Abstract
Complement C3 is a key fluid-phase protein of the immune system that covalently tags pathogenic cells and molecules for subsequent clearance. Previously, we reported that complement activation results in the formation of multiple C3b:plasma protein complexes in serum. However, it is not known if C3b attaches to any plasma protein in close proximity or preferentially binds damaged proteins. The objective of this study was to determine if C3b couples to plasma proteins in a non-native state and if this could be a potential mechanism to detect and clear damaged proteins from the blood. Using a purified in vitro system with alternative pathway proteins C3, factors B and D it was observed that guanidinium-HCl denaturation of three purified plasma proteins (albumin, alpha-1 proteinase inhibitor, vitamin D binding protein) greatly increased their capacity to form covalent complexes with C3b. However, native vitamin D binding protein, covalently attached to C3b, still retained the ability to bind its natural ligand G-actin, indicating that C3b links to plasma proteins in their native configuration but denaturation substantially increases this interaction. Serum complement activation generated a large number of C3b:plasma protein complexes that bound red blood cell membranes, suggesting a CR1-mediated clearance mechanism. Thermally denatured (60°C) serum activated the alternative pathway when added to fresh serum as evidenced by factor B cleavage and iC3b generation, but this heat-treated serum could not generate the pro-inflammatory peptide C5a. These results show that C3 recognizes and tags damaged plasma proteins for subsequent removal from the blood without triggering proinflammatory functions.
Keywords: Alternative Pathway activation, Complement C3, Extracellular chaperone, Plasma proteins, Denatured proteins, Inflammation
1. INTRODUCTION
The third component of complement (C3) is the most abundant and essential protein of the complement system in all vertebrates (Mastellos et al., 2013; Zarkadis et al., 2001). High concentrations of C3 circulate in blood plasma (adult human range: 0.7 to 1.6 mg/ml) and can increase almost two-fold during the acute phase response of inflammation. C3 is required for full activation of the classical and lectin recognition pathways and is central for the initiation and activation of the alternative pathway (Ricklin et al., 2010). During complement activation, proteolytic cleavage of C3 to C3b exposes the unstable thioester bond, which then reacts extremely fast with either a target surface or surrounding water molecules to generate surface bound or soluble C3b (Janssen et al., 2006; Pangburn and Muller-Eberhard, 1980). It is well established that C3 plays a major role in the clearance of microbial pathogens, immune complexes, apoptotic cells and tissue debris by covalently tagging targets with C3 cleavage products (C3b and iC3b) for subsequent removal via phagocytic cells expressing C3b/iC3b receptors (He et al., 2008).
Extracellular protein misfolding underlies most of the serious conditions of pathological protein deposition including systemic amyloidosis, Alzheimer’s disease, diabetes and spongiform encephalitis (Aguzzi and O'Connor, 2010; Naiki and Nagai, 2009). Both the intra and extracellular compartments can impose strains on protein structure due to fluctuations in pH, temperature and oxidative stress. Thus, it is critical to maintain correct protein structure and function in living systems, a process that is collectively known as protein homeostasis or proteostasis (Aguzzi and O'Connor, 2010; Hipp et al., 2014; Naiki and Nagai, 2009). The extracellular space is a more oxidizing environment than the intracellular compartment, and there is an additional challenge posed to protein stability in the blood due to shear stress of circulating fluids. Chaperones play a key role in protecting against such stresses either by binding and preventing aggregation of the unfolded/misfolded protein or by facilitating protein refolding or clearance from circulation. Excessive misfolding/unfolding can cause disease due to loss of normal function, gain of pathological function and/or dysfunctional accumulation in tissues (Aguzzi and O'Connor, 2010; Naiki and Nagai, 2009). Though the process of protein homeostasis has been studied extensively in the intracellular context (Hipp et al., 2014), extracellular chaperones are not as well recognized or understood and characterization of these proteins and their functions has only recently gained more attention (Wyatt et al., 2013). The number of extracellular chaperones continues to grow; currently at least seven plasma proteins have been shown to function as chaperones including clusterin, α2-macroglobulin, haptoglobin, apolipoprotein E, serum amyloid P (SAP), caseins and fibrinogen (Wyatt et al., 2013).
Recently, we have shown that upon activation of complement, C3 binds covalently via the thioester to a wide range of plasma proteins of varying abundance, molecular weight and isoelectric points (Ramadass et al., 2014). A significant proportion of C3b is complexed to plasma proteins, even with the large disparity in the percentage of water molecules to proteins in plasma (91.5% of plasma is water, 55 Molar, and only 7.5% is protein). We speculated that this could be a mechanism to neutralize the C3 thioester and limit deposition on host cells at sites of complement activation (Ramadass et al., 2014). In the present study we investigated if C3 tagging of plasma proteins facilitates their clearance, and if so does this process have a preference for non-native or unfolded plasma proteins, thus suggesting an extracellular chaperoning function for C3.
2. MATERIALS AND METHODS
2.1 Reagents
Purified cobra venom factor (CVF), human serum depleted of individual complement components (C3, factor B and factor D), and purified human complement proteins (C3, factor B and factor D) were all purchased from Complement Technology, Inc. (Tyler, TX). The following purified human proteins were all obtained from Athens Research and Technology (Athens, GA): alpha-1 proteinase inhibitor (α1PI), vitamin D binding protein (DBP), human serum albumin (HSA) and neutrophil elastase. The following reagents were purchased from Sigma-Aldrich (St. Louis, MO): 2-p-toluidinylnaphthalene-6-sulfonate (TNS), N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide (MeOSuc-AAPV-pNA) and MeOSuc-Ala-Ala-Pro-Val chloromethyl ketone (MeOSuc-AAPV-CMK). Bio-Gel P30 matrix with medium polyacrylamide beads (90–180 μm wet bead size) for size exclusion chromatography and Lowry protein assay based DC Protein Assay Kit both were purchased from Bio-Rad (Hercules, CA). Pre-diluted bovine serum albumin (BSA) protein standards were purchased from Thermo-Fisher Scientific (Pittsburgh, PA). Purified actin was purchased from Cytoskeleton, Inc. (Denver, CO). Protease inhibitor cocktail tablets were purchased from Roche (Indianapolis, IN). Blood was obtained from several healthy donors as previously described using a protocol approved by Stony Brook University Institutional Review Board in accordance with the Declaration of Helsinki (Ramadass et al., 2014). Normal human serum (NHS) was collected and pooled from several donors.
