Abstract
Besides its role in exocrine differentiation, pancreas-specific transcription factor 1a (PTF1a) is required for pancreas specification from the foregut endoderm and ultimately for endocrine cell formation. Examining the early role of PTF1a in pancreas development has been challenging due to limiting amounts of embryonic tissue material for study. Embryonic stem cells (ESCs) which can be differentiated in vitro, and without limit to the amount of experimental material, can serve as a model system to study these early developmental events. To this end, we derived and characterized a mouse ESC line with tetracycline-inducible expression of PTF1a (tet-Ptf1a mESCs). We found that transient ectopic expression of PTF1a initiated the pancreatic program in differentiating ESCs causing cells to activate PDX1 expression in bud-like structures resembling pancreatic primordia in vivo. These bud-like structures also expressed progenitor markers characteristic of a developing pancreatic epithelium. The epithelium differentiated to generate a wave of NGN3+ endocrine progenitors, and further formed cells of all three pancreatic lineages. Notably, the insulin+ cells in the cultures were monohormonal, and expressed PDX1 and NKX6.1. PTF1a-induced cultures differentiated into significantly more endocrine and exocrine cells and the ratio of endocrine-to-exocrine cell differentiation could be regulated by retinoic acid and nicotinamide signaling. Moreover, induced cultures treated with RA and Nic exhibited a modest glucose response. Thus, this tet-Ptf1a ESC-based in vitro system is a valuable new tool for interrogating the role of PTF1a in pancreas development and in directing differentiation of ESCs to endocrine cells.
Keywords: Ptf1a, pancreas development, embryonic stem cells, pluripotent stem cells, pancreatic duodenal homeobox 1 (PDX1), beta cells, insulin, endocrine, neurogenin3, acinar morphology
Integrating the transcriptional and intercellular signals in pancreas development will lead to a better overall understanding of the process and thus allow application of this knowledge to exploit the endogenous regenerative capacity of the pancreas and/or manipulate pluripotent stem cells into therapeutic β cells. Yet, we only have a partial understanding of how multiple extrinsic and intrinsic signals act together to regulate various stages of pancreas development. Moreover, investigating the role of transcription factors that are expressed very early in pancreas development and how they combine with tissue inductive events has been challenging due to difficulty in accurately determining the timing and location of their activation, and to limiting amounts of embryonic material for study. Embryonic stem cells (ESCs) are a potentially useful model system to study these early developmental events which are otherwise difficult to study and can provide insights into the gene networks involved.
PTF1a is a basic helix-loop-helix sequence-specific DNA binding subunit of a heterotrimeric transactivator complex called PTF1 whose other components include one of the common E proteins and either the vertebrate suppressor of hairless, RBPJ or its paralogue RBPJL [1]. PTF1a was originally identified in exocrine cells where it binds and activates the promoters of digestive enzymes and is required for acinar cell development [2–4]. Besides the late role of PTF1a in pancreatic exocrine differentiation, a lineage tracing study demonstrated PTF1a is required very early for pancreas specification from the foregut endoderm [5] and is expressed in the progenitors of all pancreatic cell types, including acinar, endocrine and ductal cells. This study also suggested that PTF1a is involved in fate choice decisions between duodenum and pancreas [5]. However, while the role of PTF1a in exocrine differentiation is clear, how PTF1a controls the fate of multipotent progenitors, in turn endocrine development in mammals has not been fully elucidated with different studies presenting conflicting conclusions [6]. For example, by transactivating the pdx1 promoter [7, 8], PTF1a may lead to more committed pancreatic progenitors, and/or may commit PDX1+ progenitors preferentially to pancreatic vs. the alternative duodenal fate [5]. Or, as based on a recent study in zebrafish, the dose of PTF1a may dictate endocrine/exocrine fate choice of the progenitors [9]. Other studies in mice seem to point to regulation of endocrine differentiation by the interplay of PTF1a and Delta-Notch signaling [10] or antagonist activity between PTF1a and NKX6.1 [11]. Many protocols devised to differentiate ESC into β-cells produce immature multiple-hormone expressing cells, in vitro and an in vivo environment appears essential for their maturation and amelioration of the diabetic state [12–15]. This could be due to absence/low levels of PTF1a in the cultures leading to improper specification of pancreatic fate and lack of specific transcriptional cues provided by PTF1a in normal pancreatic development. While the precise mechanism by which PTF1a influences endocrine cell specification has yet to be fully resolved, it is clear that PTF1a plays an important role in normal pancreas development and therefore is a critical gene to evaluate in ESC-based pancreatic differentiation cultures.
Hence, we established a mouse ESC line with tetracycline-inducible expression of PTF1a (tet-Ptf1a). In the present study we sought to determine the impact of ectopic PTF1a expression on pancreatic exocrine and endocrine lineage differentiation in ESCs in vitro. We found that transient PTF1a induction activated PDX1 expression resulting in PTF1a+PDX1+ progenitor cell formation and that effects on pancreatic lineage differentiation were time- and context-dependent. Notably, induction of PTF1a generated bud-like structures reminiscent of pancreatic primordia formed during development, in vivo. The results also point to the importance of achieving PTF1a expression in differentiating ESC cultures in order to optimize endocrine specification. As such, this tet-Ptf1a ESC line serves as a new tool to interrogate the role of PTF1a in pancreas development and for efficiently directing differentiation of ESCs to the endocrine lineages.
Materials and Methods
Cell Lines
The tet-Ptf1a line was generated from the parent Ainv15 ESC line [16], a gift from M. Kyba/G. Daley and available at ATCC as described previously [17].
Cell Culture
Tet-Ptf1a and Ainv15 ESCs were maintained in an undifferentiated state and cultured in differentiation media as previously described [17], with the following modifications. To initiate the formation of embryoid bodies (EBs), ESCs were resuspended at a density of three million cells per siliconized (Sigma) 60 mm petri dish (BD Biosciences). EBs were transferred to new siliconized petri dishes with fresh media every 24 hours. After 7 days of suspension culture, EBs were removed from the media and plated onto 24-well plates with 0.1% gelatinized glass coverslips at a concentration of 50 EBs per well. Plated EB cultures were maintained in DMEM-High Glucose with 100 U penicillin/streptomycin, 2mM L-glut with 1 % Serum Replacement (Gibco). To induce PTF1a expression, cultures were exposed to 1μg/ml doxycycline (Dox) renewed every 24 hrs. Retinoic acid and nicotinamide treatments were performed after Ptf1a induction using 5μM of retinoic acid (Sigma) in carrier DMSO for the three days and/or 10mM nicotinamide (Sigma) for 21 days beginning at 7 days post-plating (EB7+7).
Quantitative real-time PCR
Cells were harvested at various stages by dissolution and homogenization in 1 ml of TRIzol (Invitrogen), and RNA was isolated and purified using Qiagen RNAeasy Mini kits. QPCR was performed using Applied Biosystems gene expression assays. Assay IDs are given in Supplementary Table 1. Gapdh was used as an internal control and the comparative threshold method was used to quantify transcript abundance.
Immunofluorescent Staining
Immunostaining was performed as previously described by Kahan et al [18]. The antibodies and dilutions are listed in Supplemental Table 2. Secondary antibodies were 488, 568 and 647 Alexa Fluors of anti-goat, anti-mouse, anti-rabbit raised in either goat or donkey. Cells were counterstained with DAPI to mark nuclei. Coverslips with adherent stained cells were mounted on glass slides with Prolong® Gold Antifade reagent (Invitrogen). Images were generated using A1R-Si Nikon Confocal or a Zeiss Axiovert 200M microscope. Hormone+ cell counts were performed for each sample manually.
Flow cytometry
Cultures were dissociated by incubating in 0.25% trypsin for 3 min followed 0.05% trypsin for 15 min and heavy pipetting to make single cell suspension. Cells were then fixed in 0.1% paraformaldehyde for 10min, pelleted and resuspended in FACS buffer (2% FBS and 0.1% sodium azide in PBS). 90% methanol was used to permeabilize the cells and cells were then incubated with the primary antibody diluted in the FACS buffer for 2 hours at room temperature. After washing (with FACS buffer+0.1% Triton-X), cells were incubated in secondary antibody for 1 hour. Samples were read on a FACSCalibur flow cytometer (BD Biosciences).
Western blot analyzes
Tetptf1a and Ainv15 cells were differentiated in 60mm dishes to different time points and harvested using 1ml of NP-40 lysis buffer supplemented with Halt protease inhibitor cocktail (Thermo Scientific). 40ug of each sample lysate was run on 12% polyacrylamide gels (Biorad) and transferred onto nitrocellulose membranes (Biorad) in the cold. Following electroblotting, membranes were blocked with 5%milk in PBST and incubated overnight with the primary antibody. Horseradish peroxidase-conjugated secondary antibodies (Pierce) and chemiluminescent substrate (SuperSignal West Femto, Pierce) were used to detect the protein bands in a luminescent image analyzer (Fujifilm LAS-4000). Primary antibody: rabbit anti-PTF1A (Chris Wright), 1:6000. Secondary antibody: HRP conjugated goat anti-rabbit (1:20,000). Blots were stripped and reprobed for GAPDH (mouse anti-GAPDH HRP conjugated abcam 9482, 1:10,000).
