Abstract
Natural killer (NK) cells are an innate lymphoid cell lineage characterized by their capacity to provide rapid effector functions, including cytokine production and cytotoxicity. Here, we identify the Ikaros family member, Aiolos, as a regulator of NK-cell maturation. Aiolos expression is initiated at the point of lineage commitment and maintained throughout NK-cell ontogeny. Analysis of cell surface markers representative of distinct stages of peripheral NK-cell maturation revealed that Aiolos was required for the maturation in the spleen of CD11bhighCD27− NK cells. The differentiation block was intrinsic to the NK-cell lineage and resembled that found in mice lacking either T-bet or Blimp1; however, genetic analysis revealed that Aiolos acted independently of all other known regulators of NK-cell differentiation. NK cells lacking Aiolos were strongly hyper-reactive to a variety of NK-cell-mediated tumor models, yet impaired in controlling viral infection, suggesting a regulatory function for CD27− NK cells in balancing these two arms of the immune response. These data place Aiolos in the emerging gene regulatory network controlling NK-cell maturation and function.
Keywords: differentiation, Ikzf3, NK cell, transcription
Introduction
Natural killer (NK) cells are innate lymphocytes specialized in cytokine production and cytotoxicity toward tumors and virus-infected cells. NK cells develop in the bone marrow from hematopoietic stem cells via lymphoid precursors (reviewed by Huntington et al, 2013) and then undergo maturation and functional diversification in bone marrow, thymus and a variety of peripheral organs including the liver, spleen and lymph nodes. NK-cell peripheral maturation in the mouse spleen is characterized by the up-regulation of the markers CD43, CD11b, CD94, Ly49C and KLRG1 with concomitant down-regulation of CD27, CD51 and c-kit (Kim et al, 2002; Hayakawa & Smyth, 2006; Huntington et al, 2007b; Chiossone et al, 2009; Yu et al, 2009). Mature CD11bhighCD27−KLRG1+ NK cells are the dominant population in non-lymphoid organs except for the liver (Hayakawa & Smyth, 2006; Huntington et al, 2007b), where a distinct TRAIL+CD49b−CD11blow expressing population (termed ILC1) exists (Takeda et al, 2005; Daussy et al, 2014; Sojka et al, 2014).
There is an emerging understanding about the transcriptional circuitries controlling NK-cell development, maturation and function. Transcription factors including NF-IL3 (E4BP4) (Gascoyne et al, 2009; Kamizono et al, 2009; Firth et al, 2013; Crotta et al, 2014; Male et al, 2014; Seillet et al, 2014; Sojka et al, 2014), Tox (Aliahmad et al, 2010), Ets1 (Barton et al, 1998; Ramirez et al, 2012) and Id2 (Yokota et al, 1999; Ikawa et al, 2001; Boos et al, 2007) are required for the development of NK cells from early progenitors, whereas GATA3 is essential for thymic NK cells and modulates the function of mature NK cells (Samson et al, 2003; Vosshenrich et al, 2006). A second group of transcription factors including Blimp1 (Smith et al, 2010; Kallies et al, 2011), Zbtb32 (Beaulieu et al, 2014), MEF (Lacorazza et al, 2002), IRF2 (Lohoff et al, 2000), Eomes and T-bet (Townsend et al, 2004; Robbins et al, 2005; Kallies et al, 2011; Gordon et al, 2012; Daussy et al, 2014) are more specifically required for the later stages of NK-cell differentiation and function.
The Ikaros family of Zinc-finger transcription factors are critical regulators of many aspects of lymphopoiesis, although their importance in the NK-cell lineage remains unclear (Merkenschlager, 2010; Yoshida et al, 2010). The family members, including Ikaros, Helios and Aiolos (encoded by Ikzf1, Ikzf2 and Ikzf3, respectively), can bind DNA as either homo- or hetero-dimers and regulate gene expression in both a positive and negative manner. Ikaros is required for the appearance of common lymphoid progenitors (CLPs) and subsequent NK-cell development, although there are at present no direct data on the role of Ikaros within the NK-cell compartment. In contrast, Helios is proposed to play a role in fine-tuning NK-cell responses downstream of the NKp46 receptor through an as yet undefined mechanism (Narni-Mancinelli et al, 2012). Aiolos is expressed predominantly in B cells, with some evidence for expression in T cells (Wang et al, 1998) and most recently human NK cells (Billot et al, 2010). Aiolos-deficient mice show mildly impaired B-cell development and have a propensity to develop a Lupus-like autoimmune disease (Wang et al, 1998; Cortes & Georgopoulos, 2004). Despite also being expressed in T cells, Aiolos deficiency leaves T-cell development relatively intact (Wang et al, 1998). A function for Aiolos in other hematopoietic lineages, including NK cells is as yet unknown.
Here, we show that Aiolos is constitutively expressed by NK cells and is vital for their peripheral maturation into CD11bhighCD27− NK cells. Aiolos is dispensable for most NK-cell effector functions after culture, but is required for maximal IFN-γ expression directly ex vivo, and for the full control of viral infection. Aiolos expression is independent of T-bet and Blimp1, two factors acting at a similar point in NK-cell differentiation, but its absence, similarly to Blimp1 deficiency, results in hyper-reactivity to tumor cells. Aiolos is required for the correct expression of several hundred genes in NK cells placing it as an emerging node in the transcriptional network controlling NK-cell peripheral maturation.
Results
Aiolos is constitutively expressed in NK cells
To identify new regulators of NK-cell development, we analyzed RNAseq data derived from the pre-pro-NK (Carotta et al, 2011), NKP (Rosmaraki et al, 2001), immature (i)NK and mature (m)NK-cell stages of the mouse bone marrow (Seillet et al, 2014). These data were combined with the previously reported transcriptome of the multipotent ALP (all lymphocyte progenitor) fraction of the CLP (Revilla et al, 2012), which is the immediate precursor of the NK-cell lineage (Carotta et al, 2011). Analysis of the Ikaros family of transcription factors revealed that the expression of one member, Ikzf3, coincided precisely with NK-cell lineage commitment (Fig 1A and B). Thereafter, Ikzf3 expression was maintained throughout bone marrow NK-cell development. In contrast, the best-characterized family member, Ikzf1, was similarly expressed in ALPs and all NK-cell fractions. A third family member, Ikzf2, was predominantly expressed at the earliest stages of NK-cell maturation before down-regulation in the mNK-cell fraction (Fig 1A and B).
Figure 1. Aiolos is constitutively expressed in NK cells.

- RNA was extracted from sorted populations from Id2GFP/GFP bone marrow corresponding to the pre-pro-NK cells (ppNK), NK-cell progenitor (NKP), immature (i) and mature (m) NK cells and subjected to RNAseq analysis. The cell isolation strategy is described in detail in Materials and Methods. Read coverage for indicated Ikaros family members Ikzf1 (Ikaros), Ikzf2 (Helios) and Ikzf3 (Aiolos) is shown mapped to the exon–intron structure (below). Arrows within the genes indicate the direction of transcription.
- Graph shows the reads per kilobase per million (RPKM) mapped to the indicated genes for the cell populations described in (A). The RNAseq data from the Ly6D− all lymphocyte progenitor (ALP) and Ly6D+ B cell-biased lymphocyte progenitor (BLP) fractions of the common lymphocyte progenitor (CLP) and the pro-B-cell data have been previously reported (Revilla et al, 2012).
