Abstract
Our genome contains many G-rich sequences, which have the propensity to fold into stable secondary DNA structures called G4 or G-quadruplex structures. These structures have been implicated in cellular processes such as gene regulation and telomere maintenance. However, G4 sequences are prone to mutations particularly upon replication stress or in the absence of specific helicases. To investigate how G-quadruplex structures are resolved during DNA replication, we developed a model system using ssDNA templates and Xenopus egg extracts that recapitulates eukaryotic G4 replication. Here, we show that G-quadruplex structures form a barrier for DNA replication. Nascent strand synthesis is blocked at one or two nucleotides from the G4. After transient stalling, G-quadruplexes are efficiently unwound and replicated. In contrast, depletion of the FANCJ/BRIP1 helicase causes persistent replication stalling at G-quadruplex structures, demonstrating a vital role for this helicase in resolving these structures. FANCJ performs this function independently of the classical Fanconi anemia pathway. These data provide evidence that the G4 sequence instability in FANCJ−/− cells and Fancj/dog1 deficient C. elegans is caused by replication stalling at G-quadruplexes.
Keywords: DNA Replication, FANCJ, G4 DNA, G-quadruplex, Xenopus egg extract
Introduction
Genome stability is ensured by a large variety of specialized DNA surveillance and repair pathways. These mechanisms efficiently deal with DNA damage from exogenous sources as well as damage generated intracellularly. One of the cellular processes that can be a source of genome instability is DNA replication. Although the intrinsic error rate of this process is extremely low, its fidelity is continuously threatened, including by stable secondary structures in the DNA (Aguilera & Garcia-Muse, 2013). One particularly stable DNA structure is a G4 or G-quadruplex structure (hereafter referred to as G-quadruplex structure) (Bochman et al, 2012; Tarsounas & Tijsterman, 2013). This structure can form in DNA sequences that contain four stretches of three or more guanines (Gs) interspaced by at least one random nucleotide (Fig 1A, hereafter referred to as G4 sequence). Four Gs, one from each G-stretch, can form a planar structure stabilized by non-canonical Hoogsteen hydrogen bonds in the presence of monovalent cations, such as sodium or potassium. In vitro, G4 sequences can adopt a variety of structural conformations, depending on the length and orientation of the G-stretches and intervening loops (Fig 1B) (Burge et al, 2006; Phan et al, 2007). Because Watson and Crick base pairing in double-stranded DNA is more favorable than G4 Hoogsteen base pairing, G-quadruplex structures are preferentially formed in single-stranded DNA (Phan & Mergny, 2002). Therefore, it has been suggested that in vivo these structures form during processes that allow for temporal dissociation of duplex DNA, that is DNA replication, transcription and/or recombination (Maizels & Gray, 2013).
Figure 1. G-quadruplex structures and sequences.
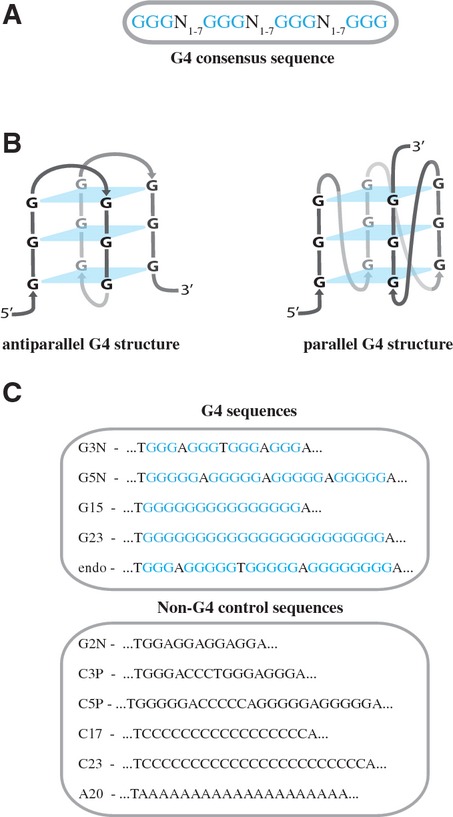
- G4 consensus sequence consisting of four stretches of at least three guanines (G) separated by 1–7 random nucleotides (N).
- Schematic representation of an antiparallel (left) and a parallel (right) G-quadruplex structure. G-planes stabilized by non-canonical Hoogsteen hydrogen bonds are shown in blue.
- G4 sequences and non-G4 control sequences used in this study.
Our genome contains over 300,000 evolutionary conserved sequences that conform to the G4 consensus sequence (Fig 1A) (Huppert & Balasubramanian, 2005; Todd et al, 2005). G-quadruplex structures have been suggested to play a role in several biological processes such as telomere maintenance (Maiti, 2010), gene regulation (Siddiqui-Jain et al, 2002), DNA replication initiation (Besnard et al, 2012; Cayrou et al, 2012; Valton et al, 2014), epigenetic regulation (Sarkies et al, 2010), and gene conversion (Cahoon & Seifert, 2009). However, which and how many of the G4 sequences present in the genome form a G-quadruplex structure at a given time in a human cell is not known. Nevertheless, a number of recent studies using fluorescently tagged G4 ligands (Rodriguez et al, 2012) and highly specific G4 antibodies (Biffi et al, 2013; Henderson et al, 2013) provide evidence that these structures are abundantly present in proliferating human cells. The assembly and disassembly of G-quadruplex structures is dynamic during the cell cycle and their number is significantly increased during DNA replication in S-phase (Biffi et al, 2013). Furthermore, DNA damage induced by a strong G4 stabilizing ligand depends on active DNA replication (Rodriguez et al, 2012), suggesting that the formation of G-quadruplex structures occurs mainly during DNA replication.
Several lines of evidence suggest that G-quadruplex structures need to be actively resolved during DNA replication. First, G-quadruplex structures form a block for DNA synthesis in primer extension assays using both prokaryotic and eukaryotic DNA polymerases in vitro (Kaguni & Clayton, 1982; Woodford et al, 1994; Weitzmann et al, 1996; Kamath-Loeb et al, 2001), indicating that polymerases cannot unwind G-quadruplex structures. Although several DNA helicases, such as BLM (Sun et al, 1998), WRN (Fry & Loeb, 1999; Kamath-Loeb et al, 2001), Pif1 (Ribeyre et al, 2009; Sanders, 2010), DOG-1/FANCJ (London et al, 2008; Wu et al, 2008), and Dna2 (Masuda-Sasa et al, 2008) can unwind G-quadruplex structures in vitro, the role of these proteins in vivo is unclear. Second, G4 sequences are found specifically enriched at chromosomal breakpoints in human cancers (De & Michor, 2011; Nambiar et al, 2011; Bose et al, 2014). This observed genome instability is thought to be a consequence of replicative stress. Third, genetic data show that dog-1 mutant animals (C. elegans) accumulate deletions that map to G4 sequences and are characterized by a specific signature that implies the collision of the replication machinery as an initiating event (Cheung et al, 2002; Youds et al, 2006; Kruisselbrink et al, 2008). Likewise, in yeast, the absence of a G4 unwinding helicase, Pif1, induces mutations that disrupt G4 sequences and this is enhanced by the addition of chemicals that stabilize G-quadruplex structures (Ribeyre et al, 2009; Paeschke et al, 2011). In addition, Pif1 deficiency induces slower movement of replication forks through genomic regions that contain G4 sequences (Paeschke et al, 2011). Altogether, these data suggest that G-quadruplex structures cause problems for the progression of the DNA replication machinery and that these problems are enhanced during replicative stress or in the absence of helicases such as Pif1 or DOG-1.