The IgG fraction of goat polyclonal anti-human DBP was purchased from DiaSorin (Stillwater, MN) and then affinity-purified in our laboratory using immobilized DBP. Chicken polyclonal anti-α1PI antibody was obtained from ProSci, Inc. (Poway, CA). Mouse monoclonal anti-human factor B (clone D33/3) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-albumin antibody was a generous gift from Dr. Berhane Ghebrehiwet, Stony Brook University. Chicken polyclonal anti-human C3 was obtained from Gallus Immunotech (Cary, NC).
2.2 Gel electrophoresis and immunoblotting
For denaturing and reducing gels, all samples were separated using 8% polyacrylamide gels with SDS at 80 V for the stacking gel and 100 V for the resolving gel. Resolved gels then were transferred onto an Immobilon PVDF membrane (Millipore, Bedford, MA) at 100 V for 75 minutes. The PVDF membrane was blocked with 5% non-fat dry milk (NFDM) in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 30 minutes, followed by primary and HRP-labeled secondary antibody incubations in 5% NFDM. Finally, blots were developed using HyGLO Quick Spray chemiluminescent detection reagent (Denville Scientific, Denville, NJ) and X-ray film. For native gels, the same procedure was carried out using 8% native polyacrylamide gel but without SDS.
2.3 In vitro complex formation
To evaluate the role of activated C3 in complex formation, the alternative pathway was assembled in vitro using the purified proteins C3 (1.3 mg/ml), factor B (200 μg/ml) factor D (1 μg/ml) and DBP (400 μg/ml) or α1PI (1 mg/ml) with 0.5 mM Mg2+ and incubated at 37°C for the specified amount of time. In control experiments, a mixture containing either (20% of the original C3 concentration (0.26 mg/ml) or no C3 was used to test C3 dependence of the complex formation. The molar ratios of the various components were maintained at physiological levels even when the exact concentrations could not be maintained due to dilution effects.
2.4 Denaturation of proteins
The protein of interest was denatured using 5 M guanidinium-HCl (GuHCl), 30 mM DTT, 10 mM EDTA, 100 mM Tris-HCl (pH 8.3) at room temperature for the time indicated on a rotary shaker. Upon denaturation, the denaturing reagents were removed by gel filtration using a Bio-Gel P-30 column. The protein concentration of denatured proteins was quantified using the Lowry assay and then tested for denaturation using functional assays.
2.5 TNS Assay
Purified DBP or purified HSA (both native or GuHCl-denatured at 0.5 μM) were incubated with 30 nM TNS at room temperature for 20 minutes to allow dye binding, fluorescence then was measured at an excitation wavelength of 322 nm and an emission wavelength of 465 nm.
2.6 Elastase Assay
Purified elastase (60 nM) was incubated at 37°C for 30 minutes with either 60 nM native α1PI, 60 nM GuHCl α1PI or 100 μM of the synthetic elastase inhibitor MeOSuc-AAPV-CMK (positive control) in assay buffer (0.1 M HEPES, pH 7.25 with 0.5 M NaCl, 0.05% Tween 20) to allow binding. The p-nitroaniline substrate MeOSuc-AAPV-pNA (100 μM) was added and the mixture was incubated at 37°C for 30 minutes. Absorbance was read at 405nm to assess elastase activity.
2.7 DBP:actin complex formation
In vitro complex was formed with either native DBP or GuHCl denatured DBP incubated with purified actin at a 1:5 molar ratio of DBP:actin for 30 minutes at room temperature. Complex formation was assessed using an 8% native polyacrylamide gel and immunoblotting for DBP to verify a shift in molecular weight upon binding actin.
2.8 Complex binding to red blood cells
Blood from normal healthy individuals was drawn into EDTA tubes and mixed well. The blood was transferred into a conical tube containing equal volume of 3% dextran, mixed thoroughly but gently and allowed to sediment in upright position for 30 minutes at room temperature until a sharp demarcation between sedimented red blood cells (RBCs) and the pink supernatant (leukocytes and platelets) was noted. The supernatant was aspirated and the RBC fraction washed 3 times with DPBS by spinning at 900 rpm for 10 minutes at 15°C. The RBCs were resuspended in DPBS and then treated with either normal human serum or CVF activated normal human serum (108 RBCs + 100 μl serum) for 2 hours at 37°C, following which the cells were spun at 1000 × g for 3 minutes, washed twice with DPBS by spinning at 1000 × g at 4°C. The cell pellet was lysed using H2O containing 1X protease inhibitor cocktail by incubating on ice for 30 minutes. The samples were spun at 3300 × g for 10 minutes at 4°C to pellet the membranes. The supernatant was collected for analysis. The membrane fraction was solubilized in 100 μl 0.5 M Tris HCl (pH 8.3) + 0.05% Triton X-100, centrifuged at 3300 × g for 10 minutes at room temperature and the soluble fraction was collected for analysis.
2.9 Complement activation using thermally denatured serum
Normal human serum (NHS) was heated to 60°C for 1 hour (Δ serum) and added to equal volume of either fresh NHS, factor B depleted serum, factor D depleted serum or C3 depleted serum and incubated at 37°C for the time indicated. The non-heated sham control received equal amount of fresh serum. The NHS+Δ serum sample was then activated using CVF, zymosan or aggregated IgG for 30 minutes. The samples were separated on an 8% SDS polyacrylamide gel to evaluate factor B cleavage and iC3b generation. C5a levels in the samples were quantified using C5a ELISA.
