Significance
Autophagy is an evolutionarily conserved catabolic process essential to maintaining cellular homeostasis through the breakdown and recycling of damaged organelles and long-lived proteins. We report that autophagy plays an essential cell-intrinsic role in maintaining the survival of a subset of innate-like cells known as invariant natural killer T (iNKT) cells. Autophagy deficiency prevents transition to a quiescent state after population expansion of thymic iNKT cells. Hence, autophagy-deficient iNKT cells accumulate mitochondria and oxygen radicals and subsequently die of apoptosis.
Keywords: CD1d, autophagy, iNKT cells, metabolism, glycolysis
Abstract
Autophagy is an evolutionarily conserved cellular homeostatic pathway essential for development, immunity, and cell death. Although autophagy modulates MHC antigen presentation, it remains unclear whether autophagy defects impact on CD1d lipid loading and presentation to invariant natural killer T (iNKT) cells and on iNKT cell differentiation in the thymus. Furthermore, it remains unclear whether iNKT and conventional T cells have similar autophagy requirements for differentiation, survival, and/or activation. We report that, in mice with a conditional deletion of the essential autophagy gene Atg7 in the T-cell compartment (CD4 Cre-Atg7−/−), thymic iNKT cell development—unlike conventional T-cell development—is blocked at an early stage and mature iNKT cells are absent in peripheral lymphoid organs. The defect is not due to altered loading of intracellular iNKT cell agonists; rather, it is T-cell–intrinsic, resulting in enhanced susceptibility of iNKT cells to apoptosis. We show that autophagy increases during iNKT cell thymic differentiation and that it developmentally regulates mitochondrial content through mitophagy in the thymus of mice and humans. Autophagy defects result in the intracellular accumulation of mitochondrial superoxide species and subsequent apoptotic cell death. Although autophagy-deficient conventional T cells develop normally, they show impaired peripheral survival, particularly memory CD8+ T cells. Because iNKT cells, unlike conventional T cells, differentiate into memory cells while in the thymus, our results highlight a unique autophagy-dependent metabolic regulation of adaptive and innate T cells, which is required for transition to a quiescent state after population expansion.
Autophagy is an evolutionarily conserved catabolic process that, by facilitating the breakdown and recycling of damaged organelles and long-lived proteins, is essential to maintaining cellular homeostasis (1). The autophagy pathway is highly regulated during development and also by environmental factors such as nutrient availability/starvation, hypoxia, and reactive oxygen species (ROS). The process is controlled by a number of autophagy-related genes (Atg) and starts with the formation of a double-membraned vesicle (the autophagosome) engulfing the cytoplasmic components to be delivered to the lysosome for degradation. Formation of the autophagosome requires the concerted action of two ubiquitin-like conjugation systems in which Atg12 is covalently linked to Atg5 and Atg8 is conjugated to phosphatidylethanolamine (2, 3). Atg7 is a necessary catalyst in both conjugation systems and is therefore essential for autophagy (3).
Due to its important role in cellular homeostasis, dysregulation of autophagy has been implicated in several pathological processes, including neurodegeneration, autoimmunity, cancer, and infection (4). Furthermore, autophagy plays an important role during immune responses by regulating pathogen handling and antigen presentation (5, 6). It is well established that different populations of αβ T lymphocytes can recognize not only peptides in the context of MHC class I and class II molecules, but also foreign and self-lipids in association with CD1 proteins (7), antigen-presenting molecules that share structural similarities with MHC class I molecules. Of the five CD1 isoforms, CD1d restricts the activity of a family of cells known as invariant natural killer T (iNKT) cells because of their semi-invariant T-cell receptor (TCR) use (7). It has been demonstrated that autophagy can deliver intracellular antigens to MHC class II containing compartments for antigen processing and loading onto MHC class II molecules (8). Considering the intersection of this compartment with the CD1 loading pathway and the observation that lysosomal storage disease models impair presentation of intracellular iNKT cell agonists to iNKT cells (9–11), we investigated the role of autophagy in lipid antigen presentation by CD1 molecules.
iNKT cells develop in the thymus in a tightly regulated selection process that requires self-lipid antigen presentation by CD4+CD8+ double-positive (DP) CD1d+ thymocytes and access of CD1d molecules to the lysosomal compartment. Concerted signaling initiated through the TCR and molecules of the SLAM family of receptors imparts to the developing iNKT cells a memory phenotype with the ability to promptly secrete a plethora of cytokines upon stimulation and to home to inflamed tissues (12). Impaired trafficking of CD1d molecules to the lysosome, lysosomal lipid storage, and lack of lipid transfer proteins have all been shown to profoundly affect iNKT cell development (7). Likewise, genetic ablation of signaling molecules, adaptors, or transcription factors downstream of the TCR/SLAM axis also leads to impaired iNKT cell development (12).
