Abstract
The non-synonymous SNP rs2228145 in the IL6R gene on chromosome 1q21.3 is associated with a wide range of common diseases, including asthma, rheumatoid arthritis, type 1 diabetes and coronary heart disease. We examined the contribution of this functional IL6R gene polymorphism rs2228145 versus other genome-wide SNPs to the variance of sIL-6R levels in blood plasma in a large population-based sample (N∼5000), and conducted an expression QTL (eQTL) analysis to identify SNPs associated with IL6R gene expression. Based on data from 2360 twin families, the broad heritability of sIL-6R was estimated at 72%, and 51% of the total variance was explained by the functional SNP rs2228145. Converging findings from GWAS, linkage, and GCTA analyses indicate that additional variance of sIL-6R levels can be explained by other variants in the IL6R region, including variants at the 3′end of IL6R tagged by rs60760897 that are associated with IL6R RNA expression.
Keywords: soluble Interleukin-6 receptor, inflammation, heritability, GWAS, gene expression, eQTL
Introduction
The interleukin-6 receptor (IL-6R) forms part of the ligand-receptor complex that mediates the activities of interleukin-6 (IL-6). So-called classical IL-6 signaling occurs in hepatocytes and some leukocyte subtypes, which express a trans-membrane form of IL-6R on their surface (membrane-bound IL-6R, or mIL-6R; Rose-John et al. 2006). A second type of IL-6 signaling called IL-6 trans-signaling is capable of stimulating a variety of different cell types as it is mediated by a soluble form of the IL-6R (soluble IL-6R, or sIL-6R; Mackiewicz et al. 1992; Taga et al. 1989). IL-6 trans-signaling allows cells lacking mIL-6R to respond to IL-6, as long as they express the trans-membrane signal transducer protein gp130 on their surface (which is thought to be expressed ubiquitously; Saito et al. 1992). On target cells, the complex of IL-6 and IL-6R binds to two molecules of gp130, thereby activating several signal transduction pathways (Taga and Kishimoto 1997), leading to cellular responses such as proliferation, differentiation and inflammatory processes. IL-6 trans-signaling plays a key role in several autoimmune diseases and inflammatory conditions, including asthma (Doganci et al. 2005), rheumatoid arthritis (Kotake et al. 1996), chronic inflammatory bowel disease (Atreya et al. 2000), some types of cancer (e.g. multiple myeloma; Becker et al. 2004; Stephens et al. 2012) and peritonitis(Hurst et al. 2001).
Two isoforms of sIL-6R have been identified in blood plasma of healthy individuals that are generated through different mechanisms (Jones et al. 2001; Muller-Newen et al. 1996). The majority of sIL-6R is thought to be produced by a process called shedding, referring to the proteolytic cleavage of mIL-6R and subsequent release of the ligand-binding ectodomain into the extracellular space (Müllberg et al. 1993). A second isoform is produced by translation of an alternatively spliced mRNA lacking a 94-bp sequence coding for part of the transmembrane domain (Horiuchi et al. 1994; Lust et al. 1992) that anchors the receptor to the cell membrane. The process of shedding is affected by a non-synonymous SNP (Asp358Ala or rs2228145 (A > C), previously also known as rs8192284) that occurs within the region encoding the proteolytic cleavage site, in exon 9 of the interleukin-6 receptor gene, IL6R, on chromosome 1q21.3 (Müllberg et al. 1994). The SNP causes a striking difference in IL-6R concentrations between carriers of different alleles, with reduced concentrations of mIL-6R and increased concentrations of sIL-6R in carriers of the minor allele (C) (Ferreira et al. 2013; Galicia et al. 2004; Lourdusamy et al. 2012; Melzer et al. 2008; Rafiq et al. 2007). Although some previous studies found no association of rs2228145 with overall IL6R RNA expression (IL6R Genetics Consortium and Emerging Risk Factors Collaboration. 2012) or expression of the RNA transcript encoding mIL-6R (Ferreira et al. 2013), a positive association has been reported for the rs2228145 C allele and expression level of the alternatively spliced mRNA (Ferreira et al. 2013; Stephens et al. 2012), and we and others found a negative association between the rs2228145 C allele and overall expression level (Revez et al. 2013).
Variants in IL6R are associated with the risk of a wide spectrum of common diseases, with the rs2228145 C allele increasing susceptibility to asthma (Ferreira et al. 2011), and decreasing susceptibility to other diseases including rheumatoid arthritis (Eyre et al. 2012), coronary heart disease (IL6R Genetics Consortium and Emerging Risk Factors Collaboration. 2012), and type 1 diabetes (Ferreira et al. 2013). Two consortia reported a protective effect of the C allele on the risk of coronary heart disease and emphasized the potential of tocilizumab, a monoclonal antibody against IL-6R used for treatment of chronic inflammatory disease, as a novel therapeutic strategy to prevent cardiovascular disease (IL6R Genetics Consortium and Emerging Risk Factors Collaboration. 2012; The Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium. 2012). Growing interest in the role of IL6R in complex disease and in therapeutic strategies targeting the IL-6R pathway highlight the value of novel insights into genetic determinants of IL-6R level. The associations with disease reported for multiple variants in the IL6R gene are generally ascribed to LD with rs2228145. Two recent study showed that some additional variance in sIL-6R level is explained by other SNPs within IL6R (Ferreira et al. 2013; Revez et al. 2013), but it is unknown how much of the variance of sIL-6R levels in total is due to variants other than rs2228145 that remain to be identified.
We describe a series of analyses conducted in a population-based sample of ∼5000 Dutch individuals (Supplementary Table S1) aimed at evaluating the contribution of the known functional polymorphism rs2228145 to the variation in sIL-6R levels, and the contribution of other genetic variants within the IL6R region and the rest of the genome. We analyzed plasma sIL-6R levels in a large sample of twins and their family members to estimate the total heritability of sIL-6R, and conducted GWA, linkage, and eQTL analyses to identify additional genetic variants influencing sIL-6R level. Using the data from twin families and the classic biometrical model, we estimate the broad heritability of sIL-6R level to be 72 % and show that rs2228145 accounts for 51% of the total variance of sIL-6R level. Results from linkage analysis corroborate this and indicate that genetic variation within the IL6R region on chromosome 1 explains 69% of the variation in sIL-6R, of which 19% is due to genetic variation other than rs2228145. Results from eQTL analysis point towards a role of genetic variants at the 3′end of IL6R contributing to the levels of sIL-6R and IL6R RNA in blood. In passing, we provide empirical evidence that different methods based on the same corpus of genetic theory, including the classic biometrical model, the twin-family model, linkage analysis at the IL6R locus, and genome-wide SNP sharing in unrelated individuals (GCTA) all converge to the same conclusion.
Materials and Methods
Subjects
Plasma sIL-6R level data were available for 8929 participants from the Netherlands Twin Register (NTR; Boomsma et al. 2006), of which 5945 individuals also had genome-wide SNP data. Data from an additional 1966 participants from the Netherlands Study of Depression and Anxiety (NESDA (Penninx et al. 2008)) were included in the eQTL analysis. Individuals using anti-inflammatory medication or medication influencing the HPA (Hypothalamic-Pituitary-Adrenal)-axis (NTR: N = 426/4.8%, NESDA: N=538/26%) at the time of blood sampling, or with a sIL-6R level > 100.000 pg/mL (N=6/0.07% of NTR subjects) were excluded from all analyses. For a detailed description of the characteristics of the subjects included in each analysis, see Supplementary Methods. NTR and NESDA studies were approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Centre, Amsterdam, an Institutional Review Board certified by the US Office of Human Research Protections (IRB number IRB-2991 under Federal-wide Assurance-3703; IRB/institute codes, NTR 03-180, NESDA 03-183). All subjects provided written informed consent.
Blood sampling
Blood sampling procedures have been described previously (Spijker et al. 2004; Willemsen et al. 2010). In short; venous blood samples were drawn in the morning after an overnight fast. For RNA measurement, heparinized whole blood from NESDA en NTR participants was transferred within 20 minutes of sampling into PAX gene Blood RNA tubes (Qiagen) and stored at -20°C. For sIL-6R measurement, EDTA plasma tubes were collected from NTR participants, and were centrifuged for 20 minutes at 2000× g at 4°C. EDTA plasma, buffy coat and red blood cells were harvested and a liquoted (0.5 ml), snap-frozen in dry ice, and stored at -30°C.
sIL-6R level
sIL-6R level was measured in EDTA plasma samples (diluted 1:100) using the Quantikine Elisa Human IL-6 sR assay of R&D systems. The inter-assay and intra-assay coefficient of variation were < 15 %. In all analyses, sIL-6R level was adjusted for sex and age by inclusion of covariates or by analysis of residualized levels. sIL-6R level was on average higher in males (4.25 × 10-8 g/mL) compared to females (4.14 × 10-8 g/mL) and increased with age (1 SD of age (14 years) was associated with an increase in sIL-6R level of 0.17 × 10-8 g/mL).
