Abstract
Background
Taxonomy that utilizes morphological characteristics has been the gold standard method to identify mosquito species. However, morphological identification is challenging when the expertise is limited and external characters are damaged because of improper specimen handling. Therefore, we explored the applicability of mitochondrial cytochrome C oxidase subunit 1 (COI) gene-based DNA barcoding as an alternative tool to identify mosquito species. In the present study, we compared the morphological identification of mosquito specimens with their differentiation based on COI barcode, in order to establish a more reliable identification system for mosquito species found in Singapore.
Methods
We analysed 128 adult mosquito specimens, belonging to 45 species of 13 genera. Phylogenetic trees were constructed for Aedes, Anopheles, Culex and other genera of mosquitoes and the distinctive clustering of different species was compared with their taxonomic identity.
Results
The COI-based DNA barcoding achieved a 100% success rate in identifying the mosquito species. We also report COI barcode sequences of 16 mosquito species which were not available previously in sequence databases.
Conclusions
Our study utilised for the first time DNA barcoding to identify mosquito species in Singapore. COI-based DNA barcoding is a useful tool to complement taxonomy-based identification of mosquito species.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-014-0569-4) contains supplementary material, which is available to authorized users.
Keywords: DNA barcode, COI, Identification, Taxonomy, Mosquitoes, Phylogeny
Background
Morphological identification is the conventional, gold standard method to identify mosquito species based on their external characters. Different mosquito species exhibit distinguishable morphological features, which are utilized in taxonomic keys such as Bram [1], Harrison and Scanlon [2] and Rattanarithikul [3-7] to identify individual species. However, morphological identification requires experienced taxonomists and the method itself is highly time-consuming, especially in the hands of the less-experienced researchers. Moreover, incomplete identification is often encountered when important morphological features such as scales and bristles are damaged as a result of improper specimen handling. In addition, similar morphological characters shared by members of species complexes make identification a difficult task based on taxonomic keys alone [8]. Furthermore, most of the taxonomic keys are limited to adult female mosquitoes and fourth instar larvae because many of the morphological characteristics are not well developed in early larval stages. Moreover, many species of the Culex subgenus Lophoceraomyia are recognised on the basis of differences in male antenna, palpus, proboscis and genitalia, making the morphological identification of their female counterparts difficult [9]. These limitations restrict the applicability of existing taxonomic keys for the identification of certain mosquito species. Therefore, there is a need for an alternative, universally-applicable method to support the existing mosquito identification methods.
DNA barcoding is a molecular method that is becoming increasingly popular for the identification of animal species, based on partial mitochondrial DNA sequences [10,11]. This method is based on the concept that every species has a unique genetic identity [11,12]. A DNA barcode is a short standardised sequence of DNA that can be used as a genetic maker for species identification [11,13]. Early studies on DNA barcoding have used the nuclear internal transcribed spacer 2 [14], cytochrome b oxidase [15,16], 12S rRNA [17,18] and nicotinamide adenine dinucleotide dehydrogenase [19,20] as target genes. In recent years, however, the mitochondrial cytochrome c oxidase subunit 1 (COI) gene has gained increasing popularity, primarily because of the ease of using a universal set of primers to amplify the gene and its ability to provide a higher sequence variation at inter-species than at intra-species level [10]. COI gene-based DNA barcoding is, therefore, an alternative species identification method that can easily be standardized to obtain comparable results from different sources.
Herbert and co-workers [10,11] proposed using a 658 base pair (bp) region of the COI gene as a universal marker to barcode animal life. Previous studies have proven that COI gene is an efficient and useful barcode for the identification of metazoans, including mosquitoes [21-26]. However, COI barcode may not be universally applicable to identify all animal species. For example, COI-based barcoding has not been promising in identifying fungi and plant species [27-29]. Likewise, COI barcoding has failed to distinguish certain mosquito species of Anopheles and Culex [21-25]. Kumar et al., [23] reported that two closely related mosquito species of the genus Ochlerotatus could not be differentiated using their COI barcode. On the other hand, the barcoding approach has other common limitations as well. The recombination within mitochondrial genes may lead to complex sequence patterns when species with divergent mitochondrial DNA genomes interbreed, resulting in inconclusive identification. Moreover, the success of DNA barcoding is dependent on the availability of representative sequences for comparison. DNA barcoding approach fails if there are insufficient reference sequences in databases for comparison and analysis [8].
In instances where COI barcode fails to accurately identify certain mosquito species, a multi locus approach has been proposed [24]. By utilizing other gene markers and combining the datasets, the accuracy of identification can be increased. These observations, therefore, testify the need to use integrated datasets, including genomic, morphological and ecological data, to further understand the species diversity of the animal kingdom [30]. In the present study, we explored the applicability of COI-based DNA barcoding in the identification of mosquito species in Singapore. We compared the identification based on morphology and COI-based barcoding of 45 mosquito species belonging to 13 genera. By using both methods, we sought to establish a more reliable and standardized identification system for mosquito species found in Singapore.