In this paper, we report that deletion of the essential autophagy gene Atg7, which has no described autophagy-unrelated functions, abrogates iNKT cell development in a T-cell–intrinsic manner, affecting survival of developing iNKT cells in the thymus. Unexpectedly, Atg7-deficient thymocytes and bone marrow-derived dendritic cells (BMDCs) showed no defect in exogenous or self-lipid antigen presentation.
Results
Early Block of iNKT Cell Development in Atg7-Deficient Mice.
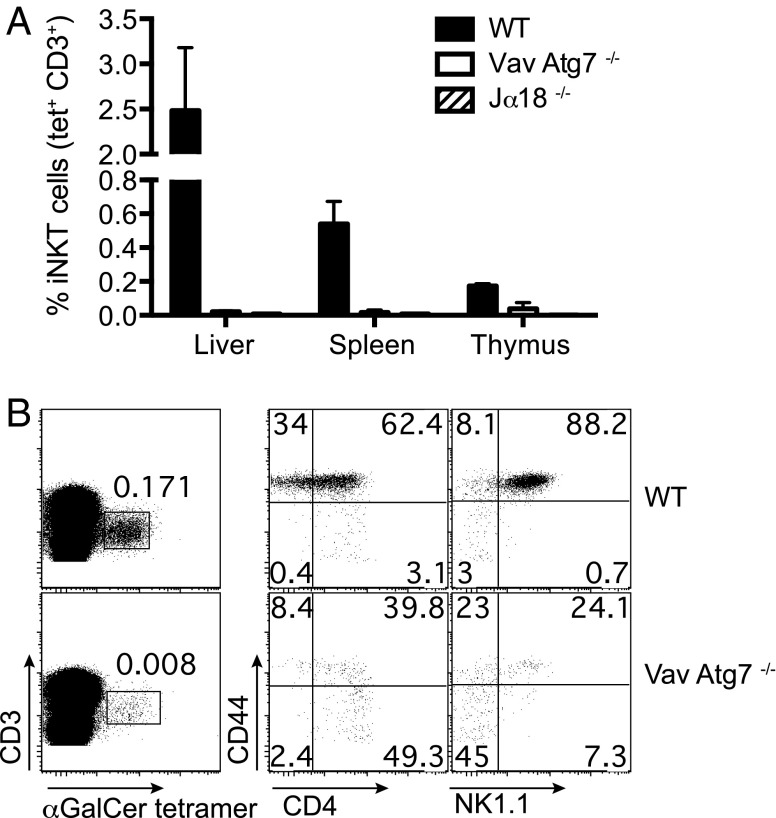
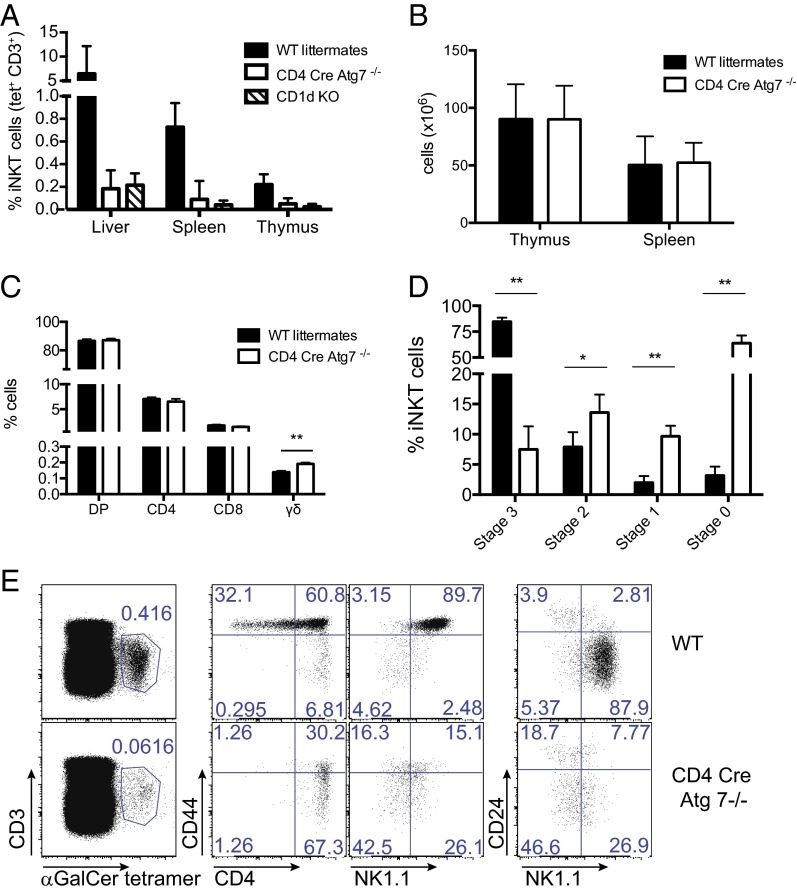
To study the role of autophagy in lipid antigen presentation, we analyzed mice in which Atg7 has been conditionally deleted in the hematopoietic system [Vav-Atg7 −/− mice (13)]. Remarkably, no mature iNKT cells could be detected by α-GalCer-CD1d tetramer staining in the liver and spleen of Vav-Atg7−/− mice (Fig. 1A). A fourfold reduction in the percentage of iNKT cells was observed in the thymus (Fig. 1A). Furthermore, the majority of remaining thymic iNKT cells displayed an immature phenotype with a reduction in the fraction of stage 3 CD44+ NK1.1+ α-GalCer-CD1d tetramer+ cells (Fig. 1B). As Vav-Atg7−/− mice have a defect in hematopoietic stem cells leading to progressive anemia, splenomegaly, lymphadenopathy, and death by 12 wk of age (14), we analyzed a second genetic mouse model in which Atg7 deficiency was restricted to the T-cell compartment by crossing Atg7flox/flox mice (15) with CD4-Cre transgenic mice. We confirmed efficient Atg7 deletion in DP and CD4+ single-positive (SP) thymocytes of CD4 Cre-Atg7−/− mice, consistent with the