Genotype data
DNA extraction has been described before (Boomsma et al. 2008). Genotyping was done on multiple chip platforms, for several partly overlapping subsets of participants. The following platforms were used: Affymetrix Perlegen 5.0, Illumina 370, Illumina 660, Illumina Omni Express 1M and Affymetrix 6.0. After array specific data analysis, genotype calls were made with the platform specific software (Genotyper, Beadstudio). In total, genotype data were available for 12,133 subjects from NTR and NESDA. The extensive genotyping quality control steps and imputation procedures are described in the Supplementary Methods. All analyses were performed on 1000G-imputed data (phase I Interim release All panel (sequence data freeze 23/11/2010), release June 2011, https://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_interim.html.
IL6R expression
RNA extraction (Spijker et al. 2004; Willemsen et al. 2010) and expression QC procedures have been described in detail previously (Jansen et al. 2014). PAXgene tubes were shipped to the Rutgers University Cell and DNA Repository (RUCDR), USA. RNA was extracted using Qiagen Universal liquid handling system (PAXgene extraction kits following the manufacturer's protocol). Total RNA was measured by spectroscopy (Trinean DropSense) to determine purity and concentration while RNA fidelity was measured by the Agilent Bioanalyzer analysis. RNA samples were hybridized to Affymetrix U219 array plates (GeneTitan), which contains 530,467 probes, each 25 bases in length. Array hybridization, washing, staining, and scanning were carried out in an Affymetrix GeneTitan System following the manufacturer's protocol. Non-uniquely mapping probes (hg19) and probes containing a polymorphic SNP based on snp137 (UCSC) were removed. Expression values were obtained using RMA normalization implemented in Affymetrix Power Tools (APT, v 1.12.0). Probes targeting IL6R (Supplementary Table S4) were selected for analysis.
Statistical analysis
The heritability of sIL-6R level in extended twin families
To estimate the broad- and narrow-sense heritability of sIL-6R and to examine the contribution of rs2228145, genetic structural equation models were fitted in M×(Neale et al. 2006) to sIL-6R data from mono- and dizygotic twins, siblings and parents. The model included additive genetic influences (A), non-additive genetic influences (D), sibling-shared environmental influences (C) and unique environment (E). These methods are outlined in full in the Supplementary Methods.
Variance explained by chromosome-wide SNPs and SNPs in the IL6R region using GCTA
The variance in sIL-6R level explained by all SNPs was estimated in GCTA (Genome-wide Complex Trait Analysis (Yang et al. 2011a; Yang et al. 2011b)) separately for each chromosome. For a full description of the methods, see Supplementary Methods.
GWA analysis
GWA analysis was performed in PLINK (Purcell et al. 2007) on dosage data. PLINK accounts for familial relations by performing a stratified association analysis with clusters based on family id using the --family option. This option implements generalized estimating equations with an independence model (Dobson 2002) and robust standard errors obtained using the sandwich correction for the family clustering (Williams 2000). The analyses included one randomly selected twin of each MZ pair.
Biometrical model
The variance due to additive effects (VA) and dominance effects (VD) of rs2228145 were estimated by applying the biometrical model (Falconer 1960) to the allele frequency estimates and the mean sIL-6R level corresponding to each genotype group. According to biometrical model, VA and VD are calculated as follows:
Where p=frequency of allele 1, q=frequency of allele 2, a= genotypic value; half the distance between the mean phenotype level of the two homozygotes, and d=dominance deviation; deviation of the mean phenotype level of heterozygotes from the midpoint of the two homozygotes.
Mean sIL-6R level per rs2228145 genotype, corrected for age and sex, were obtained in SPSS version 19. Allele frequency estimates were obtained with Sib-pair (“http://genepi.qimr.edu.au/staff/davidD/#sib-pair”) using the best linear unbiased estimator (BLUE) option, which accounts for familial relatedness (McPeek et al. 2004).
Heritability explained by rs2228145
To assess the contribution of rs2228145 to the heritability of sIL-6R level, models were fitted to the sIL-6R data from twin families with and without adjustment of sIL-6R levels for rs2228145 genotype. In the model with adjustment for rs2228145, sIL-6R level was modeled as follows:
where α=intercept, age= age at blood sampling (z-score), sex= coded as 0 for males and 1 for females, βSNPrs2228145 = additive effect of rs2228145, rs2228145 genotype = observed genotype at rs2228145 (codedas 0, 1, 2 – corresponding to the number of minor alleles) and ε=residual.
When rs2228145 genotype is not accounted for, the effect of this SNP on the resemblance of sIL-6R level among family members is included in the total genetic influences (A and D, see Supplementary Methods). When the effect of rs2228145 is incorporated (by estimating β rs2228145), sIL-6R levels are adjusted for rs2228145 genotype and the variance of residual levels (ε) is partitioned into unmeasured genetic and environmental factors (A, C, D and E). Thus, the total broad heritability (H2) of sIL-6R can be written as:
Where a2 and d2 are the proportions of the variation in sIL-6R level due to total additive genetic and non-additive genetic effects, as estimated from the twin-family data, a2residual and d2residual represent all remaining unmeasured additive and non-additive genetic effects that are not captured by rs2228145 (expressed as a proportion of the total variation in sIL-6R level): these components were estimated in a model that included adjustment of sIL-6R levels for SNP effects. a2rs2228145 and d2rs2228145 are the proportion of the total variation in sIL-6R that can be explained by additive and non-additive genetic effects of rs2228145, which can be inferred from the difference between the variance estimates from the total heritability model (without correction for rs2228145) and the variance estimates from the model with adjustment for rs2228145.
Combined linkage and association analysis
Analysis of linkage while simultaneously modeling association, as suggested by Fulker et al (1999) was conducted in QTDT (Abecasis et al. 2000). The data came from nuclear families of which both parents and offspring had data on genome-wide SNPs and sIL-6R level, where the offspring consisted of DZ twins or non-twin sib-pairs, or a single MZ twin + sibling(s). The analysis was performed for all imputed SNPs with a MAF >0.2 in the IL6R gene +/- 10 MB (IL6R gene location build 37/hg19, chr1: 154377669 - 154441926), leading to a selection of 13751 SNPs (chr1: 144377669-164441926). IBD probabilities were estimated in Merlin (Abecasis et al. 2002) using multipoint estimation, which takes into account the correlated structure of markers and is therefore suited for dense SNP data (Abecasis and Wigginton 2005). To create a centiMorgan (cM) map of the region, cM distances between SNPs were inferred from base-pair distances following the assumption that a distance of 1 million basepairs between SNPs corresponds to a distance of 1cM. The analysis was performed on residual sIL-6R levels after taking out the effects of all covariates (Supplementary Methods). First, the evidence for linkage was evaluated and next, these results were compared to the results obtained when modeling linkage and association simultaneously (using the –at option to specify the total association model - which is not a TDT test). The test for linkage while simultaneously modeling association involves comparing H0: sIL-6R = μ + βSNP and Variance (sIL-6R) = VE + VG against H1: sIL-6R = μ + βSNP and Variance (sIL-6R) = VE + VG + VA; Where μ=intercept (corrected for covariates), VE=Non-shared environmental variance, VG=Additive polygenic variance, which is estimated from the phenotypic covariance of relatives following the assumption that on average 50% of VG is shared among first degree relatives. VA=Additive major gene effect, which represents the additive effect of linkage to a major gene and is based on the pi-hat measure derived from the IBD matrix of relatives. H0=null-hypothesis. H1=Alternative hypothesis.
eQTL analysis
Inverse quantile normal transformation was applied to the individual probe data to obtain normal distributions. For each SNP-probe combination, a linear mixed model was fitted with expression level as dependent variable, and with fixed effects: genotype, sex, age, body mass index, smoking status, technical covariates (covering e.g. plate and well differences (Jansen et al. 2014)), three principle components (PCs) from the genotype data (Supplementary Methods) and five PCs from the transformed expression data. Random effects included family ID and zygosity to account for family and twin relations (Visscher et al. 2004). Cis-eQTLs are expression-associated SNPs with a distance < 1Mb to the gene, and trans-eQTLs are the complementary set of SNPs. In an initial analysis of genome-wide SNPs, no trans-eQTLs were observed. The cis-eQTL analysis yielded 36 (N probes)*2731(N SNPs) = 98316 tests. To correct for multiple testing, a conservative P value threshold of 0.01/98316 ∼ 1 × 10-7 was applied. The conditional eQTL analysis was performed using the same model and P value threshold on expression data that had been residualized for the effect of rs7512646 in advance. To examine the relationship between sIL-6R level and IL6R expression level, the Pearson correlation between sIL-6R level and expression level was computed for all probes.