2.10 C5a Sandwich ELISA
A sandwich ELISA to detect human C5a and C5a des Arg was developed as previously described in detail (Trujillo et al., 2013). Briefly, Maxi-sorb 96 well plates were coated with 500 ng of mouse monoclonal anti-human C5a (clone 295003; R&D Systems, Minneapolis, MN) capture antibody at 4°C overnight. The coating solution was removed and the wells were blocked with 300 μl of blocking solution (3% NFDM in PBS-T) for 1 hour at room temperature. After 3 washes, 100 μl of standards (78 pg/ml to 5 ng/ml of purified natural human C5a, Complement Technology, Inc.) or experimental samples were added and incubated at room temperature with shaking for 90 minutes. This was followed by 4 washes and incubation with 100 ng of the detection antibody, biotinylated mouse monoclonal anti-human C5a (clone 295009, R&D Systems) for 60 minutes at room temperature with shaking. Finally, 40 ng of HRP-conjugated streptavidin (KPL, Gaithersburg, MD) was added to each well and incubated at room temperature for 30 minutes. After 5 washes, 100 μl of HRP substrate solution (KPL) was added until color development followed by addition of 100 μl stop solution (KPL) and the absorbance was measured at 450 nm using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA).
3. RESULTS
3.1 C3b binds more avidly to non-native plasma proteins
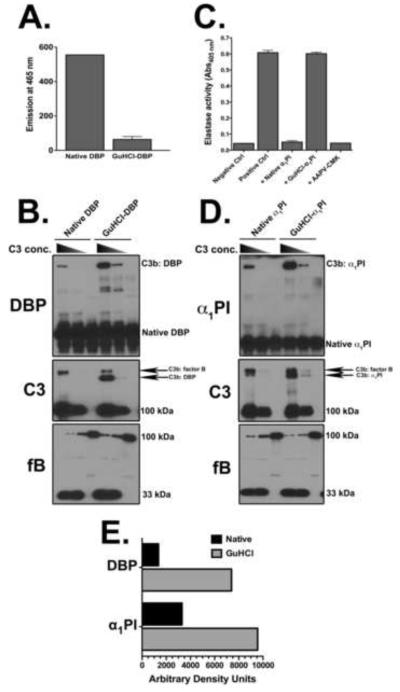
Our recent report showing that C3b covalently tags multiple plasma proteins in the fluid-phase during complement activation (Ramadass et al., 2014) coupled with the fact that C3b links to altered proteins such as beta amyloid (Aβ) (Bradt et al., 1998) prompted us to ask the question: does C3b react more avidly with misfolded/non-native plasma proteins. To test this premise, purified vitamin D binding protein (DBP) was employed as a model plasma protein as we previously described (Ramadass et al., 2014). DBP was denatured with guanidine hydrochloride (GuHCl) and dithiothreitol (DTT) for 6 hours and then analyzed with an in vitro reconstituted complement alternative pathway to evaluate complex formation with C3b. The denaturation procedure altered the native structure of DBP as confirmed by a marked reduction in the capacity to bind TNS dye (Fig. 1A). The binding of TNS to native DBP (or albumin) produces a strong increase in fluorescence emission at 465 nm (Goldschmidt-Clermont et al., 1987). Using purified alternative pathway proteins (C3, factor B, factor D) at physiological concentrations with 0.5 mM Mg2+, denatured DBP formed noticeably more complexes with C3b as compared to native DBP (Fig. 1B, C3b:DBP band in the DBP and C3 blots). Quantitative densitometry of the C3b:DBP band (DBP blot in figure 1B) showed a 5-fold increase in band intensity with denatured versus native DBP (Fig. 1E). Moreover, the intensity of the C3b:DBP complex decreased markedly when the C3 concentration was reduced by 80%, and no complexes were observed when C3 was excluded, indicating that the complexes were indeed covalent complexes of C3b:DBP, and not denatured DBP aggregates (Fig. 1B). The C3 blot in figure 1B had two higher molecular weight bands above the 110 kDa C3 α-chain: the intensity of the C3b:factor B band decreased slightly when denatured DBP was added while the C3b:DBP band showed a noticeable increase, further indicating that C3b has greater avidity for denatured proteins. The intensity of the C3b:factor D band is too faint to be observed in these experiments because of the very low factor D concentration (1 μg/ml). The bottom panel in figures 1B and 1D is a blot for factor B showing that generation of the 33 kDa Ba fragment (indicator of alternative pathway activation) correlates with C3b complex formation. Similar results were also obtained using purified human serum albumin (HSA), the most abundant protein in blood with a structure very similar to DBP. Denatured HSA formed more complexes with C3b as compared to native HSA (Supplementary Figure 1).
Figure 1.
C3b binds more avidly to denatured plasma proteins. Panel A: Fluorescence emitted by TNS binding (Ex/Em 322/465 nm) to native or guanidine hydrochloride (GuHCl) denatured DBP to measure the level of protein denaturation. Panel B: Native DBP or GuHCl DBP (both at 0.4 mg/ml) were allowed to form complexes with either 1.3 mg/ml C3 (physiological concentration), 0.26 mg/ml C3 or no C3 in the presence of factor B (fB, 0.2 mg/ml), factor D (fD, 1 μg/ml) and 0.5 mM Mg2+. All proteins were mixed at physiological (plasma) ratios with the exception of reduced or no C3 samples. The samples were incubated at 37°C for 15 minutes. Aliquots were analyzed using SDS-PAGE with β-mercaptoethanol on an 8% gel and then immunoblotted for DBP and C3 (for complex formation) and factor B (for complement activation). Panel C: Elastase inhibitory activity of α1PI was tested using the chromogenic substrate AAPV-pNA by measuring the release of pNA at 405 nm. Native and GuHCl denatured α1PI (both at 60 nM) were incubated at 37°C for 30 minutes with purified neutrophil elastase (60 nM), enzyme activity was then tested by adding 100 μM AAPV-pNA in assay buffer (0.1 M HEPES, pH 7.25 with 0.5 M NaCl, 0.05% Tween 20) and incubated at 37°C for another 30 minutes. Negative control is substrate alone without elastase, positive control is substrate and elastase only. AAPV-CMK sample is elastase pretreated with this peptide inhibitor as an additional control for inhibition of enzyme activity. Panel D: Native or GuHCl α1PI (both at 1 mg/ml) were allowed to form complexes with either 1.3 mg/ml C3, 0.26 mg/ml C3 or no C3 in the presence of factor B (fB, 0.2 mg/ml), factor D (fD, 1 μg/ml) and 0.5 mM Mg2+. All proteins were mixed at physiological (plasma) ratios with the exception of reduced or no C3 samples. The samples were incubated at 37°C for 15 minutes. Aliquots were analyzed using SDS-PAGE with β-mercaptoethanol on an 8% gel and then immunoblotted for α1PI and C3 and factor B. Panel E: Densitometry of the C3b:DBP complex bands in panel B (DBP blot) and C3b:α1PI complex bands in panel D (α1PI blot) comparing native versus GuHCl denatured proteins with the physiological (1.3 mg/ml) C3 concentration.