- NK1.1+CD49b+TCRβ− NK cells from the thymus (tNK) and spleen were analyzed for the expression of Ikzf3 by quantitative PCR. Splenic NK cells were further divided into two fractions based on the expression of CD27 as indicated. Data were normalized to Hprt and were the mean ± SEM for three samples. Sorted splenic B cells (CD19+B220+) and CD8+ T cells were positive controls. *P < 0.05
- Intracellular flow cytometry for Aiolos expression in NK cells. Left dot plots, expression of CD27 and CD11b in bone marrow and spleen NK cells, gated as NK1.1+CD49b+TCRβ−. Right histograms, expression of Aiolos in wild-type (red) NK cells gated as indicated in the left dot plots. Total Ikzf3−/− NK cells served as a negative control (blue). Numbers indicate the mean fluorescence index of Aiolos in wild-type NK cells ± SEM from three independent experiments.
Data information: Data in (A) and (B) were pooled from the sorted cells of 11 experiments and 90 Id2GFP/GFP mice.
To determine whether Aiolos expression was maintained in NK cells outside of the bone marrow, we isolated NK cells from the thymus and spleen by flow cytometry and subjected them to quantitative real-time PCR. Ikzf3 was detected in all populations of NK cells independent from their localization, with expression peaking in splenic NK cells (Fig 1C). Despite these small transcriptional changes, analysis of Aiolos protein using intracellular flow cytometry revealed strong and uniform expression throughout NK-cell differentiation (Fig 1D). Taken together, these data identify Aiolos as being constitutively expressed by NK cells from the earliest known progenitor.
Aiolos is required for peripheral NK-cell maturation
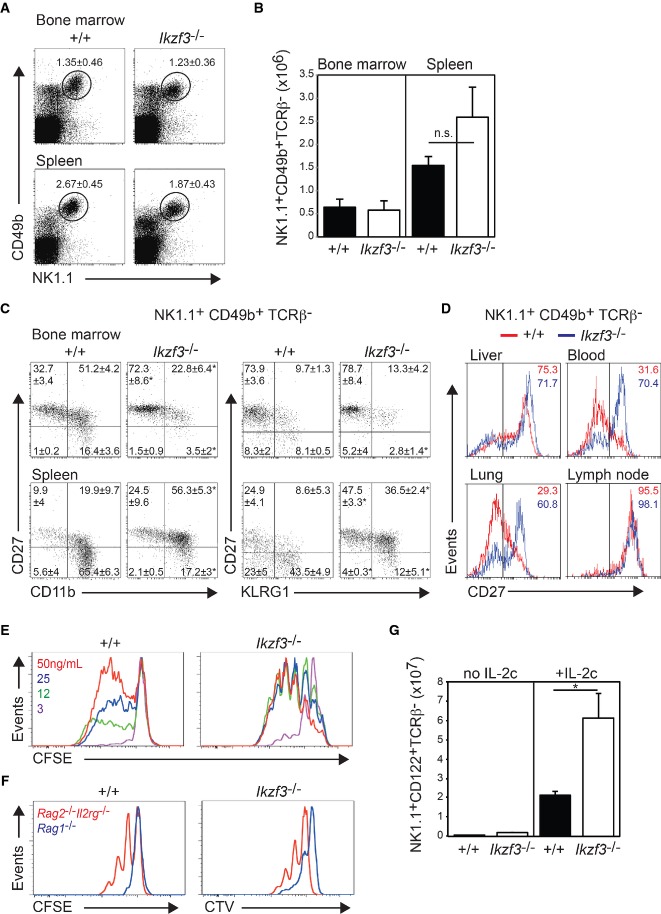
To test the functional importance of Aiolos in the NK-cell lineage, we have assessed their abundance and cell surface phenotype in Ikzf3−/− mice (Wang et al, 1998) that we have backcrossed to C57BL/6 for 10 generations. NK cells, defined as NK1.1+CD49b+TCRβ−, were present in a similar frequency and absolute number in the bone marrow and spleen of wild-type and Ikzf3−/−mice (Fig 2A and B), indicating that Aiolos was grossly dispensable for the production of NK cells.
Figure 2. Loss of Aiolos blocks peripheral NK-cell maturation.
A–D Flow cytometric analysis of the NK-cell compartment of wild-type (+/+) and Ikzf3−/− mice. Plots in (A) show total cells from the bone marrow and spleen. Values are the mean proportion of NK1.1+CD49b+ NK cells in the indicated gate ± SEM from 7 to 8 individual mice of each genotype. (B) Quantitation of the absolute numbers of bone marrow and spleen NK cells from +/+ and Ikzf3−/− mice. Numbers represent the mean ± SEM from 7 to 8 individual mice of each genotype. n.s., not significant (P > 0.05) when comparing the indicated populations. (C) Analysis of CD11b, CD27 and KLRG1 expression in gated NK cells (NK1.1+CD49b+TCRβ−) from bone marrow and spleen. Numbers are the mean proportion of NK cells in each quadrant ± SEM from 3 +/+ and 6 Ikzf3−/− mice. (D) Analysis of CD27 expression in gated NK cells from the indicated organs of +/+ and Ikzf3−/− mice. Results are representative of 4–6 independent experiments.
E, F Splenic NK cells from +/+ and Ikzf3−/− mice were labeled with CFSE or CTV and either (E) cultured for 5 days in the indicated concentration of IL-15 or (F) transferred into Rag1−/− or Rag2−/−Il2rg−/− mice and analyzed at day 7. Data are representative of two experiments.
G +/+ and Ikzf3−/− mice were treated daily for 5 days with IL-2/anti-IL-2 mAb complexes before analysis. Data are the mean number of splenic NK1.1+CD122+ TCRβ− NK cells in recipient mice ± SD from two experiments. *P < 0.05, when comparing the equivalent +/+ and Ikzf3−/− populations.
The progression of the late stages of NK-cell maturation can be visualized by flow cytometric analysis for expression of CD27 in conjunction with the auxiliary markers CD11b, KLRG1, CD51, Ly49C/I and c-kit (Kim et al, 2002; Hayakawa & Smyth, 2006; Huntington et al, 2007b). Bone marrow NK cells are predominantly CD11blowCD27+KLRG1−, while splenic NK cells first up-regulate CD11b, before differentiating through CD11bhighCD27+KLRG1− and CD11bhighCD27−KLRG1+ stages (Hayakawa & Smyth, 2006; Huntington et al, 2007b; Chiossone et al, 2009). Examination of NK cells from the bone marrow and spleen of Aiolos-deficient mice revealed an over-representation of the CD11blowCD27+ and CD11bhighCD27+ populations and a pronounced decrease in the proportion of NK cells at the CD11bhighCD27− stage (Fig 2C). While CD27 and KLRG1 are generally mutually exclusive in splenic NK cells, Aiolos deficiency resulted in a striking accumulation of CD27+KLRG1+ cells (Fig 2C). The failure to appropriately down-regulate CD27 was also observed in the lung, peripheral blood and to a lesser extent liver, while both wild-type and Ikzf3−/− NK cells in the lymph node were characteristically CD27+ (Fig 2D). In contrast, the differentiation of liver ILC1s was intact in the absence of Aiolos (data not shown).