Results
The heritability of sIL-6R level in extended twin families
To examine the overall contribution of genetic and environmental influences to the variation in sIL-6R level, genetic structural equation modeling was performed on sIL-6R level data of 4980 subjects from 2360 twin families. This approach allowed us to estimate the variance due to total heritable genetic effects (broad-sense heritability) and additive genetic effects (narrow-sense heritability). Based on the pattern of phenotypic correlations for sIL-6R level among monozygotic (MZ) twins, dizygotic (DZ) twins, siblings, and parent-offspring pairs (Table 1), the broad-sense heritability of sIL-6R level was estimated at 72% (H2=(VA +VD)/Vtotal=(0.89+0.08)/1.35) and the narrow-sense heritability was estimated at 66% (h2= VA/Vtotal=0.89/1.35). The remaining variance (28%) was ascribed to environmental factors not shared among family members (unique environment: e2=VE/Vtotal=0.38/1.35).
Table 1. Familial correlations of sIL-6R level.
| Complete pairs | Correlations, no correction for SNP effectsd | Correlations, correction for rs2228145e | |||
|---|---|---|---|---|---|
|
| |||||
| N | r | 95% CI | r | 95% CI | |
| Monozygotic twins | |||||
| MZ male twins | 208 | 0.80 | (0.7607 - 0.8388) | 0.56 | (0.4530 - 0.6416) |
| MZ female twins | 520 | 0.68 | (0.6421 - 0.7207) | 0.35 | (0.2852 - 0.4248) |
| Male siblings | |||||
| DZ male twins | 93 | 0.40 | (0.2083 - 0.5566) | 0.24 | (0.0258 - 0.4324) |
| Male sibsa | 126 | 0.24 | (0.0882 - 0.3859) | 0.27 | (0.1033 - 0.4129) |
| Female siblings | |||||
| DZ female twins | 216 | 0.37 | (0.2627 - 0.4688) | 0.25 | (0.1445 - 0.3612) |
| Female sibsb | 313 | 0.29 | (0.1784 - 0.4018) | 0.12 | (0.0116 - 0.2250) |
| Opposite-sex siblings | |||||
| DZ opposite-sex twins | 224 | 0.31 | (0.1884 - 0.4282) | 0.15 | (0.0191 - 0.2782) |
| Opposite-sex sibsc | 337 | 0.42 | (0.3276 - 0.5121) | 0.20 | (0.0748 - 0.3250) |
| Parent-offspring | |||||
| Mother-daughter | 342 | 0.33 | (0.2543 - 0.4076) | 0.18 | (0.0975 - 0.2581) |
| Mother-son | 204 | 0.43 | (0.3434 - 0.5174) | 0.29 | (0.1700 - 0.3969) |
| Father-son | 171 | 0.36 | (0.2626 - 0.4447) | 0.20 | (0.0708 - 0.3190) |
| Father-daughter | 314 | 0.31 | (0.2311 - 0.3915) | 0.18 | (0.0928 - 0.2748) |
| Spouses | |||||
| Mother-Father | 374 | 0.11 | (0.0269 - 0.2075) | 0.24 | (0.1563 - 0.3317) |
Non-twin brother-brother pairs and pairs of brother-male twin
Sister-sister and Sister-female twin
Sister-brother, brother-female twin and sister-male twin
Correlation from a saturated model with covariates age and sex
Correlation from a saturated model with covariates age, sex and rs2228145 genotype. CI =Confidence interval. Note that the correlations of sIL-6R levels adjusted for the effect of rs2228145 provide information about the proportions of residual variance due to genetic and environmental effects. The proportion of the total phenotypic variance that is due to genetic effects beyond rs2228145 is reflected in the difference between the correlations with and without adjustment for rs2228145 genotype. MZ= monozygotic, DZ= dizygotic.
Variance explained by chromosome-wide SNPs and SNPs in the IL6R region using GCTA
We next used GCTA on 1000Genomes-imputed SNP data to examine how much of the variance in sIL-6R level can be explained by all SNPs in the IL6R region (IL6R +/- 10MB), how much by other SNPs on chromosome 1 and how much by each of the other chromosomes (Table 2, Supplementary Figure S1) based on the similarity across SNPs among 2875 unrelated subjects. This method gives insight into the total contribution of additive genetic effects tagged by all genotyped and imputed SNPs together, providing an estimate of the total variance that could be identified by GWAS on this set of SNPs, given sufficient power to detect individual SNP effects. SNPs in the IL6R region with a MAF > 0.001 (N SNPs = 42268) together explained 54.7 % (SE= 2.5%) of the variance of sIL-6R levels, while the rest of chromosome 1 did not contribute to the variance. Some additional variance was captured by SNPs on chromosome 2 (5.6%, SE=2.8). When the analysis was repeated with inclusion of related subjects (including siblings, DZ twins and parent-offspring pairs, total N subjects=4846) the estimate for chromosome 2 was somewhat lower (2.8%, SE= 1.6), whereas otherwise highly similar results were obtained (Table 2).
Table 2. Variance of sIL-6R level explained by chromosome-wide SNPs estimated using GCTA.
| Unrelated subjects (N=2875) | Unrelated and Related subjects (N=4846) | ||||
|---|---|---|---|---|---|
|
|
|||||
| Chromosome | N SNPs | VG/VP | SE | VG/VP | SE |
| 1 IL6R region | 42268 | 0.547 | 0.025 | 0.533 | 0.019 |
| 1 Rest | 584291 | 0.003 | 0.025 | 0.000 | 0.015 |
| 2 | 692964 | 0.056 | 0.028 | 0.028 | 0.016 |
| 3 | 590258 | 0.000 | 0.024 | 0.009 | 0.014 |
| 4 | 605730 | 0.000 | 0.023 | 0.002 | 0.013 |
| 5 | 541738 | 0.000 | 0.023 | 0.005 | 0.013 |
| 6 | 530265 | 0.017 | 0.021 | 0.030 | 0.014 |
| 7 | 483988 | 0.000 | 0.021 | 0.009 | 0.013 |
| 8 | 447709 | 0.001 | 0.021 | 0.010 | 0.013 |
| 9 | 350386 | 0.000 | 0.021 | 0.000 | 0.012 |
| 10 | 418731 | 0.010 | 0.021 | 0.017 | 0.013 |
| 11 | 410817 | 0.000 | 0.014 | 0.005 | 0.009 |
| 12 | 393609 | 0.001 | 0.019 | 0.010 | 0.011 |
| 13 | 307137 | 0.003 | 0.017 | 0.007 | 0.011 |
| 14 | 265369 | 0.000 | 0.017 | 0.000 | 0.010 |
| 15 | 227463 | 0.013 | 0.017 | 0.000 | 0.009 |
| 16 | 242058 | 0.006 | 0.018 | 0.002 | 0.010 |
| 17 | 202299 | 0.014 | 0.017 | 0.000 | 0.010 |
| 18 | 234349 | 0.002 | 0.018 | 0.000 | 0.010 |
| 19 | 158942 | 0.003 | 0.014 | 0.001 | 0.008 |
| 20 | 180019 | 0.016 | 0.016 | 0.007 | 0.010 |
| 21 | 109887 | 0.000 | 0.013 | 0.000 | 0.007 |
| 22 | 105190 | 0.003 | 0.012 | 0.010 | 0.008 |
SE=standard error
GWA analysis
GWA analysis of sIL-6R level was conducted on imputed SNP data from 4846 subjects. The genomic inflation factor indicated no effect of population stratification (λ= 1.015). 680 Genome-wide significant hits were found (P< 5×10-8), which were all located on chromosome 1q21.3 (significant hits; chr 1: 153389207-154697624), except for four SNPs (MAF 0.01 – 0.09) on chromosomes 5, 8 and 20 (Supplementary Table S2). The top SNPs (P< 1 × 10-176) were located within IL6R and were all in LD with rs2228145.
Biometrical model
The minor (C) allele of rs2228145 occurred at a frequency of 0.39. To estimate how much of the variance in sIL-6R level is explained by rs2221845, we applied the classic biometrical model to the data. The average sIL-6R level was 5.698 (10-8 g/mL) in individuals homozygous for the minor allele (CC), 4.418 in heterozygotes (AC), and 3.238 in individuals with the AA genotype, giving an overall mean level of 4.17. The observed variance of sIL-6R levels in the total sample was 1.35. Applying the biometrical model to these data, it follows that:
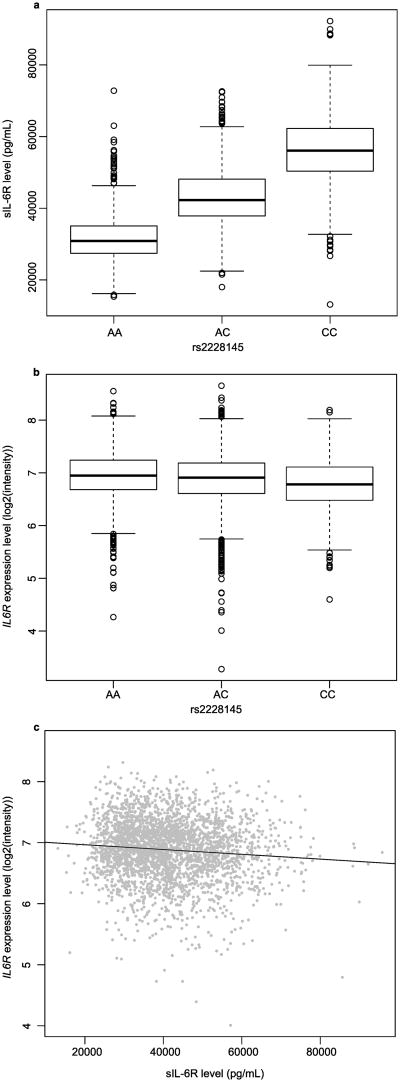
Thus, the allelic effect was almost completely additive, which illustrates that the average sIL-6R level of heterozygous individuals lies perfectly in the middle of the levels of the two homozygous groups (Figure 1A).
