. The high overall similarity of the three structures is not surprising because all of these structures correspond to the inactive form of the enzyme, lacking a bound dCTP activator. Indeed the inactivity of the dCMP-bound structure may have facilitated its particular crystallization, over dCTP, providing structure of a monophosphate deoxycytidine bound to the allosteric motif and confirming the critical role of triphosphate entity in coordinating Mg2+ ions and the consequent activation/inhibition effects (see below).
FIGURE 2.
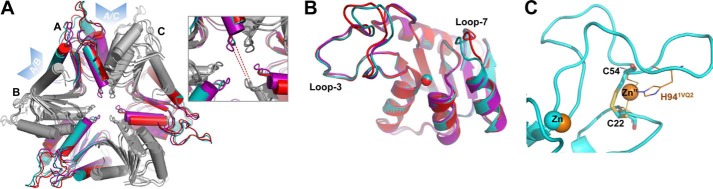
Overall structure of S-TIM5-dCD. A, superposition of the in crystallo hexameric forms (carried out in Coot). For clarity, every second protomer is shaded gray (S-TIM5-A in purple, S-TIM5-T in red, and S-TIM5-C in cyan). Blue arrowheads highlight interfaces between protomers. Inset, close-up of the middle region of the hexamer highlighting the greater compactness of the unbound S-TIM5-A structure (opposite protomers in the hexamer are ∼2.5 Å closer) as compared with the ligand-bound structures of S-TIM5-C and S-TIM5-T. B, superposition of the monomers from each structure. Zn2+ ions are displayed as spheres. C, secondary zinc binding motif in the S-TIM5-C (cyan) is substituted with a disulfide bond (represented by Cys22 and Cys54 as sticks). Secondary zinc (Zn”, orange sphere) and its binding residues (orange lines, His94 is labeled) from the structurally aligned T4 bacteriophage dCD (PDB code 1VQ2) are shown. Catalytic zinc ions are represented as overlapped cyan and orange spheres for S-TIM5-C and 1VQ2, respectively.
A unique feature of S-TIM5-dCD elucidated by these structures is the presence of a disulfide bond (formed between Cys22 and Cys54) analogously positioned in loop 3 to the second coordinated zinc ion identified in some dCDs (Fig. 2C). In most of the monomers, the electron density for the side chain of Cys22 is observed in two variant orientations corresponding to a reduced and oxidized disulfide bond. Interestingly, it was shown in T4 bacteriophage (21) that mutating His94 (Fig. 2C), which coordinates the second noncatalytic zinc, while keeping the two conserved Cys residues analogous to Cys22 and Cys54 in S-TIM5-dCD, inactivated the enzyme, suggesting that the coordinated Zn2+ ion and disulfide bond at this position are not entirely structurally interchangeable.
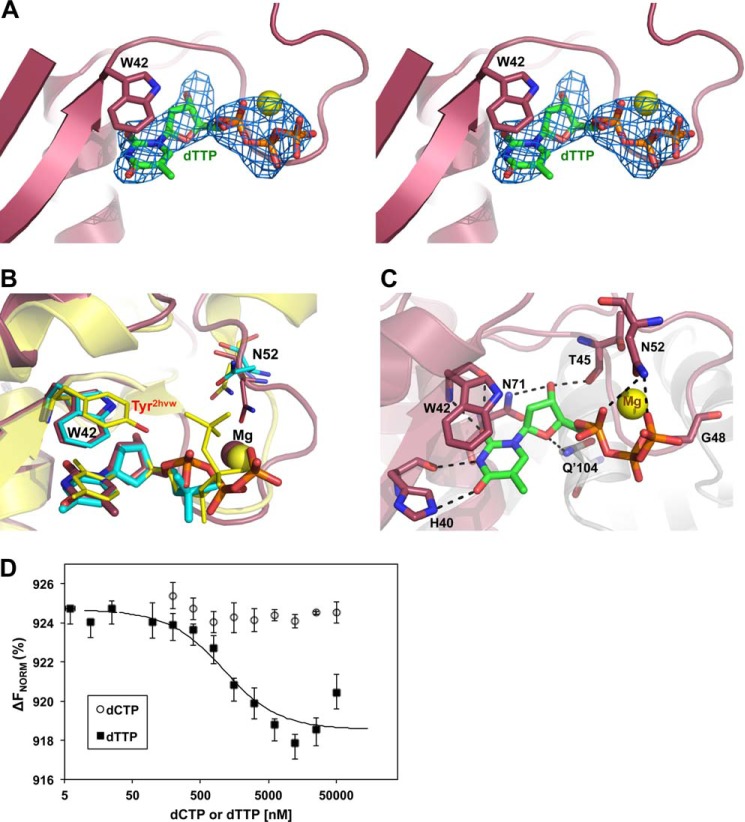
dTTP Binds Analogously to dCTP in the Allosteric Binding Site
The allosteric binding site for the dCTP activator has previously been described in the crystal structure of dCD from S. mutans complexed with dCTP·Mg2+ (PDB code 2HVW (13)) and biochemical evidence has driven the presumption that the dTTP inhibitor binds to this same site located near the AB interface (22). The S-TIM5-T structure now conclusively confirms this presumption with electron density fitting dTTP·Mg2+ clearly visualized at the allosteric binding site in the Fo − Fc map (Fig. 3A). Comparison of the S-TIM5-T and -C structures shows equivalent binding of the nucleoside component within the previously determined activator binding pocket and shows that