Methods
Mosquito collection and identification
Mosquitoes were collected from various parts of Singapore from 2003 to 2012. Adult mosquito specimens were collected using BG-sentinel traps (BioGents AG, Germany), CO2 light traps, human baited net traps and human landing catch method. Larval samples were collected using the dipping method during field surveillance activities. Laboratory strains of several mosquito species, namely Aedes (Stegomyia) aegypti (Linneaus, 1762), Anopheles (Anopheles) sinensis (Wiedemann, 1828), Culex (Culex) vishnui (Theobald, 1901), Culex (Culex) pseudovishnui (Colless, 1957) Culex (Culex) quinquefasciatus (Say, 1823), Lutzia (Metalutzia) fuscana (Wiedermann, 1820) and Culex (Culex) mimulus (Edwards, 1915) that were colonised at the Environmental Health Institute (EHI) were also included in our analysis.
Field collected larvae were reared individually to adults. The imagos were identified by experienced taxonomists at EHI according to taxonomic keys [1-7,31-35]. A reference number was assigned to each adult mosquito which then was deposited as voucher specimens in the EHI mosquito repository.
DNA extraction
In order to preserve voucher specimens for future references, DNA was extracted from the fore-, mid-, and hindlegs (n = 3) from one side of each mosquito. Legs were removed using clean, sterile forceps and were homogenised using a mixer mill (Retsch Mixer Mill MM301). Total DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. The extracted DNA was stored at -20°C until further analysis.
Polymerase Chain Reaction and DNA sequencing
A 735 bp region flanking the mitochondrial COI gene was amplified by polymerase chain reaction (PCR) using following primers: forward 5’- GGATTTGGAAATTGATTAGTTCCTT - 3’ and reverse 5’ – AAAAATTTTAATTCCAGTTGGAACAGC – 3’ [23]. The 50 μl PCR reaction consisted of 5 μl of extracted DNA, 1.5 mM MgCl2 (Promega, USA), 0.2 mM dNTPs (Promega, USA), 1x reaction buffer (Promega, USA), 1.5 U Taq DNA polymerase (Promega, USA), and 0.3 μM of each primer.
PCR reaction conditions were as follows: an initial denaturation of 95°C for 5 minutes, followed by 5 cycles of denaturation at 94°C for 40 seconds, annealing at 45°C for 1 minute and extension at 72°C for 1 minute. The amplification reaction was continued for another 35 cycles of denaturation at 94°C for 40 seconds, annealing at 51°C for 1 minute and extension at 72°C for 1 minute followed by a final extension at 72°C for 10 minutes [23].
Amplicons were visualised on 1.5% agarose gels stained with GelRed (Biotium Inc., USA). PCR products were purified using the Purelink PCR purification kit (Invitrogen Corp., USA), according to manufacturer’s recommendations. Sequencing was performed at a commercial laboratory according to the recommended protocol for BigDye Terminator Cycle Sequencing kit (Applied Biosystems, USA).
COI sequences generated in this study were deposited in the GenBank (http://www.ncbi.nlm.nih.gov) and Barcode of Life (BOLD) (http://www.boldsystems.org) databases [NCBI: KF564650 to KF564674, KF564678 to KF564740, KF564643 to KF564778 and KM609455 to KM609458].
Phylogenetic analyses and genetic distance calculation
Contiguous sequences of COI gene were generated from forward and reverse chromatograms using Lasergene 8.0 software suite (DNASTAR Inc., USA). Completed sequences were aligned using the Clustal W algorithm [36] implemented in BioEdit v7.0.5 software [37]. Separate phylogenetic trees were constructed by using the neighbour joining algorithm implemented in MEGA 6.06 software suite [38]. Parameters for phylogenetic construction included a Kimura-2 parameter substitution model with gamma distributed rates using the nearest neighbour interchange heuristic search method. Robustness of clustering was determined by bootstrap analysis with 1000 replicates. Reference DNA sequences were obtained from the GenBank and BOLD databases. The pairwise distance between individual species within Aedes, Anopheles, Culex and other genera described in this study was calculated using MEGA 6.06 software package [38].
Ethical consideration
All specimens belonging to species of mosquitoes analysed in the present study were collected as part of the vector surveillance and control programmes conducted by Environmental Health Department of National Environment Agency and Singapore Armed Forces. EHI serves as the reference centre for the identification of field collected adult and larval specimens under the above programmes. The study was approved by the Project Evaluation Committee of EHI, National Environment Agency, Singapore (Reference No. TS058).
Results and discussion
Mosquito specimens and collection habitats
In total, 128 mosquito specimens belonging to 45 species of 13 genera were analysed. Of the 128 specimens, 83 mosquitoes (65%) were collected from rural habitats. Those habitats included forested areas such as Pulau Ubin, Sungei Buloh Wetland Reserve, Pulau Tekong and military training areas. Of the remaining, 32 specimens (25%) were from urban housing estates and private houses. The majority of urban specimens were collected as larvae and were reared into adults before identification. Colonised mosquitoes (n = 13) constituted 10% of the specimens. Information about the exact locations of specimen collection is given in Additional file 1.