Electron microscopy
TetPtf1a cells grown on p60 culture dishes to EB7+28 were fixed for 45 minutes in 3% glutaraldehyde, 1% paraformaldehyde, 0.1 M cacodylate buffer solution. Fixed tissues were then prepared and sectioned as previously described [18]. Sections were then examined using a Hitachi H-7000 electron microscope.
Insulin Content and Release Assays
Insulin content was determined by acid ethanol extraction followed by a mouse C-peptide ELISA (Biovendor). For insulin release assay, cultures were grown in 60mm dishes and media was changed to DMEM-low glucose the previous night. After pre-incubation in Krebs-Ringer buffer with 3.3 mM glucose at 37°C for 120 min, the cells were then incubated in Krebs-Ringer buffer containing 25mM glucose, at 37 °C for an additional 120 min. The supernatants were collected and the C-peptide levels were normalized to total cell number in the cultures by trypsinization and counting parallel samples treated similarly.
Results
Validation of murine ESC line possessing inducible expression of Ptf1a
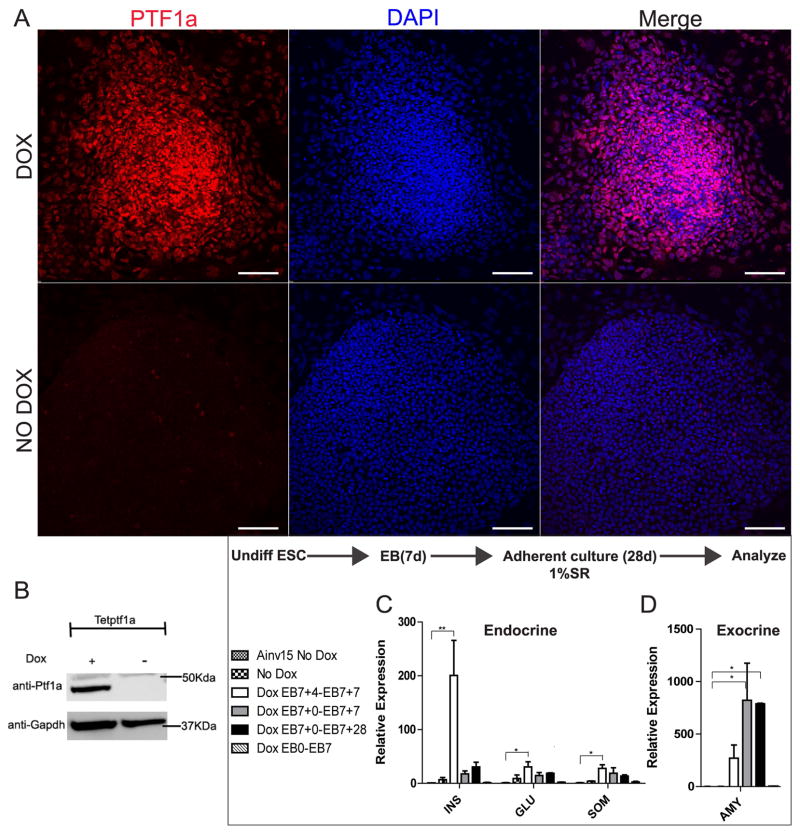
Using the tet-on system developed by Kyba et al. [16], a murine ESC line (tet-Ptf1a) with tetracycline inducible Ptf1a expression was generated [17]. To validate the cell line, tet-Ptf1a ESCs were treated with doxycycline (Dox) for 3 days. Most cells expressed PTF1a in the presence of Dox while very few cells stained positively in the absence of Dox (Fig. 1A). Similarly, Dox treatment resulted in strong PTF1a expression detected by western blot analysis (Fig. 1B). Hence, ectopic expression of Ptf1a can be induced and shutdown robustly in the tet-Ptf1a cell line.
Fig. 1.
Generation and validation of a tetracycline inducible PTF1a mESC cell line. (A) Immunofluorescent staining for PTF1a and DAPI in tet-Ptf1a ESCs induced with Dox for 3 days (upper panel) or untreated (lower panel). Most of the induced cells express PTf1a while in the absence of Dox very few cells express PTf1a. Scale bars, 100μm. (B) Western blot of whole cell protein extracts obtained from tet-Ptf1a ESCs induced with Dox for 3 days or untreated cultures, probed for PTF1a and GAPDH shows induction of PTF1a protein upon addition of Dox. (C and D) tet-Ptf1a cells were cultured as embryoid bodies (EBs) for seven days and subsequently grown in attachment culture for 28 days. Various Dox induction schemes were tested including addition for 3 days post-plating between EB7+4 to EB 7+7 (Dox EB7+4-EB7+7), 7 days post-plating between EB7+0 to EB7+7 (Dox EB7+0-EB7+7), 28 days days post-plating between EB7+0 to EB7+28 (Dox EB7+0-EB7+28) or during 7 days of EB formation (Dox EB0-EB7). Negative controls were the parental cell line Ainv15 and untreated tet-Ptf1a cultures. Cultures were grown for 28 days post-plating and analyzed for transcripts of (C) endocrine hormones such as insulin (INS), glucagon (GLU) and somatostatin (SOM), and (D) the exocrine enzyme amylase (AMY). Cultures with 3 day Dox induction between EB7+4-EB7+7 had significantly higher insulin and lower amylase when compared to other treatment groups. Error bars indicate s.e. *P<0.05, **P<0.01 determined by ANOVA with Tukey’s multiple comparison test. n=3.
Transient ectopic Ptf1a expression early in ESC differentiation significantly increases insulin transcript expression
As a starting point for Ptf1a activated pancreatic differentiation studies, we first tested several different Ptf1a induction regimes by adding Dox at different ESC differentiation stages to identify an optimal context to observe Ptf1a induced effects. We treated cultures of differentiating tet-Ptf1a cells with Dox during each of the following time intervals: 1) for 7 days of EB formation (EB0-EB7), 2) for the initial 7 days after plating EBs (EB7+0 to EB7+7), 3) for three days from EB7+4 to EB7+7, or 4) for 28 days continuously after plating (EB7+0 to EB7+28). The inducibility of the transgene was confirmed at all the time points tested. Time course analysis of Ptf1a transcript in each induction regime showed similar induction levels (Suppl Fig. 2I). The short three-day induction resulted in significantly more transcript levels of islet hormones (insulin-1, glucagon, somatostatin) when compared to other treatment regimes, uninduced (No Dox) tet-Ptf1a cells and Ainv15 control cells at the end of our culture period (EB7+28) (Fig. 1C). Ins-1 transcript levels in the three day induced cells were 200 fold higher than in Ainv15 control cells (P<0.01). Since Ptf1a is a direct transcriptional activator of many exocrine enzyme genes, we also tested whether induction of Ptf1a increased expression of a classical exocrine gene, amylase-2 (Amy2) (Fig. 1D). We observed a time-dependent activation of both endocrine and exocrine genes in ESCs induced to ectopically express Ptf1a (Fig. 1C, D). For example, whereas increased islet hormone and amylase mRNA expression was seen when Ptf1a was expressed after plating, Ptf1a over-expression during EB formation had little effect on endocrine and exocrine transcript abundance. Furthermore, prolonged (28 day) Pft1a expression led to predominantly increased Amy2 expression whereas, transient Ptf1a expression (3 day) resulted in proportionately greater hormone transcript expression. These results led us to utilize this culture protocol as the basis for further experiments.