the nucleoside is also superimposable with the deoxycytidine moiety of dCTP from the 2HVW structure (Fig. 3B). In both the S-TIM5-C and -T structures, polar contacts are made between the nucleoside of dCMP or dTTP and the side chains of His40, Asn71, and Gln′104 (from an adjacent C subunit), as well as the backbone of His40, Trp42, and Thr45 (Fig. 3C). Asn71 and Gln′104, which interact with the hydroxyl group and ring oxygen of the ribose, respectively (both interacting with the hydroxyl group in the 2HVW structure), are highly conserved residues in dCMP deaminases. These two interactions together with the stacking interaction between the nucleoside and the aromatic residue of the allosteric binding motif presumably bind the activator/inhibitor in a precise orientation necessary for function. Importantly, interaction with Gln′104 constitutes an intrasubunit interaction across the AC interface, the same interface that was presumably lost in the dimeric T4 bacteriophage R115E mutant that has reduced activity and functions independently of dCTP/dTTP activation/inhibition (6).
FIGURE 3.
dTTP bound at the allosteric binding site. A, cross-eyed stereo view of an Fo − Fc map (blue mesh) at 3 σ calculated with dTTP omitted from the S-TIM5-T structure. Trp42 is displayed as sticks, and the Mg2+ ion is a yellow sphere. B, structural alignment of the allosteric binding sites of S-TIM5-T (magenta) and 2HVW (yellow). Trp42 and dCMP from the S-TIM5-C structure are superimposed in cyan. The aromatic residue of the allosteric binding site and the bound nucleotide are represented as sticks. C, polar interactions made between dTTP (green) and the protein environment (magenta) are shown in black dashed lines. Interacting residues are labeled and represented with sticks, and the Mg2+ ion is a yellow sphere. D, MST measurements. S-TIM5-dCD binding affinities to dCTP and dTTP. Standard deviations were obtained from triplicate experiments. ΔFNorm, normalized fluorescence units calculated as percentage of bound and unbound substrate.
However, the analogous positioning of the nucleoside moieties is contrasted by the distinct conformations of the triphosphate and Mg2+ ion (Fig. 3B). The Mg2+ coordinated triphosphate of dTTP forms polar interactions with the side chain of Asn52 and the backbone of Gly48 (Fig. 3C). The triphosphate of dCTP in the 2HVW structure, which is twisted and contoured differently around the Mg2+ ion, makes these analogous contacts as well as two more interactions with nonconserved amino acids, including Tyr44 from the allosteric binding motif (Fig. 3B). Because the region of loop 3 interacting with the triphosphate of the activator/inhibitor shows low amino acid conservation (Fig. 1A), it is not clear whether the varied orientation of the triphosphate group in the S-TIM5-T and 2HVW structures is intrinsic of the different nucleotides (dTTP and dCTP, respectively) or a result of the varied protein binding environment. Only further structures, ideally a pair of structures activator and inhibitor bound from the same species, will clarify the significance of variability of triphosphate configuration.