induction of Cre expression at the DP stage (Fig. S1). Similar to previous observations in Vav-Atg7−/− mice, CD4 Cre-Atg7−/− mice also lacked mature iNKT cells in the liver or spleen, with a fivefold reduction in the percentage of iNKT cells in the thymus (Fig. 2A and Fig. S2). Total cell numbers of thymus and spleen were comparable between wild-type (WT) and CD4 Cre-Atg7−/− littermates (Fig. 2B), as were the percentages of thymic DP, CD4+, and CD8+ T cells, whereas percentages of thymic γδ T cells were significantly increased in CD4 Cre-Atg7−/− mice (Fig. 2C). These results underscore the specific defect in thymic iNKT cell development as conventional T cells were reduced only in the periphery. Indeed, despite normal splenic cell numbers (Fig. 2B), we observed reduced percentages of CD3+ T cells and of non-CD4+ T cells in CD4 Cre-Atg7−/− mice (Fig. S3A). Residual iNKT cells in the thymus were reduced both in percentages (Fig. 2D) and in absolute numbers (Fig. S3B) and displayed an immature phenotype with over 70% of the cells at stage 0 (CD24+ CD44−), around 20% between stage 1 (CD24− CD44−) and 2 (CD24− CD44+), and less than 10% reaching stage 3 CD24− CD44+ NK1.1+ (Fig. 2 D and E and Fig. S3C). Consistent with the more immature phenotype, thymic CD4 Cre-Atg7−/− iNKT cells also expressed less Bcl2, Egr2, and PLZF, the last two being transcription factors essential for iNKT cell development (Fig. S3D) (16, 17). Similar to iNKT cells, type II NKT cells (defined as α-GalCer-CD1d tetramer− TCRβ+ NK1.1+) were also reduced in frequencies in the thymus of CD4 Cre-Atg7−/− mice (Fig. S3E).