Ptf1a expression in differentiating ESCs activates Pdx1 expression and initiates formation of Pdx1+Ptf1a+ bud-like structures
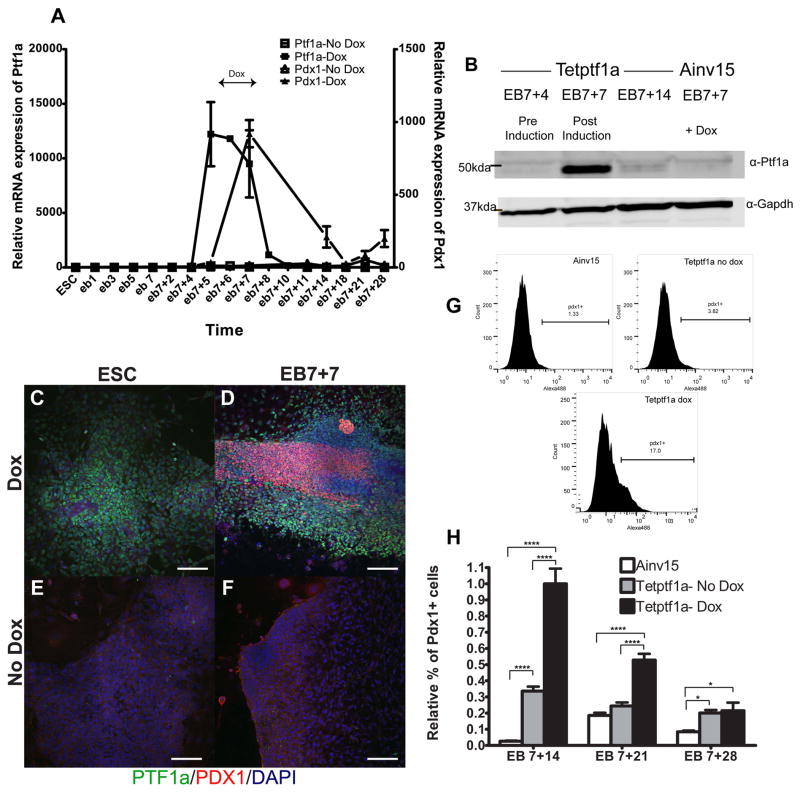
Given our findings that ectopic expression of Ptf1a in a narrow window period increased expression of both endocrine and exocrine genes and prior reports of Ptf1a transactivating the Pdx1 promoter in vivo and in pancreatic cell lines [7, 8], we sought to determine if Ptf1a was inducing expression of Pdx1 in ESC cultures. We first investigated the Ptf1a expression profile through the course of differentiation. Transcript levels (Fig. 2A) and protein (Fig. 2B) levels peaked during induction period and were very low/undetectable at other time points or in untreated differentiated tet-Ptf1a cells (Fig. 2A, 2F) and in differentiated Ainv15 cells treated with Dox (Fig. 2B) indicating that the expression of Ptf1a protein was tightly controlled by induced transgene expression. We found that transient Ptf1a expression from EB7+4 to EB7+7 resulted in extensive Pdx1 mRNA expression (Fig. 2A), and this did not occur in the absence of Dox in ESC cultures at this stage. Importantly, the effects of ectopic Ptf1a expression appeared to be context-dependent. We observed that ectopic Ptf1a activated PDX1 protein expression in EB7+4 cells but not in undifferentiated ESCs (Fig. 2C, D) or other contexts (data not shown). We then sought to perform a quantitative analysis of PDX1+ cells at different time points by flow cytometry (Fig. 2G,H). The proportion of PDX1+ cells was high in induced cultures at EB7+14 (ranging between 15–25%, Fig. 2G). Normalizing to this data value, we found 70% fewer PDX1+ cells in uninduced control cultures and ~ 98% fewer in Ainv15 cultures (Fig. 2G,H). Thus, our findings indicate that ectopic Ptf1a expression in differentiating ES cell progeny activates PDX1 expression in a context- and time-dependent manner.
Fig. 2.
Dox-induced expression of Ptf1a in tet-Ptf1a ESC differentiation cultures activates expression of Pdx1. Tet-Ptf1a cells were differentiated in adherent EB-based cultures and treated with Dox from EB7+4 to EB7+7as shown in Fig. 1 and grown to indicated time points and analyzed. (A) Gene expression profile of Ptf1a and Pdx1 in Dox-treated and untreated cultures relative to undifferentiated tet-Ptf1a ESCs. (B) Western blot of whole cell protein extracts taken from induced tet-Ptf1a and Ainv15 probed with α-ptf1a and α-gapdh. (C–F) Tet-Ptf1a cells expressed PDX1 (red) when induced with Dox in the context of EB-based cultures (D), but not in undifferentiated cells (C). Untreated ESCs and EB-based cultures did not express PTF1a (green) or PDX1 (E,F). Scale bars, 100μm. (G) Sample histograms from flow cytometry on EB7+14 populations of Dox-treated and untreated tet-Ptf1a and Ainv15. (H) FACS analysis revealed that induced cultures have more PDX1+ cells than uninduced cultures, which gradually decreases over time as cells progressively differentiate. All data were normalized to values for Dox-treated tet-Ptf1a cells grown to EB7+14. Error bars indicate s.e. *P<0.05, ****P<0.0001 determined by 2-way ANOVA with Tukey’s multiple comparison test. n=3.
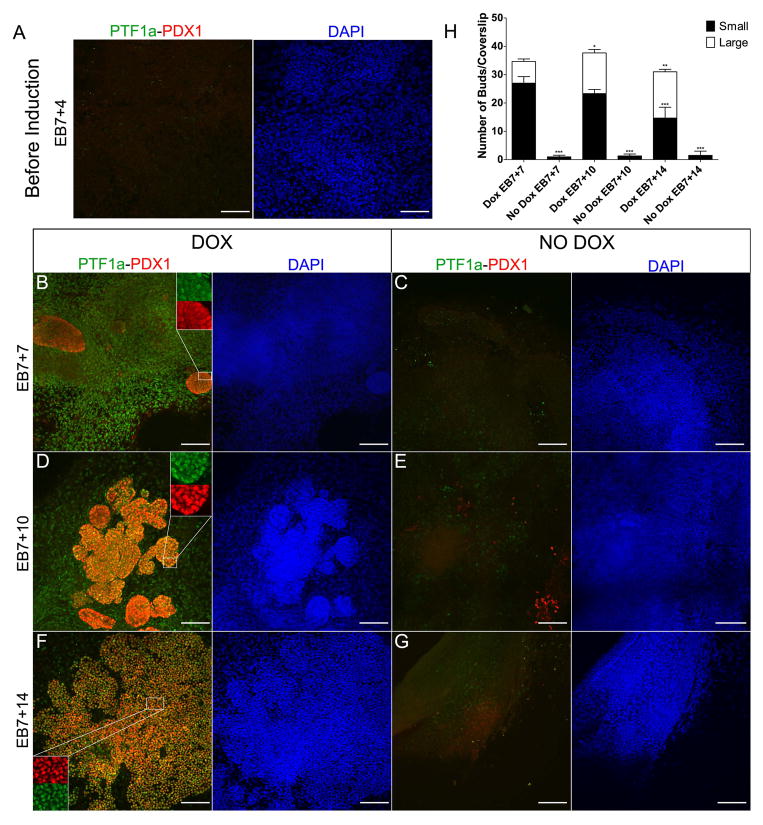
The most striking result of ectopic Ptf1a expression was the outgrowth and expansion of unique PDX1+ bud-like structures, reminiscent of morphological features occurring during in vivo development. Before induction, there were very few PDX1+ or PTF1a+ cells (Fig. 3A), but after Ptf1a induction PDX1 expression appeared in a subset of PTF1a+ cells in bud-like outgrowths (Fig. 3B) beginning within 2–3 days of ectopically expressing Ptf1a transgene. The PDX1+ cell population continued to enlarge and the bud-like structures continued to expand long after Ptf1a transgene expression was extinguished (Fig. 3D). These bud-like growths contain cells which predominantly co-express both PTF1a and PDX1, whereas neither bud-like growths nor co-expressing cells are present at any time in No Dox treated cultures (Fig. 3C, E, G). By EB7+14, the buds have grown into structures that resemble pancreatic epithelium co-expressing PTF1a and PDX1 (Fig. 3D). There were significantly more Ptf1a+Pdx1+ bud-like structures in Dox-induced cultures when compared to uninduced cells regardless of the stage of differentiation (Fig. 3H). On categorizing these buds based on size as small and large, small buds were predominant in early Dox treated cultures and their number decreased with time. In contrast, the number of large buds increased with time suggesting these bud-like structures expand over time (Fig. 3H). In cells induced to express Ptf1a, insulin+ cells are more prominently detected (starting at ~ EB7+14) than in cultures not expressing Ptf1a, gradually increasing in number until EB7+28 when PDX1 staining becomes primarily associated with insulin staining (Suppl Fig. 1). Insulin+ cells in the cultures co-express C-peptide (Fig. 5F).
Fig. 3.
Time course analysis of PTF1a-PDX1 co-expression in tet-Ptf1a cultures induced with Dox from EB7+4 to EB7+7 reveals distinctive bud-like outgrowths and expression of PDX1. (A) Before treatment with Dox, at EB7+4, neither PTF1a nor PDX1 are expressed. (B,D,F) PTF1a over-expression leads to progressive increase in PDX1+ branched epithelium. At EB7+7, simple PDX1+ clusters are seen (B). Within 3 days, PDX1+ bud-like structures have grown, and undergone branching into PDX1+ PTF1a+ epithelium (D), eventually expanding and branching extensively (F), similar to that observed during pancreatic development in vivo. Insets show magnified regions to show co-expression. These bud-like outgrowths are not observed in cultures that have not been treated with Dox (C,E,G). A few scattered PDX1+ cells without distinct epithelial morphology can be seen in E, lower right corner. Scale bars, 100μm. (H) Quantification of small (< 200 cells) and large (≥ 200 cells) Ptf1a+Pdx1+ Bud-like outgrowths at various time points in induced vs. uninduced cultures. Error bars indicate s.e. *P<0.05,**P<0.01, ***P<0.001 determined by 2-way ANOVA with Bonferroni’s multiple comparison test to Dox EB7+7. n=3.