COI-based DNA barcoding accurately identified all species of Aedes, Culex, Anopheles and other genera of mosquitoes
Accurate identification of mosquito species is instrumental in vector control programmes because only a handful of mosquito species plays an important role in disease transmission [39]. Advancement in DNA-based molecular techniques allows us to complement the taxonomical identification of mosquito species. Our analysis included 10 species of Aedes and Verrallina (n = 30), 13 species of Culex and Lutzia (n = 42), nine species of Anopheles (n = 33) and 13 species of other genera (n = 23) of mosquitoes. Other genera included Aedeomyia, Armigeres, Coquillettidia, Ficalbia, Mansonia, Toxorhynchites, Uranotaenia, and Zeugomyia.
As illustrated in Figures 1, 2, 3 and 4, COI-based phylogenetic analyses showed distinct clustering of individual species within each genus with strong bootstrap support. Clustering patterns agreed with the morphological identification, enabling the differentiation of individual species based on COI sequences. Pairwise distance analyses of COI sequences showed that inter-species “barcode gap” exceeded the proposed cut-off of 2-3% [30], supporting the ability of COI barcode to differentiate analysed species (Additional files 2, 3, 4 and 5). Furthermore, our sequences clustered with those of similar species from other endemic regions reported in the NCBI and BOLD databases. All species analysed in this study (n = 45) could, therefore, be identified based on their COI barcode, yielding a 100% compatibility between molecular and taxonomic identification, indicating that COI barcode is a useful tool to complement taxonomy for the identification of mosquito species.
Figure 1.
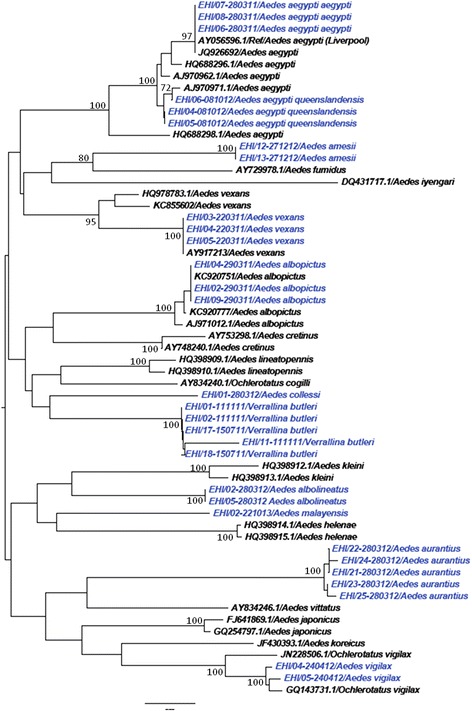
Phylogenetic tree based on COI sequences of Aedes and Verrallina spp. mosquitoes. An alignment of COI gene sequences (440 bp) was used to construct the neighbour joining tree in MEGA 6.06 software. Numbers displayed on branches are the bootstrap support obtained through 1000 replications. GenBank sequences are shown with accession numbers. Sequences starting with “EHI” were generated during this study and are highlighted in blue.
Figure 2.
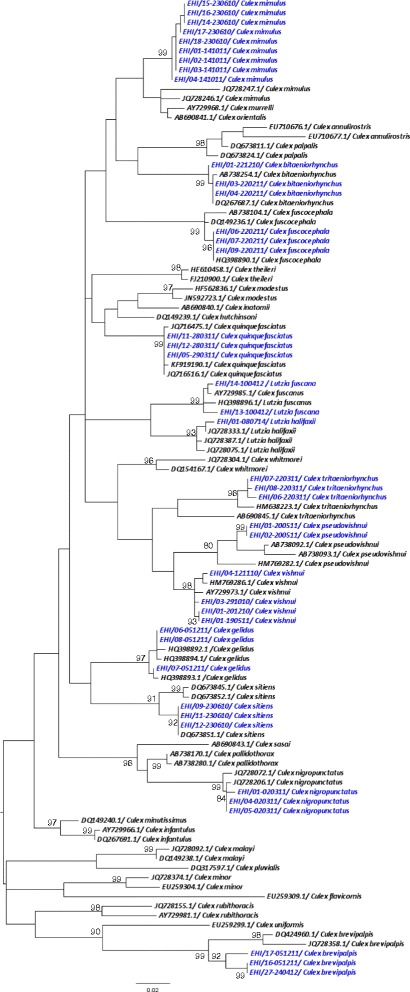
Phylogenetic tree based on COI sequences of Culex and Lutzia spp. mosquitoes. A 432 bp-region of the COI gene was used to construct the neighbour joining tree in MEGA 6.06 software. Numbers displayed on branches are the bootstrap support obtained through 1000 replications. GenBank sequences are shown with accession numbers. Sequences starting with “EHI” were generated during this study and are highlighted in blue.
Figure 3.

Phylogenetic tree based on COI sequences of Anopheles spp. mosquitoes. A 452 bp-region of the COI gene was used to construct the neighbour joining tree in MEGA 6.06 software. Numbers displayed on branches are the bootstrap support obtained through 1000 replications. GenBank sequences are shown with accession numbers. Sequences starting with “EHI” were generated during this study and are highlighted in blue.
Figure 4.