Figure 1. Associations between sIL-6R level, IL6R expression and rs2228145 genotype.
A: Boxplots of plasma sIL-6R level (pg/mL) for each rs2228145 genotype. B: Boxplots of IL6R expression level in blood as measured by expression probe 582_132 for each rs2228145 genotype (P = 3.14 × 10-19). C: Scatterplot of sIL-6R level (pg/mL) versus IL6R expression level for probe 582_132 (r = -0.092, P = 1.42 × 10-6).
Heritability explained by rs2228145
The data from twin families allowed us to examine how much of the heritability is explained by rs2228145, and how much genetic variance is left after adjusting sIL-6R levels for rs2228145. The contribution of rs2228145 to the heritability of sIL-6R level is illustrated by the drop of the correlations of sIL-6R level among family members after correcting sIL-6R level for rs2228145 genotype (Table 1). When the allelic effect of rs2228145 on sIL-6R level was taken into account in the twin family model (Table 3), the total variance of sIL-6R level dropped from 1.35 to 0.66 (difference= 0.69), illustrating that this SNP accounted for 51% of the total variance in sIL-6R level (H2SNP=0.69/1.35=0.51). Residual genetic effects not tagged by rs2228145 (VG-residual=0.26) accounted for 19% of the total variance of sIL-6R (H2residual=0.26/1.35=0.19).
Table 3. Variance of sIL-6R level due to rs2228145, total heritability of sIL-6R level, and estimates from linkage analysis.
| Observed variance due to rs2228145 genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Genotype | Frequencya | sIL-6R level (10-8 g/mL)b | Genotypic Value | Frequency* sIL-6R | (deviation from μ)2 | Frequency*squared deviation | ||||||
| CC | p2= 0.15 | 5.698 | + a | 0.15*5.698 = 0.87 | (5.698- μ)2 = 2.32 | 0.15*2.32 = 0.35 | ||||||
| AC | 2pq=0.48 | 4.418 | d | 0.48*4.418 = 2.10 | (4.418- μ)2 = 0.06 | 0.48*0.06 = 0.03 | ||||||
| AA | q2= 0.37 | 3.238 | - a | 0.37*3.238 = 1.20 | (3.238- μ)2 = 0.88 | 0.37*0.88 = 0.33 | ||||||
|
| ||||||||||||
| Total | μ=0.87+2.10+1.20 = 4.17 | VSNP=0.35+0.03+0.33 = 0.71 | ||||||||||
| (95% CI: 0.68-0.74) | ||||||||||||
|
| ||||||||||||
| Extended twin family model estimates | ||||||||||||
|
| ||||||||||||
| VsIL-6R | VA | VD | VC | VE | V/Vtotal | h2 | d2 | c2 | e2 | |||
|
| ||||||||||||
| ACDE modelc | Vtotal= 1.35 | 0.89 | 0.08 | 0.00 | 0.38 | 1.00 | 0.66 | 0.06 | 0.00 | 0.28 | ||
| ACDE + rs2228145d | Vresidual= 0.66 | 0.24 | 0.02 | 0.01 | 0.39 | Vresidual/Vtotal= 0.49 | 0.18 | 0.01 | 0.01 | 0.29 | ||
|
| ||||||||||||
| Effect of rs2228145e | VSNP= 0.69 | 0.65 | 0.06 | -0.01 | -0.01 | VSNP/Vtotal= 0.51 | 0.48 | 0.05 | 0.01 | 0.01 | ||
|
| ||||||||||||
| Linkage analysis estimates | ||||||||||||
|
| ||||||||||||
| VsIL-6R | VA-linkage | VA-polygenic | VD | VC | VE | V/Vtotal | h2 | d2 | c2 | e2 | ||
|
| ||||||||||||
| Linkage modeled at rs2228145 | Vtotal=1.39 | 0.96 | 0 | - | - | 0.43 | 1 | 0.69 | - | - | 0.31 | |
| Linkage+ association modeled at rs2228145 | Vresidual=0.67 | 0.26 | 0 | - | - | 0.41 | Vresidual/Vtotal =0.48 | 0.19 | - | - | 0.30 | |
p=minor allele frequency=0.39 and q=major allele frequency=0.61
Mean sIL-6R level, corrected for age and sex, for each genotype group.
μ= Mean sIL-6R level in the population, estimated from the genotype frequencies and corresponding sIL-6R levels for each genotype group.
ACDE model without SNP effects, in which Vtotal is decomposed into VA, VD, VC, and VE.
Model in which the additive effect of rs2228145 on sIL-6R level is modeled, and the residual variance of sIL-6R (Vresidual=Vtotal - VSNP) is decomposed into VA, VC, VD and VE.
The effect of rs2228145 was inferred from the difference between model c and model d Vtotal=Total (phenotypic) variance of age- and sex-adjusted sIL-6R levels. VSNP= Variance of sIL-6R level attributable to rs2228145, VSNP/Vtotal = Proportion of total (phenotypic) variance sIL-6R level explained by rs2228145, Vresidual/Vtotal= Proportion of total (phenotypic) variance sIL-6R level not explained by rs2228145, VA=Additive genetic variance, VD=Non-additive genetic variance, VC=Sibling-shared environmental variance, VE=Unique environmental variance, h2= VA/Vtotal, d2= VD/Vtotal, c2= VC/Vtotal, e2= VE/Vtotal.
VA-linkage =Variance due to linkage, which is based on the pi-hat measure at rs2228145, derived from the IBD matrix of relatives
VA-polygenic = Variance due to all additive genetic effects that are not captured within the linkage component (i.e. additive genetic variance that is not linked to the IL6R locus), which was estimated from the phenotypic covariance of relatives following the assumption that on average 50% of VA-polygenic is shared among first degree relatives.
The contribution of other additive SNP effects in the IL6R region
When we repeated the analysis of unrelated subjects in GCTA while correcting for the effect of rs2228145, SNPs in the IL6R region on chromosome 1 still explained 5.6% of the total variance of sIL-6R level (SE=1.8%, P = 6.0 × 10-5). This suggests that part of the residual heritability that was estimated based on twin family analysis can be explained by other variation in the IL6R region that is tagged by genotyped and imputed SNPs.
Combined linkage and association analysis
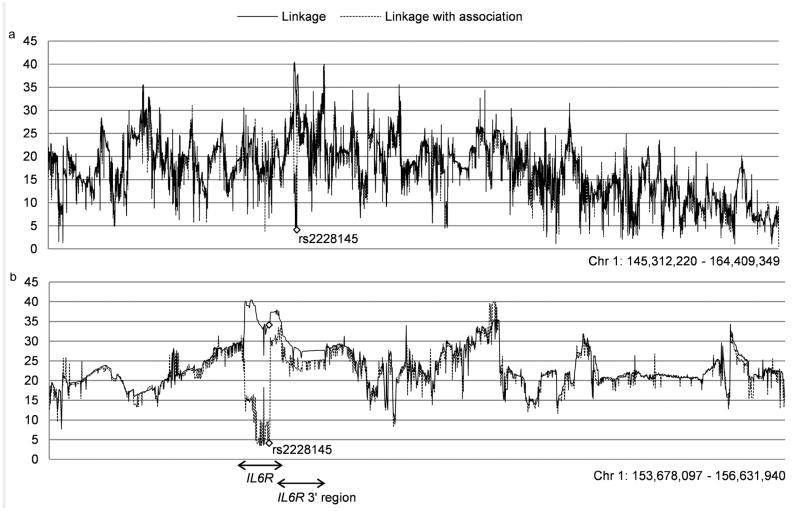
In the GWA analysis of sIL-6R levels, highly significant hits were found across the IL6R region, which may include SNPs that merely capture the effect of rs2228145 (through LD) and SNPs tagging other causal variants. To search for genetic variants that explain additional variation in sIL-6R levels beyond the effect of rs2228145, we simultaneously modeled linkage and association using data from 355 nuclear families (total N subjects = 1254) for SNPs in the IL6R region (IL6R +/- 10MB, Figure 2a). This region covers all genome-wide significant GWA hits on chromosome 1. If the linkage test is not significant while simultaneously modeling association for a SNP (while the linkage test is significant when association is not modeled simultaneously), this indicates that the respective SNP is in high LD with a causal variant. On the other hand, the linkage signal will not be fully impaired when association is modeled for SNPs that are in lower LD with a causal variant, or when multiple causal variants in partial LD contribute to variation in sIL-6R levels.