The vertebrate homologue of DOG-1 is FANCJ, also named BACH1 or BRIP1, which performs several roles in genome maintenance. FANCJ is one of the 16 genes that, when mutated, cause Fanconi anemia (FA); a human cancer-predisposition disorder characterized by cellular sensitivity to DNA interstrand crosslinking agents (Levitus et al, 2005; Levran et al, 2005; Muniandy et al, 2010). In addition to its function in interstrand crosslink repair, FANCJ has also been suggested to play a role in the processing of G-quadruplex structures, primarily based on the G4 sequence instability phenotype of DOG-1/FANCJ deficient C. elegans strains. Likewise, cells derived from human FANCJ patients accumulate gross chromosomal rearrangement more frequently near G4 sequences (London et al, 2008). In addition, FANCJ deficient human and chicken cells are sensitive to treatment with G4 stabilizing ligands and, as a consequence of this treatment, exhibit enhanced DNA damage responses (Wu et al, 2008; Schwab et al, 2013). However, direct evidence that FANCJ resolves G-quadruplex structures in cells is missing. Because cells deficient for the central FA protein FANCD2 are not sensitive to G4 ligands, it has been suggested that the function of FANCJ in maintaining G4 sequence stability is FA pathway independent. Yet, C. elegans deficient in both FANCJ and FANCD2 show a higher mutation rate at G4 sequences compared to the single FANCJ mutant (Youds et al, 2008), suggesting there might be a functional relationship between FANCJ and FANCD2.
To investigate the molecular events that take place when the replication machinery encounters a G-quadruplex structure, we employed Xenopus egg extracts to replicate exogenous G4 sequence on single-stranded DNA plasmids under physiological conditions. Using this unique model system, we show for the first time that replication stalls at a defined G-quadruplex structure. Mapping of the nascent strands at nucleotide resolution demonstrates that replication proceeds to within a few nucleotides from the G-quadruplex. After transient stalling, we observe efficient bypass and faithful replication of the G4 sequence. In addition, we show that replication stalling at G-quadruplex structures is enhanced in the absence of FANCJ. Further stabilization of the G-quadruplex by addition of a G4 stabilizing ligand increases the requirement for FANCJ. In addition to providing a framework for future studies on the mechanism of G-quadruplex structure unwinding, our data also explain the genetic instability at G4 sequences in FANCJ mutants.
Results
G-quadruplex structures form a block for DNA polymerases
To study G4 DNA replication, we generated a series of single-stranded DNA plasmids, each containing a different G4 sequence at a defined position (G4 plasmids). In addition, we generated control plasmids carrying G-rich sequences that do not conform to the G4 consensus sequence (non-G4 plasmids) (Fig 1C). The G4 sequences either consisted of 4 stretches of several guanines separated by single adenines or of a consecutive stretch of Gs. G4G3N and G4G15 are ‘minimal’ G4 sequences and can only form one G-quadruplex configuration with 3 G-planes, while G4G5N and G4G23 contain additional Gs and can form many structurally different G-quadruplex structures with up to five stacked G-planes. G-quadruplex structures were induced in the G4 plasmids by brief incubation at 80°C in the presence of physiological concentrations of potassium (Matsugami et al, 2003).
It has previously been demonstrated that G-quadruplex structures on short linear DNA templates block extension by purified polymerases (Woodford et al, 1994; Weitzmann et al, 1996; Kamath-Loeb et al, 2001). To examine whether this is also the case in our G4 sequence containing plasmids, we performed primer extension assays using these plasmid templates and the modified T7 DNA polymerase (Sequenase) (Tabor & Richardson, 1989). To this end, 5′ 32P-labeled primer B was annealed to the ssDNA plasmids approximately 200 nt 3′ of the G4 sequence and this template was incubated in Sequenase reaction mix (Fig 2A and Supplementary Fig S1A). Extension products were separated on denaturing urea-PAGE gels and visualized by autoradiography (Fig 2B). When using a non-G4 control plasmid, multiple extension products accumulated over time, but there was no specific stalling observed at the G-rich sequence (Fig 2B, left panel, and Supplementary Fig S1B). In contrast, when G4 plasmids were examined, a ˜200-nt product rapidly accumulated and persisted, indicating extension was effectively blocked close to the G4 sequence (Fig 2B, right panels, and C, and Supplementary Fig S1C and D for duplicates). The observed T7 polymerase extension stalling indicates that G-quadruplex structures are formed efficiently in all our G4 plasmid templates. This conclusion is further strengthened by the observation that stalling at a G-quadruplex is fully dependent on the presence of potassium (Supplementary Fig S1E and F). Based on the low DNA concentration used and the observation that G-rich control sequences that could theoretically form intermolecular G-quadruplexes do not stall primer extension, we assume that the majority of the G-quadruplexes are intramolecular. Importantly, these results show that G-quadruplex structures in ssDNA plasmids cannot be bypassed by T7 polymerase alone.
Figure 2. G-quadruplex structures block DNA synthesis by T7 DNA polymerase.
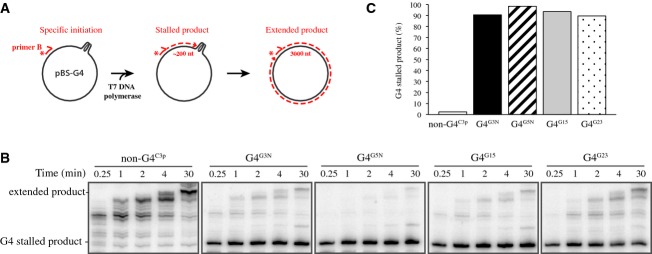
- Schematic representation of the primer extension assay. Extension is initiated from 32P-labeled (asterisk) primer B annealed to single-stranded pBluescript DNA containing a G4 sequence (left). If extension is blocked by the G-quadruplex structure, accumulation of a ˜200-nt product is expected (middle) while full extension generates a ˜3,000-nt product (right).
- G4 and non-G4 plasmid templates were subjected to primer extension by a modified T7 DNA polymerase (Sequenase). Extension was stopped after the indicated times, and reaction products were separated on 6% urea-PAGE gels and visualized by autoradiography.
