Abstract
The ba3-type cytochrome c oxidase from Thermus thermophilus is a membrane-bound proton pump. Results from earlier studies have shown that with the aa3-type oxidases proton uptake to the catalytic site and “pump site” occur simultaneously. However, with the ba3 oxidase the pump site is loaded before proton transfer to the catalytic site because the proton transfer to the latter is slower than with the aa3 oxidases. In addition, the timing of formation and decay of catalytic intermediates is different in the two types of oxidases. In the present study, we have investigated two mutant ba3 CytcOs in which residues of the proton pathway leading to the catalytic site as well as the pump site were exchanged, Thr312Val and Tyr244Phe. Even though the ba3 CytcO uses only a single proton pathway for transfer of the substrate and “pumped” protons, the amino-acid residue substitutions had distinctly different effects on the kinetics of proton transfer to the catalytic site and the pump site, respectively. The results indicate that the rates of these reactions can be modified independently by replacement of single residues within the proton pathway. Furthermore, the data suggest that the Thr312Val and Tyr244Phe mutations interfere with a structural rearrangement in the proton pathway that is rate limiting for proton transfer to the catalytic site.
Keywords: cytochrome c oxidase, electron transfer, membrane protein, respiration, electrochemical potential, redox reaction, metalloprotein, cytochrome aa3
Cytochrome c oxidases (CytcOs) are terminal enzymes of the membrane-bound respiratory chains in aerobic prokaryotic and eukaryotic organisms. This group of enzymes catalyses sequential reduction of oxygen to water and uses part of the free energy released in this reaction for generation of a transmembrane electrochemical proton gradient. Energy conservation by terminal oxidases occurs via two mechanisms. The electron donor, cytochrome c, binds on the positive (p) side of the membrane while protons used in the reduction of oxygen to water originate from opposite, negative (n) side of the membrane, which results in a charge separation across the membrane. In addition, up to now, all investigated CytcOs have been shown to energetically link electron transfer to pumping of protons across the membrane, from the n to the p side (1–5).
The ba3 CytcO consists of two main core subunits, I and II, and an additional subunit IIa. Subunit I consists of 13 transmembrane helices and holds heme b as well as the catalytic site composed of heme a3 and CuB. A fourth redox site, CuA, is located in subunit II. This site acts as the primary electron acceptor, which receives electrons from the water-soluble cytochrome c552. Subunit IIa forms one transmembrane helix, which corresponds to a helix of subunit II of e.g. the Rhodobacter sphaeroides CytcO, however, with opposite polarity (6–18). During turnover, electrons are transferred from CuA consecutively to heme b and to the catalytic site, which upon reduction binds O2 that is reduced to water. The protons involved in this reaction are transferred through specific proton-conducting pathways that span the distance between the n-side surface and the catalytic site. While the aa3-type oxidases (from e.g. R. sphaeroides, P. denitrificans or mitochondria) harbor two functional proton-conducting pathways, D and K, the ba3 oxidase presumably uses only one pathway, which partly overlaps in space with the Kpathway in the aa3 CytcOs (10, 19). This pathway is presumably used for transfer of all protons, including those used for O2 reduction and those being pumped across the membrane. The results from functional studies, combined with analyses of the ba3 CytcO structures (7–10), indicate that the entrance to the K-pathway is near a water molecule (H2O 146) and a conserved Glu in subunit II (E15(II)). The pathway is defined by a number of polar residues and water molecules that span the distance between solution and the catalytic site (Figure 1a).
Figure 1. Proton pathway and reaction scheme.
(a) Residues that comprise the K-pathway analog used for transfer of both substrate protons to the catalytic site (used for O2 reduction) and protons that are pumped across the membrane (Protein Data Bank: PDB3S8F (7). Copper and iron ions are presented as brown and orange spheres, respectively. Heme carbon atoms are shown in yellow and residues of the proton pathway are shown in green with oxygen and nitrogen atoms in red and blue, respectively. Water molecules of the proton-conducting pathway are shown as small red spheres.
(b) Schematic outline of the reaction of the four-electron reduced aa3 and ba3 CytcOs with O2. The redox centers are shown as a circle (CuA), a square (heme b or a for the ba3 and aa3 oxidases, respectively) and a merged square-circle (catalytic site, CS), where filled and empty symbols represent reduced and oxidized centers, respectively. A putative “proton-loading site” (PLS) is shown just above the catalytic site. For the ba3 CytcO the sign of absorbance changes (arrow up - increase in absorbance; arrow down, decrease in absorbance) of the different steps of the reaction are indicated in the square boxes. Protons that are pumped and transferred to the catalytic site are shown as dashed and solid lines, respectively. For the two mutant CytcOs, Y244F and T312V, two alternative scenarios (i) and (ii) are shown in the branched part of the lower reaction scheme. In scenario (ii) proton pumping does not occur because the proton at PLS is transferred to the catalytic site. This is illustrated by the blue dashed arrow next to (ii) in the third step of the lowermost scheme.