Figure 2. Results from combined linkage and association analysis of the IL6R region on chromosome 1.
The solid black line connects the X2 values from the linkage test at each SNP without modeling SNP association and the dotted black line connects the X2 values from the linkage test at each SNP when association is simultaneously modeled. The analysis was performed for all imputed SNPs with a MAF > 0.2 in IL6R +/-10MB, chr1 (build 37): 145312220-164409349 (x-axis, Figure A). Figure B zooms in to the region from basepair position 153678097 to 156631940. The arrows mark the location of the IL6R gene, and the location of an LD block stretching across the 3′UTR of IL6R and adjacent genes SHE and TDRD10, where significant hits were found in the eQTL analysis for SNPs in partial LD with rs2228145 (r2 ∼0.1).
In a model that included linkage (VA, additive variance attributable to the locus estimated using IBD), additive polygenic variance (VG, estimated from the familial resemblance in sIL-6R following the assumption that on average 50% of VG is shared among first degree relatives) and unique environment (VE), but not incorporating association, the linkage signal around rs2228145 explained 69% (VA/Vtotal) of the total variation in sIL-6R levels (X2=34.12, df=1, P = 5 × 10-9) and VG was estimated at 0 (Table 3). Comparison of the estimate of variance explained by linkage at IL6R (69%) to the broad-sense heritability (72%) and narrow-sense heritability (66%) estimated by the twin family model suggests that the entire narrow-sense heritability of sIL-6R level and nearly the entire broad-sense heritability of sIL6-R level can be captured by modeling the covariance of sIL6-R level among relatives as a function of IBD-sharing at the IL6R locus.
When the allelic effects of SNPs on the means were incorporated in the model (i.e., association was added), 19% of the total variance in sIL-6R level (VA/Vtotal) was still explained by the linkage component when modeling association at rs2228145 (similar to the estimate of residual heritability from the twin family model), although this signal was borderline significant only (X2=4.13, df=1, P = 0.042). Significant linkage was initially observed across the entire region, but when association was modeled, linkage only dropped to the level of borderline significance for SNPs in high LD with rs2228145 (r2 > 0.93 among 21 SNPs with P > 0.01) indicating that rs2228145 explains most, though not the entire linkage signal. The linkage signal was also attenuated when modeling association for SNPs within an LD block at the 3′ end of IL6R containing the IL6R 3′UTR and the adjacent genes SHE and TDRD10 (location indicated in Figure 2b). This region contains both SNPs in low LD (r2 ∼0.1) with rs2228145 and SNPs in high LD with rs2228145. When we conducted a conditional GWAS to test for association between chromosome 1 SNPs and sIL-6R levels after taking out the effect of rs2228145 (by including this SNP as a covariate in the model), significant associations were still observed for SNPs within IL6R including SNPs at the 3′end (Supplementary Table S3). These results support the presence of additional causal variants influencing sIL-6R level in the IL6R gene and 3′end.
eQTL analysis
To identify genetic variants related to IL6R RNA expression levels, eQTL analysis was performed on data from 4467 subjects for 36 probes (Supplementary Table S4) measuring IL6R RNA. A total of 341 significant associations (Supplementary Table S5) were found for 5 of these probes (4 probes targeting exon 7 and 1 probe targeting the 3′end). These associations involved 179 SNPs that were all in cis (SNP positions; chr1q21.3: 154395125-154521584). The most significant association was between rs7512646 and expression probe 582_132 targeting exon 7 (P = 2.84 × 10-22). This SNP (intronic) is in high LD with rs2228145 (r2 =0.94) and rs2228145 itself showed significant associations with all four probes targeting exon 7. Probe 308_15 targeting the 3′ end was the only probe outside exon 7 with significant eQTLs (total N associated SNPs= 92, top hit= rs4072391 located in the IL6R 3′UTR, P = 1.32 × 10-11, r2 with rs2228145=0.142). Of the 680 significant hits identified in the GWAS of sIL-6R, 157 SNPs were significantly associated with IL6R expression level. For all of these SNPs, the allele associated with higher sIL-6R levels was associated with lower IL6R expression level.
Relationship between rs2228145, IL6R RNA and sIL-6R levels
As a measure of the relationship between plasma sIL-6R levels and the abundance of IL6R RNA in whole blood, the correlation between sIL-6R level and expression level was computed for all probes targeting IL6R transcripts (Supplementary Table S6). All five expression probes with significant eQTL hits showed small but statistically significant negative correlations with sIL-6R level (e.g. probe 582_132: r = -0.092, P =1.42 × 10-6, Table 4). The negative correlations indicate that across individuals, higher sIL-6R level was associated with lower overall IL6R expression (Figure 1c). Correlations between IL6R RNA and sIL-6R levels became non-significant when expression levels were corrected for the effect of rs2228145 (e.g., probe 582_132: r = 0.019, P =0.314, Table 4), which indicates that at the population level, the relationship between IL6R expression and sIL-6R level can be explained by the effect of rs2228145. Indeed, the minor C allele of rs2228145 was associated with higher sIL-6R level and with lower IL6R expression level (Figure 1). When computed separately within rs2228145 genotype classes, correlations between IL6R expression and sIL-6R levels were not significant (Table 4).
Table 4. Correlation between sIL-6R level and IL6R expression level for all probes with significant eQTL hits.
| IL6R probe | Probe location | Overall correlation (N=2727) | Correction for rs2228145 (N=2727) | Genotype AA (N=991) | Genotype AC (N=1278) | Genotype CC (N=458) | |
|---|---|---|---|---|---|---|---|
| 323_134 | Exon 7 | r | -0.086 | 0.001 | 0.032 | 0.015 | -0.023 |
| P value | 7.48 × 10-6 | 0.955 | 0.312 | 0.601 | 0.618 | ||
| 201_497 | Exon 7 | r | -.054 | 0.011 | 0.047 | 0.009 | 0.002 |
| P value | 0.005 | 0.577 | 0.136 | 0.755 | 0.974 | ||
| 582_132 | Exon 7 | r | -0.092 | 0.019 | 0.024 | 0.005 | -0.063 |
| P value | 1.42 × 10-6 | 0.314 | 0.449 | 0.857 | 0.178 | ||
| 202_497 | Exon 7 | r | -0.071 | 0.005 | 0.072 | -0.016 | -0.017 |
| P value | 1.94 × 10-4 | 0.791 | 0.023 | 0.569 | 0.724 | ||
| 308_15 | 3′-end | r | -0.055 | -0.003 | 0.031 | -0.043 | -0.088 |
| P value | 0.004 | 0.894 | 0.335 | 0.124 | 0.059 |
r= Pearson correlation between sIL-6R level (corrected for age and sex) and expression level (corrected for technical covariates, age and sex). Colum 5 only is based on expression levels that were additionally corrected for the additive allelic effect of rs2228145. Columns 6 to 8 show the correlations between sIL-6R level and expression level computed separately within rs2228145 genotype classes. Correlations for all IL6R expression probes are given Supplementary Table S6.
Taken our eQTL design, we could not distinguish between alternative IL6R transcripts but observed a significant negative association between the rs2228145 C allele and total levels of IL6R RNA. In another eQTL analysis based on expression in peripheral blood (N=1469; Fehrmann et al. 2011), one of the probesets targeted IL6R exon 9, which is spliced out from the alternative RNA transcript that is presumed to directly code for sIL-6R. For this probeset, a negative relation between the minor allele of rs4845623 (r2 with rs2228145=0.93) and expression level was observed (P< 1 × 10-7). We also downloaded the exon-specific expression data from HapMap lymphoblastoid cell lines (GEO accession nr = GSE9703, N=162) and corresponding genotype data (Zhang et al. 2009). In this dataset, no significant association between rs2228145 and exon 9 expression level was found (possibly due to the small sample size). When we computed the ratio of exon 9 expression level over mean IL6R transcript expression, we found that the proportion of IL6R transcripts containing exon 9 decreased with each copy of the rs2228145 C allele (P < 0.01, Supplementary Figure S2), suggesting that rs2228145 is associated with the ratio of normal and alternatively spliced transcripts. Note that if the rs2228145 C allele has differential effects on alternative RNA transcripts, a negative association with total RNA levels (as assessed in our own study) will emerge if the increasing effect on levels of the alternative splice variant (Ferreira et al. 2013; Stephens et al. 2012) is smaller compared to the decreasing effect on other transcripts.