- The extension stalling products after 1 min (from B) were quantified using ImageQuant software, and the percentage of this product versus the total of products that have arrived or bypassed the G4 sequence is depicted for the various sequences used.
G-quadruplex structures are efficiently bypassed in Xenopus egg extract
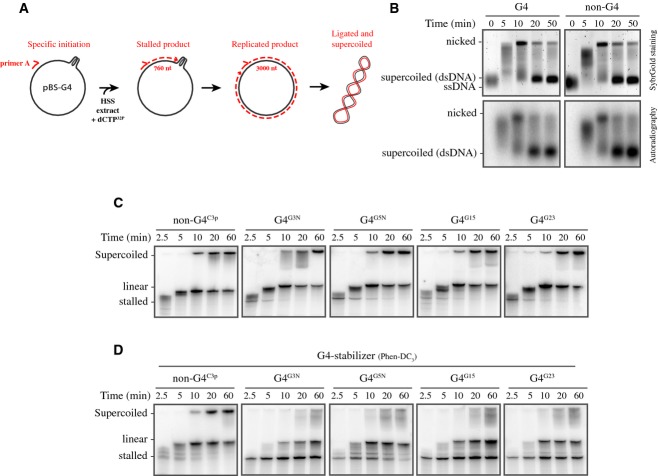
G-quadruplex structures are thought to form in ssDNA. During DNA replication, ssDNA is present at the lagging strand template but also on the leading strand template in cases of transient uncoupling of the MCM helicase from the polymerase (Pacek & Walter, 2004). In both cases, the approach of the growing 3′ end of the nascent strand to the G-quadruplex does not require unwinding of the DNA double helix. To investigate how G-quadruplex structures are resolved during eukaryotic DNA replication, we mimicked this situation by replicating primed ssDNA templates in Xenopus egg extract. A high-speed supernatant of egg cytoplasm (HSS) supports one round of efficient DNA replication of ssDNA plasmids initiated by sequence-independent priming (Mechali & Harland, 1982). The replicated plasmids are also ligated and supercoiled in this extract. At low DNA concentrations, the inherent random priming activity of the extract is inhibited (Lebofsky et al, 2011); therefore, replication can be initiated from an exogenous primer. For this, primer A, located ˜760 nt 3′ of the G4 or G-rich control sequence, was annealed to the ssDNA plasmids and these templates were incubated in HSS in the presence of 32P-α-dCTP (Fig 3A and Supplementary Fig S1A). Replication products were separated on a native agarose gel, which was subsequently stained with SybrGold (Fig 3B, top panels). In both conditions, the ssDNA plasmid template was observed at t = 0 and replication intermediates appeared within the first 5 min. Ten minutes after the start of the reaction fully replicated nicked molecules accumulated, and after 20 min, the majority of the plasmids were supercoiled. At this point, all ssDNA was converted to dsDNA indicating complete and efficient replication of the template DNA (Fig 3B, top panels). Gels were subsequently dried and autoradiography showed a very similar pattern for the nascent 32P-labeled products confirming that these are replication products (Fig 3B, bottom panels). The banding pattern for the G4 versus the non-G4 plasmid template was highly similar, suggesting that the G-quadruplex structure does not have a major effect on the replication kinetics and efficiency in HSS. Importantly, no replication was observed after depletion of PCNA, indicating the involvement of the replicative polymerases pol δ or pol ε (Supplementary Fig S2 and Mattock et al, 2001). These data show that, in contrast to the primer extension reaction with a purified polymerase, G-quadruplex structures are efficiently replicated in Xenopus egg extract.
Figure 3. G-quadruplex structures are efficiently replicated in Xenopus egg extracts.
- Schematic representation of the G4 plasmid replication assay in HSS. Replication is started from primer A located 760 nucleotides (nt) from the G-quadruplex. Stalling of replication at the G-quadruplex structure will result in the accumulation of a 760-nt product, while G4 bypass first generates a 3,000-nt product that over time is ligated and supercoiled.
- G4G3N and non-G4C3p plasmid templates were replicated in HSS. Samples collected at the indicated times were separated on agarose gels and visualized with SybrGold (top panels). Gels were subsequently dried and visualized by autoradiography (bottom panels). The bands corresponding to the ssDNA, fully replicated but still nicked, and supercoiled dsDNA are indicated.
- Non-G4C3p, G4G3N, G4G5N, G4G15, and G4G23 plasmids were replicated in HSS, samples were taken at the indicated times, separated on 6% urea-PAGE gels and visualized by autoradiography. Products stalled at the G4 sequence (‘stalled’), linear molecules resulting from denatured nicked products (‘linear’), and closed supercoiled products (‘supercoiled’) are indicated.
- Non-G4C3p, G4G3N, G4G5N, G4G15, and G4G23 plasmid templates were replicated in HSS in the presence of 5 μM of Phen-DC3. Samples collected at the indicated times were separated on urea-PAGE gels and visualized by autoradiography.
To investigate whether there is a subtle effect of the G-quadruplex on nascent strand progression, replication products were analyzed on urea-PAGE gels. Using a control non-G4 plasmid template, we observed the accumulation of ˜3,000-nt linear molecules, corresponding to fully replicated nicked molecules, and slower migrating supercoiled molecules from 10 min onwards (Fig 3C, left panel). Strikingly, when replicating various G4 plasmid templates, an additional band of around 750 nt, corresponding to a product stalled close to the G4 site, was visible from 2.5 up to 20 min depending on the replicated G4 sequence (Fig 3C, right panels, and Supplementary Fig S3). These results demonstrate that, even though G4 sequence containing plasmids are efficiently replicated, this is associated with transient stalling in close proximity of a G-quadruplex structure. In summary, Xenopus egg extract contains all factors required to resolve and bypass G-quadruplex structures, making this a suitable and powerful system to study eukaryotic G4 unwinding and replication.
G-quadruplex stabilizing agent Phen-DC3 blocks G4 replication in Xenopus egg extract
Previous reports have shown that G4 stabilizing ligands inhibit the unwinding activity of several G4 helicases in vitro and that cells are sensitive to treatment with these ligands (Piazza et al, 2010; Bharti et al, 2013). To investigate this, we assessed the capacity of the extract to bypass stabilized G-quadruplex structures by employing the G4 ligand Phen-DC3 (Piazza et al, 2010). While replication of control non-G4 plasmids showed no accumulation of products stalled at the G-rich site (Fig 3D, left panel), accumulation of stalled products was dramatically enhanced when G4 plasmids were replicated in the presence of Phen-DC3 (Fig 3D, right panels, compare with Fig 3C). Furthermore, fully replicated molecules were less abundant at later times, which indicates that replication past G-quadruplex structures was severely inhibited. This shows that increasing G4 structure stability enhances replication stalling. In contrast to this G4-specific replication stalling in extract, adding Phen-DC3 to the T7 primer extension assay in the absence of extract, induced replication stalling at several sites independent of the G4 sequence (Supplementary Fig S4A). We conclude that the plasmid contains other Phen-DC3-sensitive secondary structures that are readily resolved in Xenopus egg extract. In addition to Phen-DC3, we tested the widely used telomeric G4 stabilizer TMPyP4 and compared it to a control compound TMPyP2 (Mergny & Helene, 1998; Han et al, 2001). Although TMPyP4 slightly enhanced replication stalling at a G4 sequence in Xenopus egg extract, it induced extensive unspecific inhibition of replication, especially at high dose, which was also observed with TMPyP2 (Supplementary Fig S4B–D). This unspecific effect of TMPyP4 could be caused by the absence of telomeric context in our system. Nevertheless, we conclude that the specific enhancement of stalling at G-quadruplexes, as we have seen with Phen-DC3, could explain the hypersensitivity of cells to G4 stabilizing agents.