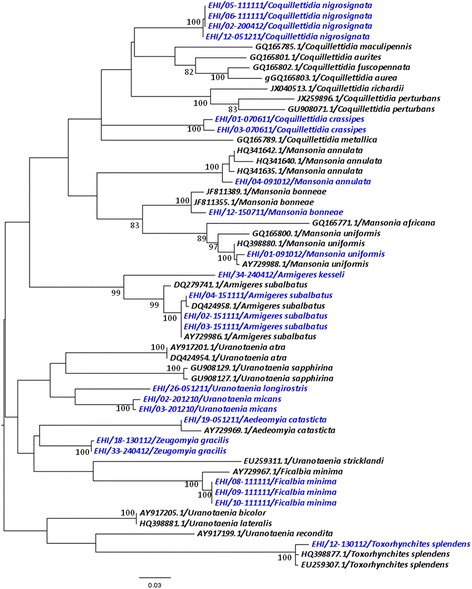
Phylogenetic tree based on COI sequences of other genera of mosquitoes. A 447 bp-region of the COI gene was used to construct the neighbour joining tree in MEGA 6.06 software. Numbers displayed on branches are the bootstrap support obtained through 1000 replications. GenBank sequences are shown with accession numbers. Sequences starting with “EHI” were generated during this study and are highlighted in blue.
COI barcode differentiated morphologically similar species of Aedes and Culex genera
It was noteworthy that several morphologically similar Aedes and Culex species could be differentiated based on the COI barcode.
One such pair was Aedes (Stegomyia) aegypti aegypti and Aedes (Stegomyia) aegypti queenslandensis (Theobald, 1901). Adult stages of these two forms are differentiated based on white scales on the abdominal terga. In Ae. aegypti queenslandensis, there are pale scales that appear as unbroken median stripes on the abdominal terga II to VII (Figure 5a). These pale scales are not seen on the abdominal terga of Ae. aegypti aegypti (Figure 5b) [33]. Given such minute differences, it is difficult to differentiate these two forms when scales on the abdominal terga are rubbed off. However, as illustrated in Figure 1, COI barcode-based phylogeny differentiated Ae. aegypti queenslandensis from Ae. aegypti aegypti. According to COI sequence analysis, the genetic distance between Ae. aegypti aegypti and Ae. aegypti queenslandensis varied from 1.5% to 1.9% (Additional file 2). This distance was lower than the “barcode gap” proposed to differentiate vertebrate (2%) and invertebrate (3%) species based on COI sequences [30]. Therefore, Ae. aegypti aegypti and Ae. aegypti queenslandensis were not considered as two subspecies. This observation agreed with previous descriptions of Ae. aegypti as a group of highly polymorphic mosquitoes. Mattingly described four morphological forms of Ae. aegypti which were distinguishable based on colour characters; a pale form (Ae. aegypti queenslandensis), an intermediate (type) form (Ae. aegypti aegypti), a dark form (Ae. aegypti formosus) and a fourth form (Ae. aegypti mascarensis) [40,41]. Based on morphology and habitat differences, Mattingly concluded that both Ae. aegypti formosus and Ae. aegypti mascarensis were clearly subspecies, but classified Ae. aegypti queenslandensis as a variety (var.) [41]. According to previous descriptions, Ae. aegypti aegypti and Ae. aegypti queenslandensis share a similar geographical distribution pattern [41]. It is known that the presence of subspecies as well as their geographic and ecological separation may affect the potential of a mosquito species to act as a vector. Aedes aegypti is considered as the primary vector of Dengue virus (DENV) in endemic regions, including Singapore [42]. Even though preliminary findings of a previous study have shown no significant differences in oral infection of DENV-2 between pale (Ae. aegypti queenslandensis) and dark (Ae. aegypti aegypti) forms of Ae. aegypti in Thailand [43], no extensive studies have so far been carried out to determine the differences in vectorial capacity between Ae. aegypti queenslandensis and Ae. aegypti aegypti. In this context, the ability of COI barcoding to differentiate Ae. aegypti queenslandensis and Ae. aegypti aegypti is especially advantageous.
Figure 5.

Morphological comparison of the abdominal terga of Ae. aegypti queenslandensis and Ae. aegypti aegypti. (a): Ae. aegypti queenslandensis: unbroken median stripes of pale scales along the abdominal terga. (b): Ae. aegypti aegypti: no median stripes of pale scales from terga II to VII.
Likewise, Aedes (Aedimorphus) vexans (Meigen, 1830) and Aedes (Ochlerotatus) vigilax (Skuse, 1889) share similar morphological features such as pale scaling on the basal underside of proboscis, dark and white scales along the costa and subcosta of wings, and a narrow abdominal segment VIII which is nearly retracted into segment VII [34]. Ae. vexans has narrow basal pale bands that span over less than 1/4 of the length of hind tarsomeres (Figure 6a) [6]. On the other hand, Ae. vigilax has broad basal pale bands covering more than 1/4 of the length of hind tarsomeres (Figure 6b) [6]. Similarly, the taxonomical differentiation of Cx. vishnui and Cx. pseudovishnui also relies on subtle differences in the hind femur. Cx. vishnui has an apical dark band on the anterior surface of hind femur which is not well contrasted due to the presence of pale scales (Figure 7a), whereas it is well-contrasted in Cx. pseudovishnui (Figure 7b) [5]. Such subtle morphological differences pose a difficulty in accurate morphological identification of those species, especially in the hands of non-experienced taxonomists. Our results showed that COI barcode is able to differentiate those morphologically similar species (Figures 1 and 2) and thus complements their taxonomic identification.