To test whether the developmental block observed in the absence of Aiolos was due to impaired proliferation or survival, we assayed NK-cell responses to IL-15, the major physiological regulator of NK-cell homeostasis (Huntington et al, 2007a). Ikzf3−/− NK cells showed enhanced proliferation responses after in vitro exposure to IL-15, with the phenotype being most pronounced in suboptimal IL-15 concentrations (Fig 2E). This enhanced proliferation occurred regardless of whether the starting NK-cell populations were derived directly ex vivo or were pre-cultured in optimal IL-15 for 5 days, suggesting that the effect was not due to the altered distribution of the mature splenic NK-cell compartments (data not shown).
To examine NK-cell proliferation in vivo, NK cells were labeled with cell division tracking membrane dyes [wild-type (CFSE), Ikzf3−/− (CTV)] and co-transferred into Rag1-deficient (T- and B-cell- deficient, NK-cell-sufficient) or Rag2/common γ chain-double-deficient (Rag2−/−Il2rγ−/−, T-, B- and NK-cell-deficient) hosts. In keeping with the in vitro proliferation data (Fig 2E), Ikzf3−/− NK cells displayed higher sensitivity to homeostatic cytokines (Rag2−/−Il2rγ−/− recipients) compared to wild-type NK cells as evident by all Ikzf3−/− NK cells having entered cell division after 7 days, whereas many wild-type NK cells had not yet divided (Fig 2F). Neither population proliferated in the presence of endogenous NK cells (Rag1−/− hosts, Fig 2F). Treatment of wild-type and Ikzf3−/− NK with IL-2/anti-IL-2 mAb complexes that promote NK-cell proliferation (Boyman et al, 2006) resulted in a significant increase in the number of Ikzf3−/− NK cells in comparison to their wild-type competitors (Fig 2G). Thus, Aiolos-deficient NK cells display hyper-responsiveness to activation via the common γ chain receptor pathway. The hyper-responsiveness observed in these assays excluded impaired cell survival or proliferation as the cause of the impaired maturation observed in Aiolos-deficient NK cells as instead suggested a differentiation block at the final step in peripheral NK-cell maturation.
Gene regulation by Aiolos
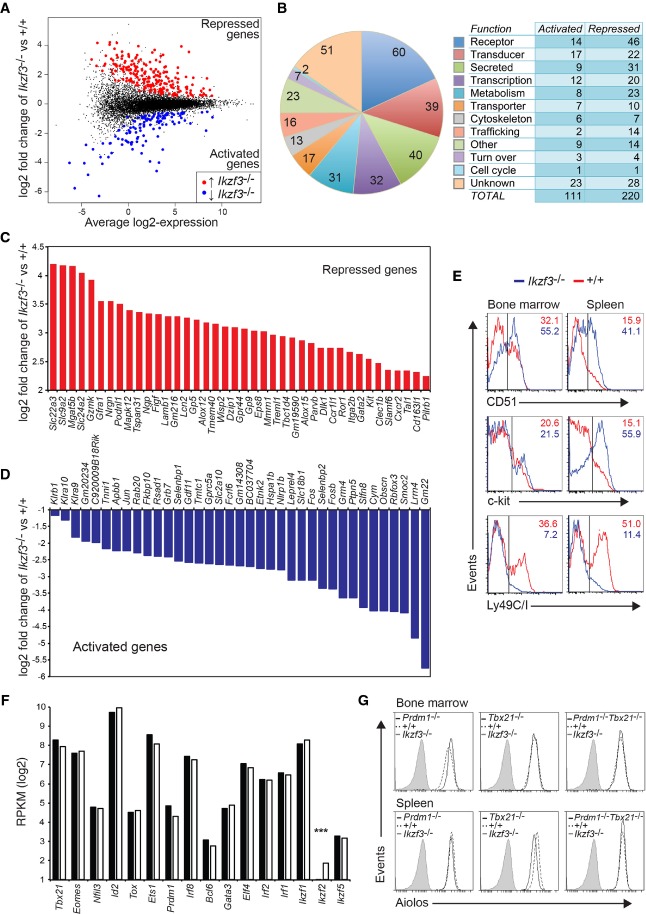
To identify the transcriptional program regulated by Aiolos in NK cells, we performed RNAseq on sorted mature NK cells from the spleen of wild-type and Ikzf3−/− mice. As Ikzf3−/− mice accumulate a CD27+KLRG1+ NK-cell population that is not presented in wild-type mice, we have analyzed NK1.1+NKp46+CD11bhigh mature NK cells regardless of their expression of CD27 and KLRG1. This strategy also excludes any potential contamination by ILC1s, which are CD11blow (Daussy et al, 2014; Sojka et al, 2014). A stringent analysis of this data revealed 331 differentially expressed transcripts, including 220 whose expression increased in the absence of Aiolos and 111 genes that required Aiolos for full expression (Fig 3A–D and Supplementary Table S1; false discovery rate < 0.05). The differentially expressed genes encoded proteins belonging to diverse functional categories, with a relatively large number of transcripts coding for proteins with no known function (15%, 51/331 genes). Flow cytometry confirmed the differential expression of several of the candidates including Itgav (encoding CD51), Kit (encoding c-kit) and Klra9 (encoding Ly49I and recognized by the Ly49C/I antibody), and Cd38, Icos and Csf2rb (encoding CD131; Fig 3E and data not shown). Other differentially expressed genes relevant to NK-cell biology included increased expression of Gzmk, Gzmc, Gzmm and Tnf and mildly reduced Gzmb, while several additional NK receptors including Klrb1 and Klra10 were reduced in expression.
Figure 3. Gene regulation by Aiolos in NK cells.
A–D NK1.1+NKp46+CD244+CD11bhighTCRβ− NK cells were sorted from the spleen of wild-type (+/+) and Ikzf3−/− mice and subjected to RNAseq. Data are the mean of two independent experiments. (A) Scatter plot of differential expression. Genes with significantly increased (red) or decreased (blue) expression in the absence of Aiolos are indicated (false discovery rate < 0.05). The full list of differentially expressed genes is shown in Supplementary Table S1. (B) Left, pie chart showing the proportion of differentially expressed genes that fall into 1 of 12 functional categories. Right, number of Aiolos activated (upregulated in Ikzf3−/−) and repressed (downregulated in Ikzf3−/−) genes in each functional category. (C) Differential expression of the genes whose expression was most increased in the absence of Aiolos (n = 41). (D) Differential expression of the genes whose expression was most decreased in the absence of Aiolos (n = 37).
E Expression of Itgav (encoding CD51), Kit (encoding c-kit) and Klra9 (encoding Ly49I and recognized by the Ly49C/I antibody) in NK1.1+CD49b+TCRβ− NK cells from the bone marrow and spleen of +/+ and Ikzf3−/− mice. Data are representative of 4–6 independent experiments. Numbers indicate the proportion of cells that were positive for the indicated markers.
F Expression of the major transcription factors implicated in NK-cell differentiation in +/+ and Ikzf3−/− NK cells from the RNAseq data described in (A). Graph shows the mean reads per kilobase per million (RPKM). ***P < 0.005.
G NK1.1+CD49b+TCRβ− NK cells from the bone marrow or spleen of mice lacking Blimp1 (encoded by Prdm1), T-bet (encoded by Tbx21) or both proteins were analyzed for the expression of Aiolos by intracellular flow cytometry. Ikzf3−/− NK cells act as a background control.