The reaction of the reduced ba3 CytcO with oxygen has been studied previously upon laser flash photolysis of CO from the ba3-CO complex in the presence of O2. The results from these experiments showed that at neutral pH the general reaction sequence is similar to that observed with the aa3 CytcOs. However, notable differences were observed in the timing of the individual electron and proton-transfer reactions (Figure 1b) (20–24), which may be related to the 0.5 H+/e− pumping stoichiometry of the ba3 oxidase (25, 26) (compared to 1 H+/e− in the aa3 oxidases studied to date). In brief, in both the aa3 (here data with the R. sphaeroides CytcO are discussed for comparison, (5, 27)) and ba3 CytcOs the reaction is initiated by oxygen binding to the reduced heme a3 with a time constant of 5–10 μs (at 1 mM O2). The binding of O2 is followed in time by electron transfer from heme b (heme a in aa3 CytcO) to the catalytic site with a time constant of ~15 μs or ~40 μs in the ba3 and aa3 CytcOs, respectively (Table 1). This electron transfer results in formation of a state that is called PR. In the next step, a proton is taken up from solution and the electron at CuA equilibrates with heme b/a with a time constant of ~60 μs and ~90 μs with the ba3 and aa3 CytcOs, respectively. However, while with the aa3 CytcO this proton is transferred to the catalytic site to form state F, with the ba3 CytcO the proton is transferred to a site that is located at a distance from the catalytic site, suggested to be the so-called proton-loading site (PLS) (23). With the aa3 CytcO, the fourth electron, accompanied by proton uptake, is transferred to the catalytic site with a time constant of ~1 ms forming the oxidized CytcO (both the PR → F and F → O reactions are linked in time to proton pumping). With the ba3 CytcO the 60-μs proton uptake is followed in time by proton uptake to form the F state with a time constant of ~0.8 ms at neutral pH, which is significantly slower than with the aa3 CytcO (c.f. ~100 μs). This reaction approximately overlaps in time with transfer of the last electron to the catalytic site and formation of the oxidized CytcO (state O). In other words, the F state is not significantly populated with the ba3 CytcO. At pH >8 the two processes are separated in time such that formation of F and O are resolved separately1. Thus, the main difference between the reaction sequences for the aa3 and ba3 CytcOs is that while with the former proton uptake to the PLS and the catalytic site are synchronized, with the ba3 CytcO the two processes are separated in time; proton transfer to PLS occurs before proton transfer to the catalytic site. Furthermore, while with the aa3 CytcO two protons are pumped during O2 reduction, with the ba3 CytcO only one proton is pumped.
Table 1.
Rate constants of the absorbance changes during reaction of reduced wild-type and, T312V and Y244F mutant CytcOs with O2 (pH 7.5). 4e and 3e refer to the four and three-electron reduced CytcOs, respectively. Rate constants were typically determined from at least four experiments. The standard error of the mean was ≤10%. pT, proton transfer; eT, electron transfer.
| Observed absorbance changes | ΔA560 decrease ΔA610 increase# (s−1) | ΔA560; ΔA830 increase (s−1) | 1st proton uptake (s−1) | ΔA610 decrease (s−1) | ΔA560 slow decrease (s−1) | 2nd proton uptake (s−1) |
|---|---|---|---|---|---|---|
| Reaction | formation of state P | fractional eT from CuA to heme b | pT to PLS | formation of state F | final oxidation | see Fig 1b |
| WT 4e# | 68000 (15 μs) | 16000 (62 μs) | 17000 (60 μs) | 1300 (0.8 ms) | 900 (1.1 ms) | 1100 (0.9 ms) |
| T312V 4e | 66000 (15 μs) | 13000 (80 μs) | 8300 (120 μs) | 1200 (0.8 ms) | 170 (6 ms) | 170 (6 ms) |
| Y244F 4e | 44000 (22 μs) | 8500 (120 μs) | 7600 (130 μs) | 900 (1.1 ms) | 190 (5.3 ms) | 190 (5.3 ms) |
| WT 3e | 30000 (30 μs) | no¤ | 14000 (70 μs) | 1000 (1 ms) | no | 1000 (1 ms) |
| T312V 3e | 32000 (30 μs) | no | 8300 (120 μs) | 950 (1.0 ms) | no | 290 (3.4 ms) (30%) |
| Y244F 3e | 29000 (34 μs) | no | 6700 (150 μs) | 900 (1.1 ms) | no | 190 (5.3 ms) (30%) |
| T312S 4e& | 33000 (30 μs) | 6000 (170 μs) | 6000 (170 μs) | 30 (33 ms) | 30 (33 ms) | 25 (40 ms) |
| T315V 4e& | 68000 (15 μs) | 13000 (77 μs) | 8500 (120 μs) | ~1000 (~1 ms) | 200 (5 ms) | 200 (5 ms) |
For the four-electron reduced CytcO (the wild-type and both mutant CytcOs) the absorbance decrease at 560 nm (oxidation of heme b) was about a factor of two faster than the increase in absorbance at 610 nm (formation of PR). This is a relatively small dissimilarity in rates and a detailed discussion of any functional differences would require more detailed studies, which is outside of the scope of the present manuscript. As done previously (21) we consider these two events to be synchronous until further studies are done.
no - not observed
The data are from (24).
To elucidate the mechanism by which the ba3 CytcO couples O2 reduction to proton pumping, we searched for structural modifications in the K pathway analog of the ba3 CytcO (10) that could result in specific changes in the kinetics of proton uptake during the reaction steps associated with O2 reduction. Two such structural modifications are the replacement of Thr312 by a Val residue (T312V) and Tyr244 by a Phe residue (Y244F). The turnover activities of these mutant CytcOs were <1% and ~15%, respectively, of that of the wild-type CytcO (~300 s−1). The proton-pumping stoichiometry of the Y244F mutant CytcO was similar to that of the wild-type CytcO (the activity of the T312V was too low to allow measurements of proton pumping) (10).
In this work, we show that in these mutant CytcOs, the first proton was taken up only slightly slower than with the wild-type CytcO, but the second proton uptake was significantly slowed such that the oxidized state (O) was formed slower than state F resulting in a temporal separation of the two events. The data indicate that even though the substrate and pumped protons are transferred through a single pathway, the T312V and Y244F substitutions have distinctly different effects on the relative rates of the electron and proton-transfer reactions, which offers insights into the mechanism by which these processes are regulated.
Materials and Methods
Bacterial growth, enzyme purification and characterization
Thermus thermophilus HB8 strain YC 1001 (with a deletion of the cba gene and a plasmid with the ba3 gene with a 6-His-tag at the N-terminus of subunit I) was used for production of cytochrome ba3 CytcO. Mutations were introduced as described previously (10, 28). Purification of the recombinant ba3-CytcO was performed as described in (23). The ba3-CytcO variants at a concentration of 100–150 μM in 5 mM HEPES, pH 8.0, 0.05% DDM were kept at 4°C.