The hypothesis was further tested using another abundant plasma protein, α1-proteinase inhibitor (α1PI). Purified α1PI was denatured with GuHCl/DTT for 20 hours (α1PI required a longer denaturation time to lose functional capacity), and then tested for C3b complex formation using the in vitro assembled alternative pathway. To verify loss of native structure and function, the ability of α1PI to inhibit neutrophil elastase (its primary physiological target) was determined (Fig. 1C). α1PI covalently binds to the active site serine in elastase and abolishes its enzymatic activity. Using the chromogenic elastase substrate AAPV-pNA, it was noted that the GuHCl-α1PI completely lost elastase inhibitory activity, thus confirming that GuHCl denaturation abolished functional activity (Fig. 1C). Similar to the results using DBP or HSA, denatured α1PI formed more intense complexes with C3b as compared to native α1PI (Figs. 1D, 1E). The intensity of the denatured C3b:α1PI also was decreased dramatically when less C3 was used, and no complexes were seen when C3 was omitted (Fig 1D). These results indicate that C3b reacts more avidly with denatured proteins versus their native counterparts.
3.2 C3b binds plasma proteins in their native state, though it binds more readily to plasma proteins in their non-native form.
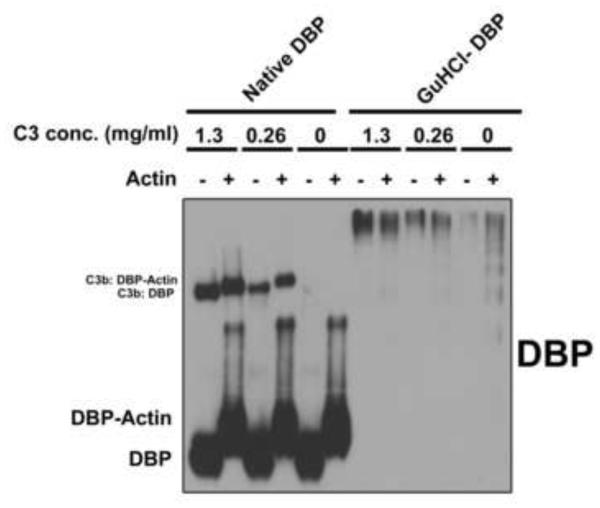
Our previous paper (Ramadass et al., 2014) and the results from figure 1 show that only a small percentage of the total target protein form complexes with C3b. The increased avidity of C3 for misfolded/unfolded proteins probably is due to an increased accessibility of potential thioester binding sites. Although figure 1 shows that denaturation substantially increases the amount of C3b complexes, these remain a fraction of the total. Thus, it is not clear if upon activation C3 binds basally to native plasma proteins or only to a small proportion of the protein pool that is in a non-native state. To examine this possibility, C3b:DBP complexes were first formed in vitro using native DBP and then tested for the ability of DBP to bind actin while covalently coupled to C3b. DBP is a known actin monomer binding protein and it binds G-actin in 1:1 molar ratio with high affinity (Kd of 10−9 M), and denaturation of DBP completely abolishes this function (Chun, 2012; Goldschmidt-Clermont et al., 1987). Purified C3b:DBP complexes were allowed to react with a 5-fold molar excess of purified G-actin and samples then were evaluated using an 8% non-denaturing gel to maintain the non-covalent DBP-actin complexes (Fig. 2). Results demonstrate that DBP in the C3b:DBP complex still retains the ability to bind actin as shown by the shift in molecular weight in samples where actin was added. This indicates that C3 can recognize and attach to proteins in their native state. Figure 2 also shows that GuHCl denatured DBP formed large aggregates (both with or without C3b attached) that did not migrate into the gel under non-denaturing conditions. This is in contrast to denaturing conditions where the SDS-PAGE migration pattern of native and denatured DBP was similar (Fig. 1B). The results from figures 1 and 2 suggest that C3 attaches to plasma proteins in their native state during complement activation, but denaturation substantially increases this interaction.
Figure 2.
Native DBP complexed with C3b retains the capacity to bind actin. Native DBP or GuHCl denatured DBP (both 0.4 mg/ml) were allowed to form complexes with either 1.3, 0.26 mg/ml C3 or no C3, in the presence of fB (0.2 mg/ml), fD (1 μg/ml) and 0.5 mM Mg2+ at physiological ratios. The samples were incubated at 37°C for 15 minutes. The complexes once formed in vitro were then treated with a 5-fold molar excess of purified G-actin and allowed to bind at room temperature for 30 minutes. Mixtures were then separated on an 8% native (non-denaturing) gel and immunoblotted for DBP.