The surprisingly few NK-cell-associated genes that were differentially expressed suggested that Aiolos regulates a molecular program distinct from that previously implicated in NK-cell biology. In keeping with this conclusion, analysis of the expression of a panel of transcription factors known to control NK-cell differentiation revealed no significant differences between the wild-type and Ikzf3−/− genotypes (Fig 3F). In addition, we observed little evidence of compensation by other family members, as both Ikaros (Ikzf1) and Pegasus (Ikzf5) were equivalently expressed between the genotypes. The expression of one family member, Helios (Ikzf2), was increased in Ikzf3−/− NK cells, although its overall concentration was very low (Fig 3F). Taken together, these data suggest that Aiolos regulates a transcriptional program in maturing NK cells that is, at least in the absence of exogenous stimulation, largely independent of the characteristic NK-cell surface receptors, transcription factors and effector molecules.
Ikzf3 expression in NK cells is independent of T-bet and Blimp1
The maturation defect we have observed in Aiolos-deficient NK cells resembled that previously reported for mice deficient in T-bet (encoded by Tbx21) or Blimp1 (encoded by Prdm1) (Robbins et al, 2005; Kallies et al, 2011). Thus, it was possible that the defective NK-cell maturation in the absence of T-bet or Blimp1 was due to reduced Aiolos expression. Flow cytometric analysis of mice lacking either Blimp1, T-bet or both proteins showed that these factors were dispensable for normal Aiolos expression (Fig 3G).
Aiolos is intrinsically required in NK cells
Aiolos-deficient mice display spontaneously activated B cells and development of a lupus-like syndrome with age (Wang et al, 1998), raising the possibility that the impaired NK-cell maturation is secondary to this autoimmune syndrome. To rigorously test whether the maturation phenotype of Aiolos-deficient NK cells was intrinsic to the NK-cell lineage, we generated chimeric mice by reconstituting lethally irradiated Ly5.1 mice with a mixture of congenically marked bone marrow from wild-type (Ly5.1+) and Ikzf3−/− (Ly5.2+) mice. Examination of the bone marrow and spleen of these mice revealed that Aiolos-deficient NK cells were present at normal frequencies in the chimeric mice (Fig 4A). Importantly, the Ikzf3−/− NK cells maintained their immature CD27+ phenotype, with decreased Ly49C and increased c-kit and CD51, indicating that the requirement of Aiolos for the peripheral maturation of NK cells was cell intrinsic (Fig 4B).
Figure 4. Cell intrinsic requirement for Aiolos during NK-cell maturation.
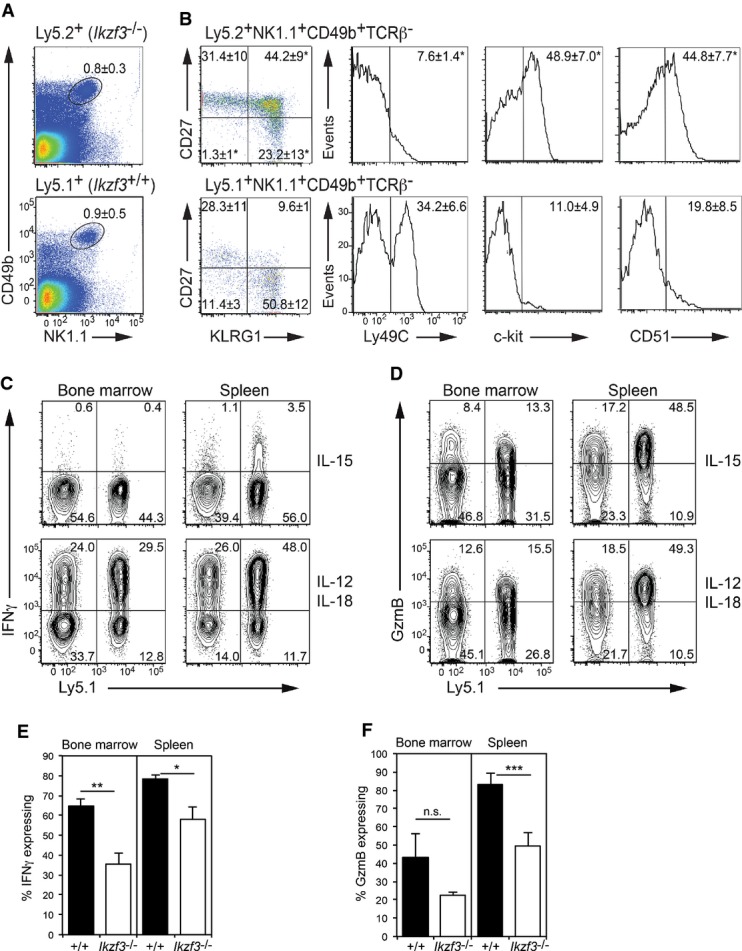
A C57Bl/6 Ly5.1+ mice were lethally irradiated and reconstituted with a 1:1 ratio of C57Bl/6 Ly5.1+ and Ikzf3−/− (Ly5.2+) bone marrow. Splenocytes from these mice were analyzed 8 weeks after reconstitution for the indicated markers. Numbers are the mean proportion of NK1.1+CD49b+ cells ± SEM from the Ikzf3−/− (Ly5.2+, upper plot) or wild-type (Ly5.1+, lower plot).
B NK1.1+CD49b+TCRβ− NK cells as in (A) were gated as either wild-type (Ly5.1+) or Ikzf3−/− (Ly5.2+) and analyzed for the markers as indicated. Numbers in the dot plots show the mean proportion of NK cells in each quadrant ± SEM. Numbers in the histograms are the proportion of marker expressing cells ± SEM. Results are representative of at least three experiments with each containing 3–4 mice.
C, D Gated NK1.1+CD49b+TCRβ− NK cells from mixed bone marrow chimeras, reconstituted with a 1:1 ratio of C57Bl/6 Ly5.1+ and Ikzf3−/− (Ly5.2+) bone marrow, were examined for (C) IFN-γ and (D) GzmB expression by intracellular staining and flow cytometry. Numbers in the plots show the proportion of NK cells in each quadrant. Prior to analysis, the cells were stimulated with either IL-15 or IL-12 and IL-18 for 5 h.
E, F Graphs show the mean proportion of IL-12- and IL-18-stimulated NK cells expressing (E) IFN-γ and (F) GzmB from the C57Bl/6 Ly5.1+ (+/+) and Ikzf3−/− (Ly5.2+) fractions. Data are the means ± SEM from three mice of each genotype. P-values compare the indicated genotypes. n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.005.
To further assess the function of Aiolos-deficient NK cells in this competitive setting, NK cells from the bone marrow or spleen of chimeric mice were stimulated ex vivo for 5 h by either IL-15 or the combination of IL-12 and IL-18, and the production of IFN-γ and GzmB was determined by intracellular flow cytometry (Fig 4C–F). Collectively. these data showed that Ikzf3−/− NK cells were capable of expressing these two important mediators of NK-cell function, but did so at a modestly reduced frequency compared to wild-type (Ly5.1+) cells.
Aiolos is dispensable for cytokine production and cytotoxicity in vitro
To test the functional properties of Aiolos-deficient NK cells, we measured lytic activity of splenic NK cells either directly ex vivo (Fig 5A) or after culture in IL-15 (Fig 5B). Wild-type and Ikzf3−/− NK cells were equivalent in their ability to lyse NKG2D-ligand expressing MHC-I-deficient targets (Fig 5A and B). Similarly, NK cells cultured in IL-15 alone or with the cytotoxicity-promoting factor IL-21 were also able to lyse target cells in the absence of Aiolos (Fig 5B).