Figure 6.
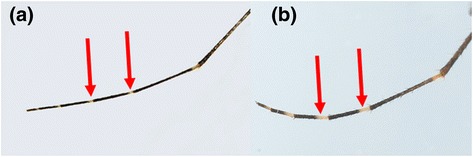
Morphological comparison of the hind tarsa of Ae. vexans and Ae. vigilax . (a): Ae. vexans: pale basal bands in less than a quarter of the length of hind tarsomeres. (b): Ae. vigilax: pale basal bands covering more than a quarter of the length of hind tarsomeres.
Figure 7.
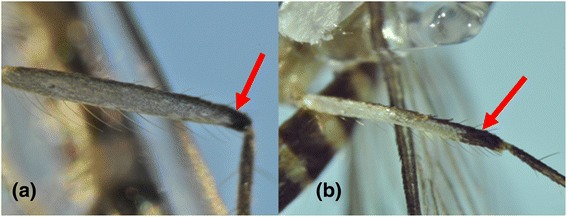
Morphological comparison of the hind femur of Cx. vishnui and Cx. pseudovishnui. (a): Cx. vishnui: hind femur with apical dark band not well contrasted with pale scales on the hind femur. (b): Cx. pseudovishnui: hind femur with apical dark band well contrasted with pale scales on the hind femur.
Moreover, morphological identification of mosquitoes becomes a challenge when the morphological characterisation is stage-specific. For example, even though the adult mosquitoes of Lt. fuscana and Lutzia (Metalutzia) halifaxii (Theobald, 1903) can be distinguished based on differences of their abdominal terga (Figures 8a, b), their larval stages cannot be distinguished from each other [1,5]. Another example is Aedes (Stegomyia) albopictus (Skuse, 1894) and Aedes (Stegomyia) malayensis (Colless, 1962). Even though the adult mosquitoes can easily be distinguished based on differences of their abdominal terga area (Figures 9a, b) and supraalar (Figures 10a, b) [6], their larval stages are morphologically similar. As Ae. albopictus and Ae. malayensis coexist in the same habitats [32], larvae of both species are often found mixed during larval surveillance activities. Accurate identification of these two species is important as only Ae. albopictus is a vector of human pathogens such as DENV and Chikungunya virus. In contrast, adults of Cx. vishnui and Cx. pseudovishnui share many similar external characteristics (Figures 7a, b), which make them difficult to separate from each other. However, they are distinguishable at the larval stage. Nonetheless, rearing of larvae into adults as a requirement of taxonomic differentiation of certain species is time and resource intensive. Our findings showed that COI barcoding could differentiate these mosquito species (Figures 1 and 2) and thus be utilized to overcome such challenging scenarios. Therefore, complementing morphological identification with molecular characterisation has the potential to enhance vector surveillance capacity.
Figure 8.

Morphological comparison of the abdominal terga of Lt. fuscana and Lt. halifaxii. (a): Lt. fuscana: yellowish scales on abdominal terga. (b): Lt. halifaxii: entirely dark abdominal terga.
Figure 9.
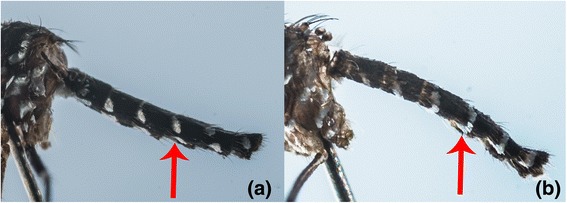
Morphological comparison of the abdominal terga of Ae. albopictus and Ae. malayensis . (a): Ae. albopictus: dorsal white bands separated from the lateral spots on abdominal terga. (b): Ae. malayensis: dorsal white bands connected to the lateral pale patches on abdominal terga.
Figure 10.
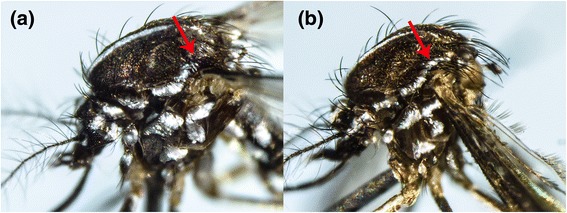
Morphological comparison of the thorax of Ae. albopictus and Ae. malayensis. (a): Ae. albopictus: pale patches of scales do not extended towards the scutellum on the thorax. (b): Ae. malayensis: pale scales extended towards the scutellum on the thorax.
DNA barcoding enhanced the identification of Anopheles sinensis
An interesting phenomenon was observed when field-caught An. sinensis mosquitoes were subsequently colonised in the laboratory. According to taxonomic keys, An. sinensis is identified based on morphological characteristics such as apical pale bands on hind tarsomeres and the wing venation [2,4,31]. We observed differences in the wing venation of field-caught and laboratory colonised An. sinensis adults in Singapore. Reid [31] previously reported that approximately 75% of An. sinensis specimens had pale fringe spots at vein CuA. However, as shown in Figures 11a and b, our specimens of An. sinensis had both pale and dark fringe spots at the end of vein CuA. Nevertheless, all COI barcode sequences of An. sinensis specimens clustered together in the phylogenetic tree (Figure 3), regardless of their morphological differences at the end of vein CuA. Therefore, despite a polymorphic nature of wing venation that may impede taxonomic identification, COI barcoding enabled us to confirm that those specimens were indeed An. sinensis. These observations also indicated that existing taxonomic keys for the identification of An. sinensis need to be revised further.