3.3 C3b:plasma protein complexes bind to red blood cell membranes.
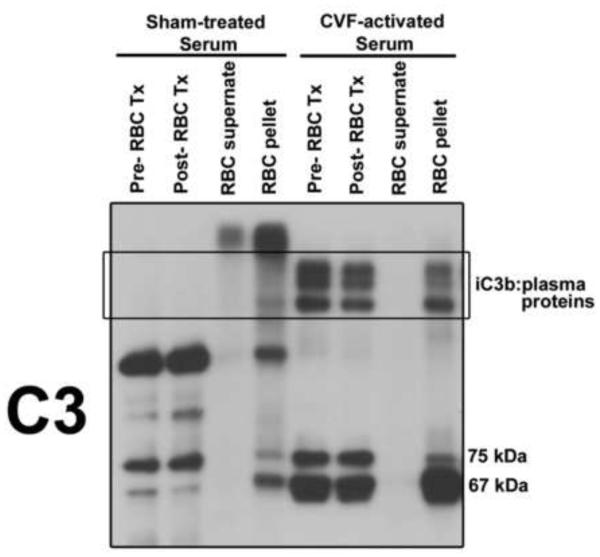
Since C3b binds to plasma proteins, and more specifically to proteins in their non-native state, we wanted to determine if these complexes bind RBCs as a potential clearance mechanism. RBCs are the most abundant cell in circulation and primate RBCs express large amount of complement receptor CR1 that is used to transport C3-opsonized complexes to the liver and spleen for clearance (Zipfel and Skerka, 2009). To test if RBCs bind C3b:plasma protein complexes, cells were purified from the blood of healthy individuals and treated with either normal human serum or complement activated (using CVF) serum. RBCs were separated from the serum, washed, lysed and the supernatant and pellet (membranes) were examined by immunoblotting for C3b:plasma protein complexes. Figure 3 shows that as expected, there were no C3b:plasma protein complexes in sham-treated serum either before (pre-RBC Tx) or after incubation with RBCs (post-RBC Tx). However, there were prominent complexes (degradation of C3b by factor I in the serum generated iC3b:plasma protein complexes, note increase in the 67 kDa fragment of iC3b) in CVF-activated serum both before and after incubation with RBCs (Fig. 3). The RBC membranes (pellet) from cells treated with CVF-activated serum also contained iC3b:plasma proteins and these were not seen in the RBC supernatant or pellet from cells treated with negative control sham-activated serum (Fig. 3). These results indicate that C3b:plasma protein complexes can bind RBC membranes and this can potentially function as a mechanism to clear complexes from the circulation.
Figure 3.
iC3b:plasma protein complexes bind to red blood cells (RBCs). Isolated human RBCs were treated for 30 minutes at 37°C with either with sham-treated NHS or serum activated with CVF. Serum aliquots were removed before (pre-RBC Tx) and after (post-RBC Tx) RBC treatment. RBCs were lysed with H2O and the membrane fraction (pellet) was separated from the supernatant. All samples were separated using SDS-PAGE on an 8% gel and immunoblotted for C3.
3.4 Thermally denatured proteins activate the complement alternative pathway but do not generate C5a.
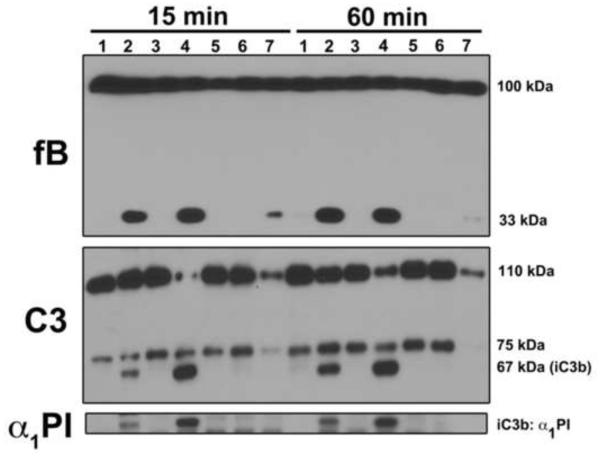
The increased capacity of C3b to bind non-native proteins led us to determine if these proteins could also activate the alternative pathway of complement to trigger clearance. To test this, normal human serum (NHS) was heated to 60°C for 1 hour to thermally denature proteins. Heated serum then was added to either fresh NHS, factor B depleted serum, factor D depleted serum or C3 depleted serum followed by a subsequent incubation at 37°C for 15 or 60 minutes. Factor B cleavage (increase in 33 kDa band) and iC3b generation (increase in 67 kDa band) was then evaluated by immunoblotting to assess alternative pathway activation (Fig. 4). Controls included 60°C-heated serum treated with EDTA to inhibition activation or CVF as a positive control for alternative pathway activation. As expected, complement could not be activated in 60°C-heated serum (not shown), but when added to fresh NHS for either 15 or 60 minutes at 37°C, the heat-treated serum triggered cleavage of C3 and factor B (Fig. 4, lanes 2, C3 and factor B blots). These samples also caused the generation of C3b:plasma protein complexes as seen in the α1PI blot (Fig. 4, lanes 2). Addition of CVF to the 60°C-heated serum + fresh NHS mixture induced robust complement activation (Fig. 4, lanes 4). In contrast, factor B and C3 cleavage, as well as α1PI:iC3b complex formation, was not observed in untreated NHS incubated at 37°C for 15 or 60 minutes (lanes 1) or in NHS treated with heated serum containing 10 mM EDTA to inhibit complement activation (lanes 3). Likewise, factor B (lanes 5), factor D (lanes 6) or C3-depleted (lanes 7) serum samples treated with heated serum show no evidence of complement activation or protein complex formation.
Figure 4.
Thermally denatured proteins activate the complement system via the alternative pathway. NHS was heated to 60°C for 1 hour (Δ serum) and then added to equal volume of either fresh NHS (lanes 2), factor B depleted serum (lanes 5), factor D depleted serum (lanes 6) or C3 depleted serum (lanes 7) and incubated for either 15 or 60 minutes at 37°C. The negative control was untreated NHS (lanes 1) or NHS that received equal amount of Δ serum in the presence of 10 mM EDTA (lanes 3) to prevent complement activation. The positive control was a mixture of Δ serum plus fresh serum treated with CVF (lanes 4) to activate the alternative pathway. Samples were separated using an 8% SDS-PAGE gel and blotted for factor B (alternative pathway activation and generation of the 33 kDa Ba fragment), C3 (110 kDa α-chain cleavage and 67 kDa iC3b generation) and α1PI (iC3b:α1PI complex formation).