The optical absorbance spectra of both structural variants T312V and Y244F in their oxidized, reduced and in CO-ligated forms were essentially the same as those of the wild-type CytcO.
Flow-flash experiments
The cytochrome ba3 sample was prepared in a Thunberg cuvette as described in (29, 30). Briefly, air was replaced by nitrogen on a vacuum line after which the samples were reduced upon addition of 2–3 mM sodium ascorbate and 2 μM of the redox mediator phenazine methosulphate (PMS) in 100 mM HEPES-NaOH or 100 mM KCl (for measurements of proton uptake) and 0.05 % dodecyl-β-D-maltoside (DDM). Then, nitrogen was exchanged for carbon monoxide about one hour before initiation of the experiment. In order to obtain the partially reduced enzyme (3 electrons/enzyme) the samples were supplemented with 30 μM EDTA, air was replaced with nitrogen and then carbon monoxide on the vacuum line. The samples were kept over night in order to reach the three-electron reduced state and 10–30 μM FeSO4 was added. In order to test whether or not the presence of EDTA and FeSO4 had any specific effect on the studied reactions, in part of the experiments ascorbate and PMS were added to fully reduce the CytcO (with 4 electrons) after completing the experiments with the three-electron reduced CytcO, and the reaction with O2 was monitored. No differences were observed compared to the data obtained with the four-electron reduced CytcO prepared from the oxidized CytcO as described above.
Flow-flash experiments were performed using a locally modified stopped-flow apparatus (Applied Photophysics, DX-17MV) as described in (31). Briefly, the enzyme-containing solution was mixed with an oxygen-saturated solution at a ratio of 1:5 resulting in a final oxygen concentration of ~1 mM. About 30 ms after mixing the reaction of the enzyme with oxygen was initiated by flash-photolysis of the enzyme-CO complex (10 ns; 200 mJ; 532 nm, Nd-YAG laser, Quantel). The oxidation kinetics were monitored at different wavelengths (see Figure legends). The concentration of the reactive enzyme was calculated from the amplitude of the flash-induced absorbance increase at 445 nm using an absorption coefficient of 67 mM−1cm−1 (20).
Proton-uptake measurements
Proton uptake during oxidation of the fully or partially reduced enzyme with oxygen was measured using the pH indicator dye cresol red (pKa=8.3) at a concentration of 33 μM (after mixing). The sample buffer was exchanged for 100 mM KCl, 0.05% DDM, pH adjusted to ~8 with 50 mM KOH using gel filtration on a pre-packed Sephadex G-25 column (PD-10; Pharmacia). The pH during measurement was found to be 7.5–7.7. Traces were collected also in the presence of buffer (100 mM HEPES-NaOH at pH 7.5, 0.05% DDM) and they were subtracted from those obtained in the buffer-free solution in order to remove possible contribution of the hemes (about 12 traces were averaged). To estimate the number of protons taken up per enzyme molecule, the exhaust solution from the stopped-flow apparatus (in the absence of buffer) was collected, its pH was adjusted to 8.0 and absorbance changes corresponding to a given proton concentration were determined by additions of well-defined amounts of hydrochloric acid.
Determination of the heme concentration
The concentration of heme b was determined from the absorbance spectrum of the reduced ba3 CytcO using the absorption coefficient ε(560–590) = 26 mM−1cm−1 (28) or from a reduced minus oxidized spectrum with ε(560–658) = 21 mM−1cm−1. Ferro-heme as3 was quantified using ε(613–658) = 6.3 mM−1cm−1 in the reduced minus oxidized spectrum (32).
Results
Four-electron reduced enzyme
A solution of the fully reduced ba3 CytcO with CO bound to heme a3 was mixed with an oxygen-saturated solution after which the CO ligand was dissociated by means of a laser flash approximately 30 ms after mixing. Upon removal of the CO ligand, O2 binds to heme a3, which initiates the reaction. Figure 2ab shows absorbance changes at 560 nm and 610 nm for the T312V and Y244F mutant CytcOs, compared to those obtained with the wild-type CytcOs. The rate constants are summarized in Table 1. At 560 nm the absorbance changes are dominated by heme b. With the wild-type CytcO, after flash-photolysis of CO at t=0, a decrease in absorbance is seen (Figure 2a), which is associated with oxidation of heme b upon electron transfer to the catalytic site forming intermediate PR with a rate constant of 6.8·104 s−1 (τ≅15 μs). In the T312V and Y244F mutant CytcOs this reaction rate was about the same, ~6.6·104 s−1 (τ≅15 μs) or slightly slower ~4.4·104 s−1 (τ≅22 μs), respectively. The same process is also seen at 610 nm where the PR state displays maximum absorbance (Figure 2b) (see comment in Table 1).
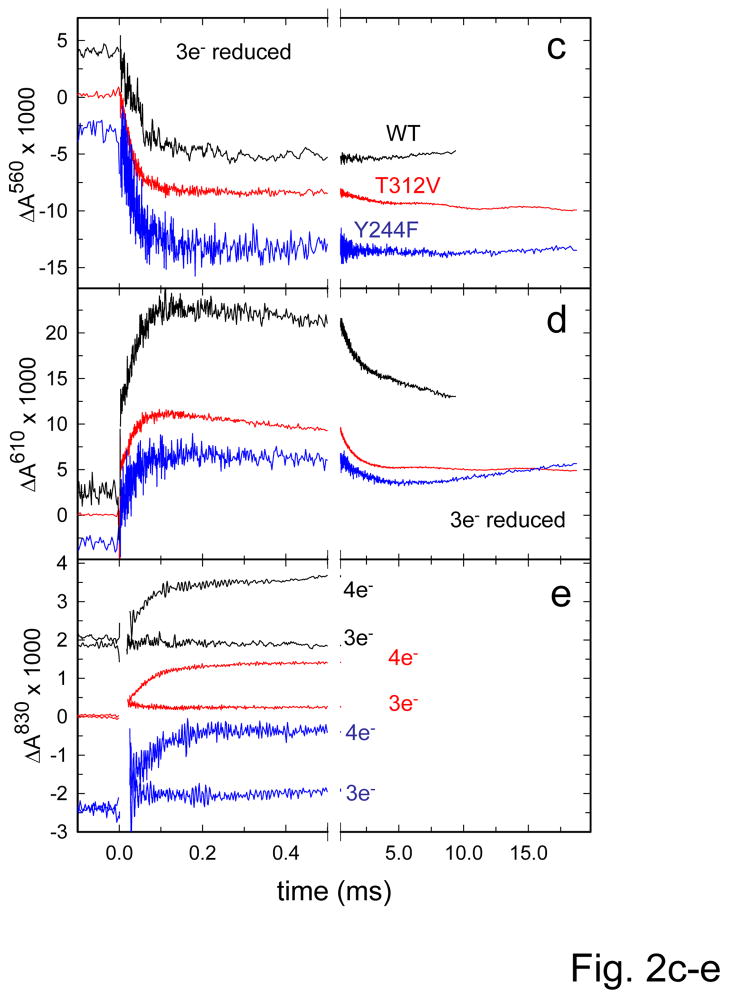
Figure 2. Absorbance changes associated with reaction of the reduced wild-type, T312V and Y244F mutant ba3 oxidases withO2.