Figure 5. In vitro effector functions in the absence of Aiolos.

A, B Cytotoxic activity of freshly isolated (A) or cultured (B) NK cells of the indicated genotypes against RMAS-Rae1β tumor cells. DX5+ NK cells were cultured in either IL-15 or IL-15 and IL-21 for 5 days before being subjected to a standard 51Cr release assay. Data are the mean proportion of specific lysis of triplicate measurements ± SEM and are representative of at least three experiments.
C NK cells sorted as in (A) were expanded in 50 ng/ml IL-15 for 5 days prior to culture for 2 days in the indicated cytokines. Cells were then washed and equal numbers seeded into fresh cytokine for 24 h, after which the supernatants were harvested and examined for cytokine production by bead array. Data are the mean ± SEM from at least three experiments. P-values compare the indicated genotypes. *P < 0.05.
As longer-term exposure to the cytokines including IL-12, IL-18 and IL-21 alters the functional maturation of NK cells (Brady et al, 2010), we cultured Ikzf3−/− and wild-type NK cells with these factors and measured cytokine secretion by bead arrays. While these data overall indicated that Aiolos played no essential role in cytokine expression in response to these factors, we did observe a consistent drop in the secretion of the inflammatory chemokines, Ccl3 (MIP1α), Ccl4 (MIP1β) and Ccl5 (RANTES) in all conditions tested (Fig 5C).
In vivo NK-cell functions in Aiolos-deficient mice
To test NK-cell functions in vivo, we infected the wild-type and Ikzf3−/− mice with mouse cytomegalovirus (MCMV), which in C57Bl/6 mice is controlled by activities mediated by Ly49H+ NK cells (Voigt et al, 2003). We examined NK-cell dynamics and viral load in the organs of mice for up to 18 days after infection with MCMV. As in resting mice, the numbers of both total and Ly49H+ NK cells were similar in the spleen and liver of infected wild-type and Ikzf3−/− mice (data not shown). Moreover, analysis of infected +/+ (Ly5.1+): Ikzf3−/− (Ly5.2+) bone marrow chimeras showed equal contribution of wild-type and Aiolos-deficient NK cells (both total and Ly49H+) to the MCMV response (Supplementary Fig S1A and B). Interestingly, analysis of the viral titers from infected wild-type and Ikzf3−/− mice showed significantly increased MCMV load at the initial site of viral replication, the liver, in the Aiolos-deficient mice (Fig 6A). In keeping with the characteristic viral spread to other organs at later time points, we observed increased viral load at day 10 in the spleen, lung and salivary gland. The increased viral load did not appear to be mediated by defects in the clonal expansion of Aiolos-deficient T cells as the chimeric analysis showed that CD4+ and CD8+ (both total and virus-specific) T cells contributed equally to the immune response to MCMV (Supplementary Fig S1C–E).
Figure 6. In vivo functions of Aiolos-deficient NK cells.

A Wild-type (+/+) and Ikzf3−/− mice were infected with MCMV. Organs were removed at the indicated times post-infection and viral load determined by plaque assay. Data are the mean ± SEM combined from two independent experiments and at least four mice per genotype. *P < 0.05, **P < 0.01. Dotted line indicates the limit of detection of the assay.
B +/+ and Ikzf3−/− mice were inoculated intravenously with B16B6 melanoma cells at the indicated doses. The number of lung metastases was measured at day 14. Each dot represents an individual mouse. Horizontal line shows the mean number of lung metastases ± SEM for each group. *P < 0.05.
C–E Groups of 5 +/+ and Ikzf3−/− mice were inoculated subcutaneously with RMA-S tumor cells at the indicated doses. Tumor volume was determined at the indicated days after injection and shown as the means ± SEM of mice in each group. Data in (E) are representative of two independent experiments. Asterisks indicate the groups that are statistically different between the genotypes using a Mann–Whitney rank sum test (*P < 0.05).
F Left plots, NKp46+CD122+TCRβ− NK cells were analyzed from the lungs and spleen of mice injected with 40,000 B16F10 melanoma cells 14 days before. Data are the mean number of NK cells ± SEM from four experiments. Right plots, expression of CD27 and KLRG1 in NK cells from the lungs and spleen gated as in the left plots.
NK cells are also known for their ability to kill some types of tumor cells. To examine whether Aiolos deficiency also impacts on tumor control, we injected wild-type and Ikzf3−/− mice with tumor cells that are controlled in an NK-cell-dependent manner (Hayakawa et al, 2002; Smyth et al, 2002) and measured tumor burden. Surprisingly, Aiolos-deficient NK cells were superior in reducing the number of lung metastases that arise after intravenous injection of B16B6 melanoma cells (Fig 6B). Aiolos-deficient NK cells were also superior to their wild-type counterparts in their ability to control the growth of RMAS tumor cells across a spectrum of inoculation doses (Fig 6C–E). As the cytotoxic function of freshly isolated NK cells per se did not seem to be altered (Fig 5A and B), this result suggested that NK cells in the absence of Aiolos showed either enhanced activation or recruitment to the site of tumor growth. To examine this possibility in more detail, we examined the frequency and phenotype of NK cells recruited to the lung of tumor bearing mice. Ikzf3−/− NK cells were recruited to the lung at a similar frequency to wild-type cells, but maintained their aberrant CD27+KLRG1+ phenotype, suggesting that it was this subset that is providing the superior tumor control in the absence of Aiolos (Fig 6F).
Discussion
In this study, we identified Aiolos as an important player in the transcriptional network governing NK-cell differentiation. NK cells constitutively express Aiolos from a very early point in their development, although the requirement for Aiolos was largely restricted to the peripheral differentiation of NK cells from CD11blowCD27+ to the CD11bhighCD27− mature NK cells. Aiolos-deficient NK cells accumulated at an intermediate CD27+KLRG1+ stage that also displayed aberrant CD51 and c-kit expression and lacked Ly49C/I. Aiolos-deficient NK cells displayed relatively intact, cytotoxic function and production of cytokines after in vitro culture, but showed reduced production of IFN-γ, inflammatory chemokines and MCMV control in vivo. Most surprisingly, Aiolos-deficient NK cells displayed enhanced ability to control tumor cells that are usually subject to NK-cell-mediated regulation, suggesting a negative regulatory role for Aiolos in NK-cell activation.
The Ikaros family of Zinc-finger transcription factors plays multiple roles in the adaptive immune system; however, to date, Aiolos has not been implicated in NK-cell biology. Aiolos is expressed throughout B-cell differentiation, where it is under the control of Pax5 (Schebesta et al, 2007; Pridans et al, 2008). Aiolos-deficient B cells show a complex phenotype characterized by mildly impaired early B-cell differentiation and the late appearance of a lupus-like syndrome (Wang et al, 1998; Cortes & Georgopoulos, 2004). This B-cell expression pattern and function is strikingly similar to that reported here for NK cells, which constitutively express Aiolos, while Ikzf3−/− NK cells showed impaired differentiation and hyper-responsiveness to activation stimuli in vivo, in this case tumor cells. Aiolos deficiency does not appear to have a significant impact on T-cell development or function (Wang et al, 1998), although Ikzf1+/−Ikzf3−/− mice show an increased prevalence of lymphomas, suggesting a redundancy between these two family members in some aspects of lymphopoiesis (discussed below) (Cortes et al, 1999).