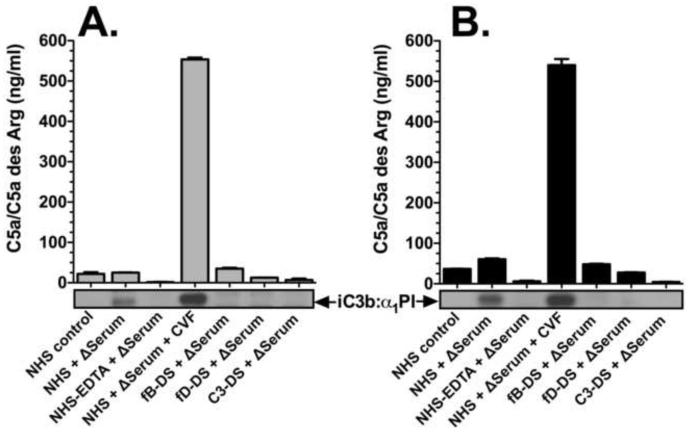
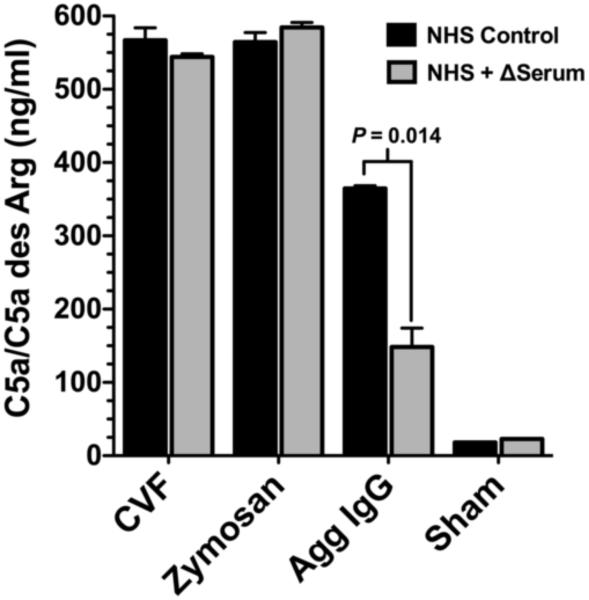
Although significant levels of factor B cleavage and iC3b generation were observed in NHS treated with heated serum, there was no C5a generation in these samples serum after 15 (Fig. 5A) or 60 minutes (Fig. 5B). Addition of CVF to the NHS + heated serum sample induced C5a generation (Figs. 5A and 5B) indicating that the addition of thermally denaturated serum did not alter the ability of NHS to form a C5 convertase and generate C5a. However, CVF is the cobra snake’s homologue of mammalian C3b and can function independent of C3 to generate C5a (Vogel and Fritzinger, 2010). To determine if C3-dependent complement activators can generate C5a in fresh NHS + heated serum, samples were treated with zymosan or aggregated IgG (HAGG). Figure 6 shows that zymosan, HAGG and CVF treatments could all induce C5a generation in both the NHS alone and NHS + heated serum. The 50% decrease in C5a levels in the NHS + heated serum treated with HAGG was due the half the level of active C2 present in that sample (enzymatic activity of C2 is destroyed by heat treatment). This data indicates that thermally denatured plasma proteins activate the alternative pathway of complement to generate C3 and factor B cleavage fragments, but do not result in C5a production. This is consistent with the observation that complement is involved in the non-inflammatory clearance of apoptotic cells by preventing downstream effects of complement activation (Ricklin et al., 2010).
Figure 5.
Thermally denatured proteins do not generate C5a. The level of C5a measured by ELISA in the same serum samples analyzed in Figure 4. Panel A: serum incubated for 15 minutes at 37°C. Panel B: serum incubated for 60 minutes at 37°C. The α1PI blot showing iC3b:α1PI complex formation is displayed for comparison below the X-axis.
Figure 6.
C3 dependent complement activators generate C5a in thermally denatured serum. Untreated NHS (Control) or NHS plus 60°C heated serum (Δ Serum) were treated with either CVF, zymosan, HAGG or buffer (Sham) for 60 minutes at 37°C and C5a levels were quantified by ELISA.
4. DISCUSSION
Prompt clearance of damaged proteins from the circulation is essential to prevent potential pathological consequences triggered by molecules with altered non-native structure. This report demonstrates that chemically or thermally denatured plasma proteins activate the alternative pathway of complement and are covalently tagged by C3b, marking the complexes for subsequent clearance by cells expressing C3b/iC3b receptors. Moreover, complement activation induced by non-native protein structure does not proceed to the level of C5 cleavage, thus avoiding both generation of the potent pro-inflammatory mediator C5a and activation of the terminal (membrane attack) pathway. This shows that denatured proteins can activate complement without triggering an inflammatory reaction, perhaps in much the same manner as apoptotic cells activate complement and are cleared without triggering C5a-mediated inflammation (Flierman and Daha, 2007; Ricklin et al., 2010). Thus, the complement system can differentiate between tagging waste products for clearance (apoptotic cells, denatured proteins) and more serious disturbances such as infections with pathogenic microorganisms that require rapid activation of the immune system. The alternative pathway of complement is well-positioned for rapid recognition of damaged plasma proteins, however, it is not known if denatured plasma proteins can also activate the classical or lectin complement pathways. Additional studies will be needed to investigate this possibility.