Fig. 1.
Early block of iNKT cell development in Vav-Atg7−/− mice. (A) Frequencies (mean ± SD) of liver, splenic, and thymic iNKT cells (CD3+ α-GalCer-CD1d tetramer+ cells) after gating on live cells, singlets, and excluding B cells. n = 4 mice/group (WT, Vav-Atg7−/−, and Jα18−/− mice, one experiment representative of three). (B) Representative FACS dot plots depicting thymic iNKT cells (Left) and their maturation after staining with CD44, CD4, and NK1.1 antibodies (Right). Top, WT. Bottom, Vav-Atg7−/− mice.
Fig. 2.
Early block of iNKT cell development in CD4 Cre-Atg7−/− mice. (A) Frequencies (mean ± SD) of liver, splenic, and thymic iNKT cells (CD3+ α-GalCer-CD1d tetramer+ cells) after gating on live cells, singlets, and excluding B cells. n = 33 mice/group (8 for CD1d KO; 16 livers). (B) Absolute numbers of live cells in the thymus and spleen of WT and CD4 Cre-Atg7−/− mice. (C) Frequencies of DP, CD4+, CD8+, and γδ T cells in the thymus of WT and CD4 Cre-Atg7−/− mice. **P = 0.002. (D) Frequency of iNKT cell subsets identified by CD24, CD44, and NK1.1 expression (according to ref. 71) in the thymus of WT and CD4 Cre-Atg7−/− mice. *P = 0.02; **P < 0.0003. (E) Representative FACS dot plots depicting thymic iNKT cells (Left) and their maturation after staining with CD44, CD4, CD24, and NK1.1 antibodies (Right). Top, WT. Bottom, CD4 Cre Atg7−/− mice.
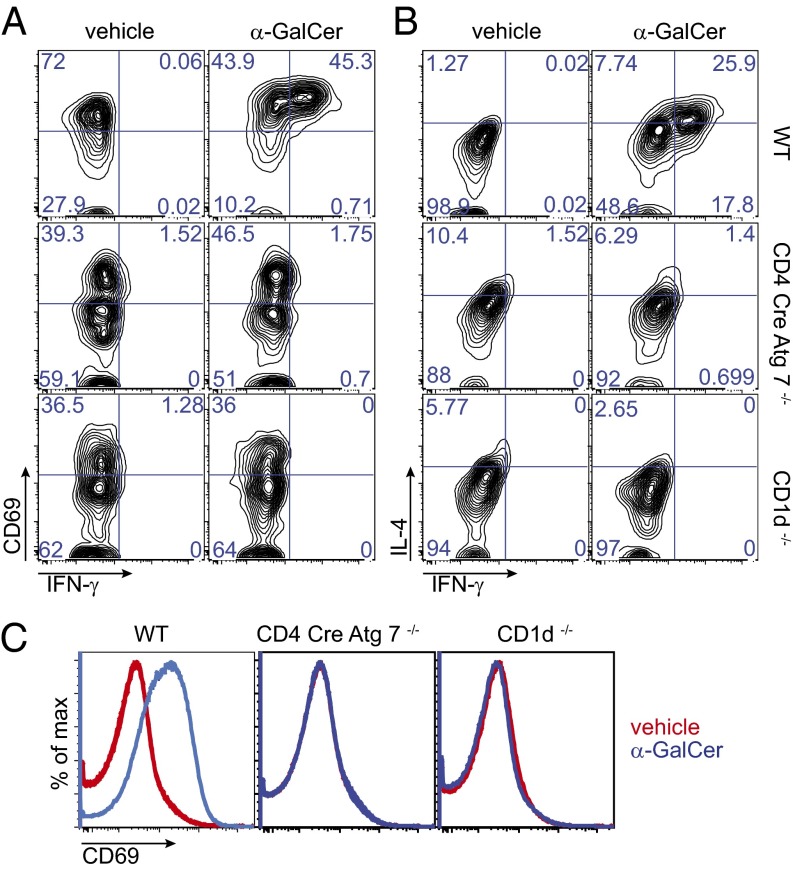
In agreement with the lack of mature splenic iNKT cells upon in vivo injection of the strong glycolipid agonist α-GalCer, we failed to detect a population of activated (CD69 bright) IFNγ+ IL4+ α-GalCer-CD1d tetramer+ cells in the spleen of CD4 Cre-Atg7−/− and CD1d−/− mice, which in contrast was clearly detectable in the spleen of WT mice (Fig. 3 A and B). Likewise, iNKT cell-dependent B-cell activation (as depicted by CD69 expression) was observed only in WT, and not in CD4 Cre-Atg7−/− or CD1d−/− mice (Fig. 3C).