Peripheral maturation of NK cells is a multi-step process. The most immature NK cells in the mouse spleen express the marker combination CD11blowCD27+. During maturation, these cells first become CD11bhighCD27+ before silencing CD27 and up-regulating KLRG1 to undergo their final maturation (Hayakawa & Smyth, 2006; Huntington et al, 2007b; Chiossone et al, 2009). Aiolos-deficient NK cells show an accumulation of CD27+KLRG1+ NK cells, a normally transient state of differentiation between the CD27+KLRG1− and CD27−KLRG1+ maturation stages. The developmental staging of these cells, however, is problematic as Aiolos-deficient NK cells also have altered expression of several of other maturation markers, including a lower CD11b and Ly49C/I and increased expression of the immature markers c-kit and CD51, which together do not indicate a simple differentiation block between the CD27+ and CD27− states and instead suggest a more general dysregulation of maturation (Hayakawa & Smyth, 2006; Huntington et al, 2007b; Chiossone et al, 2009). In keeping with this conclusion, only 11% (37 of 331) of the Aiolos-regulated genes were also previously reported to be differentially expressed between CD11blowCD27+ and CD11bhighCD27− NK cells (Chiossone et al, 2009).
Our transcriptome analysis showed that, in addition to Aiolos, NK cells also expressed Ikaros (Ikzf1) and Helios (Ikzf2). Ikaros family members are known to homo- and hetero-dimerize with each other, raising the possibility that Ikaros and Helios provided some compensatory function in the absence of Aiolos (Morgan et al, 1997; Cortes et al, 1999). Ikaros, like Aiolos, is constitutively expressed in NK cells, however, due to the absence of NK cells in Ikzf1-null mice, resulting from a deficiency in CLPs, no function for Ikaros in committed NK cells has been defined. Moreover, we observed no additional NK-cell functional or developmental deficiencies in Ikzf1+/−Ikzf3−/− mice (R.P.L. Thong, S.L. Nutt, unpublished observations). Helios is known to be silenced in mature NK cells in an NKp46-dependent manner (Narni-Mancinelli et al, 2012), and its expression was increased in the absence of Aiolos, although only to a relatively low expression level. It will now take the development of new mouse models to address the possible redundancy of Ikaros family members in NK cells.
The phenotypic consequence of Aiolos deficiency in NK cells reported in this study most closely resembles mice lacking T-bet or Blimp1 (Robbins et al, 2005; Kallies et al, 2011). In each case, NK cells accumulated at the CD27+ stage and over-expressed c-kit; however, in contrast to T-bet- and Blimp1-deficient NK cells, Ikzf3−/− NK cells up-regulated KLRG1. Thus, it appears that while T-bet or Blimp1 deficiency results in a block in further development, the loss of Aiolos results in aberrant marker expression and failure to normally transition between the CD27+ and CD27− cell states. Such a role of Ikaros family members as drivers of cell differentiation between stable developmental stages has been recently proposed for Ikaros in pre-B cells (Ferreiros-Vidal et al, 2013).
Despite the commonalities in the knockout phenotypes, we found no evidence that Aiolos played a role in the regulation of the expression of T-bet or Blimp1. Similarly, loss of Blimp1, T-bet or both factors did not alter Aiolos expression. This contrasts with Blimp1 that is downstream of T-bet in the same process (Kallies et al, 2011). The novel function of Aiolos was also highlighted by our global transcriptome analysis, with few NK-cell-specific genes and no known NK-cell transcription factors being differentially expressed in the absence of Aiolos. Thus, Aiolos acts in peripheral NK-cell maturation independently of the major known regulators of the process.
The existence of a final maturation program in late NK-cell differentiation, as well as the tissue specific distribution of CD27+ and CD27− NK cells, has been recognized for a number of years; however, our understanding of the functions of these subsets remains limited to the modest differences observed in vitro assays (Hayakawa & Smyth, 2006; Hayakawa et al, 2006; Huntington et al, 2007b; Chiossone et al, 2009). CD27+KLRG1− NK cells undergo enhanced proliferation in response to IL-15 in vitro and under lymphopenic conditions in vivo, compared to the CD27−KLRG1+ NK cells, as well as being more efficient at performing some NK-cell effector functions in vitro (Hayakawa et al, 2006; Huntington et al, 2007b; Chiossone et al, 2009). The enhanced tumor control reported here for Aiolos-deficient mice and previously for mice lacking Blimp1 (Kallies et al, 2011) may stem from the preponderance of CD27+KLRG1− NK cells in these mice, which we showed efficiently home to tumor containing organs. In contrast, Aiolos deficiency resulted in a mild defect in IFN-γ expression and reduced ability to control the MCMV virus. These observations are likely to be linked as the optimal host response to MCMV is known to require NK-cell-produced IFN-γ (Loh et al, 2005; Orange et al, 1995). Taken together, these findings may suggest that the coordination of the two best-characterized NK-cell effector functions, cytokine secretion and cytotoxicity, relies on the normal peripheral maturation of the CD27+ and CD27− populations. An alternative scenario is that the CD27−KLRG1+ population may possess a negative regulatory function whose absence results in enhanced NK-cell lytic activity from the more proliferative CD27+KLRG1− cells. Further studies are required to test these possibilities.
Materials and Methods
Mice
Mixed background Ikzf3-deficient (Wang et al, 1998) mice were obtained from Dr. Katia Georgopoulos and backcrossed to C57BL/6 for 10 generations. Prdm1GFP (Kallies et al, 2004), Id2GFP (Jackson et al, 2011) and Tbx21−/− (Szabo et al, 2002) mice have been previously described. Prdm1GFP/GFP mice were generated as described (Kallies et al, 2011). Ikzf3−/−: Ly5.1 mixed bone marrow chimeras were produced by reconstituting lethally irradiated C57BL/6-Ly5.1 recipients with a mixture of bone marrow cells isolated from Ikzf3−/− and Ly5.1 mice. Chimeric mice were analyzed after a minimum of 8 weeks post-reconstitution. All animal experimentation was performed with the approval of the Animal Ethics Committees of the Walter and Eliza Hall Institute or the University of Western Australia and according National Health and Medical Research Council (NHMRC) of Australia guidelines.
Antibodies and flow cytometry
The following anti-mouse mAbs were used for flow cytometric analysis: NK1.1 (PK136), Ly5.2 (104), Ly5.1 (A20), CD27 (LG.7F9/LG.3A10), KLRG1 (2F1), CD49b (HMα2), CD51 (RMV-7), CD122 (TM-β1), CD244 (244F4), NKp46 (29A1.4) CD11b (Mac-1, M1/70), c-kit (2B8), TCRβ (H57-597), CD4 (GK1.5), CD8 (53-6.7), IFN-γ (XMG1.2), Ly49H (3D10) and Ly49C/I (SW5E6). Intracellular staining for Aiolos (8B2) was performed using the Foxp3 staining kit (eBioscience). Anti-GzmB (GB12) was purchased from Invitrogen, and the remaining reagents were from BD Biosciences or eBioscience. Viable cells were identified by propidium iodide or cytox blue exclusion. Cells were analyzed on a FACS Canto cytometer (BD Biosciences), and cell sorting was performed on MoFlo (Becton Coulter) or Aria cytometers (BD Biosciences). Data were processed using FlowJo and Weasel software.