G4 sequences are not mutated after replication in Xenopus egg extract
It has been shown that G4 sequences give rise to genome alterations in a variety of species, including bacteria, yeast, and C. elegans (Kruisselbrink et al, 2008; Cahoon & Seifert, 2009; Ribeyre et al, 2009). To determine whether replication of G4 sequences in Xenopus egg extract induces mutations, G4 replication products were sub-cloned and sequenced. None of the analyzed replication products showed mutations or deletions at the G4 sequence (Supplementary Fig S5). The observation that replication through G-quadruplex is non-mutagenic indicates that the mechanism of bypass does not involve G4 skipping via template switching or any other mechanism that would lead to deletion of the G4 sequence. This supports a model in which, after initial replication stalling, the G-quadruplex structure is unwound and faithfully replicated.
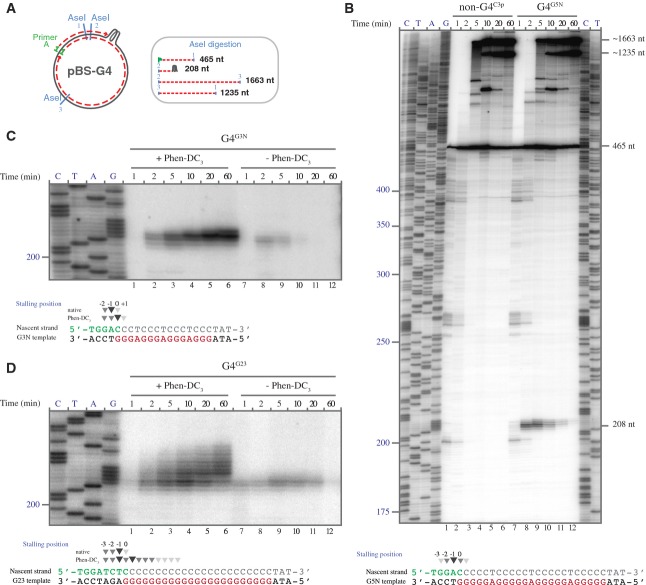
Replication in Xenopus egg extract stalls at the site of the G4
To investigate the mechanism by which G-quadruplex structures are resolved and bypassed, we set out to determine where the replication machinery stalls with respect to the G-quadruplex. To this end, ssDNA plasmids were replicated in egg extract starting from primer A located 760 nt 3′ of the G4 site. Replication products were digested with AseI and separated on high-resolution sequencing gels (Fig 4A and B). When a non-G4 plasmid was replicated, a band of ˜465 nt accumulated after 1 min, corresponding to a product that starts from the primer and ends at the first AseI site (Fig 4A and B, lane 1). Over time, two new bands appeared, one corresponding to a digestion product from the second to the third AseI site (1,663 nt), followed by a band corresponding to the digested product from the third to the first AseI site after ligation (1,235 nt) (Fig 4A and B, lanes 2–6). The order at which these bands appeared confirmed that replication started at the primer and passed the three AseI sites sequentially. When replicating the G4G5N template, the same 465-nt band was visible after 1 min but, shortly thereafter, a cluster of bands around 208 nt accumulated (Fig 4B, lane 7–9). The size of these bands corresponds to products starting at the second AseI site and ending at the G4 sequence (Fig 4A), indicating that these products are a result of replication stalling at the G-quadruplex structure. Using a sequencing ladder starting from the second AseI site, we mapped the 3′ ends of the stalled nascent strands at nucleotide resolution. Of note, we used wild-type pBluescript vector as a template for the sequencing ladder because the Sequenase enzyme cannot synthesize through a G-quadruplex structure. The 3′ end of the major stalled product was at the −1 position, 1 nucleotide before the G4 sequence, while minor stalling products appeared at the −3, −2, 0 and +1 positions, with respect to the G4 sequence (Fig 4B, bottom).
Figure 4. Mapping the sites of replication stalling at G-quadruplex structures.
A Schematic representation of a G4 template showing AseI restriction sites. Products formed after AseI digestion and/or replication stalling at the G-quadruplex structure are depicted on the right. Of note, AseI will only cut in double-stranded DNA and thus replication past the site is required.
B G4G5N and non-G4C3P plasmids were replicated in HSS. Samples collected at the indicated times were extracted, AseI digested, separated on a high-resolution urea-PAGE sequencing gel and visualized by autoradiography (top). Sequencing ladder generated by extension of primer A on pBluescript allows the mapping of the replication products. Stalling positions are depicted on top of the G4G5N sequence (bottom) and are numbered such that the first nucleotide within the G4 sequence is numbered 0, the last nucleotide 3′ to the G4 sequence is numbered -1 and so forth.
C, D G4G3N (C) and G4G23 (D) plasmids were replicated in HSS, in the presence or absence of Phen-DC3, and analyzed as in (B). The sections of the sequencing gels containing the stalled replication products are depicted. Replication stalling sites were mapped and depicted on the relevant G4 sequences below the gels. Stalling positions were numbered as in (B).
Next, we mapped stalling sites for two additional G4 plasmid templates, G4G3N and G4G23, both under native conditions and in the presence of Phen-DC3 (Fig 4C and D, and Supplementary Fig S6). In the presence of Phen-DC3, replication stalls 1 nucleotide before, but also at multiple positions within the G4 sequence (Fig 4C and D, lanes 2–6, and Supplementary Fig S6A and B). This is most striking for the G4G23 template and indicates that Phen-DC3 induces multiple G4 conformations during replication. Importantly, a non-G4 control plasmid did not result in replication stalling at the G-rich sequence in the presence of Phen-DC3 (Supplementary Fig S6C). Under native conditions, the major replication stalling product for both G4G3N and G4G23 was at the −1 position while minor bands accumulated at the −2, and 0 positions (Fig 4C, lanes 8 and 9, and Fig 4D, lanes 7–11) similar to what we observed with G4G5N (Fig 4B, lanes 7–9). Interestingly, the 3′ positions of these stalling products are very similar to the 3′ ends of deletions found at G4 sequences in dog-1 deficient C. elegans (Kruisselbrink et al, 2008). Altogether, these data indicate that, in this system, the replication machinery transiently stalls when it is in close proximity to the G-quadruplex. After a brief period, the stalled intermediate disappears indicating the block is relieved and replication proceeds. We consider two possible options for this relieve: one, the gradual incorporation of flanking nucleotides destabilizes the G-quadruplex structure leading to its unfolding, or two, specialized enzymes are recruited to resolve the replication barrier.