Fig. 3.
Lack of iNKT cell activation in the spleen of CD4 Cre-Atg7−/− mice injected with α-GalCer. Four mice/group were injected i.v. with 1 μg of α-GalCer or vehicle, and intracellular staining of splenocytes was performed after 3 h. Shown are representative FACS dot plots depicting the expression of CD69 and IFN-γ (A) or IL-4 and IFN-γ (B) by gated splenic iNKT cells. Contour dot plots display 8,000 cells (WT vehicle), 2,948 cells (WT α-GalCer); 329 cells (CD4 Cre-Atg7−/− vehicle), and 286 cells (CD4 Cre-Atg7−/− α-GalCer). A small population of cells binding α-GalCer CD1d tetramers but not secreting cytokines is also found in the spleen of CD1d-deficient mice (156 cells in vehicle and 151 cells in α-GalCer–injected mice). (C) Expression of CD69 on B220-gated B cells in vehicle (red) or α-GalCer–injected (blue) WT, CD4 Cre-Atg7−/−, and CD1d-deficient mice (overlays of two mice/group/treatment, one for CD1d-deficient mice).
Normal Lipid Antigen Presentation by Atg7-Deficient Cells.
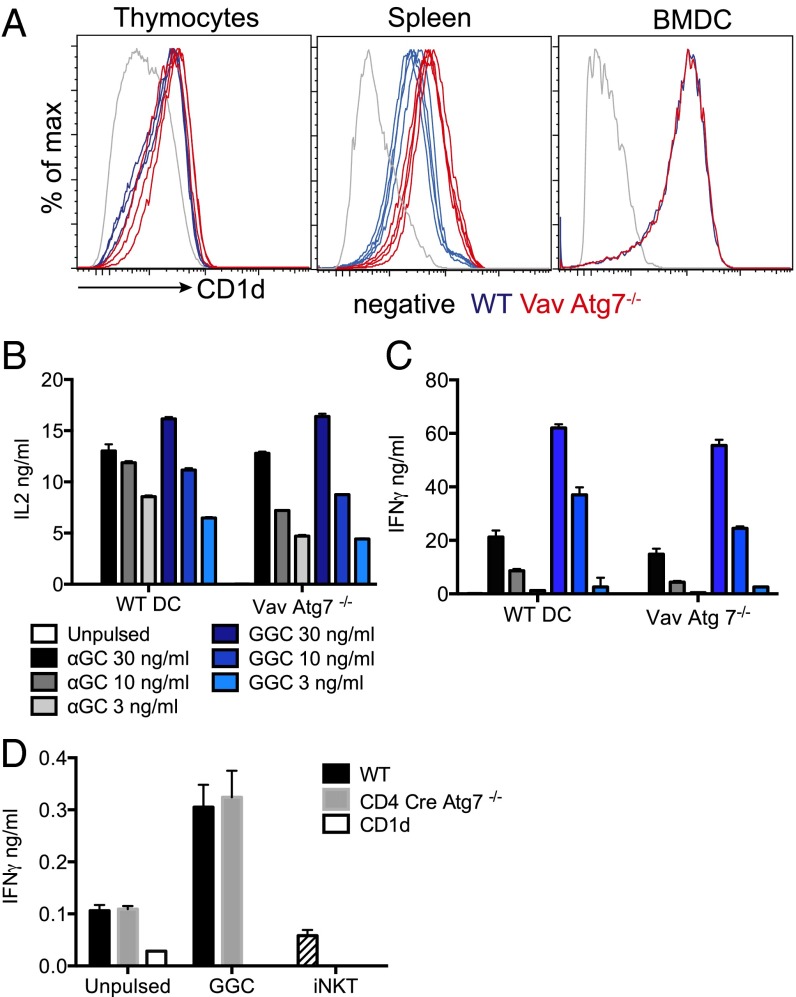
For iNKT cell selection to take place in the thymus, thymocytes need to express CD1d molecules, which need to traffic through the lysosomes where loading of self-lipids takes place supported by lipid transfer proteins (18, 19). We reasoned that Atg7 deficiency could influence CD1d expression, the lipid repertoire, and/or the capacity of selecting thymocytes to present lipids on CD1d molecules, potentially impairing iNKT cell-positive selection. However, the results of our experiments were not consistent with this possibility, as we showed that CD1d expression in thymocytes and splenic B-cell and BMDCs from Vav-Atg7−/− mice was normal or even increased (Fig. 4A). Furthermore, BMDC from WT and Vav-Atg7−/− mice were equally potent in stimulating the murine hybridoma DN32 (Fig. 4B) and human iNKT cells (Fig. 4C) when pulsed with different concentrations of α-GalCer and of the disaccharide Gal(α1→2)GalCer (GalGalCer), whose presentation is strictly dependent on uptake and processing by α-galactosidase in the lysosome (10). As lysosomal storage disease models impair presentation of GalGalCer and access of CD1d to the lysosomes (9–11), these results suggest normal trafficking of CD1d molecules and lack of lipid storage in Vav-Atg7−/− cells. Finally, we also observed comparable autoreactivity of human iNKT cells to WT and Vav-Atg7−/− cells (Fig. 4D). However, we could not determine autoreactivity of murine iNKT cells as the release of cytokines was below the detection limit of our assay.