Proteins in extracellular fluids are exposed to multiple stresses that can alter normal structure and trigger misfolding and/or aggregation, potentially resulting in pathological protein deposition in tissues (Wyatt et al., 2013). The mechanism by which extracellular chaperones function appears to be different than intracellular chaperones (that largely utilize ATP-dependent re-folding) and involves binding misfolded proteins and inhibiting their aggregation and/or enhanced clearance from fluids (Hipp et al., 2014; Wyatt et al., 2013). The work described herein demonstrates that complement C3 displays enhanced recognition of non-native protein structures and covalently tags the denatured protein for subsequent clearance, thus effectively functioning as an extracellular chaperone. However, C3 can also covalently attach to plasma proteins in a native configuration, this may serve to intercept excess C3b in the fluid-phase and limit deposition on host cells during episodes of complement activation (Ramadass et al., 2014). The increased binding of C3b to denatured proteins most likely is due to increased accessibility of potential thioester binding sites. Disparity in binding of C3b was noted previously where activation of C3 led to 35% incorporation of nascent C3b into C3b:C4b complexes in the fluid phase, while only 12% of C3b was incorporated into C3b:IgG complexes (Meri and Pangburn, 1990). Further studies showed that the C3b attachment site is Ser-1217 in human C4 whereas Thr144 was the major acceptor site for C3b in the CH1 domain of IgG (Kim et al., 1992; Sahu and Pangburn, 1994). Thus the thioester binding sites vary from protein to protein, and studies using model peptides have also shown that Tyr residues react with the thioester in C3b 11-fold greater than threonine residues, 47-fold better than serine and 50-fold better than hydroxyl groups in carbohydrates (Sahu and Pangburn, 1995). In the current study, a large proportion of denatured DBP did not react with C3b and still migrated at its native molecular weight (56 kDa) on SDS-PAGE (Fig. 1B). GuHCl denatured DBP clearly was aggregated since it did not migrate into a native gel (Fig. 2) but was dissociated by SDS into DBP monomers (Fig. 1B). A plausible explanation is that C3 has access only to a small fraction of nucleophile acceptor groups on the exterior of DBP aggregates, while the majority of denatured protein is located in the aggregate interior and is unable to react with the thioester in C3. So the level of aggregation may limit the extent of C3b binding to denatured protein targets. In an in vivo system, denaturation and tagging can proceed simultaneously and hence the proportion of C3 tagged denatured protein could be higher.
Numerous previous reports have shown that C3 is found in amyloid plaques in brain tissue from Alzheimer’s disease patients (Bradt et al., 1998; Emmerling et al., 1997; Veerhuis et al., 1995) and C3b binds to beta amyloid (Aβ) and results in alternative pathway activation (Bradt et al., 1998). In addition to interacting with C3, beta amyloid can activate the classical complement pathway by binding C1q (Jiang et al., 1994; Rogers et al., 1992). It has been reported that beta amyloid-induced activation of complement does not proceed further than C3 cleavage (Cadman and Puttfarcken, 1997; Veerhuis et al., 1995), but others have shown activation with C5a generation and terminal pathway formation (Bradt et al., 1998). Inhibition of C3 activation in transgenic mice containing the human amyloid precursor protein (hAPP) using soluble complement receptor-related protein y (sCrry, a murine complement inhibitor) leads to 2- to 3-fold higher Aβ deposition in the brain and is accompanied by a prominent accumulation of degenerating neurons when complement is inhibited (Wyss-Coray et al., 2002). C3 deficiency also accelerates the brain Aβ deposition in a mouse model of Alzheimer’s disease (Maier et al., 2008). Furthermore, beta amyloid attached to C3b binds to CR1 on RBCs for subsequent clearance (Rogers et al., 2006). These results indicate that complement activation products can protect against beta amyloid-induced neurotoxicity and may reduce the accumulation or promote the clearance of amyloid. These studies with beta amyloid deposition in the brain are consistent with our findings herein that indicate that C3 is an effective extracellular chaperone that recognizes non-native plasma proteins and tags them for subsequent clearance from the blood.
C3 is highly expressed in physiological fluids and has a central role in the activation of complement, but this fluid-phase innate immune effector also functions broadly to maintain homeostasis. Very recent reports have shown that C3 also functions inside cells to regulate T-cell homeostasis (Liszewski et al., 2013) and sense intracellular pathogens (Tam et al., 2014). Moreover, covalent attachment of C3b to antigens on apoptotic cells is needed for proper intracellular processing of apoptotic cell fragments and modulation of T-cell responses to self-antigens (Baudino et al., 2014). Thus, the functional roles of this versatile protein are expanding and we believe that the results of this study, coupled with the prior reports discussed above, indicate that C3 functions as an extracellular chaperone to tag non-native proteins in extracellular fluids for subsequent clearance.
Supplementary Material
HIGHLIGHTS.
C3 interacts more avidly with denatured plasma proteins versus their native forms
Plasma proteins covalently tagged with C3b bind red blood cell membranes
Heated (60°C) serum activates the alternative pathway but does not generate C5a
C3 may serve to detect and clear damaged proteins from the blood without producing C5a
ACKNOWLEDGEMENTS
This worked was supported by U.S. National Institutes of Health grants GM063769 (R.R.K.) and AI060866 and AI084178 (B.G.).
Abbreviations
- α1PI
alpha 1 proteinase inhibitor
- CVF
cobra venom factor
- DBP
vitamin D binding protein
- HAGG
heat-aggregated human IgG
- HSA
human serum albumin
- NHS
normal human serum
- TNS
2-p-toluidinylnaphthalene-6-sulfonate fluorescent dye
- Δ serum
NHS heated at 60°C for 60 minutes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
The authors declare that no conflict of interest exists.