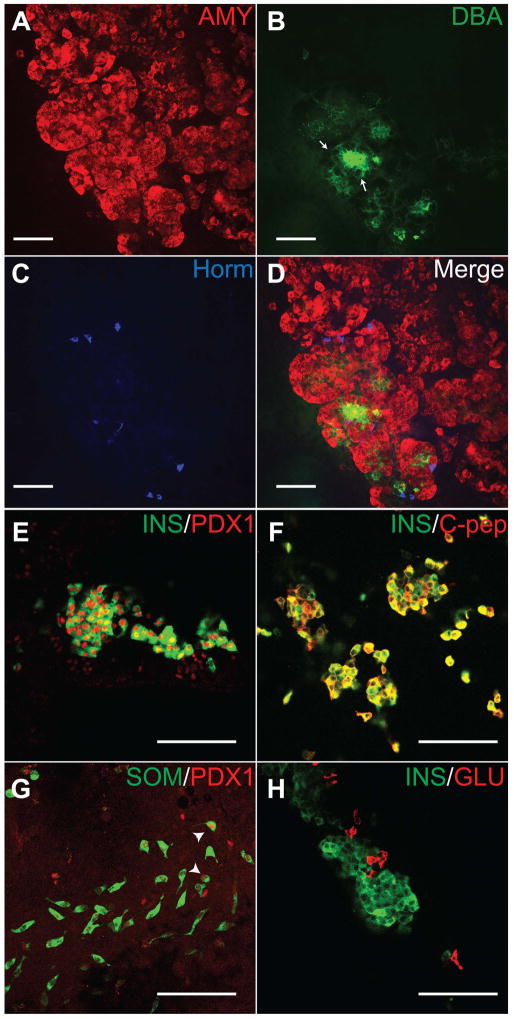
Fig. 5.
Pancreatic progenitors in induced cultures can further differentiate into all the three pancreatic lineages. Cultures were grown to EB7+28 and analyzed. (A) Acinar cells as indicated by Amylase expression (A) are arranged in typical lobular acinar-like structures, with DBA-staining duct cells (B) present amongst the lobules. Arrows show DBA staining. (C) Hormone+ cells (antibody cocktail against insulin, somatostatin and glucagon) are scattered between the acinar lobules. On further characterizing individual islet cell types, insulin+ cells expressed nuclear PDX1 (E) and co-expressed C-peptide (F). Abundant somatostatin+ cells co-expressing PDX1 were present (G). Nuclear PDX1 expression in somatostatin+ cells is indicated by arrowheads. Glucagon and insulin were not co-expressed (H). Scale bars, 100μm.
Key markers of pancreatic developmental program are expressed in Ptf1a-induced cultures
To support the hypothesis that the bud-like structures developing in Ptf1a-induced ESCs represents pancreatic differentiation, other markers of pancreas development were investigated at various stages. Induced and uninduced cultures were co-stained with PDX1 and early pancreatic endoderm markers such as HNF6, FOXA2 and MNX1 immediately after induction (EB7+7) to determine if the emerging PDX1+ bud-like clusters correspond to early budding and commitment to the pancreatic fate [19, 20]. We found the majority of cells contained within these bud-like regions of the Ptf1a-induced cultures co-expressed PDX1 and FOXA2, PDX1 and HNF6, and PDX1 and MNX1 (Suppl Fig. 2E, G, H). FOXA2 and HNF6 were expressed in a larger domain than was observed for PDX1, and PDX1+ cells emerged from within these FOXA2 and HNF6 regions (Suppl Fig. 2E, G). Apart from a few PDX1+MNX1+(Suppl Fig. 2H, inset) bud-like structures, few isolated MNX1+(Suppl Fig 2H, arrow) cells were seen, possibly of neuronal lineage [21]. HNF4a+ cells were rarely observed, and did not co-express PDX1; one such area is shown (Suppl Fig. 2F), and the HNF4a+PDX1− cells could represent a population of liver progenitors [22]. In contrast, uninduced cells did not express PDX1, and had very little FOXA2 and HNF6 (Suppl Fig. 2A, C), and more widespread MNX1 and HNF4a at the same stage (Suppl Fig. 2B,D). SOX9 was expressed in most PDX1+ cells (Fig 4A) as is the case in the pancreatic epithelium during early pancreas development [23]. These observations demonstrate transient Ptf1a over-expression gives rise to bud-like outgrowths that express many early pancreatic endoderm markers. By acting at the early pancreas specification stage, ectopic Ptf1a expression may potentiate further pancreatic lineage differentiation.
Fig. 4.
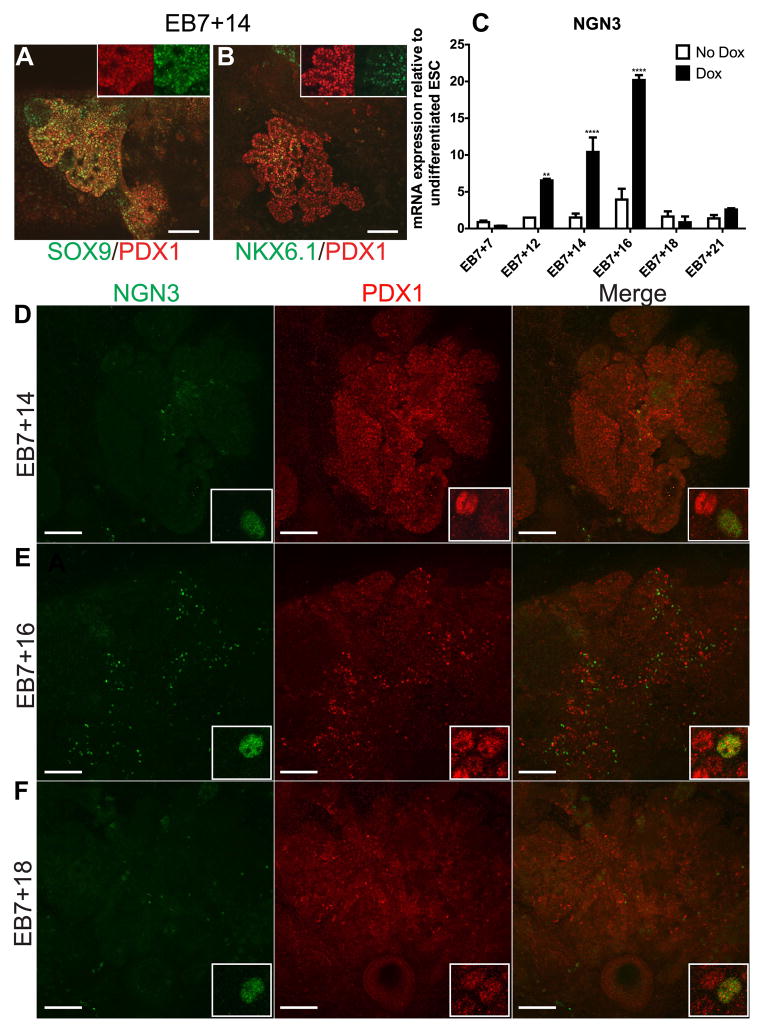
Ptf1a-induced cultures express characteristic markers of developing pancreatic epithelium, especially a wave of transient Neurogenin3. (A) Majority of the PDX1+ cells in the EB7+14 bud-like structures express SOX9 with the exception of some cells at budding tip regions. (B) NKX6.1 is predominantly restricted to PDX1+ cells at core of the bud-like structures and never at the periphery or tip. Insets show magnified regions. (C) Time course analysis of Ngn3 transcript levels in Dox-treated and untreated cultures relative to undifferentiated tet-Ptf1a ESCs shows that Ptf1a expression increased Ngn3 expression in ESC cultures. Error bars indicate s.e. **P<0.01, ****P<0.0001 determined by 2-way ANOVA with Dunnet’s multiple comparison test to No Dox tet-Ptf1a EB7+7. n=3–4. (D–F) A wave of NGN3 protein expression occurs in induced cultures peaking at EB7+16 (E vs. D, F) and corresponds with relative mRNA levels. There are still detectable numbers of Ngn3+ cells at EB7+14 and EB7+18 (D,F). Ngn3 appears to be primarily expressed in interior/trunk regions of branching structures and in low Pdx1 expressing cells as indicated in the insets. Scale bars, 100μm.
Downstream pancreatic markers, including NKX6.1 (epithelial-endocrine progenitor), and Ngn3 (endocrine committed) were then evaluated. A time course analysis of NKX6.1 staining revealed its expression within a subset of PDX1+ cells. NKX6.1 expression was widespread at EB7+10 (Suppl Fig. 3A–C), became restricted to the central cells of branching-like epithelium by EB7+14 (Fig. 4B; Suppl Fig. 3D–F), and eventually was present only in Insulin+ PDX1+ cells (Suppl Fig. 3G–I). This pattern of NKX6.1 expression is similar to that normally seen in the developing pancreas in vivo [11, 24].