Insights into the Mechanism of dTTP Allosteric Inhibition
The mechanism of dCTP activation/dTTP inhibition has not been defined, although it is known that a Mg2+ (or less effectively Ca2+ or Mn2+) ion is essential for allosteric control of dCD activity (14). The S-TIM5-C structure, which captures dCMP in the allosteric site, demonstrates that this monophosphate is not able to coordinate Mg2+, reinforcing the importance of the triphosphate entity in activation and inhibition. It has been suggested that interaction of the dCTP phosphate groups may provide the necessary forces to induce structural changes required to stably bind the substrate molecule and facilitate activity (13); however, to understand the allosteric mechanism, it is imperative to identify how interactions between dCD and the activator or inhibitor allow these similar molecules to prompt drastically different activity results. Comparison of the nucleotides from the S-TIM5-C and -T structures shows highly similar positioning of the nucleoside. Cytidine and thymidine, however, contain three chemical differences: the identity of the functional group attached to C4, the protonation state of N3, and the methylation of C5 in thymidine. The methyl group seems to be readily accommodated in the S-TIM5-T structure with no steric interference (Fig. 3A). The C4 functional group, O in thymidine and NH2 in cytidine, interacts with the nonconserved amino acid preceding the allosteric binding motif (His40 in S-TIM5-dCD) (Fig. 3C). Indeed the potency of dTTP varies from species to species with apparent correspondence to the ability of this residue preceding the allosteric motif to interact with the C4 oxygen of thymidine. S-TIM5 dCD activity was completely abolished at a 1:1 ratio of dTTP to dCTP (Fig. 1B, line e). Whereas the amino acid preceding the allosteric binding motif is a His, for which the NH1 group is at H-bonding distance with the O moiety of dTTP (Fig. 3C), the equivalent residue in S. mutans, Thr, does not support such a polar interaction, and a similar 1:1 dTTP to dCTP ratio resulted only in around 25% reduction of activity (13). Both chick embryo and human dCD (which has a weak activity even in the absence of the activator (14)) have an Ile residue preceding the allosteric motif, and the effect of dTTP on their activity has been reported as similar (5) with a 5:2 dCTP to dTTP ratio resulting only in a 50% reduction of activity at pH 8 (7). Most significant, however, with respect to allosteric control is the proximity of the nucleoside N3 to the backbone of the allosteric binding motif. Protonation of N3 in deoxythymidine and not deoxycytidine would facilitate the hydrogen bonding of deoxythymidine but not deoxycytidine with the main chain carbonyl of the residue preceding the allosteric motif. The carbonyl oxygen of the main chain of His40 in S-TIM5-dCD is at an ideal H-bonding distance of 2.8 Å from N3. Concurring with this hypothesis is the finding that S-TIM5-dCD dCMP deamination activity is completely lost at pH 5.0 (Fig. 1C) because at low pH the N3 group of deoxycytidine may also be protonated similar to deoxythymidine. Further supporting the notion that N3 protonation is important in defining inhibitor or activator identity is a study assessing the effect of phosphorylated pyrimidine analogues on dCMP deaminase activity in the presence of dCTP (23). Although there are many variables when comparing the 13 analogues investigated in that study (including several containing C5 methyl groups and several with the O moiety at C4), the two best inhibitors were the only two analogues having a protonated N3 (23).
To compare the affinity of dTTP and dCTP to S-TIM5-dCD, MST measurements were conducted (Fig. 3D). Because it is difficult to delineate between the affinity of substrate or activator/inhibitor, affinity measurements were carried out between S-TIM5-dCD and each dTTP and dCTP in buffer containing MgCl2 but in the absence of the dCMP substrate. Although the substrate surely influences the affinity of dCTP/dTTP, the MST measurements clearly show a higher intrinsic affinity of dTTP for the apo enzyme. The kDa of dTTP binding was calculated to be 960 nm, whereas over the same concentration range of dCTP no binding was observed.