REFERENCES
- Aguzzi A, O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 2010;9:237–48. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Baudino L, Sardini A, Ruseva MM, Fossati-Jimack L, Cook HT, Scott D, Simpson E, Botto M. C3 opsonization regulates endocytic handling of apoptotic cells resulting in enhanced T-cell responses to cargo-derived antigens. Proc. Natl. Acad. Sci. USA. 2014;111:1503–8. doi: 10.1073/pnas.1316877111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradt BM, Kolb WP, Cooper NR. Complement-dependent proinflammatory properties of the Alzheimer's disease beta-peptide. J. Exp. Med. 1998;188:431–8. doi: 10.1084/jem.188.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman ED, Puttfarcken PS. Beta-amyloid peptides initiate the complement cascade without producing a comparable effect on the terminal pathway in vitro. Exp. Neurol. 1997;146:388–94. doi: 10.1006/exnr.1997.6540. [DOI] [PubMed] [Google Scholar]
- Chun RF. New perspectives on the vitamin D binding protein. Cell Biochem. Funct. 2012;30:445–56. doi: 10.1002/cbf.2835. [DOI] [PubMed] [Google Scholar]
- Emmerling MR, Spiegel K, Watson MD. Inhibiting the formation of classical C3-convertase on the Alzheimer's beta-amyloid peptide. Immunopharmacology. 1997;38:101–9. doi: 10.1016/s0162-3109(97)00067-2. [DOI] [PubMed] [Google Scholar]
- Flierman R, Daha MR. The clearance of apoptotic cells by complement. Immunobiology. 2007;212:363–70. doi: 10.1016/j.imbio.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Williams MH, Galbraith RM. Altered conformation of Gc (vitamin D-binding protein) upon complexing with cellular actin. Biochem. Biophys. Res. Commun. 1987;146:611–7. doi: 10.1016/0006-291x(87)90572-9. [DOI] [PubMed] [Google Scholar]
- He JQ, Wiesmann C, van Lookeren Campagne M. A role of macrophage complement receptor CRIg in immune clearance and inflammation. Mol. Immunol. 2008;45:4041–7. doi: 10.1016/j.molimm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–6. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Jiang H, Burdick D, Glabe CG, Cotman CW, Tenner AJ. beta-Amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J. Immunol. 1994;152:5050–9. [PubMed] [Google Scholar]
- Kim YU, Carroll MC, Isenman DE, Nonaka M, Pramoonjago P, Takeda J, Inoue K, Kinoshita T. Covalent binding of C3b to C4b within the classical complement pathway C5 convertase. Determination of amino acid residues involved in ester linkage formation. J. Biol. Chem. 1992;267:4171–6. [PubMed] [Google Scholar]
- Liszewski MK, Kolev M, Le Friec G, Leung M, Bertram PG, Fara AF, Subias M, Pickering MC, Drouet C, Meri S, Arstila TP, Pekkarinen PT, Ma M, Cope A, Reinheckel T, Rodriguez de Cordoba S, Afzali B, Atkinson JP, Kemper C. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity. 2013;39:1143–57. doi: 10.1016/j.immuni.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M, Peng Y, Jiang L, Seabrook TJ, Carroll MC, Lemere CA. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J. Neurosci. 2008;28:6333–41. doi: 10.1523/JNEUROSCI.0829-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastellos DC, Deangelis RA, Lambris JD. Complement-triggered pathways orchestrate regenerative responses throughout phylogenesis. Semin. Immunol. 2013;25:29–38. doi: 10.1016/j.smim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri S, Pangburn MK. A mechanism of activation of the alternative complement pathway by the classical pathway: protection of C3b from inactivation by covalent attachment to C4b. Eur. J. Immunol. 1990;20:2555–61. doi: 10.1002/eji.1830201205. [DOI] [PubMed] [Google Scholar]
- Naiki H, Nagai Y. Molecular pathogenesis of protein misfolding diseases: pathological molecular environments versus quality control systems against misfolded proteins. J. Biochem. 2009;146:751–6. doi: 10.1093/jb/mvp119. [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Muller-Eberhard HJ. Relation of putative thioester bond in C3 to activation of the alternative pathway and the binding of C3b to biological targets of complement. J. Exp. Med. 1980;152:1102–14. doi: 10.1084/jem.152.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass M, Ghebrehiwet B, Smith RJ, Kew RR. Generation of Multiple Fluid-Phase C3b:Plasma Protein Complexes during Complement Activation: Possible Implications in C3 Glomerulopathies. J. Immunol. 2014;192:1220–30. doi: 10.4049/jimmunol.1302288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, Civin WH, Brachova L, Bradt B, Ward P, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1992;89:10016–20. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, Cao P, Kolody H, Vedders L, Kolb WP, Sabbagh M. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol. Aging. 2006;27:1733–9. doi: 10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Sahu A, Pangburn MK. Covalent attachment of human complement C3 to IgG. Identification of the amino acid residue involved in ester linkage formation. J. Biol. Chem. 1994;269:28997–9002. [PubMed] [Google Scholar]
- Sahu A, Pangburn MK. Tyrosine is a potential site for covalent attachment of activated complement component C3. Mol. Immunol. 1995;32:711–6. doi: 10.1016/0161-5890(95)98933-f. [DOI] [PubMed] [Google Scholar]
- Tam JC, Bidgood SR, McEwan WA, James LC. Intracellular sensing of complement C3 activates cell autonomous immunity. Science. 2014;345 doi: 10.1126/science.1256070. 1256070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo G, Habiel DM, Ge L, Ramadass M, Cooke NE, Kew RR. Neutrophil Recruitment to the Lung in Both C5a- and CXCL1-Induced Alveolitis Is Impaired in Vitamin D-Binding Protein-Deficient Mice. J. Immunol. 2013;191:848–56. doi: 10.4049/jimmunol.1202941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerhuis R, van der Valk P, Janssen I, Zhan SS, Van Nostrand WE, Eikelenboom P. Complement activation in amyloid plaques in Alzheimer's disease brains does not proceed further than C3. Virchows Archiv. 1995;426:603–10. doi: 10.1007/BF00192116. [DOI] [PubMed] [Google Scholar]
- Vogel CW, Fritzinger DC. Cobra venom factor: Structure, function, and humanization for therapeutic complement depletion. Toxicon. 2010;56:1198–222. doi: 10.1016/j.toxicon.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Wyatt AR, Yerbury JJ, Ecroyd H, Wilson MR. Extracellular chaperones and proteostasis. Annu. Rev. Biochem. 2013;82:295–322. doi: 10.1146/annurev-biochem-072711-163904. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T, Yan F, Lin AH, Lambris JD, Alexander JJ, Quigg RJ, Masliah E. Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer's mice. Proc. Natl. Acad. Sci. USA. 2002;99:10837–42. doi: 10.1073/pnas.162350199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkadis IK, Mastellos D, Lambris JD. Phylogenetic aspects of the complement system. Dev. Comp. Immunol. 2001;25:745–62. doi: 10.1016/s0145-305x(01)00034-9. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009;9:729–40. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.