(a,b) Fully reduced CytcO (with four electrons) and (c–e) three-electron reduced CytcO. Absorbance changes are shown at 560 nm (a and c, main contribution from heme b), 610 nm (b and d, formation and decay of PR) and 830 nm (e, main contribution from CuA). In e traces are shown also for the 4-electron reduced CytcO for comparison. Traces plotted in black, red and blue correspond to wild-type, T312V and Y244F CytcOs, respectively. All trace were normalized to 1 μM reactive enzyme. Conditions: 100 mM HEPES-NaOH (pH 7.5), 0.05% DDM. The O2 concentration was 1 mM after mixing. The reaction was initiated by a laser flash at t=0.
The subsequent increase in absorbance at 560 nm (re-reduction of heme b) occurs upon electron transfer from CuA to heme b, which is also seen as an absorbance increase at 830 nm, attributed to oxidation of CuA (not shown). This reaction displayed rate constants of ~1.3×104 s−1 (τ≅80 μs) and ~0.85×104 s−1 (τ≅120 μs) for the T312V and Y244F mutant ba3 CytcOs, respectively (Figure 2a) being slightly slower than with the wild-type CytcO (~1.6×104 s−1, τ≅60 μs). With the wild-type CytcO this electron transfer is also linked in time to proton uptake from solution (1.4–1.7×104 s−1 (τ≅60–70 μs), as measured by monitoring absorbance changes of a pH-sensitive dye, cresol red at 575 nm (Figure 3). With the T312V and Y244F mutant CytcOs the proton uptake was slowed by approximately a factor of two to 0.83 × 104 s−1 (τ≅120 μs) and 0.76 × 104 s−1 (τ≅130 μs), respectively. With the wild-type CytcO this proton is presumably transferred to a protonloading site (PLS) that is located at a distance from the catalytic site (see Discussion).
Figure 3. Absorbance changes associated with proton uptake.
Proton uptake was followed as a function of time during reaction with O2 of the four (black) and tree-electron (green) reduced wild-type (a), T312V (b) and Y244F (c) ba3 CytcO. Measurements were done at 575 nm with 33 μM cresol red (after mixing). The traces are differences between those measured in unbuffered and buffered solutions. The reaction solutions contained 100 mM KCl (pH 7.5) (unbuffered solution) or 100 mM HEPES-NaOH (pH 7.5) (buffered solution), 0.05% DDM. Trace amplitudes were normalized to 1 μM reactive enzyme (see Methods).
The decrease in absorbance at 610 nm with a rate constant of ~1.3×103 s−1 (τ≅0.8 ms) with the wild-type CytcO is associated with decay of the PR state and presumably formation of the F state. This decrease displayed about the same rate constants with the T312V and Y244F mutant CytcOs, 1.2·103 s−1 (τ≅0.8 ms) and 0.9·103 s−1 (τ≅1.1 ms), respectively (Figure 2b, Table 1).
The final decrease in absorbance at 560 nm, associated with formation of the oxidized state displayed a rate constant of 900 s−1 (τ≅1.1 ms) with the wild-type CytcO. At neutral pH this absorbance decrease occurs approximately with the same rate as the slower proton uptake (~1.1×103 s−1, τ≅0.9 ms) and the decrease in absorbance at 610 nm (~1.3×103 s−1, τ≅0.8 ms), associated with decay of the PR state (Table 1). However, at higher pH values the 560 nm decrease in absorbance (oxidation of the CytcO) is significantly slowed. This reaction occurs after the decay at 610 nm (PR decay), accompanied by simultaneous proton uptake, where the two latter are essentially unaffected by an elevated pH. In other words, the 560 nm and 610 nm absorbance changes represent processes that at neutral pH by coincidence display approximately the same rate constants, while at high pH, only the PR decay (610 nm) and proton uptake remain linked in time (21–23). With the T312V and Y244F mutant CytcOs the oxidation of the CytcO and proton uptake were significantly slowed to ~170 s−1 (τ≅5.8 ms) and 190 s−1 (5.2 ms) for the two mutant CytcOs, respectively (Figure 3), and occurred after the 610-nm absorbance decrease (0.8 ms and 1.1 ms, respectively). In other words, in contrast to the wild-type CytcO, with both mutant CytcOs the final oxidation of the CytcO (absorbance decrease at 560 nm) was accompanied by (a slowed) proton uptake with the same time constant as the oxidation process (Figures 2a and 3). We also note that in the mutant CytcOs formation of state F was not linked in time to proton uptake from solution even though the PR → F reaction requires proton transfer to the catalytic site (see Discussion).