Ngn3 is expressed transiently in islet progenitors appearing within the PDX1+ pancreatic epithelium and is critical to initiate endocrine specification of the secondary transition [19]. Hence, we investigated whether Ptf1a over-expression and PDX1+ pancreatic epithelium-like cells demonstrated Ngn3 gene activation. First, we found Ngn3 mRNA levels in Ptf1a-induced cultures were transiently increased ~20fold over undifferentiated ESCs, whilst uninduced cultures did not show a similar increase (Fig. 4C). In concert with transient Ngn3 transcript expression, immunoflourescent staining of cultures demonstrated transient NGN3 protein expression within the interior/trunk regions of PDX1+ branching-like epithelium in the Ptf1a-induced cultures with NGN3+ cells most prominent at ~EB7+16 (Fig. 4E). Also, the tip marker CPA1 was not co-expressed with NGN3 (Suppl Fig. 4). It was generally observed that NGN3 was expressed in PDX1lo cells (Fig. 4, insets), and it is interesting to note that several other studies have reported this observation in vivo [25, 26]. In contrast, NGN3+ cells were not detected in uninduced cells at any stage (data not shown). The appearance of NGN3+ cells among the PTF1a-expressing PDX1+ cell population demonstrates the capacity of ESC-derived pancreatic epithelial-like buds for endocrine specification.
Ptf1a-induced cultures can further generate adult pancreatic cell types
Given the increased number of both exocrine and endocrine progenitors (CPA+ cells and NGN3+ cells) in the Ptf1a-induced cultures, we sought to determine if adult pancreatic cell types are produced. Large numbers of Amy+ cells were found in Dox-treated cultures (Fig. 5A). In particular, the amylase expressing tissue generated by transient Ptf1a-induction exhibited tissue morphology similar to the “grape-like” or acinar structures typically seen in more mature pancreatic tissue. These tissues contained polarized epithelial cells possessing zymogen granules, a basement membrane and a central lumen thus forming “acinar” units (Suppl Fig. 5B, C). Dolichos Biflorus Agglutinin (DBA), a marker of duct epithelium [27] was found intertwined with the Amy expressing cell clusters (Fig. 5B,D). Hormone+ cells, expressing insulin, glucagon or somatostatin were also found embedded within these regions (Fig. 5C,D). H&E sections of the Ptf1a-induced cultures depict regions possessing epithelial morphology with areas containing insulin+ cells (Suppl Fig. 6). In cultures where Ptf1a was not induced, such hormone-positive and enzyme-expressing cells are rare (data not shown). In summary, all pancreatic lineages, including acinar, endocrine and ductal cells are generated upon Ptf1a-induction in differentiating ESC cultures.
Insulin+ cells in Ptf1a-induced cultures possess some typical features of mature β cells
Individual hormone-expressing cells were further investigated by staining and quantified in late stage cultures. Cells individually expressing insulin, glucagon and somatostatin were observed (Fig. 5E, G, H). Insulin+ cells almost uniformly co-expressed PDX1 and C-peptide (Fig. 5E, F). Large populations of these insulin/C-peptide co-staining cells formed clusters with apparent direct cell-cell contact that was not generally observed in uninduced or wild-type ESC cultures. These insulin+ cells co-express NKX6.1 (Suppl Fig. 3G–I) but do not co-express glucagon (97% of insulin+ cells did not express glucagon and none expressed somatostatin) (Fig. 5H). These insulin+ cells were then examined further for ultrastructural morphology. Electron microscopic evaluation of these late stage cells showed electron-dense secretory granules with peripheral halos similar to those seen in β cells (Suppl Fig. 5D). Thus, the insulin+ cells generated after further differentiation of ESC-derived PDX1+PTF1a+ progenitors express many characteristic β cell features.
Treatment with Nicotinamide and Retinoic Acid enhances endocrine differentiation in Ptf1a induced cultures
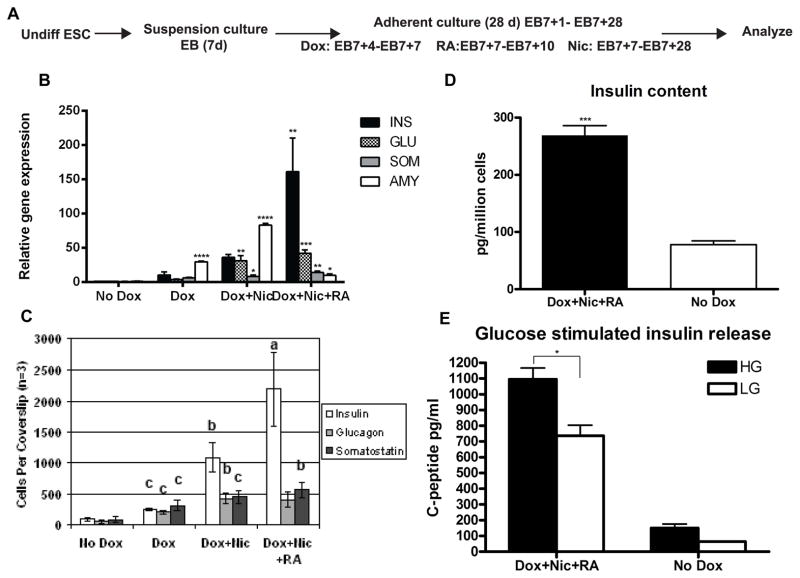
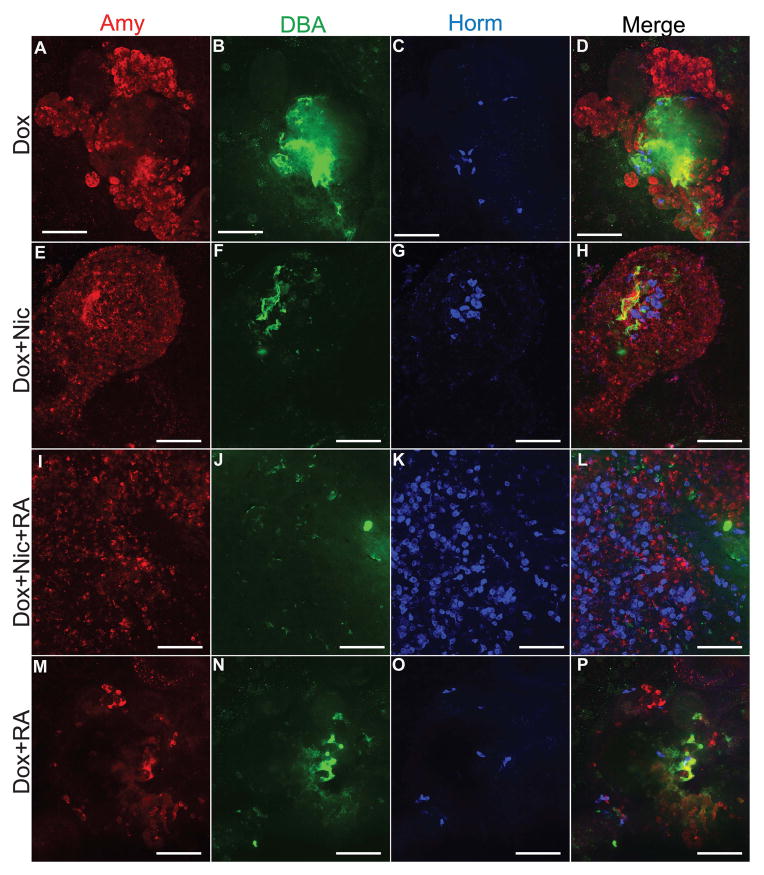
Although β cells appeared to form spontaneously in cultures comprised of PDX1+PTF1a+ progenitors, we desired to increase the yield of β cells as well as test whether ESC-derived pancreatic progenitors in vitro are responsive to defined differentiation factors. Nicotinamide (Nic) has been shown to enhance further differentiation and glucose responsiveness of fetal pancreatic tissue and possibly stimulate proliferation of β cells [28, 29], and to promote ESC differentiation to insulin-secreting cells [30] possibly in part due to its ability to inhibit histone deacetylases [31] and/or its anti-inflammatory effects [32]. Retinoid signaling contributes at multiple stages to pancreas differentiation in the embryo [33, 34], and from ESCs [35]. Retinoic acid (RA) has been shown to induce generation of NGN3+ cells and further promote their differentiation to β cells in a mouse pancreatic explant assay [36]. Based on these data, we treated tet-Ptf1a cells sequentially with Dox to induce Ptf1a expression, followed immediately by RA and Nic treatment according to the protocol described in the methods and Fig. 6A. We then compared these cultures to those which had been treated with No Dox, Dox alone, or Dox+Nic. Whereas PTF1a induction alone modestly increased islet hormone and amylase transcripts (Fig. 6B) and the number of endocrine cells (Fig. 6C), addition of Nic to induced tet-Ptf1a cells increased insulin+ cells significantly (~3–4 fold or ~1000 cells per coverslip) over that obtained by PTF1a induction alone (Fig. 6C). Treatment with both RA and Nic resulted in further increase in insulin transcript expression (Fig. 6B) and the number of insulin+ cells (~2300 per coverslip);1.15% of total cells compared to 0.04% in No Dox cultures (Fig. 6C). Comparatively, RA+Nic treatment dramatically reduced amylase gene expression in PTF1a-induced cultures (Fig. 6B), thus suggesting a shift in the differentiation of progenitors away from exocrine fate. This observation is corroborated by immunoflourescent staining for amylase, DBA and hormones shown in Fig. 7. Whereas induction of PTF1a alone led to more Amy+ cells, more DBA+ cells, and a modest increase in hormone+ cells (Fig. 7A, C), addition of both RA and Nic to Ptf1a over-expressing cells greatly enhanced the number of hormone+ cells (Fig. 7I–L). Further staining for insulin specifically and amylase yielded similar results; combined RA+Nic treatment led to better insulin expression and endocrine differentiation (Suppl Fig. 7E vs. C, and G, H). On the other hand, neither Nic nor RA alone significantly increased the number of hormone+ cells (Fig. 7E–H, M–P). To confirm the authenticity of insulin+ cells, the cultures were co-stained with C-peptide showing nearly all insulin+ cells expressed C-peptide (Suppl Fig. 7H). Importantly, treatment of other ESC lines with RA+Nic did not considerably increase insulin expression (Suppl Fig. 7F) indicating that prior Ptf1a expression preconditions cells to have the ability to respond to these morphogens. These findings suggest that Nic and RA regulate the in vitro development of Ptf1a-induced ESCs with the major effects being a shift toward endocrine differentiation and concomitant reduction of acinar and ductal differentiation.