Conditional eQTL analysis
When the eQTL analysis was conducted after adjusting expression levels for the effect of the most significant eQTL SNP from the initial analysis (rs7512646), significant associations (Supplementary Table S7) were observed between probe 308_15 (targeting the 3′ end of IL6R transcripts) and 80 SNPs located in an LD block covering the 3′UTR of IL6R and the adjacent 3′region. This region was also highlighted by the analysis of linkage and association of sIL-6R levels (location indicated in Figure 2b). The most significantly associated conditional eQTL SNPs with a P value of 1.89 × 10-9 were rs60760897, rs60255122, rs61698846 and rs61275241 (Located in TDRD10, r2 with rs2228145=0.133, MAF=0.21), followed by rs4072391 (P = 2.10 × 10-9, located in the IL6R 3′UTR, r2 with rs2228145=0.142; these SNPs are in high LD with each other and for each of the SNPs, the minor allele was associated with higher expression level and with lower sIL-6R level (P value (GWAS) for rs60760897= 1.49 × 10-121, and P value (conditional GWAS) for rs60760897= 8.04 × 10-4). All the initially observed associations for exon 7 expression probes were no longer significant after correcting for the most significant eQTL, suggesting that genetic variation tagged by rs2228145and variation tagged by rs60760897 (IL6R 3′region) are both independently associated with IL6R expression level.
Discussion
Based on the analysis of three different types of familial relations in extended twin families (monozygotic twins, sibling pairs/dizygotic twins, and parent-offspring pairs), the broad heritability of sIL-6R level was estimated at 72% and the narrow-sense heritability was estimated at 66%. Linkage analysis closely recaptured this, with 69% of the variance in sIL-6R levels explained by all variation in the IL6R region that is captured by IBD. Both estimates are similar to the heritability of sIL-6R levels reported by a previous study conducted in middle-aged male twins (h2=0.68) (Raggi et al. 2010). The estimate of variance explained by additive SNP effects in the IL6R region in unrelated subjects (GCTA) was slightly lower at 54.7%, which suggests that the heritability of sIL-6R cannot be fully ascribed to additive effects of currently genotyped and imputed SNPs. Whereas the variance explained by all SNPs in unrelated subjects (in which LD extends over relatively short distances) specifically provide information about effects tagged by a given set of SNPs under an additive allelic model, linkage and twin analyses, which are based on IBD-sharing among close relatives, capture more variation contributing to the similarity of family members. Estimates from linkage may include variation in a region that is not (fully) captured by additive SNP effects, such as rare variants segregating in families, structural variation and effects of interacting loci (epistasis). Overall, the three different methods show the expected convergence.
After establishing the heritability, we examined the contribution of the functional IL6R polymorphism rs2228145, a known candidate SNP for sIL-6R (Galicia et al. 2004; Lourdusamy et al. 2012; Melzer et al. 2008; Rafiq et al. 2007; Reich et al. 2007), to the population variance in plasma sIL-6R levels. Using the classic biometrical model and twin family analysis, we showed that rs2228145 explained 51% of the total variance of sIL-6R level, corresponding to 71% of the broad heritability (51/72=0.71). The estimate of variance explained by this SNP in our sample is comparable to an estimate previously reported for subjects of European descent (49% explained by rs2228145 (Reich et al. 2007)), and is larger compared to several other previous reports (33% explained by rs2228145 in African Americans (Reich et al. 2007), 29% explained by rs2228145 (Ferreira et al. 2013), 30% explained by rs4129267; a SNP in LD (Revez et al. 2013), and 20% explained by rs4537545; another SNP in LD (Rafiq et al. 2007)). The variation between studies might be related to differences in the lab protocol (e.g. differences related to sIL-6R detection assay and dilution of samples) and to differences between study populations. Whereas our study and the studies by Reich et al (2007) and Rafiq et al (2007) were conducted on population-based cohorts, the studies by Ferreira et al (2013) and Revez et al (2013) included patient populations. In contrast to all other studies, we excluded individuals using anti-inflammatory medication, which could have led to a slightly healthier population compared to previous studies, and it could be hypothesized that the variance explained by rs2228145 may vary with health status, as the levels of sIL-6R may rise 2-3 fold within a person during inflammation (Scheller et al. 2013). Although the allele frequencies of rs2228145 differ between individuals of European versus African descent, variation related to ancestry only seems explain the different estimates observed in the study by Reich et al (2007), as all other studies focused on individuals of European descent and reported similar allele frequencies for rs2228145.
Functional studies have indicated that amino-acid mutations at the position encoded by rs2228145 can influence the production of sIL-6R through shedding of membrane-bound receptors (Müllberg et al. 1994). The effects of rs2228145 and other SNPs on IL6R expression are less well characterized. To gain insight into the regulatory impact of genetic variation on IL6R expression, we studied the relation between IL6R expression level and genome-wide SNPs. We found two clusters of SNPs within the IL6R region that were associated with expression level, one of which included rs2228145, suggesting that this SNP also influences IL6R expression, though the signal may also come from another causal variant in LD. A previous analysis of gene expression in multiple samples and tissues revealed no significant association between rs2228145 genotype and overall expression of IL6R RNA transcripts (possibly due to limited sample size; IL6R Genetics Consortium and Emerging Risk Factors Collaboration. 2012). Two other studies found that the rs2228145 C allele was associated with higher levels of an alternative IL6R mRNA splice variant (Ferreira et al. 2013; Stephens et al. 2012), which lacks a 94-bp sequence encoding part of the trans-membrane domain that anchors the membrane-bound IL-6R to the cell membrane and is presumed to directly code for the sIL-6R. In our study and in the study by Revez et al (2013), the rs2228145 C allele was associated with lower overall IL6R RNA level and our analysis of the HAPMAP expression data suggested that rs2228145 (or a variant in LD) has an effect on the relative abundance of alternative IL6R RNA transcripts.
Though the opposite effects of rs2228145 on sIL-6R levels and overall IL6R expression level may appear contradictory, this finding is not unexpected given that rs2228145 has a strong effect on alternative splicing. As the C allele is associated with increased splicing of exon 9 (Stephens et al. 2012), this allele is likely to decrease the level of the full-length RNA transcript. It can also be hypothesized that rs2228145 may have secondary effects on IL6R expression by impacting on feedback systems that control IL6R expression. In hepatocytes, a positive feedback circuit has been identified through which increased membrane-bound receptor-mediated classic IL-6 signaling triggers microRNA (miRNA)-mediated regulatory pathways that stimulate increased IL6R expression (Hatziapostolou et al. 2011). The rs2228145 C allele has been shown to impair classical IL-6 signaling, as indicated by reduced levels of mIL-6R and decreased IL-6 responsiveness of CD4+ T cells and monocytes from C allele carriers (Ferreira et al. 2013). As the rs2228145 C allele weakens classical IL-6 signaling, positive feedback on IL6R expression may be reduced in individuals with the C allele, leading to lower RNA expression levels compared to individuals with the A allele. The associations between SNPs and variation in RNA and soluble IL-6 receptor abundance observed in our study provide guidance for functional studies into the mechanistic relationships among polymorphisms in IL6R, IL6R RNA expression and IL-6 receptor levels.
While the variance in sIL-6R levels that can be explained by rs2228145 is very large compared to single SNP effect sizes generally observed for quantitative traits, our study showed that other genetic effects also make an important contribution to the heritability of sIL-6R levels. Linkage analysis and analysis of the variance explained by chromosome wide SNP effects in unrelated subjects (GCTA) indicated that the remaining heritability of sIL-6R level appears to be primarily accounted for by other variation in the IL6R region on chromosome 1. Linkage analysis showed that genetic effects in this region not tagged by rs2228145 account for 19% of the total variance in sIL-6R levels. We therefore tried to identify other SNPs in the IL6R region that explain additional variance in this clinically important soluble cytokine receptor.
Evidence for association with sIL-6R level and IL6R expression was seen for SNPs located in the IL6R 3′ region including the IL6R 3′UTR, which is an important region of regulatory control. Genetic variation in the 3′UTRs of genes can affect transcript levels in several ways (Kwan et al. 2008), including effects on mRNA stability, translation efficiency, and by affecting regulatory control by miRNAs. Previous studies have shown that the IL6R 3′UTR contains binding sites for several miRNAs and that IL6R transcript levels are (down)regulated by binding of these miRNAs to their 3′UTR target in IL6R mRNA (Gong et al. 2012; Hatziapostolou et al. 2011; Jia et al. 2012; Zhu et al. 2010). Alterations in components of this regulatory pathway have been reported in cancer tissues (Gong et al. 2012; Hatziapostolou et al. 2011; Jia et al. 2012; Zhu et al. 2010) and in synovial fibroblasts from patients with rheumatoid arthritis (de la Rica et al. 2013). Our novel finding that SNPs in the IL6R 3′ region are associated with IL6R expression level generates novel hypotheses about the role of genetic variation in regulatory pathways of IL6R expression, and into the contribution of such pathways to complex disease susceptibility.