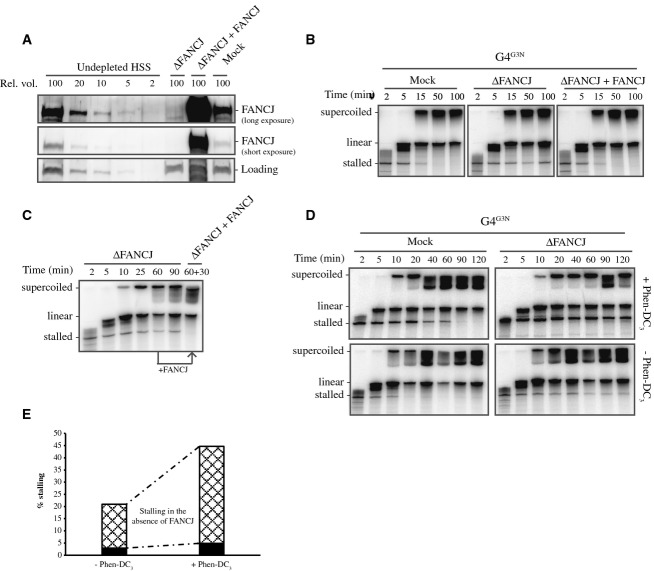
Depletion of FANCJ causes persistent replication stalling at G-quadruplex structures
The purified FANCJ helicase unwinds G-quadruplex in vitro, and FANCJ deficiency leads to G4-associated deletions in vivo (Youds et al, 2006; Kruisselbrink et al, 2008; London et al, 2008; Wu et al, 2008) suggesting this helicase plays a role in unwinding G-quadruplex. To examine the role of FANCJ in G4 processing during replication, we generated an antibody against xlFANCJ (Supplementary Fig S7A). Immunodepletion with this antibody removed the vast majority of FANCJ from HSS (Fig 5A). Importantly, FANCJ depletion had no effect on replication of a non-G4 plasmid template (Supplementary Fig S7B). We then replicated the G4G3N template in mock-depleted and FANCJ-depleted extracts. At early time points, the replication stalling product at the G4 site accumulated with similar kinetics in both extracts, which indicates that in the absence of FANCJ, G-quadruplexes still form a similar replication barrier. Consistent with this, the position of the stalled 3′ end did not change in the absence of FANCJ (Supplemental Fig S7C). However, while the stalling band readily disappeared as the G4 was bypassed in the mock-depleted extract, stalled products persisted in the FANCJ-depleted extract (Fig 5B). This indicates that a significant fraction of the G-quadruplex structures is not resolved in the absence of FANCJ. Furthermore, addition of the full-length xlFANCJ recombinant protein (Supplementary Fig S7D) to a FANCJ-depleted extract fully reversed the persistent stalling at G-quadruplex structures (Fig 5A and B, right panel) showing that this effect is caused specifically by the depletion of FANCJ. Replication of the G4G23 and G4G5N plasmid templates in FANCJ-depleted extracts also resulted in persistent replication stalling (Supplementary Fig S7E). These results strongly suggest that FANCJ plays a specific role in G4 unwinding and thereby facilitates eukaryotic G4 DNA replication.
Figure 5. Depletion of FANCJ results in persistent stalling at G-quadruplex structures.
- Mock-depleted, FANCJ-depleted, and FANCJ-depleted HSS supplemented with recombinant xlFANCJ were analyzed by Western blotting using xlFANCJ antibody. A dilution series of undepleted extract was loaded on the same blot to determine the degree of depletion. A relative volume of 100 corresponds to 0.7 μl of HSS. A non-specific band cross-reacting with FANCJ antibody is used as a loading control (‘Loading’).
- Mock-depleted, FANCJ-depleted, and FANCJ-depleted HSS supplemented with recombinant xlFANCJ were used to replicate the G4G3N plasmid template starting from primer A. Replication products were extracted, separated on 6% urea-PAGE gels, and visualized by autoradiography. Products stalled at the G4 sequence (‘stalled’), linear molecules resulting from denatured nicked products (‘linear’), and closed supercoiled products (‘supercoiled’) are indicated.
- G4G3N was replicated in FANCJ-depleted HSS starting from primer A. After 60 min, xlFANCJ or buffer was added followed by an additional 30-min incubation. Replication products were extracted, separated on 6% urea-PAGE gels, and visualized by autoradiography.
- Mock-depleted and FANCJ-depleted HSS were used to replicate G4G3N starting from primer A in the absence or presence of a low concentration (0.75 μM) of Phen-DC3. Replication products were extracted, separated on 6% urea-PAGE gels, and visualized by autoradiography.
- The G4 stalled and bypassed products of the 90-min time point in (D) were quantified using ImageQuant software, and the percentage of stalling versus bypassed products was plotted for the various conditions. Black bars represent percentage of stalling in the mock-depleted samples, the sum of black and dashed bars represent the percentage of stalling in the FANCJ-depleted samples. Therefore, dashed bars represent percentage of stalling as a result of FANCJ depletion only.
The persistence of stalling products at the G4 site in the absence of FANCJ could be a result of extended pausing of the replication machinery, but could also result from disassembly of the replication machinery or even breakage of the parental stand. To investigate this, recombinant FANCJ was added to a replication reaction in FANCJ-depleted extract after 60 min. While a stalled product was present at this 60-min time point, this product disappeared after an additional 30-min incubation in the presence of FANCJ (Fig 5C). This indicates that the replication machinery is still present, or rapidly reassembles after stalling, and that replication past these G-quadruplex structures is critically dependent on FANCJ. This is in agreement with a recent report that showed efficient replication restart of hydroxyurea-stalled forks in FANCJ deficient cells (Schwab et al, 2013). In addition, we did not observe any mutations or deletions when sequencing G4 replication products after FANCJ depletion arguing against a fork collapse and repair mechanism (Supplementary Fig S5).
Finally, we showed that increasing the stability of G-quadruplex structures by the addition of a low dose of Phen-DC3 enhanced the requirement of FANCJ (Fig 5D and E). In summary, these findings demonstrate that FANCJ plays a direct role in G4 unwinding during DNA replication and that this helicase is likely required for the resolution of particularly stable G-quadruplex structures.