The amplitudes of the pH dye absorbance changes, associated with proton uptake, were recalculated into proton-concentration changes by titration of the buffer capacity of the sample (see “Materials and Methods”). The total stoichiometry of proton uptake was approximately 2 H+ per CytcO for both the wild-type and mutant CytcOs, with approximately equal contributions of the two components.
Three-electron reduced enzyme
To monitor proton uptake that is required for (and triggered by) formation of the F state without interference from the next reaction step, we studied the reaction of the three-electron reduced CytcO with O2 (Figure 2c–e) (22, 23). In this state CuA is oxidized and all other redox sites are reduced. As seen previously with the wild-type CytcO (22, 23), after O2 binding to heme a3 and the initial absorbance decrease at 560 nm (Figure 2c), associated with oxidation of heme b, no further absorbance changes were observed, i.e. heme b was not re-reduced because CuA was initially oxidized. This conclusion was confirmed by the absence of any absorbance changes at 830 nm (reflecting redox changes at CuA, Figure 2e). As with the wild-type CytcO, with the T312V and Y244F mutant CytcOs the rate constants of heme b oxidation were about a factor of two slower for the three-electron reduced CytcO compared to the four-electron reduced variant, 3.2×103 s−1 (τ≅30 μs) and 2.9×103 s−1 (τ≅34 μs), respectively. In this case these absorbance changes coincided in time with those observed at 610 nm (increase in absorbance), i.e. oxidation of heme b and formation of PR occurred over the same time scales. Formation of the F state with the T312V and Y244F mutant CytcOs was observed as a decrease in absorbance at 610 nm with rate constants of 0.95 × 103 s−1 (τ≅1 ms) and 0.90 × 103 s−1 (τ≅1.1 ms), respectively (Figure 2d). Generally, the rates were very similar to those observed with the four-electron reduced enzyme.
The net proton uptake during reaction of the three-electron reduced CytcO with O2 is shown in Figure 3. With the wild-type ba3 CytcO, two components were observed with time constants of 70 μs and 1 ms, respectively, and the net stoichiometry was similar to that observed with the four-electron reduced CytcO (23). With the T312V and Y244F mutant CytcOs the rapid proton-uptake phase displayed rate constants of 0.83·104 s−1 (τ≅120 μs) and 0.67·103 s−1 (150 μs), respectively. This kinetic component displayed the same amplitude and about the same rate as for the four-electron reduced CytcOs. The second proton-uptake component displayed significantly smaller amplitudes (~30%) than with the wild-type three-electron reduced CytcO, and for the T312V mutant CytcO the rate was slightly faster than that observed with the four-electron reduced CytcO (see Table 1).
Discussion
Results from recent studies of the ba3 CytcO from T. thermophilus indicate that the reaction sequence is similar to that of the well-studied aa3 CytcOs (20, 22). However, there are significant differences in the timing of the proton-transfer reactions as outlined in the Introduction. Briefly, in the wild-type ba3 CytcO, first an electron is transferred from heme b to the catalytic site forming state PR with a time constant of ~15 μs. Then, a proton is taken up from solution with a time constant of ~60 μs. However, in contrast to the aa3 CytcO, the proton is not transferred to the catalytic site to form state F, but presumably to the PLS. Over a slower time scale the PR state decays, accompanied by proton uptake to the catalytic site with a time constant of ~1 ms. With the wild-type CytcO, at neutral pH the decay of the PR state occurs approximately at the same time as formation of the oxidized CytcO and therefore state F is not observed (22) (see also (20)). However, at high pH the two events, PR → F and F → O, are separated in time because the latter is significantly slower than the former.
In the wild-type ba3 CytcO the last step, i.e., the F → O reaction is not accompanied by any net proton uptake when measured in solution. Because formation of the oxidized CytcO (state O) does require proton transfer to the catalytic site, we proposed that during the F → O reaction the proton residing at PLS is released simultaneously with proton uptake to the catalytic site. With the T312V and Y244F mutant CytcOs heme b was oxidized with the same time constant (T312V, τ ≅ 15 μs) or slightly slower (Y244F, τ ≅ 22 μs) than with the wild-type CytcO. The increase in absorbance at 560 nm (electron transfer from CuA to heme b) and the accompanying first proton uptake (to the PLS in the wild-type CytcO) were both slowed from ~60 μs to ~120–130 μs with the mutant CytcOs (Figures 2a and 3, Table 1), which is a relatively small effect. In the next step, the absorbance decrease at 610 nm displayed about the same time constant with the T312V and Y244F mutant CytcOs as with the wild-type CytcO, which indicates that the PR state decayed (and presumably F was formed) with the same time constants of approximately 1.0 ms (Table 1).
The main differences in the reaction sequences of the wild-type and mutant CytcOs were observed at this point in time. While with the wild-type CytcO the absorbance decay at 610 nm coincides in time with proton uptake (c.f. Figures 2b and 3a), in the T312V and Y244F mutant CytcOs no net proton uptake was observed on this time scale (c.f. Figures 2b and 3bc), which is surprising given that formation of state F requires proton transfer to the catalytic site. The absence of any net proton uptake from solution could be explained by:
release of the proton at PLS (taken up earlier, c.f. first proton uptake) to the p side over the same time scale as proton uptake to the catalytic site from the n side of the membrane (to form state F). This scenario would imply proton pumping over the same time scale as with the wild-type CytcO, but the proton release to the p side would now be linked to formation of the F state rather than formation of the O state (as with the wildtype CytcO (Figure 1b)).
internal transfer of the proton initially taken up to the PLS (τ≅120–130 μs) from the PLS to the catalytic site (to form F) over a time scale of ~1 ms without any additional proton uptake from solution.
While the first scenario (i) is compatible with proton pumping, the second (ii) is not, at least during the reaction steps investigate here. The relatively active Y244F mutant CytcO (activity 15% of that of the wild-type CytcO) pumps protons (10), which is compatible with scenario (i) above. The very low activity of the T312V mutant CytcO (<1%) did not allow for a determination of the pumping stoichiometry (10). Such a low activity due to a mutation in the proton pathway suggests that at some point in the reaction cycle, proton transfer is dramatically slowed, which most likely would reduce the pumping stoichiometry (27, 33) because there would be sufficient time for the proton at PLS to be transferred to the catalytic site. In this case scenario (ii) would explain the behavior of the T312V mutant CytcO (see Figure 1b).