SNP rs4129267 (r2 with rs2228145=0.97) has been identified as a risk variant for asthma; the minor allele that increases sIL-6R level is associated with increased asthma risk (OR 1.09; Ferreira et al. 2011). We looked at the effect of the top 3′ eQTL hits for IL6R (rs60760897 and SNPs in LD (r2 >= 0.3)) on asthma in data from the Australian Asthma Genetics Consortium (Ferreira et al. 2011) (AAGC, N=2110 cases, N=3857 controls) and the GABRIEL consortium (Moffatt et al. 2010) (N=10,365 cases, N=16,110 controls). rs60760897 was not associated with asthma risk (Tables S9 and S10) but a suggestive association signal was found for SNPs in modest LD (r2=0.46) with rs60760897 (rs4478801; GABRIEL: OR=0.95, P =0.014 and AAGC: OR=0.94, P=0.045). The minor G allele of rs448801 that showed a trend of lower asthma risk in the AAGC and GABRIEL study was associated with lower sIL-6R level in our study (GWAS P = 1.27 × 10-145) and with higher IL6R expression (eQTLP = 4.87 × 10-8).
Compared to previous reports that common SNPs together generally explain less than 50% of the total variation of complex traits (Yang et al. 2011b), it seems remarkable that a single common variant in the IL6R gene alone accounts for such a large part of the variation in sIL-6R levels. What makes the level of circulating sIL-6R different from other quantitative traits such as height and BMI? An important factor is that sIL-6R can be produced through two mechanisms (receptor cleavage/shedding and translation of an alternatively spliced mRNA), and that the IL6R SNP rs2228145 has a major impact on both mechanisms. In contrast, it seems likely, although this is actually unknown, that many complex traits result from the integration of numerous different processes that each make a small contribution to the endpoint, in which case there could be many genetic variants in different pathways that each contribute to a small portion of the total variation in the trait.
In conclusion, we have shown that sIL-6R levels are highly heritable and that variants in IL6R other than the well-known functional SNP make an important contribution to the heritability. Our findings shed novel light on the effects of rs2228145 and SNPs at the IL6R 3′end on IL6R expression. At the same time, we demonstrated that results from different methods based on the classic biometrical model are in agreement and converge to the same conclusion.
Supplementary Material
Figure ∷ S1 Variance of sIL-6R level explained by genome-wide SNPs in unrelated individuals
Figure ∷ S2 Association between rs2228145 genotype and the ratio of IL6R exon 9 expression over mean IL6R expression
Table S1: Characteristics of subjects included in the analyses
Table S2: GWA results and information for all 680 SNPs reaching genome-wide significance in the GWAS of sIL-6R level
Table S3: SNP information and Conditional GWAS results of all 293 SNPs reaching genome wide significance in the GWAS of sIL-6R level after correction for the effect of rs2228145
Table S4: Location (Build 37/hg19) and sequence of expression probes targeting IL6R transcripts (Affymetrix U219 array)
Table S5: SNP information and eQTL P values for all 341 significant eQTL associations
Table S6: Correlations between sIL-6R level and IL6R expression level for all expression probes
Table S7: SNP information and conditional eQTL P values for all 80 significant eQTL associations after correction for the effect of rs2228145
Table S8: Results for the association with asthma for rs60760897 and SNPs in LD in the Australian Asthma Genetics consortium
Table S9: Results for the association with asthma for SNPs in LD with rs60760897 in the GABRIEL consortium
Supplementary Methods: Additional Supplementary Methods.
Acknowledgments
This work was supported by: Genotype/phenotype database for behavior genetic and genetic epidemiological studies (ZonMW Middelgroot [911-09-032]); Genetics of Mental Illness: A lifespan approach to the genetics of childhood and adult neuropsychiatric disorders and comorbid conditions [ERC-230374]; multiple grants for genotyping and expression data, funded by Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL) [184.021.007]; the National Instiute for Mental Health (NIMH) [1RC2 MH089951-01: Integration of Genomics &Transcriptomics in Normal Twins & Major Depression, and 1RC2MH089995-01: Genomics of Developmental Trajectories in Twins]; and Nederlandse organisatie voor Wetenschappelijk Onderzoek (NWO) [NWO/SPI 56-464-14192].
Footnotes
AAGC – Australian Asthma Genetics Consortium. For a full list of authors, see Supplementary material.
Conflict of interest: All authors declare that they have no conflict of interest.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77(5):754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya R, Mudter J, Finotto S, Mullberg J, Jostock T, Wirtz S, Schutz M, Bartsch B, Holtmann M, Becker C, Strand D, Czaja J, Schlaak JF, Lehr HA, Autschbach F, Schurmann G, Nishimoto N, Yoshizaki K, Ito H, Kishimoto T, Galle PR, Rose-John S, Neurath MF. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat Med. 2000;6(5):583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, Nikolaev A, Burg J, Strand S, Kiesslich R, Huber S, Ito H, Nishimoto N, Yoshizaki K, Kishimoto T, Galle PR, Blessing M, Rose-John S, Neurath MF. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21(4):491–501. doi: 10.1016/j.immuni.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, Posthuma D, van Beijsterveldt TC, Hudziak JJ, Bartels M, Willemsen G. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9(6):849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, Kluft C, Smit G, Nolen WA, Zitman FG, Smit JH, Hoogendijk WJ, van DR, de Geus EJ, Penninx BW. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16(3):335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- de la Rica L, Urquiza JM, Gomez-Cabrero D, Islam AB, Lopez-Bigas N, Tegner J, Toes RE, Ballestar E. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J Autoimmun. 2013;41:6–16. doi: 10.1016/j.jaut.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Dobson A. An introduction to generalized linear models. Chapman & Hall/CRC; London: 2002. [Google Scholar]
- Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ, Haddad e, Lehr HA, Schmitt E, Bopp T, Kallen KJ, Herz U, Schmitt S, Luft C, Hecht O, Hohlfeld JM, Ito H, Nishimoto N, Yoshizaki K, Kishimoto T, Rose-John S, Renz H, Neurath MF, Galle PR, Finotto S. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. J Clin Invest. 2005;115(2):313–325. doi: 10.1172/JCI22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, Zhernakova A, Stahl E, Viatte S, McAllister K, Amos CI, Padyukov L, Toes RE, Huizinga TW, Wijmenga C, Trynka G, Franke L, Westra HJ, Alfredsson L, Hu X, Sandor C, de Bakker PI, Davila S, Khor CC, Heng KK, Andrews R, Edkins S, Hunt SE, Langford C, Symmons D, Concannon P, Onengut-Gumuscu S, Rich SS, Deloukas P, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Arlsetig L, Martin J, Rantapaa-Dahlqvist S, Plenge RM, Raychaudhuri S, Klareskog L, Gregersen PK, Worthington J. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS. Introduction to quantitative genetics. Ronald Press Co; New York: 1960. [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, Weersma RK, Hofstra RM, Buurman WA, Rensen S, Wolfs MG, Platteel M, Zhernakova A, Elbers CC, Festen EM, Trynka G, Hofker MH, Saris CG, Ophoff RA, van den Berg LH, van Heel DA, Wijmenga C, Te Meerman GJ, Franke L. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7(8):e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le SP, Danoy P, Baltic S, Nyholt DR, Jenkins M, Hayden C, Willemsen G, Ang W, Kuokkanen M, Beilby J, Cheah F, de Geus EJ, Ramasamy A, Vedantam S, Salomaa V, Madden PA, Heath AC, Hopper JL, Visscher PM, Musk B, Leeder SR, Jarvelin MR, Pennell C, Boomsma DI, Hirschhorn JN, Walters H, Martin NG, James A, Jones G, Abramson MJ, Robertson CF, Dharmage SC, Brown MA, Montgomery GW, Thompson PJ. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RC, Freitag DF, Cutler AJ, Howson JM, Rainbow DB, Smyth DJ, Kaptoge S, Clarke P, Boreham C, Coulson RM, Pekalski ML, Chen WM, Onengut-Gumuscu S, Rich SS, Butterworth AS, Malarstig A, Danesh J, Todd JA. Functional IL6R 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases. PLoS Genet. 2013;9(4):e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64(1):259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004;5(6):513–516. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, Zhuang SM. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene. 2012 doi: 10.1038/onc.2012.318. [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147(6):1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S, Koyanagi Y, Zhou Y, Miyamoto H, Tanaka Y, Waki M, Matsumoto A, Yamamoto M, Yamamoto N. Soluble interleukin-6 receptors released from T cell or granulocyte/macrophage cell lines and human peripheral blood mononuclear cells are generated through an alternative splicing mechanism. Eur J Immunol. 1994;24(8):1945–1948. doi: 10.1002/eji.1830240837. [DOI] [PubMed] [Google Scholar]
- Hurst SM, Wilkinson TS, McLoughlin RM, Jones S, Horiuchi S, Yamamoto N, Rose-John S, Fuller GM, Topley N, Jones SA. Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001;14(6):705–714. doi: 10.1016/s1074-7613(01)00151-0. [DOI] [PubMed] [Google Scholar]