Role of FANCJ in G4 unwinding is independent of the classical FA pathway
To investigate the role of the Fanconi anemia (FA) pathway in processing G-quadruplex (Youds et al, 2008; Kitao et al, 2011), we examined the monoubiquitylation of FANCD2, a central event in the FA pathway. No differences were observed in FANCD2 monoubiquitylation levels when replicating G4 versus non-G4 sequences (Fig 6A). To further study a potential role of FANCD2 in G4 replication, the protein was immunodepleted from HSS. FANCD2 depletion did not co-deplete FANCJ and did not affect replication efficiency of ssDNA control templates (Fig 6B and C). We then replicated a G4 template in a mock- and FANCD2-depleted extract and found that FANCD2 depletion did not enhance replication stalling (Fig 6C). These data argue against a direct function for FANCD2 in G4 DNA processing and provide evidence that the role of FANCJ in G4 unwinding is independent of its role in the classical FA pathway.
Figure 6. Role of FANCJ in promoting G-quadruplex structure replication is independent of FANCD2.
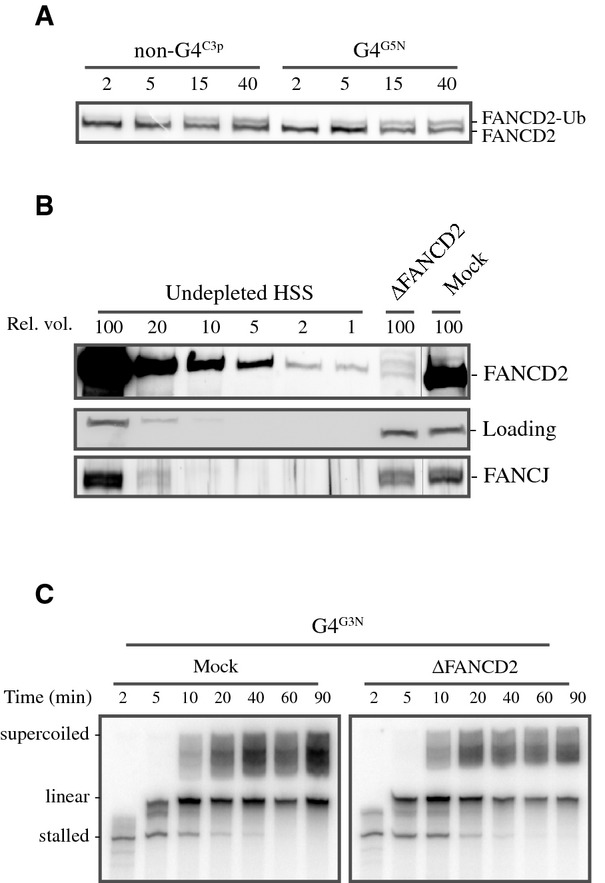
- G4G3N and non-G4C3p plasmids were replicated in HSS. Samples were taken at various time points and analyzed by Western blotting using xlFANCD2 antibody.
- Mock-depleted and FANCD2-depleted HSS were analyzed by Western blotting using xlFANCJ and xlFANCD2 antibodies. A dilution series of undepleted extract was loaded on the same blot to determine the degree of depletion. A relative volume of 100 corresponds to 0.7 μl of HSS. A non-specific band cross-reacting with FANCJ antibody is used as loading control (‘Loading’).
- Mock-depleted and FANCD2-depleted HSS from (B) were used to replicate the G4G3N template starting from primer A. Replication products were extracted, separated on 6% urea-PAGE gels, and visualized by autoradiography.
Discussion
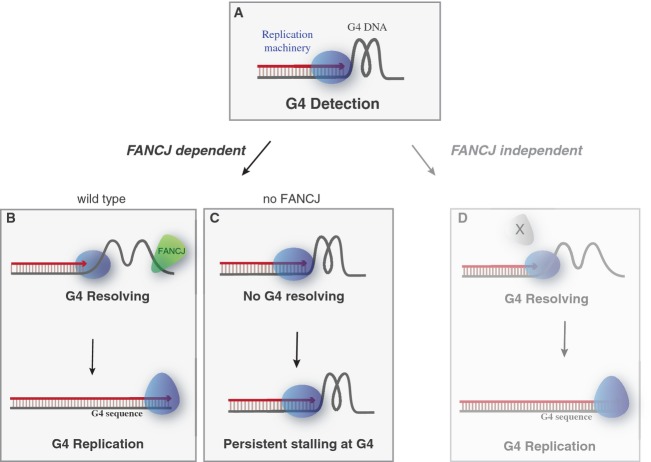
G-quadruplex structures are mutagenic in animal cells, and a number of models have proposed a replication block as a cause for this genetic instability. However, whether, and to what extent, these structures block replication directly is not known. Moreover, how these structures are resolved is poorly understood at the molecular level. In this study, we use a cell-free system based on Xenopus egg extracts to investigate G4 resolving and replication. We show that G-quadruplex structures have the intrinsic capacity to transiently block DNA replication even under non-compromised conditions (Fig 7A). Our data demonstrate that FANCJ is strictly required for the unwinding of stable G-quadruplexes (Fig 7B). Depletion of the FANCJ helicase results in persistent replication stalling at G-quadruplex structures (Fig 7C), arguing that in the absence of FANCJ temporal stalls become persistent. However, our data also suggest that additional G-quadruplex structures are present and resolved independently of FANCJ (Fig 7D). This could be mediated by other specialized helicases such as BLM, WRN, RTEL1, or XPD, or by a polymerase incorporating new nucleotides stepwise leading to destabilization of the structure. We also demonstrate that DNA synthesis is readily resumed after a short period to faithfully replicate the G4 containing DNA (Fig 7B and D).
Figure 7. Model for G4 DNA recognition and unwinding during DNA replication.
(A) When the DNA replication machinery encounters a G-quadruplex structure, it temporarily stalls right at the G4 site (G4 detection). Stable G-quadruplex structures depend on FANCJ for their resolving (B), while others are resolved through FANCJ-independent mechanisms (D) after which DNA synthesis proceeds and the G4 sequence is faithfully replicated. In the absence of FANCJ (C), the G-quadruplex structures are not resolved leading to persistent replication stalling at the G4 sequence.
In yeast, Pif1 helicase deficiency causes DNA replication to slow down in regions that contain sequences with G4 forming potential (Lopes et al, 2011; Paeschke et al, 2011). In contrast to a general replication slow-down, we here show that G-quadruplex causes stalling of nascent strand progression right at the G4 site demonstrating that G-quadruplex structures form a direct replication challenge. However, stalling is only transient, and replicated molecules do not show a high incidence of mutations, indicating that there is a mechanism in place that efficiently resolves G-quadruplex structures. This is consistent with G4 sequences being conserved in our genome. However, we could envision that in conditions of replication stress, these structures are more problematic which could explain the higher incidence of translocation junctions near G4 sequences observed in cancer cells (De & Michor, 2011; Bose et al, 2014). Furthermore, failure to properly resolve G-quadruplex structures could deregulate the propagation of epigenetic histone marks during replication leading to a loss of epigenetic memory and misregulation of gene expression (Sarkies et al, 2010).