Fig. 4.
Normal CD1d expression and lipid antigen presentation in Vav Cre-Atg7−/− mice. (A) CD1d expression at the cell surface of thymocytes, splenic B cells, and BMDCs of WT (blue) and Vav Cre-Atg7−/− mice (red). Overlays of four mice/group are shown (one sample for BMDCs). Gray lines represent unstained controls. (B and C) BMDCs were pulsed with the indicated concentrations of α-GalCer and GalGalCer (GGC) or left unpulsed and used to stimulate the murine DN32 hybridoma (B) or human iNKT cells (C). IL-2 (B) and IFN-γ (C) released in the supernatant by activated iNKT cells were measured by ELISA after 36 h (mean ± SD). (D) Human iNKT cells were incubated with thymocytes unpulsed or GGC-pulsed, and IFN-γ released in the supernatant was measured by ELISA after 36 h (mean ± SD). Hatched bars (iNKT group) represent spontaneous IFN-γ release in the absence of stimulation.
All together, these results argue against a contribution of Atg7-dependent autophagy in modulating CD1d-dependent lipid antigen presentation to developing iNKT cells.
Atg7 Deficiency Regulates iNKT Cell Development in a T-Cell–Intrinsic Manner.
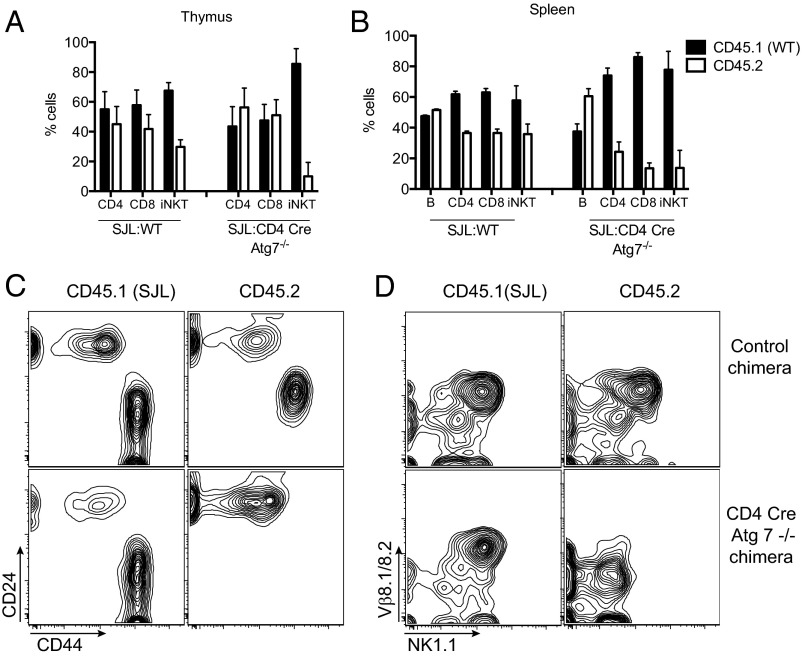
The above results suggested that a T-cell–intrinsic defect may be determining the block in iNKT cell development in CD4 Cre-Atg7−/− mice. To confirm and extend these findings, we set up bone marrow chimeras. CD4 Cre-Atg7−/− or WT bone marrow cells (CD45.2) were mixed 1:1 with B6.SJL bone marrow (CD45.1) and transplanted into lethally irradiated CD45.1 B6.SJL hosts. The peripheral blood of recipient mice was analyzed after 8 wk to assess reconstitution, which was comparable between control (SJL:WT) and CD4 Cre-Atg7−/− chimeras (Fig. S4A). Mice were killed at 10 wk after reconstitution to analyze thymus and spleen. The overall distribution of cell populations in thymus and spleen was comparable between the two types of chimeras (Fig. S4 B and C). We next analyzed the contribution of each donor population toward reconstitution on the basis of expression of the congenic marker CD45. In the thymus of SJL:WT control chimeras, we observed a moderate bias toward CD45.1 T cells, likely due to radio-resistant recipient cells (20) (Fig. 5A). In the thymus of the CD4 Cre-Atg7−/− chimeras, despite