As already noted above, with the wild-type ba3 CytcO there is no net proton uptake during the final step of oxidation of heme b and formation of the oxidized (O) enzyme. This observation was previously explained in terms of proton uptake from the n side to the catalytic site that coincides in time with release of a proton to the p side from the PLS (22). In contrast, the behavior of the T312V and Y244F mutant CytcOs was different in that we did observe proton uptake over the time scale of O formation (τ ≅ 5–6 ms), but, as outlined above, in these mutant CytcOs there was no net proton uptake upon F formation. Taken together, these two observations indicate that the PLS in the two mutant enzymes is not protonated in state F. A net proton uptake is observed during the F → O reaction because this reaction does not coincide with proton release from PLS (as it does with the wild-type CytcO).
The sequence of events in the variants were further investigated using the threeelectron reduced CytcO (Figure 2c–e) in which the reaction with O2 stops upon forming the F state. With the wild-type ba3 CytcO this reaction is accompanied by proton uptake with time constants of ~70 μs and 1 ms, where the first proton has been suggested to be transferred to the PLS and the second to the catalytic site to form the end product, state F (22, 23). With the T312V and Y244F mutant CytcOs only the first proton uptake displayed about the same rate and amplitude as observed with the wild-type CytcO, while the second proton-uptake phase displayed a significantly smaller amplitude (30 % of that observed with the four-electron reduced CytcO) (Figure 3bc). These observations are consistent with the explanation above, i.e., that, in the major fraction of the mutant enzyme population, the proton needed to form state F is either taken internally from within the CytcO (scenario ii), or it is accompanied by simultaneous proton release to the p side (scenario i).
The identity of the PLS, which is assumed to be the same for all heme-copper oxidases, is so far not known. Nevertheless, the number of candidates is limited (4, 34). For the ba3 CytcO there is data suggesting that propionate A of heme a3, possibly together with nearby sites at a hydrogen-bonding distance, may be involved in proton gating (8, 10, 34–36), see also (4, 37). For example, data from studies of CO photolysis from heme a3 suggest that changes in the ligation state at the catalytic may be linked to changes in structure and protonation state of residues near the heme a3 propionate A (35, 38).
In the ba3 oxidase both substrate and pumped protons are transferred through a single pathway. The data suggests that transfer of the first proton to the PLS is faster than that to the catalytic site. In the wild-type CytcO, after protonation of PLS, the proton to the catalytic site must be transferred from solution and not from the PLS in order to allow pumping. One possibility to regulate the proton flow is by means of a local structural change (e.g. relocation of a side chain) that would allow alternating access to the PLS and the catalytic site respectively. In addition, proton transfer to and from the PLS must be controlled such that the PLS is protonated only from the n-side and deprotonated only to the p-side.
Structural modeling of the K-pathway analog of the ba3 oxidase indicates that a water molecule is hydrogen bonded to Thr312 and interacts with Tyr244 (10). The Y244F mutation must not necessarily destabilize this water molecule, but in the T312V mutant CytcO it is likely to be lost. Yet, the data from this study suggest that the effect of the T312V and Y244F mutations is not to significantly modify the proton conductivity of the pathway because the rate of the first proton uptake was only a factor of two slower than that of the wild-type CytcO. Yet, proton uptake that followed in time after this first proton uptake was slower with both mutant CytcOs. This observation suggests that the effect of the mutations is not to slow proton transfer per se (c.f. the rate of the first proton uptake was not significantly affected). We speculate that the mutations instead interfere with structural changes that follow in time after the initial proton transfer to the PLS and that these changes are rate limiting for the second proton transfer. These structural changes are suggested to involve a segment of the proton pathway “below” the catalytic site because residues T312 and Y244 are located in this segment (Figure 1a). The structural modifications at these sites are suggested to control proton transfer to the catalytic site and the PLS, respectively. In addition, these changes would have to involve also a segment of the protein around the PLS to control proton transfer to and from this site. A link between structural changes below and above the hemes during turnover is expected because the enzyme must control the proton flux to the catalytic site and to/from the PLS. A comparable scenario has been observed, for example, with the R. sphaeroides aa3 CytcO where the flux of protons to the catalytic site and the PLS, respectively, is presumably controlled by Glu286. Any structural changes at Glu286, “below” the catalytic site would be linked to changes at a putative PLS, located “above” the hemes. Furthermore, proton pumping has been shown to be uncoupled from O2 reduction upon mutation of residues close the n-side surface of the CytcO, i.e. at a significant distance from Glu286 (33, 39–42).
The modulation of the structural changes by the T312V and Y244F mutations would result in slowed uptake of the second proton to form state F such that in the mutant CytcO this proton uptake occurs over the same time scale as proton release to the p side (scenario i above for the Y244F). Alternatively, a mutation that interferes with the structural changes may lead to slowed release of the proton from PLS, such that the proton is transferred back to the catalytic site as outlined in case ii above (possibly for the T312V mutant CytcO). It should be mentioned that observation of proton pumping without a membrane potential (or with a small potential present) as is the case in standard pumping measurements, does not directly imply that proton pumping also takes place in the presence of a transmembrane proton electrochemical gradient. It is possible that any structural changes that lead to alteration in the temporal sequence of the proton pump, may reveal their full effect only in the presence of a transmembrane potential (i.e. the Y244F mutant CytcO may display a lower pumping stoichiometry in vivo).