During our study, we observed some differences in the degree in which various G4 sequences stall DNA replication (Fig 3). It has been shown extensively that the sequence is an important determinant of G-quadruplex stability (Guedin et al, 2010; Bharti et al, 2013). Moreover, this notion is in accordance with previous genetic data in C. elegans deficient for dog-1, showing that some G4 sequences are more mutagenic than others (Kruisselbrink et al, 2008). It would be interesting to perform a thorough investigation into the effect of G-content, loop length and composition, and G-quadruplex conformation on replication stalling in Xenopus egg extract. Another aspect that is worth exploring is the influence of chromatinization of DNA, which is currently not addressed in our assay.
Purified FANCJ helicase can unwind G-quadruplexes in vitro (Wu et al, 2008) but whether it also performs this function under physiological conditions has been difficult to address. Also, many proteins have been shown to have G-quadruplex unfolding ability when tested with purified proteins and model substrates. We now provide direct evidence that FANCJ is required to efficiently replicate past G-quadruplex structures in a system that recapitulates vertebrate G4 replication, which strongly suggests that FANCJ resolves these structures. Of great interest, the 3′ ends of the stalled replication products in the absence of FANCJ are at a very similar position compared to the 3′ ends of the G4-induced deletions found in dog-1 deficient C. elegans (Cheung et al, 2002; Kruisselbrink et al, 2008). This almost exact overlap provides strong support for the hypothesis that a stalled nascent strand is a determinant in deletion induction in this genetic system. Genetic instability caused by persistent stalling at G4 sequences in the absence of FANCJ could, at least in part, explain the identification of FANCJ as a cancer susceptibility gene (Seal et al, 2006).
Although previous data and our results indicate that G-quadruplex are efficiently resolved during DNA replication, persistent G-quadruplex structures have been shown to induce DNA damage and mutations, both of which are linked to DNA replication (Kruisselbrink et al, 2008; Rodriguez et al, 2012). Whether this DNA damage is induced when replication is stalled at the G-quadruplex or whether it is only generated in the following cell cycle as recently suggested by Koole et al ( 2014) remains to be determined. In our hands, persistent stalling after depletion of FANCJ does not lead to breaks or extensive mutations. This is consistent with a recent report that showed that the formation of DSBs is not enhanced upon fork stalling after treatment of FANCJ deficient cells with hydroxyurea (Schwab et al, 2013).
G4 ligands have been suggested to act as anticancer drugs based on their effect on specific gene promoters and telomeres (Salvati et al, 2007; Balasubramanian et al, 2011). Alternatively, inducing DNA damage by enhancing replication stalling at G-quadruplex structures could also be a strategy to target rapidly dividing cancer cells. Accordingly, it has been shown that G4 ligands induce synthetic lethality in cells deficient for various DNA damage repair pathways (Rodriguez et al, 2012). To optimize these synthetic lethal strategies, it will be important to determine which G4 ligand most potently stalls the progression of the replication machinery and deficiency of which repair protein or helicase could enhance this effect. Our system is highly suited to investigate these aspects.
The unique approach described here represents a powerful tool to further unravel the mechanism of G-quadruplex structure resolution and to elucidate signaling events leading to this.
Materials and Methods
Preparation of ssDNA plasmids containing G4 and non-G4 sequences
G4 sequences or control G-rich sequences were cloned into the pBS-SK(−) phagemid vector using HindIII and BamHI restriction enzymes. Subsequently, single-stranded DNA (ssDNA) templates were generated by viral replication using a helper phage as described previously (Jupin & Gronenborn, 1995; Trower, 1996). Consequently, ssDNA plasmids used in this study were identical except for a small region of 17–30 nucleotides (nt) containing the control G-rich or G4 sequences (Fig 1C).
Primer A (GGGTTCGTGCACACA) and B (TAATGTGAGTTAGCT) were annealed to the ssDNA templates 760 nt or 208 nt, respectively, from the G4 sequence or control G-rich sequence (Supplementary Fig S1A). In order to prevent 5′-to-3′ DNA degradation in extract, primers were synthetized with phosphorothioate bonds connecting the twelve most 5′ nucleotides. Primers without phosphorothioate bonds were used for Fig 4C and D and for Supplementary Figs S3 and S6. Where indicated, primers were 32P-γ-dATP radioactively labeled at the 5′-ends using T4 polynucleotide kinase (PNK).
G-quadruplex structures were induced as follows: 21.9 fmol/μl (20 ng/μl) of ssDNA plasmid was incubated with 175 fmol/μl primer A in EB buffer (10 mM Tris pH 8) containing 50 mM KCl for 5 min at 80°C. Subsequently, the annealing mix was slowly cooled down to room temperature in approximately 2 h allowing the G-quadruplex structure to form and the primer to anneal. Where indicated, the G4 stabilizing compound Phen-DC3 (Piazza et al, 2010; Bharti et al, 2013) was added to the mix at 50°C and incubated for 30 min after which the mix was further cooled down to room temperature.
Primer extension reaction
Pre-labeled primer B (105.6 fmol/μl) was annealed to ssDNA template (18.3 fmol/μl) in Sequenase reaction buffer provided by the Sequenase kit supplemented with 50 mM KCl. The mix was incubated for 2 min at 80°C and slowly cooled down to room temperature. DTT (6.7 mM), dNTPs (0.9 mM), and T7 DNA polymerase (3.3 units) (Sequenase, USB, Cleveland, OH, USA) were added to the annealing reaction following manufacturer instructions. The reaction was carried out at room temperature. At the indicated time points, the primer extension reaction was stopped by addition of formamide loading dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF) and the products were separated on 6% urea-PAGE gels for 30 min at 15 W.
Xenopus egg extract and replication of ssDNA plasmids
Preparation of high-speed supernatant (HSS) from Xenopus laevis unfertilized eggs was performed as previously described (Mechali & Harland, 1982; Walter et al, 1998; Lebofsky et al, 2009). All HSS replication reactions in this study were performed using low ssDNA concentrations (3 ng/μl) to prevent random priming events.
HSS was supplemented with 3 ng/μl nocodazole, ATP regeneration mix (18.9 mM phosphocreatine, 1.9 mM ATP and 4.7 ng/μl creatine phosphokinase), and 32P-α-dCTP. At t = 0, the primed template was added to the Xenopus egg extract and the replication reaction was carried out at room temperature. At the indicated time points, 1 μl of the reaction was stopped with 5 μl stop solution I (8 mM EDTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 0.1% bromophenol blue, and 80 mM Tris pH 8) and 4 μl of the reaction was stopped with 50 μl stop solution II (0.5% SDS, 10 mM EDTA, 50 mM Tris pH 7.5). Samples in stop solution I were separated on native agarose gels. Samples in stop solution II were treated with RNase A and Proteinase K (0.5 mg/ml) and phenol–chloroform extracted prior to nascent strand analysis.