Fig. 6.
Treatment of Ptf1a-induced cultures with Nicotinamide (Nic) and Retinoic acid (RA) increases endocrine differentiation. (A) Schematic representation of the differentiation protocol. After induction of Ptf1a, cultures were treated with Retinoic acid for 3 days (EB7+7 to EB7+10) and/or Nicotinamide for the remainder of the culture period (EB7+7 to EB7+28). (B) Transcript analyses show treatment with Nic and RA increases insulin, glucagon and somatostatin expression and decreases amylase expression, while treatment with Nic alone increases amylase expression. Gene expression was calculated relative to uninduced tet-Ptf1a cell cultures. Error bars indicate s.e. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 determined by 2-way ANOVA with Tukey’s multiple comparison test. n=3. (C) Hormone+ cells were counted manually. Hormone cell expression increases significantly with growth factor treatments. a = P<0.05 over all other tested conditions, b = P<0.05 over No Dox controls and Dox treated cultures, c = P<0.05 over No Dox controls. The overall number of cells per coverslip in the different conditions was similar at ~ 2×105 cells. (D) Insulin content in Nic and RA treated Ptf1a-induced cultures were significantly higher than uninduced cultures. Error bars indicate s.e. ***P=0.0007, determined by unpaired two-tailed t test with Welch’s correction. n=2–4. (E) Glucose stimulated insulin release assay on Nic and RA treated Ptf1a-induced cultures showed a significant difference in the amount of C-peptide released under conditions of low (3.3mM) and high (25mM) glucose (LG vs. HG) compared to uninduced cells. Error bars indicate s.e. *P<0.05, determined by 2-way ANOVA with Tukey’s multiple comparison test. n=3.
Fig. 7.
Immunofluorescence images showing changes in differentiation status with Nic and RA treatment. Cell cultures were examined at EB7+28. The acinar-like lobular morphology and DBA expression present in Dox only-treated cell cultures (A, B) are lost by the addition of Nic and/or RA (I, J). Ptf1a-induced cell cultures treated with both Nic and RA have more hormone+ cells compared to cells treated with Dox alone (C vs. K). Adding Nic alone did not increase hormone+ cells significantly (E, G). Treatment with RA alone did enhance differentiation into amylase+ or hormone+ cell types (M, O). Scale bars, 100μm.
To correlate phenotypic endocrine differentiation with functional assessments, insulin content and glucose-stimulated insulin release were evaluated by C-peptide ELISAs. C-peptide content of cells subjected to Ptf1a over-expression followed by differentiation with Nic and RA treatment was significantly higher than in uninduced controls (Fig. 6D). Glucose-stimulated C-peptide release assays showed that the Dox+RA+Nic cultures were marginally responsive to glucose, releasing significantly more C-peptide after exposure to high glucose than low glucose (Fig. 6E). Moreover, C-peptide secretion into the media under both basal and stimulated conditions was significantly greater for Dox+RA+Nic treated cells compared to untreated, differentiated cells (Fig. 6E).
Discussion
Efficient differentiation of ESCs to insulin-expressing β cells in vitro has remained elusive, due to incomplete understanding of the transcriptional networks and their timing of activation, and the difficulty in reproducing suitable niches in a dish. In this regard, directed differentiation by induced expression of many pancreatic genes has been studied. Adenoviral mediated PTF1a and MIST1 expression in mouse ESCs was used to generate acinar cells [37]. Besides the late role of PTF1a in exocrine differentiation, it has been established that PTF1a is required very early in development for specification of the pancreas from the foregut endoderm [5, 38]. However, a study exploiting this early function of PTF1a to facilitate differentiation of ESCs to the pancreatic lineage has not been performed. Here, we demonstrate transient ectopic expression of PTF1a alone (without the requirement for PDX1) in ESCs drives the whole pancreatic program exhibiting many of the characteristic morphological and molecular signatures of pancreas development, in vitro.
One of the salient features of this system is the activation of Pdx1 expression by PTF1a that has not been previously reported in ESCs. PTF1a-induced Pdx1 activation is context-dependent and is consistent with prior studies that showed that PTF1a binds to Area III of the Pdx1 promoter that is only active early in development [7, 8]. These studies are also consistent with prior mouse studies showing that PTF1a is a crucial determinant of pancreatic fate besides PDX1 as most PDX1+ progenitors that lack PTF1a expression fail to differentiate into pancreatic cells and acquire a duodenal fate [5]. Ineffectiveness of current protocols of pancreatic differentiation of human and mouse ESCs could be attributed to suboptimal PTF1a co-expression in PDX1+ cells and, consequently resulting in some PDX1+ cells giving rise to intestinal, bile duct and/or stomach cells in culture. This model system provides a means of generating progenitors that express both PTF1a and PDX1, and thus committing the cultures to pancreatic fate, by the ectopic expression of one factor i.e. PTF1a alone.
Notably, the developmental phenotype observed with transient PTF1a induction was different from a similar cell line with PDX1 induction [39]. Ectopic PDX1 expression led to significantly lower endocrine and exocrine gene expression than did PTF1a induction and did not result in the bud-like structures seen in Ptf1a-induced cultures. Other studies that have used either transient or biphasic PDX1 expression in mouse ESCs [40] or constitutive PDX1 expression in human ESCs [41] have reported some endocrine differentiation but no morphological changes. The distinctive PDX1+PTF1a+ bud-like structures differ from islet-like clusters (ILCs) reported by many groups [42] in that they are of epithelial nature and closely resemble pancreatic buds that arise from the foregut endoderm. These structures are unique to Ptf1a-induced cultures, and might be derived from direct downstream genes targets of Ptf1a since they appear immediately after induction. There is strong co-expression of PDX1 and PTF1a along with NKX6.1 in these epithelial structures, and hence mark a fully specified pancreas progenitor unlike other reports of PDX1+ buds [43]. NKX6.1 expression domain emerges as a subset of PDX1+ population, and gets restricted to the endocrine lineage over time. NGN3+ cells arise within the PDX1 domain, some of which were PDX1lo cells, in agreement with previous reports in mouse embryos [25, 26]. All three pancreatic cell types are produced which are organized into striking acinar-like cell clusters expressing amylase and DBA+ duct-like cells with interspersed hormone+ cells, as observed in mature pancreas. More importantly, the insulin+ cells in these cultures were monohormonal in contrast to those generated by popular β-cell differentiation protocols, despite their higher efficiency [12–15]. Finally, the Ptf1a-induced ESC-derived tissue is responsive to endocrine promoting effects of Nic/RA signaling.
Although it may seem intuitive that an endoderm population would be a better starting material than EB-based cultures to evaluate pro-pancreatic effects of PTF1a, we observed that a highly enriched definitive endoderm population, generated from Nodal/Activin/BMP4/ILV treatments, was not a sufficient context to observe the pancreatic and endocrine differentiation promoting effects of ectopic Ptf1a expression (data not shown). This suggests that a more committed foregut endoderm context and/or an additional “inductive” cell type(s) may be necessary. Hence, we conclude an EB-based system which is more heterogeneous than a pure endoderm population may contain the correct combination of cells and extrinsic signals to be able to respond to Ptf1a.
The cell culture model described here possesses many features reminiscent of pancreatic organogenesis in vivo, and therefore would make an excellent model system to study Ptf1a-initiated pancreatic development. This model system could potentially be used for assessing Ptf1a dose control on endocrine vs. exocrine fate decisions, and investigating early events of pancreas specification activated by PTF1a that are difficult to access in an embryo, including the transcriptional networks surrounding PTF1a (i.e. direct and indirect targets). In summary, this work establishes Ptf1a as a sufficient initiator of pancreatic differentiation from ESCs leading to the subsequent formation of acinar, endocrine and ductal cell types after progressing through developmental stages that are phenotypically and morphologically similar to those that occur in vivo.