Taken together, although the key structural elements (e.g. the PLS, catalytic site) are the same in the wild-type and mutant CytcOs, the structural changes required to control the access to these sites might be perturbed, which would result in uncontrolled release either to the p side or proton transfer from the PLS to the catalytic site. For the T312V mutant CytcO, this conclusion is further supported by the differences in effects of the mutation observed for the CytcO turnover (<3e−/s and for oxidation of the mutant CytcO where the slowest component displayed a time constant of ~6 ms (because during oxidation of the CytcO four electrons are transferred to O2, the time constant for transfer of one electron is 6 ms/4 = 1.5 ms, i.e. the corresponding turnover rate would be ~670 electrons·s−1, i.e. a factor of >200 faster than the actual turnover rate). Apparently, there must be rate-limiting events, which take place during turnover, but not during oxidation of the reduced CytcO (c.f. the structural changes controlling the proton pump). In an earlier study, we investigated the reaction of reduced T315V and T312S mutant CytcOs with O2 (24). At the time of that study, the sequence of proton-transfer events in the wild-type ba3 CytcO was not well understood. Now, we can offer more reliable mechanistic speculations in the framework of more recent data, including those from the present study. The residue Thr315 is located about 5 Å below Thr312 in the K-pathway analog. In the case of the T315V CytcO the pumping stoichiometry could be measured (the activity was 6 % of that of the wild-type CytcO) (10) and the mutant CytcO was found not to pump protons. Qualitatively, the T315V mutant CytcO displayed the same behavior as the T312V mutant CytcO (see Table 1) studied here (c.f. scenario ii), i.e. the 610 nm absorbance decrease occurred before final oxidation of the CytcO without any accompanying proton uptake from solution.
With the T312S mutant CytcO, which pumps protons (the activity was 13 % of that of the wild-type CytcO) the formation of state PR and proton transfer to PLS were only slightly slowed (by less than a factor of 3), while uptake of the second proton was slowed dramatically to ~25 s−1 (24). However, in contrast to the results with the T312V mutant CytcO from the present work, with the T312S mutant CytcO state F was not observed. Instead the 610 nm absorbance decayed approximately over the same time scale as oxidation of the CytcO (see Table 1). We conclude that with the T312S mutant CytcO (back)proton transfer from PLS to the catalytic site could not take place and the PLS remained protonated (similar to the scenario observed with the Y244F mutant CytcO, scenario (i) above). In order to form the fully oxidized CytcO a net of two protons must be taken up to the catalytic site. However, after the initial proton transfer to PLS in the T312S mutant CytcO a net uptake of only one proton was observed over the time scale of oxidation of the CytcO, which would suggest that formation of the oxidized CytcO was accompanied by release of the PLS proton, i.e. proton pumping. Again, these earlier data support the model discussed above because the T312S mutant CytcO was found to pump protons (10). Also, the turnover rate for the T312S mutant CytcO (~40 s−1) is only a factor of 2.5 slower than the oxidation rate (for the T312V mutant CytcO the difference was a factor of >200, as outlined above) because with this mutant CytcO the slowest component displayed a rate constant of 25 s−1 (for four electrons, see above) i.e. corresponding to a turnover rate of ~100 electrons/s.
In summary, the results from the present study indicate that proton transfer to the catalytic site and the PLS of the ba3 CytcO can be modulated independently of each other by means of site-directed structural modifications of residues Y244 and T312 within the K proton pathway analog. Furthermore, the data suggest that in the wild-type CytcO proton transfer through this proton pathway to the PLS and the catalytic site is controlled by means of structural rearrangements that involve residues Y244 and T312.
Acknowledgments
Funding: These studies were supported by grants from the Swedish Research Council and by grant HL 16101 from the National Institutes of Health. IS was supported by a grant from Stockholm University.
Abbreviations
- CytcO
cytochrome c oxidase
- n side
negative side of the membrane
- p side
positive side of the membrane
- R
the four-electron reduced CytcO
- A
reduced CytcO with O2 bound to heme a3
- PR
the “peroxy” state formed after transfer of a third electron to the catalytic site
- F
the ferryl state formed at the catalytic site after protonation of PR
- O
the oxidized CytcO
- DDM
n-Dodecyl β-D-maltoside
References
- 1.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: Proton transfer through the respiratory complexes. Annual Review of Biochemistry. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belevich I, Verkhovsky MI. Molecular mechanism of proton translocation by cytochrome c oxidase. Antioxid Redox Signal. 2008;10:1–29. doi: 10.1089/ars.2007.1705. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson-Miller S, Babcock GT. Heme/copper terminal oxidases. Chem Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 4.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Brzezinski P, Gennis RB. Cytochrome c oxidase: exciting progress and remaining mysteries. J Bioenerg Biomembr. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soulimane T, Than ME, Dewor M, Huber R, Buse G. Primary structure of a novel subunit in ba3-cytochrome oxidase from Thermus thermophilus. Protein Science. 2000;9:2068–2073. doi: 10.1110/ps.9.11.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High resolution structure of the ba3 cytochrome c oxidase from thermus thermophilus in a lipidic environment. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the heme-Cu oxidases. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 10.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci U S A. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 12.Svensson-Ek M, Abramson J, Larsson G, Törnroth S, Brzezinski P, Iwata S. The X-ray Crystal Structures of Wild-Type and EQ(I-286) Mutant Cytochrome c Oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 13.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Qin L, Ferguson-Miller S. Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump. Proc Natl Acad Sci U S A. 2011;108:1284–1289. doi: 10.1073/pnas.1012846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO Journal. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoyama H, Muramoto K, Shinzawa-Itoh K, Hirata K, Yamashita E, Tsukihara T, Ogura T, Yoshikawa S. A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump. Proc Natl Acad Sci U S A. 2009;106:2165–2169. doi: 10.1073/pnas.0806391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radzi Noor M, Soulimane T. Bioenergetics at extreme temperature: Thermus thermophilus ba 3- and caa 3-type cytochrome c oxidases. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:638–649. doi: 10.1016/j.bbabio.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 20.Siletsky SA, Belevich I, Jasaitis A, Konstantinov AA, Wikström M, Soulimane T, Verkhovsky MI. Time-resolved single-turnover of ba3 oxidase from Thermus thermophilus. Biochim Biophys Acta. 2007;1767:1383–1392. doi: 10.1016/j.bbabio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Von Ballmoos C, Lachmann P, Gennis RB, Ädelroth P, Brzezinski P. Timing of electron and proton transfer in the ba 3 cytochrome c oxidase from Thermus thermophilus. Biochemistry. 2012;51:4507–4517. doi: 10.1021/bi300132t. [DOI] [PubMed] [Google Scholar]
- 22.Von Ballmoos C, Ädelroth P, Gennis RB, Brzezinski P. Proton transfer in ba3 cytochrome c oxidase from Thermus thermophilus. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:650–657. doi: 10.1016/j.bbabio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Von Ballmoos C, Gennis RB, Ädelroth P, Brzezinski P. Kinetic design of the respiratory oxidases. Proc Natl Acad Sci U S A. 2011;108:11057–11062. doi: 10.1073/pnas.1104103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smirnova I, Reimann J, Von Ballmoos C, Chang HY, Gennis RB, Fee JA, Brzezinski P, Ädelroth P. Functional role of Thr-312 and Thr-315 in the proton-transfer pathway in ba3 cytochrome c oxidase from thermus thermophilus. Biochemistry. 2010;49:7033–7039. doi: 10.1021/bi100749p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannt A, Soulimane T, Buse G, Becker A, Bamberg E, Michel H. Electrical current generation and proton pumping catalyzed by the ba3- type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 26.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, Daldal F, Blanke SR, Gennis RB. Adaptation of aerobic respiration to low O2 environments. Proceedings of the National Academy of Sciences. 2011;108:14109–14114. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namslauer A, Brzezinski P. Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase. FEBS Lett. 2004;567:103–110. doi: 10.1016/j.febslet.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Hunsicker-Wang L, Pacoma RL, Luna E, Fee JA. A homologous expression system for obtaining engineered cytochrome ba 3 from Thermus thermophilus HB8. Protein Expr Purif. 2005;40:299–318. doi: 10.1016/j.pep.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Ädelroth P, Ek M, Brzezinski P. Factors Determining Electron-Transfer Rates in Cytochrome c Oxidase: Investigation of the Oxygen Reaction in the R. sphaeroides and Bovine Enzymes. Biochim Biophys Acta. 1998;1367:107–117. doi: 10.1016/s0005-2728(98)00142-x. [DOI] [PubMed] [Google Scholar]
- 30.Ädelroth P, Svensson-Ek M, Mitchell DM, Gennis RB, Brzezinski P. Glutamate 286 in cytochrome aa3 from Rhodobacter sphaeroides is involved in proton uptake during the reaction of the fully-reduced enzyme with dioxygen. Biochemistry. 1997;36:13824–13829. doi: 10.1021/bi9629079. [DOI] [PubMed] [Google Scholar]
- 31.Brändén M, Sigurdson H, Namslauer A, Gennis RB, Ädelroth P, Brzezinski P. On the role of the K-proton transfer pathway in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2001;98:5013–5018. doi: 10.1073/pnas.081088398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Munck E. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc Natl Acad Sci U S A. 1988;85:5779–5783. doi: 10.1073/pnas.85.16.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brzezinski P, Johansson AL. Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase. Biochimica et Biophysica Acta - Bioenergetics. 2010;1797:710–723. doi: 10.1016/j.bbabio.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Chang HY, Choi SK, Vakkasoglu AS, Chen Y, Hemp J, Fee JA, Gennis RB. Exploring the proton pump and exit pathway for pumped protons in cytochrome ba 3 from Thermus thermophilus. Proc Natl Acad Sci U S A. 2012;109:5259–5264. doi: 10.1073/pnas.1107345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutsoupakis C, Soulimane T, Varotsis C. Probing the Q-Proton Pathway of ba3-Cytochrome c Oxidase by Time-Resolved Fourier Transform Infrared Spectroscopy. Biophys J. 2004;86:2438–2444. doi: 10.1016/S0006-3495(04)74300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fee JA, Case DA, Noodleman L. Toward a chemical mechanism of proton pumping by the B-type cytochrome c oxidases: Application of density functional theory to cytochrome ba3 of Thermus thermophilus. J Am Chem Soc. 2008;130:15002–15021. doi: 10.1021/ja803112w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaila VRI, Sharma V, Wikström M. The identity of the transient proton loading site of the proton-pumping mechanism of cytochrome c oxidase. Biochimica et Biophysica Acta - Bioenergetics. 2011;1807:80–84. doi: 10.1016/j.bbabio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Koutsoupakis C, Soulimane T, Varotsis C. Ligand Binding in a Docking Site of Cytochrome c Oxidase: A Time-Resolved Step-Scan Fourier Transform Infrared Study. J Am Chem Soc. 2003;125:14728–14732. doi: 10.1021/ja036107e. [DOI] [PubMed] [Google Scholar]
- 39.Pfitzner U, Hoffmeier K, Harrenga A, Kannt A, Michel H, Bamberg E, Richter OMH, Ludwig B. Tracing the D-pathway in reconstituted sitedirected mutants of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39:6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Han H, Pawate A, Gennis RB. Decoupling mutations in the D-Channel of the aa3-Type cytochrome c oxidase from Rhodobacter sphaeroides suggest that a continuous hydrogen-bonded chain of waters is essential for proton pumping. Biochemistry. 2010;49:4476–4482. doi: 10.1021/bi100344x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson AL, Carlsson J, Högbom M, Hosler JP, Gennis RB, Brzezinski P. Proton uptake and pKa changes in the uncoupled Asn139Cys variant of cytochrome c oxidase. Biochemistry. 2013;52:827–836. doi: 10.1021/bi301597a. [DOI] [PubMed] [Google Scholar]
- 42.Johansson AL, Högbom M, Carlsson J, Gennis RB, Brzezinski P. Role of aspartate 132 at the orifice of a proton pathway in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2013;110:8912–8917. doi: 10.1073/pnas.1303954110. [DOI] [PMC free article] [PubMed] [Google Scholar]