an equivalent reconstitution of CD4+ and CD8+ T cells by WT (CD45.1) and knockout (CD45.2) cells, iNKT cells were almost entirely derived from WT CD45.1 donor cells (Fig. 5A). Overall, thymic Atg7-deficient iNKT cell reconstitution was 5.95-fold less efficient than WT iNKT cell reconstitution (Table 1). Furthermore, although the iNKT cells derived from WT donor cells reached full maturity, as shown by the expression of CD44, Vβ8.1/8.2, and NK1.1 (Fig. 5 C and D), the few iNKT cells derived from the CD4 Cre-Atg7−/− bone marrow precursors were arrested in their development at the CD24high CD44dim/neg NK1.1− immature stage (Fig. 5 C and D).
Fig. 5.
Atg7 deficiency causes a T-cell–intrinsic block of iNKT cell maturation. CD45.2 BM cells from WT or CD4 Cre-Atg7−/− were mixed 1:1 with CD45.1 SJL (WT) BM cells and transplanted into lethally irradiated CD45.1 SJL WT recipients (n = 4/group). (A) Frequency (mean ± SD) of CD45.1 (black bars) and CD45.2 (white bars) cells among CD4+, CD8+, and iNKT cells in the thymus of control (SJL:WT) and SJL:CD4 Cre-Atg7−/− chimeras, 9 wk after engraftment. (B) Frequency (mean ± SD) of CD45.1 (black bars) and CD45.2 (white bars) cells among CD4+, CD8+, and iNKT cells in the spleen of control (SJL:WT) and SJL:CD4 Cre-Atg7−/− chimeras, 9 wk after engraftment. (C and D) Representative FACS dot plots depicting maturation of thymic iNKT cells (after gating on CD3+ α-GalCer-CD1d tetramer+ cells) as assessed by expression of CD24 and CD44 (C) or Vβ8.1/8.2 and NK1.1 (D). Top, control chimeras. Bottom, CD4 Cre-Atg7−/− chimeras.
Table 1.
Reconstitution efficiency of the bone marrow chimeras
| Cell type | Thymus | Spleen |
| B | — | 1.488 |
| CD4 | 1.58 | 0.554 (2.68) |
| CD8 | 1.48 (1.06) | 0.27 (5.49) |
| iNKT | 0.26 (5.95) | 0.28 (5.2) |
Reconstitution efficiency of each indicated population in SJL:CD4 Cre-Atg7−/− chimeras compared with control SJL:WT chimeras was calculated as follows: (% of population of interest from CD45.2 Atg7−/− donor/% of population of interest from CD45.1 WT donor)/(% of population of interest from CD45.2 WT donor/% of population of interest from CD45.1 WT donor). In parentheses are the ratios of each population against CD4 cells (thymus) or B cells (spleen), indicating that iNKT cell reconstitution in the Atg7-deficient compartment is 5.95-fold less efficient in the thymus and 5.2-fold less efficient in the spleen.