Monophosphate Nucleotides Bound in the Catalytic Site Highlight Aspects of Substrate Selectivity
Clear electron density corresponding in size and shape to monophosphate pyrimidine nucleotides were identified both in the catalytic sites of the S-TIM5-T and S-TIM5-C structures. Whereas the monophosphate species in the catalytic site of S-TIM5-T structure clearly corresponds to dTMP, the catalytic site of S-TIM5-C structure may contain either dCMP substrate or dUMP product because dCMP in the crystallization solution would provide the necessary combination of substrate and activator for the ready formation of dUMP product. Differentiating between dCMP or dUMP in a static structure at 2.6 Å is dubious. Hence, although the S-TIM5-C structure models the catalytic site with dUMP (because analysis of the electron density appeared to indicate direct coordination of the substrate to the catalytic zinc ion rather than a water bridged positioning as was the case for dTMP in the S-TIM5-T structure), we cannot discount the possibility that dCMP is actually present or a mixture of the two.
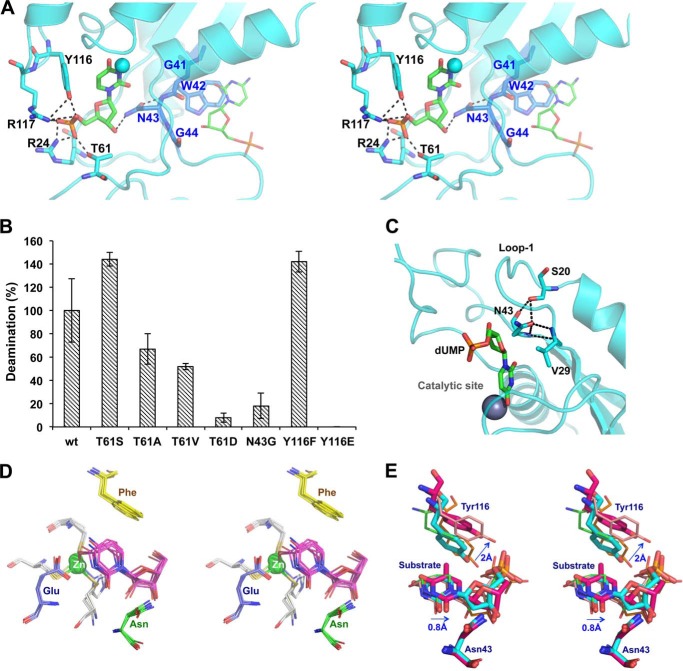
The dUMP and dTMP positioning and bonding networks within the catalytic site are similar to each other and to that of the DHOMP substrate analogue found in the 2HVW and 1VQ2 structures (Table 2). Apart from interactions made with the catalytic motif, the nucleoside sugar interacts with Asn43 from the allosteric motif (Fig. 4A). The GXNG allosteric motif (where X represents an aromatic residue) creates a barrier between the allosteric activator/inhibitor binding pocket and the substrate-binding pocket (Fig. 4A). Although the aromatic residue Trp in S-TIM5-dCD is oriented into the allosteric binding pocket, forming stacking interactions with either the inhibitor or activator, Asn forms part of the substrate-binding pocket. Although only dCDs harbor the allosteric motif, this particular Asn residue is highly conserved in all zinc-dependent cytidine deaminases (alignment not shown), and indeed a N43G mutation in S-TIM5-dCD results in a dramatic loss of activity, confirming the essentiality of this residue for activity (Fig. 4B). Interestingly Asn43 forms part of a hydrogen bonding triad together with two other residues highly conserved in dCDs: Ser20 and the backbone of Val29 (Fig. 4C). Ser20 and Val29 are located at either end of loop 1, and this triad may be essential in forming the closed conformation of this loop facilitating activity on the smaller substrates because this triad is conserved in the bifunctional dCTP-dCMP enzyme from chlorovirus and in CDAs but not in APOBECs.
FIGURE 4.