- IL6R Genetics Consortium and Emerging Risk Factors Collaboration. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet. 2012;379(9822):1205–1213. doi: 10.1016/S0140-6736(11)61931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Batista S, Brooks AI, Tischfield JA, Willemsen G, van GG, Hottenga JJ, Milaneschi Y, Mbarek H, Madar V, Peyrot W, Vink JM, Verweij CL, de Geus EJ, Smit JH, Wright FA, Sullivan PF, Boomsma DI, Penninx BW. Sex differences in the human peripheral blood transcriptome. BMC Genomics. 2014;15(1):33. doi: 10.1186/1471-2164-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HY, Wang YX, Yan WT, Li HY, Tian YZ, Wang SM, Zhao HL. MicroRNA-125b Functions as a Tumor Suppressor in Hepatocellular Carcinoma Cells. Int J Mol Sci. 2012;13(7):8762–8774. doi: 10.3390/ijms13078762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;15(1):43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- Kotake S, Sato K, Kim KJ, Takahashi N, Udagawa N, Nakamura I, Yamaguchi A, Kishimoto T, Suda T, Kashiwazaki S. Interleukin-6 and soluble interleukin-6 receptors in the synovial fluids from rheumatoid arthritis patients are responsible for osteoclast-like cell formation. J Bone Miner Res. 1996;11(1):88–95. doi: 10.1002/jbmr.5650110113. [DOI] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40(2):225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- Lourdusamy A, Newhouse S, Lunnon K, Proitsi P, Powell J, Hodges A, Nelson SK, Stewart A, Williams S, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Lovestone S, Dobson R. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 2012;21(16):3719–3726. doi: 10.1093/hmg/dds186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lust JA, Donovan KA, Kline MP, Greipp PR, Kyle RA, Maihle NJ. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine. 1992;4(2):96–100. doi: 10.1016/1043-4666(92)90043-q. [DOI] [PubMed] [Google Scholar]
- Mackiewicz A, Schooltink H, Heinrich PC, Rose-John S. Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J Immunol. 1992;149(6):2021–2027. [PubMed] [Google Scholar]
- McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 2004;60(2):359–367. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, Rafiq S, Simon-Sanchez J, Lango H, Scholz S, Weedon MN, Arepalli S, Rice N, Washecka N, Hurst A, Britton A, Henley W, van de LJ, Li R, Newman AB, Tranah G, Harris T, Panicker V, Dayan C, Bennett A, McCarthy MI, Ruokonen A, Jarvelin MR, Guralnik J, Bandinelli S, Frayling TM, Singleton A, Ferrucci L. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von ME, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllberg J, Oberthur W, Lottspeich F, Mehl E, Dittrich E, Graeve L, Heinrich PC, Rose-John S. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994;152(10):4958–4968. [PubMed] [Google Scholar]
- Müllberg J, Schooltink H, Stoyan T, Gunther M, Graeve L, Buse G, Mackiewicz A, Heinrich PC, Rose-John S. The soluble interleukin-6 receptor is generated by shedding. Eur J Immunol. 1993;23(2):473–480. doi: 10.1002/eji.1830230226. [DOI] [PubMed] [Google Scholar]
- Muller-Newen G, Kohne C, Keul R, Hemmann U, Muller-Esterl W, Wijdenes J, Brakenhoff JP, Hart MH, Heinrich PC. Purification and characterization of the soluble interleukin-6 receptor from human plasma and identification of an isoform generated through alternative splicing. Eur J Biochem. 1996;236(3):837–842. doi: 10.1111/j.1432-1033.1996.00837.x. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical Modeling. Department of Psychiatry Virginia Commonwealth University; Richmond, VA: 2006. [Google Scholar]
- Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, Cuijpers P, De Jong PJ, Van Marwijk HW, Assendelft WJ, Van Der MK, Verhaak P, Wensing M, De GR, Hoogendijk WJ, Ormel J, van DR. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17(3):121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq S, Frayling TM, Murray A, Hurst A, Stevens K, Weedon MN, Henley W, Ferrucci L, Bandinelli S, Corsi AM, Guralnik JM, Melzer D. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun. 2007;8(7):552–559. doi: 10.1038/sj.gene.6364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggi P, Su S, Karohl C, Veledar E, Rojas-Campos E, Vaccarino V. Heritability of renal function and inflammatory markers in adult male twins. Am J Nephrol. 2010;32(4):317–323. doi: 10.1159/000319449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, Choy E, Hu D, Tamraz B, Pawlikowska L, Wassel-Fyr C, Huntsman S, Waliszewska A, Rossin E, Li R, Garcia M, Reiner A, Ferrell R, Cummings S, Kwok PY, Harris T, Zmuda JM, Ziv E. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet. 2007;80(4):716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revez JA, Bain L, Chapman B, Powell JE, Jansen R, Duffy DL, Tung JY, Penninx PM, Visscher PM, de Geus EJ, Boomsma DI, Hinds DA, Martin NG, Montgomery GW, Ferreira MA. A new regulatory variant in the interleukin-6 receptor gene associates with asthma risk. Genes Immun. 2013;14(7):441–446. doi: 10.1038/gene.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006;80(2):227–236. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- Saito M, Yoshida K, Hibi M, Taga T, Kishimoto T. Molecular cloning of a murine IL-6 receptor-associated signal transducer, gp130, and its regulated expression in vivo. J Immunol. 1992;148(12):4066–4071. [PubMed] [Google Scholar]
- Scheller J, Garbers C, Rose-John S. Interleukin-6: From basic biology to selective blockade of pro-inflammatory activities. Semin Immunol. 2013 doi: 10.1016/j.smim.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Spijker S, van de Leemput JC, Hoekstra C, Boomsma DI, Smit AB. Profiling gene expression in whole blood samples following an in-vitro challenge. Twin Res. 2004;7(6):564–570. doi: 10.1375/1369052042663878. [DOI] [PubMed] [Google Scholar]
- Stephens OW, Zhang Q, Qu P, Zhou Y, Chavan S, Tian E, Williams DR, Epstein J, Barlogie B, Shaughnessy JD., Jr An intermediate-risk multiple myeloma subgroup is defined by sIL-6r: levels synergistically increase with incidence of SNP rs2228145 and 1q21 amplification. Blood. 2012;119(2):503–512. doi: 10.1182/blood-2011-07-367052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, Matsuda T, Hirano T, Kishimoto T. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell. 1989;58(3):573–581. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- The Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379(9822):1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Benyamin B, White I. The use of linear mixed models to estimate variance components from data on twin pairs by maximum likelihood. Twin Res. 2004;7(6):670–674. doi: 10.1375/1369052042663742. [DOI] [PubMed] [Google Scholar]
- Willemsen G, de Geus EJ, Bartels M, van Beijsterveldt CE, Brooks AI, Estourgie-van Burk GF, Fugman DA, Hoekstra C, Hottenga JJ, Kluft K, Meijer P, Montgomery GW, Rizzu P, Sondervan D, Smit AB, Spijker S, Suchiman HE, Tischfield JA, Lehner T, Slagboom PE, Boomsma DI. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13(3):231–245. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- Williams RL. A note on robust variance estimation for cluster-correlated data. Biometrics. 2000;56(2):645–646. doi: 10.1111/j.0006-341x.2000.00645.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011a;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de AM, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT1, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common. SNPs Nat Genet. 2011b;43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Bleibel WK, Wisel SA, Huang RS, Wu X, He L, Clark TA, Chen TX, Schweitzer AC, Blume JE, Dolan ME, Cox NJ. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum Genet. 2009;125(1):81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, Li X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277(18):3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure ∷ S1 Variance of sIL-6R level explained by genome-wide SNPs in unrelated individuals
Figure ∷ S2 Association between rs2228145 genotype and the ratio of IL6R exon 9 expression over mean IL6R expression
Table S1: Characteristics of subjects included in the analyses
Table S2: GWA results and information for all 680 SNPs reaching genome-wide significance in the GWAS of sIL-6R level
Table S3: SNP information and Conditional GWAS results of all 293 SNPs reaching genome wide significance in the GWAS of sIL-6R level after correction for the effect of rs2228145
Table S4: Location (Build 37/hg19) and sequence of expression probes targeting IL6R transcripts (Affymetrix U219 array)
Table S5: SNP information and eQTL P values for all 341 significant eQTL associations
Table S6: Correlations between sIL-6R level and IL6R expression level for all expression probes
Table S7: SNP information and conditional eQTL P values for all 80 significant eQTL associations after correction for the effect of rs2228145
Table S8: Results for the association with asthma for rs60760897 and SNPs in LD in the Australian Asthma Genetics consortium
Table S9: Results for the association with asthma for SNPs in LD with rs60760897 in the GABRIEL consortium
Supplementary Methods: Additional Supplementary Methods.