Figure 11.
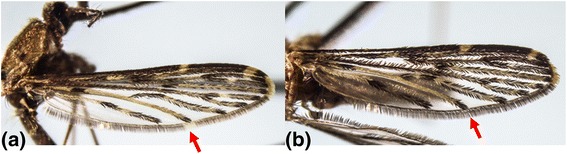
Morphological comparison of the vein CuA of An. sinensis. (a): Pale fringe spot at vein CuA. (b): Dark fringe spot at vein CuA.
In summary, our findings suggested that COI-based DNA barcode can effectively be used when morphological traits of certain species do not clearly distinguish one species from another [44]. DNA barcodes also allow taxonomists to re-confirm the reference voucher specimens. For instance, the COI barcode of a voucher specimen initially identified as Culex (Culex) bitaeniorhynchus (Giles. 1901) in our samples clustered with Cx. vishnui in the phylogenetic analysis. This prompted us to re-visit the voucher specimen which was then correctly re-identified as Cx. vishnui after a thorough morphological analysis. Therefore, COI-based molecular characterisation has immense potential to be used as a complementary tool for the identification of mosquito species.
Conclusions
DNA barcoding is a useful tool to complement taxonomy for the identification of mosquito species. In the present study, we demonstrated that mitochondrial COI gene-based DNA barcoding was comparable to morphological identification for the differentiation of 45 mosquito species analysed. In our analyses, COI barcode was even able to differentiate several mosquito species that were difficult to distinguish morphologically. However, empirical evidence has shown that COI-based barcoding is not successful all the time [21-23]. Firstly, the limited availability of sequences to be used as references for comparison has restricted its usage on species identification. In the present study, we provided COI gene sequences of 16 mosquito species which were not available previously in sequence databases (Additional file 1). We believe that those new sequences would contribute to the on-going global effort to standardise DNA barcoding as a molecular means of species identification by the Consortium for the Barcode of Life (CBOL) [8]. Secondly, the cut-off limit of “barcode gap” for species differentiation still remains controversial. Both of these limitations have implications on the identification of new species based on barcoding alone. Therefore, COI-based DNA barcoding may not always be useful on its own, but would rather be an alternative tool to complement morphological identification. The use of integrated datasets and multi locus analyses will further enhance the molecular identification. Although the taxonomic keys have been developed to identify different genera of mosquitoes from various geographical settings, little progress has been made to classify mosquito species based on phylogenetic relationships [45,46]. The phylogeny-assisted DNA barcode analyses enable us to refine the taxonomic identification and further understand the genetics and evolution of mosquito species in endemic habitats.
Acknowledgements
We thank our colleagues at the Environmental Health Institute (EHI) and Environmental Health Department of National Environment Agency, Singapore as well as Headquarters Medical Corps of Singapore Armed Forces for collecting and processing of specimens analysed in this study. We are greatly obliged to Mr Png Ah Bah, Mdm Foo Siew Yoong, and other staff at EHI for their assistance in identification and mounting of the specimens. We also thank Mr. Deng Lu for taking photos of mosquito specimens and Dr Indra Vythilingam for her supervisory support.
Abbreviations
- BOLD
Barcode of Life
- CBOL
Consortium for the Barcode of Life
- COI
Cytochrome c oxidase subunit 1
- DENV
Dengue virus
- EHI
Environmental Health Institute
- NCBI
National Centre for Biotechnology Information
- PCR
Polymerase chain reaction
Additional files
Location of specimen collection and sequence accession information of mosquito specimens (n = 128) included in the study.
Pairwise nucleotide distance of mitochondrial cytochrome C oxidase subunit 1 ( COI ) gene of Aedes and Verrallina species.
Pairwise nucleotide distance of mitochondrial cytochrome C oxidase subunit 1 ( COI ) gene of Anopheles species.
Pairwise nucleotide distance of mitochondrial cytochrome C oxidase subunit 1 ( COI ) gene of Culex and Lutzia species.
Pairwise nucleotide distance of mitochondrial cytochrome C oxidase subunit 1 ( COI ) gene of species belonging to other genera.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SGLP, SCP, LPC and RL collected mosquitoes from the field. SGLP and RL identified mosquitoes. CHT and SCP optimized conditions for DNA extraction and PCR. AC, HCH and LPC executed DNA extraction, PCR and performed sequence analysis. LCN, KSL, HCH, SGLP and AC wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Abigail Chan, Email: abigail_chan@nea.gov.sg.
Lee-Pei Chiang, Email: lipasein@yodenza.com.
Hapuarachchige C Hapuarachchi, Email: chanditha_hapuarachchi@nea.gov.sg.
Cheong-Huat Tan, Email: tan_cheong_huat@nea.gov.sg.
Sook-Cheng Pang, Email: pang_sook_cheng@nea.gov.sg.