Monophosphate nucleotides bound at the catalytic sites. A, cross-eyed stereo view of the catalytic site of S-TIM5-C (cyan) with dUMP (green sticks) bound. Interacting residues (sticks), polar interactions (black dashed lines), and catalytic zinc ion (cyan sphere) are shown. Highlighted in blue sticks are the residues of the GWNG motif (with blue labels) separating the catalytic from allosteric sites, and shown with green lines is the dCMP bound at the allosteric site. B, deamination activity of S-TIM5-dCD containing substitutions to the conserved Tyr116, Asn43, and nonconserved Thr61, which interact with the bound substrate. C, the highly conserved hydrogen bonding triad (black dashed lines) formed in dCDs by Ser20, Val29, and Asn43 (shown as sticks) is highlighted on the cyan cartoon structure of S-TIM5-C. dUMP in the catalytic site is shown as green sticks, and the catalytic zinc is a gray sphere. D, cross-eyed view of superposition of CDAs with substrate/inhibitor-bound catalytic sites (PDB codes 1ZAB, 1JTK, 2FR6, 2FR5, 4EG2, 1ALN, and 1MQ0). Highlighted are the catalytic site zinc (green sphere), zinc-coordinating residues (gray), catalytic Glu (blue), substrate/inhibitor (magenta), the conserved phenylalanine from an adjacent subunit (yellow), and conserved asparagines (green). E, cross-eyed stereo view of superposition of dCDs with substrate/inhibitor-bound catalytic sites (S-TIM5-T in magenta sticks, S-TIM5-C in cyan sticks, 2HVW in pink lines, and 1VQ2 in orange lines). A CDA (2FR5 in green) from D is included for reference. Arrows and distances refer to relative movement of S-TIM5-T compared with S-TIM5-C. Residue numbering is for S-TIM5.
The phosphate group of both dUMP and dTMP makes polar interactions with the conserved Tyr116, as well as nonconserved Thr61, Arg117, and Arg24 residues (Fig. 4A). The positioning of the Tyr116 residue is significantly different between the two S-TIM5 structures. Highly conserved in the entire zinc-dependent cytidine deaminase family (as either tyrosine in dCDs and APOBECs or phenylalanine in CDAs) and implicated as important for orienting the cytidine substrate of CDAs by base stacking (24) (Fig. 4D), this residue is pushed back by at least 2 Å in the dTMP-bound structure (Fig. 4E). Whereas dTMP itself is slightly distanced from the active site compared with dUMP, probably as a result of the water bridging its interaction with the catalytic zinc ion, it appears that the presence of the methyl group prevents analogous positioning and stacking interaction of the tyrosine with respect to dUMP/dCMP (Fig. 4E). It is also of note that in substrate/inhibitor-bound CDA structures, the position of this phenylalanine residue is spatially tightly conserved (Fig. 4D), whereas much greater variability in the position of the corresponding tyrosine residue is noted for substrate/inhibitor-bound structures of dCDs (Fig. 4E).
Because unlike in the CDAs, the conserved Tyr116 of dCDs also interacts with the phosphate group of the substrate, we created a series of mutants to assess the relative importance of phosphate binding and substrate stacking interactions by Tyr116. S-TIM5-dCD Y116F, which is able to stack but not form a polar bond, was found to be almost 1.5 times as active as the WT protein, whereas for Y116E, which lacks the stacking ability, activity was totally abolished (Fig. 4B). These results indicate that the stacking ability of the conserved aromatic residue on loop 7 is essential probably for the correct positioning of the substrate for the deamination reaction, although the relative importance of otherwise binding the substrate through polar interactions is variable.
Further probing the role of the accurate positioning and interactions of the phosphate group of substrate in the deamination activity of S-TIM5-dCD, mutations to Thr61 (Fig. 4A) were made. We found that serine substitution increased the activity, substitutions with hydrophobic alanine or valine reduced the activity, and a phosphomimetic aspartic acid significantly reduced the activity (Fig. 4B). These results allow the conclusion that although binding of the phosphate group of the substrates by nonconserved residues of the catalytic cavity is helpful but not essential for the deamination activity, substrate docking with proper coulomb interactions with regard to the catalytic residues, the zinc ion, and the water molecule is crucial. Mutating S-TIM5-dCD Thr61 to the significantly larger aspartic acid probably leads to clashes with the dCMP substrate, preventing correct positioning with respect to the catalytic site and so resulting in drastically decreased activity.