Identification and sorting of NK-cell progenitor populations by flow cytometry
Pre-pro-NK cells (ppNK, Lin−Sca-1highc-kitlow/−Id2-GFP+CD127+CD135−) (Carotta et al, 2011) and NK-cell progenitors (NKP, Lin−CD122+NK1.1−CD49b−Id2-GFP+), immature NK cells (iNK, Lin−CD122+NK1.1+CD49b−Id2-GFP+) and mature NK cells (mNK, Lin−CD122+NK1.1+CD49b+Id2-GFP+) were defined as described (Seillet et al, 2014).
Cell isolation and culture
Unless otherwise indicated, splenic NK cells were enriched using DX5 magnetic beads (Miltenyi) and cultured in IL-15 (50 ng/ml, R&D Systems) for 5 days. The cells were then used or re-cultured in the presence of IL-15 and either IL-12 (2 ng/ml), IL-18 (50 ng/ml) or IL-21 (100 ng/ml), all from R&D Systems. Lung and liver lymphocytes were purified from single cell suspensions by centrifugation on Histopaque (1.077 g/ml, Sigma-Aldrich) for 20 min at 400 g at room temperature.
Intracellular cytokine stains
DX5 bead-enriched NK cells were cultured in 50 ng/ml IL-15 or 5 ng/ml IL-12 and 50 ng/ml IL-18 for 5 h in the presence of GolgiPlug, stained for relevant surface molecules, fixed and permeabilized using the Cytofix/Cytoperm reagent (BD Biosciences) and analyzed for GzmB and IFN-γ.
Cytokine bead array
NK cells were expanded in IL-15 for 5 days before being transferred into either IL-15/IL-21, IL-15/IL-12 or IL-12/IL-18. After another 2 days in culture, cell numbers were determined and equal numbers of cells seeded in the same conditions. Supernatants were collected after an additional 24 h and assayed for cytokines using the Bio-Rad Bioplex cytokine bead assay (Mouse 23-Plex Panel).
mRNA analysis
Total RNA was prepared from flow cytometrically purified mouse NK cells using an RNeasy kit (Qiagen) and subjected to quantitative real-time PCR. Primer sequences for Ikzf3 (Pridans et al, 2008) and Hprt (Kallies et al, 2006) were as described. mRNA isolated from pre-pro-NK cells was subjected to RNAseq analysis and will be described in detail elsewhere (S. Carotta, unpublished). The ALP, BLP, pro-B cell (Revilla et al, 2012; GEO repository, accession number GSE 38046) and NKP, iNK and mNK-cell (Seillet et al, 2014) RNAseq data were previously reported.
Approximately 5 × 105 NK1.1+NKp46+CD244+CD11bhighTCRβ− mature NK cells were sorted from the spleen of wild-type (+/+) and Ikzf3−/− mice, and two biological replicates were generated and subjected to 100 bp single end sequencing on an Illumina HiSeq2000 at the Australian Genome Research Facility (Melbourne, Australia). More than 30 million reads were generated for each replicate and aligned to the GRCm38/mm 10 build of the Mus musculus genome using the Subread aligner (Liao et al, 2013). Genewise counts were obtained using featureCounts (Liao et al, 2014). Reads overlapping exons in annotation build 38.1 of NCBI RefSeq database were included. Genes were filtered from downstream analysis if they failed to achieve a CPM (counts per million mapped reads) value of at least 0.5 in at least two libraries. Counts were converted to log2 counts per million, quantile normalized and precision weighted with the voom function of the limma package (Law et al, 2014). A linear model was fitted to each gene (Smyth, 2004), and empirical Bayes moderated-t P-values were computed relative to a fold change cutoff of 1.2-fold by using treat (McCarthy & Smyth, 2009). Genes were called differentially expressed if they achieved a false discovery rate of < 0.05.
Cytotoxicity assays
The cytotoxicity of NK cells was assessed using RMAS-Rae1β tumor cell lines as described (Hayakawa et al, 2002; Brady et al, 2004).
Proliferation assays
Division tracking
Splenic NK1.1+CD49b+TCRβ− NK cells were labeled with CFSE (carboxyfluorescein succinimidyl ester, Molecular Probes) or CTV (Cell Tracker Violet; Molecular Probes) at a concentration of 5 μM and cultured for 5 days in various concentrations of IL-15. Alternatively, NK-cell numbers were expanded in vitro in IL-15, CTV/CFSE labeled and transferred into Rag1−/− or Rag2−/−Il2rg−/− mice as described (Sathe et al, 2014).
IL-2/anti-IL-2mAb complexes
1.5 μg IL-2 (Peprotech) and 10 μg anti-IL-2 (S4B6, prepared in house) were incubated together at 37°C for 30 min and then injected IP. Mice were treated daily for 5 days.
Mouse cytomegalovirus (MCMV) infection
Mice were injected (IP) with 1 × 104 plaque forming units of salivary gland-propagated virus stock of MCMV-K181 diluted in PBS containing 0.5% fetal bovine serum and analyzed as described (Sumaria et al, 2009). Viral titers were quantified by plaque assay on monolayers of permissive cells as described (Allan & Shellam, 1984).
In vivo tumor model
Groups of 5 wild-type or Ikzf3−/− mice were inoculated subcutaneously with RMA-S tumor cells (Hayakawa et al, 2002) or intravenously with B16B6 or B16F10 melanoma cells (Takeda et al, 2011) at the indicated doses. RMA-S tumor growth was examined every second day with calipers. The number of lung metastases in mice injected with B16B6 cells was measured at day 14.
Statistics
Data were analyzed using a two-tailed, Student's t-test (paired or unpaired as appropriate) or a Mann–Whitney rank sum test. P-values < 0.05 were considered significant.
Acknowledgments
We wish to thank K. Georgopoulos for the Ikzf3-deficient mice and J. Leahy and K. Elder for technical assistance. This work was supported by program and project grants from the NHMRC (1049407 to NDH 1027472 to SC and GTB 575500 and APP1054925 to SLN). AK, GTB and SLN were supported by Australian Research Council Future Fellowships, and NDH, GKS, SC and MJS by a NHMRC Fellowships. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS.