Supplementary Material
Suppl Fig 1. Time course staining of Dox-treated and untreated cells for Insulin and Pdx1 expression. Uninduced tet-Ptf1a cell cultures (A, C, E, G) are grown to the time point indicated and compared to Dox-treated (induced) tet-Ptf1a cell cultures (B, D, F, H). All cell cultures were stained for PDX1 (red) and insulin (green). The dashed line outlines the border of the plated EBs. Arrows point out the budding structures in the induced cultures. Arrowheads point to Insulin+ cells. Scale bars, 100μm.
Suppl Fig 2. Fluorescence micrographs showing expression of key pancreatic markers in the cultures post-induction and time course analysis of Ptf1a transcript levels in various induction regimes. (A–H) Differences in marker expression between uninduced and induced cultures at EB7+7. PDX1+ clusters are absent in uninduced (No Dox) cultures; There is low expression of FOXA2(A) and HNF6 (C) but more abundant expression of HNF4a (B) and MNX1(D). After Dox addition, PDX1+ bud-like structures appeared in the cultures. PDX1 expression was induced in a subset of FOXA2+ (E) and HNF6+ (G) populations. Some PDX1+ cells co-expressed MNX1 (H, cell cluster inside rectangle) and are distinct from HNF4a expressing cells (F), suggesting induction of Ptf1a activated pancreas specification and expression of key markers of pancreatic endoderm. Scale bars, 100μm. (I) Ptf1a mRNA levels over the course of differentiation in different induction schemes tested showed similar induciblity of the transgene. Error bars indicate s.e. n=3.
Suppl Fig 3. Co-expression pattern of Pdx1 and Nkx6.1 in the induced cultures. (A–C) At EB7+10, a broad subset of Pdx1+ cells mostly in the interior region of bud-like structures co-express Nkx6.1. (D–F) By EB7+14, Nkx6.1 is predominantly restricted to Pdx1+ cells at core of the bud-like structures and never at the periphery or tip, unlike Ptf1a. Insets show magnified regions to show coexpression. (G–I) Insulin+ cells by EB7+28 express both Pdx1 and Nkx6.1. Arrows point out coexpressing cells. Arrowhead points to a cell shown in inset at 60x magnification. Scale bars, 50μm
Suppl Fig 4. Co-staining for Cpa1 and Ngn3 in induced cultures at EB7+14. (A) Branching-like epithelium at 20x showing Cpa1+ cells in the periphery and arrows pointing to Ngn3+ cells embedded in interior regions. (B) Higher magnification (60x) image of another field of view indicating Cpa1 and Ngn3 are expressed in distinct cells in core regions. Scale bars, 100μm.
Suppl Fig 5. Electron micrographs of EB7+28 tet-Ptf1a cell cultures. (A) Representative image of uninduced cultures shows cells having non-specific structural morphology (2000x). (B) Representative image of Dox-induced culture demonstrating an acinar-like structure within dotted lines (3500x). (C) 10000x image of the edge of the same acinar structure. The chevron is pointing to a basement membrane, the arrowhead identifies a zymogen granule, and the arrow highlights the presence of abundant rough endoplasmic reticulum. (D) Representative image of a cell with secretory granules containing an electron-dense core and clear halo typical of mature insulin-containing granules (arrows; 10000x).
Suppl Fig 6. H&E staining and insulin immunohistochemistry (IHC) of Dox-treated tet-Ptf1a cell cultures in comparison to mouse pancreas. (A) H&E staining of induced tet-Ptf1a cell cultures showing glandular epithelial structures embedded in mesenchyme within dotted lines. (B) Contiguous section shows insulin staining within that structure. (C, D) H&E and insulin staining of a mouse pancreas sections, respectively. Scale bars, 100μm.
Suppl Fig 7. Differentiating ESCs ectopically expressing PTF1a respond to treatment with nicotinamide (Nic) and retinoic acid (RA) resulting in increased insulin+ cells to a greater degree than differentiating wild type ESCs (Ainv15) treated with or without factors. (A, B) tet-Ptf1a and Ainv15 cells differentiate spontaneously into few insulin+ cells in the absence of doxycycline (Dox). Induction of PTF1a increases differentiation into insulin+ cells (C), but has no effect on Ainv15 cells. Treatment with Nic and RA further enhances insulin expression (E), whereas Ainv15 cultures treated with Nic and RA exhibit much less insulin expression (F). Insulin+ cells derived from Nic+RA treated tet-Ptf1a cells co-express C-peptide (H). Scale bars, 100μm.
Acknowledgments
The authors would like to thank the following individuals for providing access to important reagents: Chris Wright for anti-Pdx1 antibody; Ray Macdonald for anti-Ptf1a antibody, Samuel Pfaff for anti-hlxb9 antibody, BCBC for anti-Ngn3 antibody and Michael Kyba for Ainv15 cell line. We would also like to acknowledge the CMN Waisman Core facility for training and use of their A1R-Si Nikon Confocal and FACS Calibur machines. Financial support for this project was provided by the following funding: NIH ARRA DK-78889-1A2, JDRF Research Grant #2007-75, ADA Innovative Award, Department of Surgery Surgical Associates Award, Vilas Associate Professorship Award, and ROTRF/JDRF Award #71612962.
Footnotes
Author Contributions: GN designed and performed experiments, wrote manuscript and analyzed data. RKV designed and performed experiments, reviewed and edited manuscript and analyzed data. JSO designed the experiments, analyzed data, and wrote and edited manuscript.
The authors declare no conflict of interest directly related to the subject matter of the manuscript.
References
- 1.Beres TM, Masui T, Swift GH, et al. PTF1 Is an Organ-Specific and Notch-Independent Basic Helix-Loop-Helix Complex Containing the Mammalian Suppressor of Hairless (RBP-J) or Its Paralogue, RBP-L. Molecular and Cellular Biology. 2006;26(1):117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krapp A, Knöfler M, Frutiger S, et al. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. The EMBO journal. 1996;15(16):4317–4329. [PMC free article] [PubMed] [Google Scholar]
- 3.Krapp A, Knöfler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Development. 1998;12 (23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockell M, Stevenson BJ, Strubin M, et al. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Molecular and Cellular Biology. 1989;9(6):2464–2476. doi: 10.1128/mcb.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawaguchi Y, Cooper B, Gannon M, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 6.Gittes GK. Developmental biology of the pancreas: A comprehensive review. Developmental Biology. 2009;326(1):4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Wiebe PO, Kormish JD, Roper VT, et al. Ptf1a Binds to and Activates Area III, a Highly Conserved Region of the Pdx1 Promoter That Mediates Early Pancreas-Wide Pdx1 Expression. Molecular and Cellular Biology. 2007;27(11):4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyatsuka T, Matsuoka T-a, Shiraiwa T, et al. Ptf1a and RBP-J cooperate in activating Pdx1 gene expression through binding to Area III. Biochemical and Biophysical Research Communications. 2007;362(4):905–909. doi: 10.1016/j.bbrc.2007.08.076. [DOI] [PubMed] [Google Scholar]
- 9.Dong PDS, Provost E, Leach SD, et al. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes & Development. 2008;22(11):1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahnfelt-Rønne J, Jørgensen MC, Klinck R, et al. Ptf1a-mediated control of Dll1 reveals an alternative to the lateral inhibition mechanism. Development. 2012;139(1):33–45. doi: 10.1242/dev.071761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaffer AE, Freude KK, Nelson SB, et al. Nkx6 Transcription Factors and Ptf1a Function as Antagonistic Lineage Determinants in Multipotent Pancreatic Progenitors. Developmental Cell. 2010;18 (6):1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotech. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 13.Rezania A, Bruin JE, Riedel MJ, et al. Maturation of Human Embryonic Stem Cell–Derived Pancreatic Progenitors into Functional Islets Capable of Treating Pre-existing Diabetes in Mice. Diabetes. 2012 doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotech. 2006;24(11):1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 15.Nostro MC, Sarangi F, Ogawa S, et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138(5):861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyba M, Perlingeiro RCR, Daley GQ. HoxB4 Confers Definitive Lymphoid-Myeloid Engraftment Potential on Embryonic Stem Cell and Yolk Sac Hematopoietic Progenitors. Cell. 2002;109(1):29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 17.Vincent R, Treff N, Budde M, et al. Generation and characterization of novel tetracycline-inducible pancreatic transcription factor-expressing murine embryonic stem cell lines. Stem cells and development. 2006;15(6):953–962. doi: 10.1089/scd.2006.15.953. [DOI] [PubMed] [Google Scholar]