DISCUSSION
The first dTTP inhibitor-bound dCD crystal structure reported here has confirmed and defined the nature of dTTP binding in the allosteric site, and the similarity to dCTP binding highlights the subtlety of activation and inhibition. The absence of coordinated magnesium by the dCMP bound in the allosteric site of the S-TIM5-C structure reiterates the importance of the triphosphate entity, which, when magnesium is coordinated, probably positions loop 3 around the substrate facilitating activity. Whether the orientation of the triphosphate intrinsically varies between dTTP and dCTP in the allosteric site of dCDs is still not clear in the absence of a pair of structures from the same species, activator- and inhibitor-bound. Still, the mechanism of activation or inhibition must be related to the interactions particularly made by the dCTP or dTTP base moiety with the allosteric binding pocket, with this being the chemical difference between the two nucleotides, which may define varied conformations of the triphosphate or directly affect activity. Analysis of the dTTP structure suggests that dTTP may have a higher affinity than dCTP to the allosteric site, leading to its inhibitory effect, and indeed, at least in the absence of the dCMP substrate, this is supported by MST measurements. A clear and definitive mechanistic description of the allosteric mechanism, however, remains elusive, partially because of the variability of sequences around the allosteric site, which clouds distinction between intrinsic differences in dCTP or dTTP binding or insignificant variations of interactions as a result of species variability, particularly around the triphosphate.
Although allosteric regulation is distinctive to the dCDs, defining substrate binding and specificity should be of broader interest across studies into all members of the zinc-dependent cytidine deamination family. Because, for example, a substrate-bound form of any APOBEC protein has remained elusive, structures of substrate-bound CDAs have been used to model substrates into apo APOBEC structures. To this end, the dTMP- and dUMP-bound structures of S-TIM5-dCD will certainly be of interest in future APOBEC substrate modeling efforts. Indeed the drastic decrease in activity by mutation of S-TIM5-dCD Thr61 to the significantly larger aspartic acid has been similarly observed for APOBEC3G T218E and AID T27E mutants whose cytidine deamination activity was greatly reduced without affecting DNA binding or steady state subcellular localization (25). Although the threonines in A3G and AID are themselves homologous, they do not spatially align with Thr61 from S-TIM5-dCD. Still being located on the solvent exposed loop 3 surrounding the active site, it is possible that there is a similar molecular basis for phosphorylation-dependent inhibition of their deamination activity, presumably via clashes with the target deoxycytidine preventing proper alignment necessary for deamination or preventing the presumable base flipping of target deoxycytidine into the catalytic site of APOBEC/AID proteins.
Acknowledgments
We thank the European Synchrotron Radiation Facility for the allocation of synchrotron radiation beamtime and the staff for assistance at Beamlines ID23-1 and ID14-4. We thank Prof. Debbie Lindell for providing S-TIM5 genome and Rina Benhamou for cloning and preliminary purification and crystallization of the S-TIM5 dCMP deaminase protein.
This work was supported by Israel Science Foundation Grant 1354/14.
The atomic coordinates and structure factors (codes 4P9E, 4P9D, and 4P9C) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- dCD
- deoxycytidylate deaminase
- CDA
- cytidine deaminase
- APOBEC
- apolipoprotein B mRNA editing enzyme catalytic polypeptide-like
- MST
- microscale thermophoresis
- DHOMP
- 3,4-dihydrouridine 5-monophosphate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Mathews C. K. (2006) DNA precursor metabolism and genomic stability. FASEB J. 20, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 2. Rampazzo C., Miazzi C., Franzolin E., Pontarin G., Ferraro P., Frangini M., Reichard P., Bianchi V. (2010) Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat Res. 703, 2–10 [DOI] [PubMed] [Google Scholar]