Author contributions
MLH, NDH, RPLT, JB, YH, CEA, PF, MAD-E, GTB, AK, SC and MJS designed and performed the research and analyzed the data, WS and GKS performed the bioinformatic analysis, and SLN analyzed the data and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Aliahmad P, de la Torre B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan JE, Shellam GR. Genetic control of murine cytomegalovirus infection: virus titres in resistant and susceptible strains of mice. Arch Virol. 1984;81:139–150. doi: 10.1007/BF01309303. [DOI] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat Immunol. 2014;15:546–553. doi: 10.1038/ni.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot K, Parizot C, Arrouss I, Mazier D, Debre P, Rogner UC, Rebollo A. Differential aiolos expression in human hematopoietic subpopulations. Leuk Res. 2010;34:289–293. doi: 10.1016/j.leukres.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- Brady J, Hayakawa Y, Smyth MJ, Nutt SL. IL-21 induces the functional maturation of murine NK cells. J Immunol. 2004;172:2048–2058. doi: 10.4049/jimmunol.172.4.2048. [DOI] [PubMed] [Google Scholar]
- Brady J, Carotta S, Thong RP, Chan CJ, Hayakawa Y, Smyth MJ, Nutt SL. The interactions of multiple cytokines control NK cell maturation. J Immunol. 2010;185:6679–6688. doi: 10.4049/jimmunol.0903354. [DOI] [PubMed] [Google Scholar]
- Carotta S, Pang SH, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 2011;117:5449–5452. doi: 10.1182/blood-2010-11-318956. [DOI] [PubMed] [Google Scholar]
- Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- Cortes M, Wong E, Koipally J, Georgopoulos K. Control of lymphocyte development by the Ikaros gene family. Curr Opin Immunol. 1999;11:167–171. doi: 10.1016/s0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- Cortes M, Georgopoulos K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J Exp Med. 2004;199:209–219. doi: 10.1084/jem.20031571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, Nisoli I, Brady HJ, Wack A. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J Immunol. 2014;192:2677–2688. doi: 10.4049/jimmunol.1302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, Yoh K, Takahashi S, Prinz I, de Bernard S, Buffat L, Walzer T. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, Spivakov M, Srivastava P, Petretto E, Fisher AG, Merkenschlager M. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–1782. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, Schluns KS, Kubo M, Rothman PB, Vivier E, Sun JC. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J Exp Med. 2013;210:2981–2990. doi: 10.1084/jem.20130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y, Kelly JM, Westwood JA, Darcy PK, Diefenbach A, Raulet D, Smyth MJ. Cutting edge: tumor rejection mediated by NKG2D receptor-ligand interaction is dependent upon perforin. J Immunol. 2002;169:5377–5381. doi: 10.4049/jimmunol.169.10.5377. [DOI] [PubMed] [Google Scholar]
- Hayakawa Y, Huntington ND, Nutt SL, Smyth MJ. Functional subsets of mouse natural killer cells. Immunol Rev. 2006;214:47–55. doi: 10.1111/j.1600-065X.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- Huntington ND, Puthalakath H, Gunn P, Naik E, Michalak EM, Smyth MJ, Tabarias H, Degli-Esposti MA, Dewson G, Willis SN, Motoyama N, Huang DC, Nutt SL, Tarlinton DM, Strasser A. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol. 2007a;8:856–863. doi: 10.1038/ni1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, Smyth MJ, Tarlinton DM, Nutt SL. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol. 2007b;178:4764–4770. doi: 10.4049/jimmunol.178.8.4764. [DOI] [PubMed] [Google Scholar]
- Huntington ND, Nutt SL, Carotta S. Regulation of murine natural killer cell commitment. Front Immunol. 2013;4:14. doi: 10.3389/fimmu.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JT, Hu Y, Liu R, Masson F, D'Amico A, Carotta S, Xin A, Camilleri MJ, Mount AM, Kallies A, Wu L, Smyth GK, Nutt SL, Belz GT. Id2 expression delineates differential checkpoints in the genetic program of CD8alpha+ and CD103+ dendritic cell lineages. EMBO J. 2011;30:2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, Nutt SL. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, Nutt SL. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117:1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Loh J, Chu DT, O'Guin AK, Yokoyama WM, Virgin H. Natural killer cells utilize both perforin and gamma interferon to regulate murine cytomegalovirus infection in the spleen and liver. J Virol. 2005;79:661–667. doi: 10.1128/JVI.79.1.661-667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Duncan GS, Ferrick D, Mittrucker HW, Bischof S, Prechtl S, Rollinghoff M, Schmitt E, Pahl A, Mak TW. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med. 2000;192:325–336. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, Wack A, Brady HJ. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211:635–642. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 2010;584:4910–4914. doi: 10.1016/j.febslet.2010.09.042. [DOI] [PubMed] [Google Scholar]
- Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E, Jaeger BN, Bernat C, Fenis A, Kung S, De Gassart A, Mahmood S, Gut M, Heath SC, Estelle J, Bertosio E, Vely F, Gastinel LN, Beutler B, Malissen B, Malissen M, Gut IG, Vivier E, Ugolini S. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335:344–348. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- Orange JS, Wang B, Terhorst C, Biron CA. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J Exp Med. 1995;182:1045–1056. doi: 10.1084/jem.182.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL. Identification of Pax5 target genes in early B cell differentiation. J Immunol. 2008;180:1719–1728. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla IDR, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, Busslinger M. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins SH, Tessmer MS, Van Kaer L, Brossay L. Direct effects of T-bet and MHC class I expression, but not STAT1, on peripheral NK cell maturation. Eur J Immunol. 2005;35:757–765. doi: 10.1002/eji.200425797. [DOI] [PubMed] [Google Scholar]
- Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31:1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, Buer J, Grosveld F, Godin I, Di Santo JP. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- Sathe P, Delconte RB, Souza-Fonseca-Guimaraes F, Seillet C, Chopin M, Vandenberg CJ, Rankin LC, Mielke LA, Vikstrom I, Kolesnik TB, Nicholson SE, Vivier E, Smyth MJ, Nutt SL, Glaser SP, Strasser A, Belz GT, Carotta S, Huntington ND. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun. 2014;5:4539. doi: 10.1038/ncomms5539. [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, Busslinger M, Smyth MJ, Belz GT, Carotta S. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192:2667–2676. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- Smith MA, Maurin M, Cho HI, Becknell B, Freud AG, Yu J, Wei S, Djeu J, Celis E, Caligiuri MA, Wright KL. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185:6058–6067. doi: 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Crowe NY, Pellicci DG, Kyparissoudis K, Kelly JM, Takeda K, Yagita H, Godfrey DI. Sequential production of interferon-gamma by NK1.1(+) T cells and natural killer cells is essential for the antimetastatic effect of alpha-galactosylceramide. Blood. 2002;99:1259–1266. doi: 10.1182/blood.v99.4.1259. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumaria N, van Dommelen SL, Andoniou CE, Smyth MJ, Scalzo AA, Degli-Esposti MA. The roles of interferon-gamma and perforin in antiviral immunity in mice that differ in genetically determined NK-cell-mediated antiviral activity. Immunol Cell Biol. 2009;87:559–566. doi: 10.1038/icb.2009.41. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, Smyth MJ. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- Takeda K, Nakayama M, Sakaki M, Hayakawa Y, Imawari M, Ogasawara K, Okumura K, Smyth MJ. IFN-gamma production by lung NK cells is critical for the natural resistance to pulmonary metastasis of B16 melanoma in mice. J Leukoc Biol. 2011;90:777–785. doi: 10.1189/jlb.0411208. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HR, Yokoyama WM, Scalzo AA. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci USA. 2003;100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, Corcuff E, Guy-Grand D, Rocha B, Cumano A, Rogge L, Ezine S, Di Santo JP. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, Andrikopoulos K, Liang L, Pillai S, Morgan BA, Georgopoulos K. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol. 2010;22:154–160. doi: 10.1016/j.coi.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wei M, Mao H, Zhang J, Hughes T, Mitsui T, Park IK, Hwang C, Liu S, Marcucci G, Trotta R, Benson DM, Jr, Caligiuri MA. CD94 defines phenotypically and functionally distinct mouse NK cell subsets. J Immunol. 2009;183:4968–4974. doi: 10.4049/jimmunol.0900907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.