Where indicated, agarose gels were SybrGold stained (Molecular Probes, Eugene, OR, USA) for visualization of non-radiolabeled DNA.
Nascent strand analysis
To determine G4 stalling, extracted products were separated on 6% urea-PAGE gels, which, after drying, were visualized by autoradiography. To assess the stalling efficiency, replication stalling products were quantified, and their relative amount with respect to the total lane intensity was plotted.
Where indicated, replication products were analyzed at nucleotide resolution by separation in 5% urea-polyacrylamide sequencing gels prepared in 0.8× GTG buffer (USB, Cleveland, OH, USA). Briefly, extracted products were digested for 3 h at 37°C with AseI restriction enzyme. Reaction was stopped by addition of formamide loading dye, and products were separated on sequencing gels for 200 min at 55 W. After drying, the products were visualized by autoradiography. Sequencing ladders were generated from dsDNA pBluescript template and primer B using the Thermo Sequenase TM Cycle Sequencing kit (USB, Cleveland, OH, USA) following manufacturer instructions.
Antibodies and immunodepletions
FANCJ antibody was prepared using amino acids 69–249 of Xenopus laevis FANCJ as antigen. Antiserum was raised in rabbits, and antibody specificity was confirmed by Western blotting. Antibodies against FANCD2 were described previously (Knipscheer et al, 2009).
To deplete Xenopus egg extracts of FANCJ, one volume of protein A-sepharose Fast Flow (PAS) (GE Healthcare, Piscataway, NJ, USA) was bound to 3 volumes of α-xlFANCJ serum or pre-immune serum through an overnight incubation at 4°C. Beads were washed twice with 500 μl PBS, once with 500 μl ELB (10 mM HEPES–KOH at pH 7.7, 50 mM KCl, 2.5 mM MgCl2, and 250 mM sucrose), twice with 500 μl ELB + 0.5 M NaCl, and finally twice with 500 μl ELB. Three rounds of depletion for 20 min at 22°C were performed using one volume of pre-cleared HSS mixed with 0.2 volumes of the antibody-bound sepharose. Extracts were collected and immediately used for DNA replication experiments following the same procedure as described above. When indicated, recombinant xlFANCJ was added to the depleted extract. FANCD2-immunodepleted HSS extracts were prepared as described previously (Knipscheer et al, 2009). The level of depletion was determined by Western blot analysis of a dilution series of undepleted HSS next to the depleted extract.
Acknowledgments
This work was supported by the Netherlands organization for Scientific Research (VIDI 700.10.421 to PK), a Dutch Cancer Society Fellowship (to PK), a La Caixa fellowship to SSB, a European research Council grant (203379, DSBrepair to MT), and a ZonMW/NGI-Horizon/Zenith grant (to MT). We thank JC Walter for the α-xlPCNA antibody and suggestions on the manuscript, the Hubrecht animal caretakers for animal support, and the other members of the Knipscheer laboratory feedback. Furthermore, we would like to thank A. Nicolas, M.P. Teulade-Fichou, and C. Guetta for generously providing Phen-DC3.
Author contributions
PCB, SSB, MT, and PK designed the experiments. PCB and SSB conducted the experiments. WK, JvH, and MT designed and produced ssDNA plasmids. JD produced the FANCJ antibody. PCB, MT, and PK wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Aguilera A, Garcia-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discovery. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Bharti SK, Sommers JA, George F, Kuper J, Hamon F, Shin-Ya K, Teulade-Fichou MP, Kisker C, Brosh RM., Jr Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem. 2013;288:28217–28229. doi: 10.1074/jbc.M113.496463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose P, Hermetz KE, Conneely KN, Rudd MK. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS ONE. 2014;9:e101607. doi: 10.1371/journal.pone.0101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Gregoire D, Coulombe P, Danis E, Mechali M. Genome-scale identification of active DNA replication origins. Methods. 2012;57:158–164. doi: 10.1016/j.ymeth.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Langley DR, Rangan A, Hurley LH. Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Jr, Sen D, Lansdorp PM. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2013;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupin I, Gronenborn B. Abundant, easy and reproducible production of single-stranded DNA from phagemids using helper phage-infected competent cells. Nucleic Acids Res. 1995;23:535–536. doi: 10.1093/nar/23.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni LS, Clayton DA. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci USA. 1982;79:983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- Kitao H, Nanda I, Sugino RP, Kinomura A, Yamazoe M, Arakawa H, Schmid M, Innan H, Hiom K, Takata M. FancJ/Brip1 helicase protects against genomic losses and gains in vertebrate cells. Genes Cells. 2011;16:714–727. doi: 10.1111/j.1365-2443.2011.01523.x. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, van Oijen AM, Walter JC. DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Res. 2011;39:545–555. doi: 10.1093/nar/gkq739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, Arwert F, Mathew CG, Zdzienicka MZ, Hiom K, De Winter JP, Joenje H. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, Ott J, Petrini J, Schindler D, Hanenberg H, Auerbach AD. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S. Human telomeric G-quadruplex. FEBS J. 2010;277:1097. doi: 10.1111/j.1742-4658.2009.07468.x. [DOI] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G: G tetrad and a G(:A):G(:A):G(:A): G heptad of GGA triplet repeat DNA. J Biol Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- Mattock H, Jares P, Zheleva DI, Lane DP, Warbrick E, Blow JJ. Use of peptides from p21 (Waf1/Cip1) to investigate PCNA function in Xenopus egg extracts. Exp Cell Res. 2001;265:242–251. doi: 10.1006/excr.2001.5181. [DOI] [PubMed] [Google Scholar]
- Mechali M, Harland RM. DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell. 1982;30:93–101. doi: 10.1016/0092-8674(82)90015-0. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Helene C. G-quadruplex DNA: a target for drug design. Nat Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–948. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Mergny JL. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D'Incalci M, Blasco M, Chiorino G, Bauwens S, Horard B, Gilson E, Stoppacciaro A, Zupi G, Biroccio A. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shin-ya K, Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J Cell Biol. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989;264:6447–6458. [PubMed] [Google Scholar]
- Tarsounas M, Tijsterman M. Genomes and G-quadruplexes: for better or for worse. J Mol Biol. 2013;425:4782–4789. doi: 10.1016/j.jmb.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trower MK. Preparation of ssDNA from phagemid vectors. Methods Mol Biol. 1996;58:363–366. doi: 10.1385/0-89603-402-X:363. [DOI] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Weitzmann MN, Woodford KJ, Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J Biol Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- Woodford KJ, Howell RM, Usdin K. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J Biol Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, O'Neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Barber LJ, Ward JD, Collis SJ, O'Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.