- 3. Maley F., Maley G. F. (1990) A tale of two enzymes, deoxycytidylate deaminase and thymidylate synthase. Prog. Nucleic Acids Res. Mol. Biol. 39, 49–80 [DOI] [PubMed] [Google Scholar]
- 4. Maley G. F., Maley F. (1982) Allosteric transitions associated with the binding of substrate and effector ligands to T2 phage induced deoxycytidylate deaminase. Biochemistry 21, 3780–3785 [DOI] [PubMed] [Google Scholar]
- 5. Maley G. F., Lobo A. P., Maley F. (1993) Properties of an affinity-column-purified human deoxycytidylate deaminase. Biochim. Biophys. Acta 1162, 161–170 [DOI] [PubMed] [Google Scholar]
- 6. Keefe R. G., Maley G. F., Saxl R. L., Maley F. (2000) A T4-phage deoxycytidylate deaminase mutant that no longer requires deoxycytidine 5′-triphosphate for activation. J. Biol. Chem. 275, 12598–12602 [DOI] [PubMed] [Google Scholar]
- 7. Maley G. F., Maley F. (1968) Regulatory properties and subunit structure of chick embryo deoxycytidylate deaminase. J. Biol. Chem. 243, 4506–4512 [PubMed] [Google Scholar]
- 8. Jansen R. S., Rosing H., Schellens J. H., Beijnen J. H. (2011) Deoxyuridine analog nucleotides in deoxycytidine analog treatment: secondary active metabolites? Fundam Clin. Pharmacol. 25, 172–185 [DOI] [PubMed] [Google Scholar]
- 9. Conticello S. G. (2008) The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bransteitter R., Prochnow C., Chen X. S. (2009) The current structural and functional understanding of APOBEC deaminases. Cell Mol. Life Sci. 66, 3137–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johansson E., Fanø M., Bynck J. H., Neuhard J., Larsen S., Sigurskjold B. W., Christensen U., Willemoës M. (2005) Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J. Biol. Chem. 280, 3051–3059 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Maley F., Maley G. F., Duncan G., Dunigan D. D., Van Etten J. L. (2007) Chloroviruses encode a bifunctional dCMP-dCTP deaminase that produces two key intermediates in dTTP formation. J. Virol. 81, 7662–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou H. F., Liang Y. H., Li L. F., Su X. D., Dong Y. H. (2008) Crystal structures of Streptococcus mutans 2′-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP × Mg2+. J. Mol. Biol. 377, 220–231 [DOI] [PubMed] [Google Scholar]
- 14. Almog R., Maley F., Maley G. F., Maccoll R., Van Roey P. (2004) Three-dimensional structure of the R115E mutant of T4-bacteriophage 2′-deoxycytidylate deaminase. Biochemistry 43, 13715–13723 [DOI] [PubMed] [Google Scholar]
- 15. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 16. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sabehi G., Shaulov L., Silver D. H., Yanai I., Harel A., Lindell D. (2012) A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc. Natl. Acad. Sci. U.S.A. 109, 2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moore J. T., Silversmith R. E., Maley G. F., Maley F. (1993) T4-phage deoxycytidylate deaminase is a metalloprotein containing two zinc atoms per subunit. J. Biol. Chem. 268, 2288–2291 [PubMed] [Google Scholar]
- 21. McGaughey K. M., Wheeler L. J., Moore J. T., Maley G. F., Maley F., Mathews C. K. (1996) Protein-protein interactions involving T4 phage-coded deoxycytidylate deaminase and thymidylate synthase. J. Biol. Chem. 271, 23037–23042 [DOI] [PubMed] [Google Scholar]
- 22. Maley F., Maley G. F. (1982) Studies on identifying the allosteric binding sites of deoxycytidylate deaminase. J. Biol. Chem. 257, 11876–11878 [PubMed] [Google Scholar]
- 23. Liou J. Y., Krishnan P., Hsieh C. C., Dutschman G. E., Cheng Y. C. (2003) Assessment of the effect of phosphorylated metabolites of anti-human immunodeficiency virus and anti-hepatitis B virus pyrimidine analogs on the behavior of human deoxycytidylate deaminase. Mol. Pharmacol. 63, 105–110 [DOI] [PubMed] [Google Scholar]
- 24. Vincenzetti S., Cambi A., Maury G., Bertorelle F., Gaubert G., Neuhard J., Natalini P., Salvatori D., De Sanctis G., Vita A. (2000) Possible role of two phenylalanine residues in the active site of human cytidine deaminase. Protein Eng. 13, 791–799 [DOI] [PubMed] [Google Scholar]
- 25. Demorest Z. L., Li M., Harris R. S. (2011) Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J. Biol. Chem. 286, 26568–26575 [DOI] [PMC free article] [PubMed] [Google Scholar]