Ruth Lee, Email: ruth_lee@nea.gov.sg.
Kim-Sung Lee, Email: lks9@np.edu.sg.
Lee-Ching Ng, Email: ng_lee_ching@nea.gov.sg.
Sai-Gek Lam-Phua, Email: lam-phua_sai_gek@nea.gov.sg.
References
- 1.Bram RA. Contributions to the mosquito fauna of Southeast Asia II: the Genus Culex in Thailand (Diptera: Culicidae) Contrib Am Entomol Inst. 1967;2:No. 1. [Google Scholar]
- 2.Harrison BA, Scanlon JE. The subgenus Anopheles in Thailand (Diptera: Culicidae) Contrib Am Entomol Inst. 1975;12:No. 1. [Google Scholar]
- 3.Rattanarithikul R, Harrison BA, Panthusiri P, Peyton EL, Coleman RE. Illustrated keys to the mosquitoes of Thailand III. Genera Aedeomyia, Ficalbia, Mimomyia, Hodgesia, Coquillettidia, Mansonia, and Uranotaenia. Southeast Asian J Trop Med Public Health. 2006;37 Suppl 1:1–85. [PubMed] [Google Scholar]
- 4.Rattanarithikul R, Harrison BA, Harbach RE, Panthusiri P, Coleman RE. Illustrated keys to the mosquitoes of Thailand. IV. Anopheles. Southeast Asian J Trop Med Public Health. 2006;37 Suppl 2:1–128. [PubMed] [Google Scholar]
- 5.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Jones JW, Coleman RE. Illustrated keys to the mosquitoes of Thailand. II. Genera Culex and Lutzia. Southeast Asian J Trop Med Public Health. 2005;36 Suppl 2:1–97. [PubMed] [Google Scholar]
- 6.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE, Richardson JH. Illustrated keys to the mosquitoes of Thailand. VI. Tribe Aedini. Southeast Asian J Trop Med Public Health. 2010;41 Suppl 1:1–225. [PubMed] [Google Scholar]
- 7.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE. Illustrated keys to the mosquitoes of Thailand V. Genera Orthopodomyia, Kimia, Malaya, Topomyia, Tripteroides, and Toxorhynchites. Southeast Asian J Trop Med Public Health. 2007;38 Suppl 2:1–65. [PubMed] [Google Scholar]
- 8.Jinbo U, Kato T, Ito M. Current progress in DNA barcoding and future implications for entomology. Entomol Sci. 2011;14:107–124. doi: 10.1111/j.1479-8298.2011.00449.x. [DOI] [Google Scholar]
- 9.Sirivanakarn S. A revision of the subgenus Lophoceraomyia of the genus Culex in the Oriental region (Diptera: Culicidae) Contrib Am Entomol Inst (Ann Arbor) 1977;13:1–245. [Google Scholar]
- 10.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert PD, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rueanghiran C, Apiwathnasorn C, Sangthong P, Samung Y, Ruangsittichai J. Utility of a set of conserved mitochondrial cytochrome oxidase subunit I gene primers for Mansonia annulata identification. Southeast Asian J Trop Med Public Health. 2011;42:1381–1387. [PubMed] [Google Scholar]
- 13.Paz A, Crawford AJ. Molecular-based rapid inventories of sympatric diversity: a comparison of DNA barcode clustering methods applied to geography-based vs clade-based sampling of amphibians. J Biosci. 2012;37:887–896. doi: 10.1007/s12038-012-9255-x. [DOI] [PubMed] [Google Scholar]
- 14.Merget B, Koetschan C, Hackl T, Forster F, Dandekar T, Muller T, Schultz J, Wolf M. The ITS2 Database. J Vis Exp. 2012;61:pii: 3806. doi: 10.3791/3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sevilla RG, Diez A, Norn M, Mouchel O, Jerome M, Verrezagnis V, Van Pelt H, Favrery L, Krey G, Bautista JM. Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol Ecol Notes. 2007;7:730–734. doi: 10.1111/j.1471-8286.2007.01863.x. [DOI] [Google Scholar]
- 16.Shen Y-Y, Chen X, Murphy RW. Assessing DNA barcoding as a tool for species identification and data quality control. PLoS ONE. 2013;8:e57125. doi: 10.1371/journal.pone.0057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vences M, Thomas M, van der Meijden A, Chiari Y, Vieites DR. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front Zool. 2005;2:5. doi: 10.1186/1742-9994-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu KH, Li C, Qi J. Ribosomal RNA as molecular barcodes: a simple correlation analysis without sequence alignment. Bioinformatics. 2006;22:1690–1701. doi: 10.1093/bioinformatics/btl146. [DOI] [PubMed] [Google Scholar]
- 19.Rach J, Desalle R, Sarkar IN, Schierwater B, Hadrys H. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc Biol Sci. 2008;275:237–247. doi: 10.1098/rspb.2007.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster BL, Emery AM, Webster JP, Gouvras A, Garba A, Diaw O, Seye MM, Tchuente LAT, Simoonga C, Mwanga J. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Negl Trop Dis. 2012;6:e1882. doi: 10.1371/journal.pntd.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurito M, de Oliveira TM, Almiron WR, Sallum MAM. COI barcode versus morphological identification of Culex (Culex) (Diptera: Culicidae) species: a case study using samples from Argentina and Brazil. Mem Inst Oswaldo Cruz. 2013;108:110–122. doi: 10.1590/0074-0276130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cywinska A, Hunter FF, Hebert PD. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- 23.Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:DBCDSO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Bourke BP, Oliveira TP, Suesdek L, Bergo ES, Sallum M. A multi-locus approach to barcoding in the Anopheles strodei subgroup (Diptera: Culicidae) Parasit Vectors. 2013;6:111. doi: 10.1186/1756-3305-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H. Identifying the main mosquito species in China based on DNA barcoding. PLoS ONE. 2012;7:e47051. doi: 10.1371/journal.pone.0047051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamis FM, Masiga DK, Mohamed SA, Salifu D, de Meyer M, Ekesi S. Taxonomic identity of the invasive fruit fly pest. Bactrocera invadens: concordance in morphometry and DNA barcoding. PLoS One. 2012;7:e44862. doi: 10.1371/journal.pone.0044862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, Chaudhary LB, Datt B, Bag SK, Singh PK, Nair NK. Universal plant DNA barcode loci may not work in complex groups: a case study with Indian Berberis species. PLoS ONE. 2010;5:e13674. doi: 10.1371/journal.pone.0013674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dentinger BT, Didukh MY, Moncalvo J-M. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina) PLoS ONE. 2011;6:e25081. doi: 10.1371/journal.pone.0025081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinoff D. DNA barcoding evolves into the familiar. Conser Biol. 2006;20:1548–1549. doi: 10.1111/j.1523-1739.2006.00542.x. [DOI] [PubMed] [Google Scholar]
- 31.Reid J. Studies from the Institute for Medical Research Malaya, No. 31. Government of Malaysia. Kettering: Staples Printer Ltd; 1968. Anopheline mosquitoes of Malaya and Borneo. [Google Scholar]
- 32.Huang YM. A redescription of Aedes (Stegomyia) scutellaris malayensis Colless and the differentiation of the larva from that of Aedes (S.) albopictus (Skuse) Proc Entomol Soc Wash. 1971;73:1–8. [Google Scholar]
- 33.McClelland G. A worldwide survey of variation in scale pattern of the abdominal tergum of Aedes aegypti (L.) (Diptera: Culicidae) Trans R Entomol Soc Lond. 1974;126:239–259. doi: 10.1111/j.1365-2311.1974.tb00853.x. [DOI] [Google Scholar]
- 34.Reinert JF. Contributions to the Mosquito Fauna of Southeast Asia. XV. Genus Aedes Meigen, Subgenus Ayurakitia Thurman. Contrib Am Entomol Inst. 1972;9:No. 2. [Google Scholar]
- 35.Apiwathnasorn C. A List of Mosquito Species in Southeast Asia. Museum and Reference Centre: SEAMED-TROPMED National Centre of Thailand, Faculty of Tropical Medicine, Mahidol University; 1986. [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higa Y, Toma T, Tsuda Y, Miyagi I. A multiplex PCR-based molecular identification of five morphologically related, medically important subgenus Stegomyia mosquitoes from the genus Aedes (Diptera: Culicidae) found in the Ryukyu Archipelago, Japan. Jpn J Infect Dis. 2010;63:312–316. [PubMed] [Google Scholar]
- 40.Mattingly PF. The Culicine Mosquitoes of the Indomalayan Area. Part VI. Genus Aedes Meigen, subgenus Stegomyia Theobald (Groups A, B and D) London: British Museum (Natural History); 1965. [Google Scholar]
- 41.Mattingly P. Genetical aspects of the Aedes aegypti problem. I. Taxonomy and bionomics. Ann Trop Med Parasitol. 1957;51:392–408. [PubMed] [Google Scholar]
- 42.Chan YC, Ho BC, Chan KL. Aedes aegypti (L.) and Aedes albopictus (Skuse) in Singapore City. 5. Observations in relation to dengue haemorrhagic fever. Bull World Health Organ. 1971;44:651–657. [PMC free article] [PubMed] [Google Scholar]
- 43.Wasinpiyamongkol L, Thongrungkiat S, Jirakanjanakit N, Apiwathnasorn C. Susceptibility and transovarial transmission of dengue virus in Aedes aegypti: a preliminary study of morphological variations. Southeast Asian J Trop Med Public Health. 2003;34:131–135. [PubMed] [Google Scholar]
- 44.Frezal L, Leblois R. Four years of DNA barcoding: current advances and prospects. Infect Genet Evol. 2008;8:727–736. doi: 10.1016/j.meegid.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Harbach RE. Classification within the cosmopolitan genus Culex (Diptera: Culicidae): the foundation for molecular systematics and phylogenetic research. Acta Trop. 2011;120:1–14. doi: 10.1016/j.actatropica.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Foster PG, Bergo ES, Bourke BP, Oliveira TM, Nagaki SS, SantAna DC, Sallum MAM. Phylogenetic analysis and DNA-based species confirmation in Anopheles (Nyssorhynchus) PLoS ONE. 2013;8:e54063. doi: 10.1371/journal.pone.0054063. [DOI] [PMC free article] [PubMed] [Google Scholar]