- 18.Kahan BW, Jacobson LM, Hullett DA, et al. Pancreatic Precursors and Differentiated Islet Cell Types From Murine Embryonic Stem Cells. Diabetes. 2003;52(8):2016–2024. doi: 10.2337/diabetes.52.8.2016. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen MC, Ahnfelt-Rønne J, Hald J, et al. An Illustrated Review of Early Pancreas Development in the Mouse. Endocrine Reviews. 2007;28(6):685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 20.Pan FC, Wright C. Pancreas organogenesis: From bud to plexus to gland. Developmental Dynamics. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 21.Arber S, Han B, Mendelsohn M, et al. Requirement for the Homeobox Gene Hb9 in the Consolidation of Motor Neuron Identity. Neuron. 1999;23(4):659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 22.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and Development of the Liver. Developmental Cell. 2010;18(2):175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Seymour PA, Freude KK, Tran MN, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proceedings of the National Academy of Sciences. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, Law AC, Rajagopal J, et al. A Multipotent Progenitor Domain Guides Pancreatic Organogenesis. Developmental Cell. 2007;13(1):103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Oliver-Krasinski JM, Kasner MT, Yang J, et al. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. The Journal of Clinical Investigation. 2009;119(7):1888–1898. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson SB, Schaffer AE, Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134(13):2491–2500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi H, Spilde TL, Li Z, et al. Lectin as a marker for staining and purification of embryonic pancreatic epithelium. Biochemical and Biophysical Research Communications. 2002;293(2):691–697. doi: 10.1016/S0006-291X(02)00278-4. [DOI] [PubMed] [Google Scholar]
- 28.Otonkoski T, Beattie GM, Mally MI, et al. Nicotinamide is a potent inducer of endocrine differentiation in cultured human fetal pancreatic cells. The Journal of Clinical Investigation. 1993;92(3):1459–1466. doi: 10.1172/JCI116723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandler S, Andersson A. Stimulation of cell replication in transplanted pancreatic islets by nicotinamide treatment. Transplantation. 1988;46(1):30–31. doi: 10.1097/00007890-198807000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Vaca P, Berná G, Araujo R, et al. Nicotinamide induces differentiation of embryonic stem cells into insulin-secreting cells. Experimental Cell Research. 2008;314(5):969–974. doi: 10.1016/j.yexcr.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 31.Kruszewski M, Szumiel I. Sirtuins (histone deacetylases III) in the cellular response to DNA damage—Facts and hypotheses. DNA Repair. 2005;4(11):1306–1313. doi: 10.1016/j.dnarep.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Caton P, Kieswich J, Yaqoob M, et al. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54(12):3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]
- 33.Martín M, Gallego-Llamas J, Ribes V, et al. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Developmental Biology. 2005;284(2):399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 34.Shen C-N, Marguerie A, Chien C-Y, et al. All-trans retinoic acid suppresses exocrine differentiation and branching morphogenesis in the embryonic pancreas. Differentiation. 2007;75(1):62–74. doi: 10.1111/j.1432-0436.2006.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Micallef SJ, Janes ME, Knezevic K, et al. Retinoic Acid Induces Pdx1-Positive Endoderm in Differentiating Mouse Embryonic Stem Cells. Diabetes. 2005;54(2):301–305. doi: 10.2337/diabetes.54.2.301. [DOI] [PubMed] [Google Scholar]
- 36.Öström M, Loffler KA, Edfalk S, et al. Retinoic Acid Promotes the Generation of Pancreatic Endocrine Progenitor Cells and Their Further Differentiation into β-Cells. PLoS ONE. 2008;3(7):e2841. doi: 10.1371/journal.pone.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rovira M, Delaspre F, Massumi M, et al. Murine Embryonic Stem Cell–Derived Pancreatic Acinar Cells Recapitulate Features of Early Pancreatic Differentiation. Gastroenterology. 2008;135(4):1301–1310. e5. doi: 10.1053/j.gastro.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afelik S, Chen Y, Pieler T. Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes & Development. 2006;20(11):1441–1446. doi: 10.1101/gad.378706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent RK, Odorico JS. Reduced serum concentration is permissive for increased in vitro endocrine differentiation from murine embryonic stem cells. Differentiation. 2009;78(1):24–34. doi: 10.1016/j.diff.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Bernardo AS, Cho CHH, Mason S, et al. Biphasic Induction of Pdx1 in Mouse and Human Embryonic Stem Cells Can Mimic Development of Pancreatic β-Cells. STEM CELLS. 2009;27(2):341–351. doi: 10.1634/stemcells.2008-0310. [DOI] [PubMed] [Google Scholar]
- 41.Lavon N, Yanuka O, Benvenisty N. The Effect of Overexpression of Pdx1 and Foxa2 on the Differentiation of Human Embryonic Stem Cells into Pancreatic Cells. STEM CELLS. 2006;24(8):1923–1930. doi: 10.1634/stemcells.2005-0397. [DOI] [PubMed] [Google Scholar]
- 42.Jiang J, Au M, Lu K, et al. Generation of Insulin-Producing Islet-Like Clusters from Human Embryonic Stem Cells. STEM CELLS. 2007;25(8):1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 43.Micallef SJ, Li X, Janes ME, et al. Endocrine cells develop within pancreatic bud-like structures derived from mouse ES cells differentiated in response to BMP4 and retinoic acid. Stem Cell Research. 2007;1(1):25–36. doi: 10.1016/j.scr.2007.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Fig 1. Time course staining of Dox-treated and untreated cells for Insulin and Pdx1 expression. Uninduced tet-Ptf1a cell cultures (A, C, E, G) are grown to the time point indicated and compared to Dox-treated (induced) tet-Ptf1a cell cultures (B, D, F, H). All cell cultures were stained for PDX1 (red) and insulin (green). The dashed line outlines the border of the plated EBs. Arrows point out the budding structures in the induced cultures. Arrowheads point to Insulin+ cells. Scale bars, 100μm.
Suppl Fig 2. Fluorescence micrographs showing expression of key pancreatic markers in the cultures post-induction and time course analysis of Ptf1a transcript levels in various induction regimes. (A–H) Differences in marker expression between uninduced and induced cultures at EB7+7. PDX1+ clusters are absent in uninduced (No Dox) cultures; There is low expression of FOXA2(A) and HNF6 (C) but more abundant expression of HNF4a (B) and MNX1(D). After Dox addition, PDX1+ bud-like structures appeared in the cultures. PDX1 expression was induced in a subset of FOXA2+ (E) and HNF6+ (G) populations. Some PDX1+ cells co-expressed MNX1 (H, cell cluster inside rectangle) and are distinct from HNF4a expressing cells (F), suggesting induction of Ptf1a activated pancreas specification and expression of key markers of pancreatic endoderm. Scale bars, 100μm. (I) Ptf1a mRNA levels over the course of differentiation in different induction schemes tested showed similar induciblity of the transgene. Error bars indicate s.e. n=3.
Suppl Fig 3. Co-expression pattern of Pdx1 and Nkx6.1 in the induced cultures. (A–C) At EB7+10, a broad subset of Pdx1+ cells mostly in the interior region of bud-like structures co-express Nkx6.1. (D–F) By EB7+14, Nkx6.1 is predominantly restricted to Pdx1+ cells at core of the bud-like structures and never at the periphery or tip, unlike Ptf1a. Insets show magnified regions to show coexpression. (G–I) Insulin+ cells by EB7+28 express both Pdx1 and Nkx6.1. Arrows point out coexpressing cells. Arrowhead points to a cell shown in inset at 60x magnification. Scale bars, 50μm
Suppl Fig 4. Co-staining for Cpa1 and Ngn3 in induced cultures at EB7+14. (A) Branching-like epithelium at 20x showing Cpa1+ cells in the periphery and arrows pointing to Ngn3+ cells embedded in interior regions. (B) Higher magnification (60x) image of another field of view indicating Cpa1 and Ngn3 are expressed in distinct cells in core regions. Scale bars, 100μm.
Suppl Fig 5. Electron micrographs of EB7+28 tet-Ptf1a cell cultures. (A) Representative image of uninduced cultures shows cells having non-specific structural morphology (2000x). (B) Representative image of Dox-induced culture demonstrating an acinar-like structure within dotted lines (3500x). (C) 10000x image of the edge of the same acinar structure. The chevron is pointing to a basement membrane, the arrowhead identifies a zymogen granule, and the arrow highlights the presence of abundant rough endoplasmic reticulum. (D) Representative image of a cell with secretory granules containing an electron-dense core and clear halo typical of mature insulin-containing granules (arrows; 10000x).
Suppl Fig 6. H&E staining and insulin immunohistochemistry (IHC) of Dox-treated tet-Ptf1a cell cultures in comparison to mouse pancreas. (A) H&E staining of induced tet-Ptf1a cell cultures showing glandular epithelial structures embedded in mesenchyme within dotted lines. (B) Contiguous section shows insulin staining within that structure. (C, D) H&E and insulin staining of a mouse pancreas sections, respectively. Scale bars, 100μm.
Suppl Fig 7. Differentiating ESCs ectopically expressing PTF1a respond to treatment with nicotinamide (Nic) and retinoic acid (RA) resulting in increased insulin+ cells to a greater degree than differentiating wild type ESCs (Ainv15) treated with or without factors. (A, B) tet-Ptf1a and Ainv15 cells differentiate spontaneously into few insulin+ cells in the absence of doxycycline (Dox). Induction of PTF1a increases differentiation into insulin+ cells (C), but has no effect on Ainv15 cells. Treatment with Nic and RA further enhances insulin expression (E), whereas Ainv15 cultures treated with Nic and RA exhibit much less insulin expression (F). Insulin+ cells derived from Nic+RA treated tet-Ptf1a cells co-express C-peptide (H). Scale bars, 100μm.