In the spleen of control SJL:WT chimeric mice, we again observed a slight bias toward CD45.1 donor T cells, whereas the B-cell compartment was equally reconstituted (Fig. 5B). In the spleen of CD4 Cre-Atg7−/− chimeric mice, as expected, we observed an equal contribution of the two donor populations toward reconstitution of the B-cell compartment because the Cre transgene is not active in B cells. In contrast, the contribution of CD4 Cre-Atg7−/−-deficient cells toward reconstitution of the T-cell compartment was markedly reduced and iNKT cells were almost entirely derived from WT donor cells (Fig. 5B). Overall, we calculated that reconstitution of Atg 7-deficient splenic CD4+, CD8+, and iNKT cells was 2.68-, 5.49-, and 5.2-fold less efficient than that of their WT counterparts, respectively (Table 1).
The phenotype of the chimeric mice thus resembles that of the CD4 Cre-Atg7−/− mice, demonstrating that Atg7 deficiency has a T-cell–intrinsic effect on the development/survival of the iNKT cell compartment and on the survival of peripheral T cells.
Normal Rearrangement of Vα14-Jα18 Gene Segments.
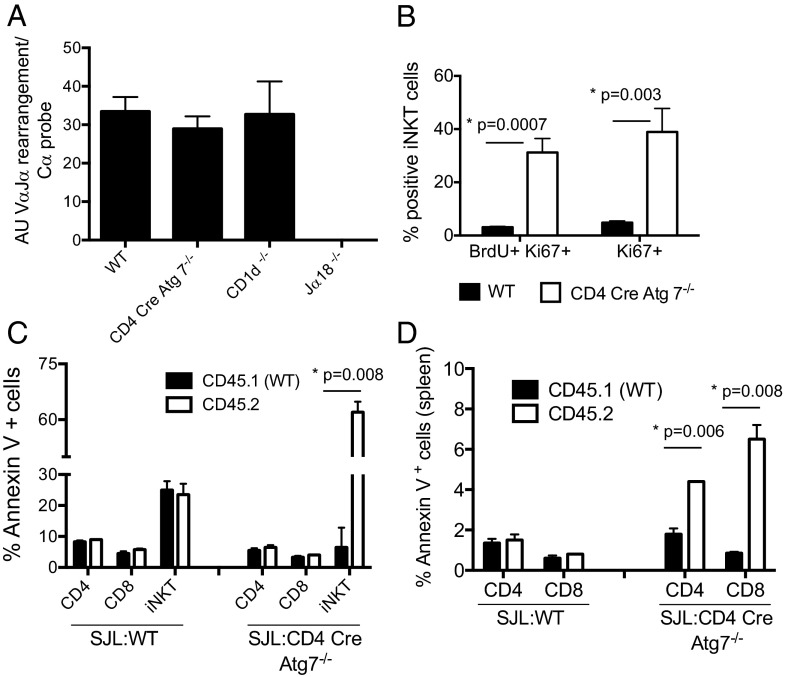
Rearrangement of Vα14-to-Jα18 gene segments in DP thymocytes is necessary for iNKT TCR-positive selection by CD1d self-lipid complexes (7). The process is temporally regulated during ontogeny and requires adequate survival of the DP population (21). We therefore measured by qPCR Vα14–Jα18 rearrangement in DP thymocytes, sorted after exclusion of α-GalCer-CD1d tetramer+ cells, and found equal rearrangement in WT, CD4 Cre Atg7−/−, and CD1d−/−, whereas no rearrangement was detected in control Jα18−/− mice, as expected (Fig. 6A). These results suggest that the reduced numbers of iNKT cells in Atg7-deficient mice are not due to reduced life span of DP thymocytes potentially impairing the late Vα14–Jα18 rearrangement.
Fig. 6.
(A) Equivalent VαJ14–Jα18 rearrangement in thymocytes from WT and CD4 Cre-Atg7−/− mice. The VαJ14–Jα18 rearrangement was determined by qPCR as described in Materials and Methods on DP thymocytes sorted after depletion of α-GalCer-CD1d tetramer+ cells. Data from two independent sorts: n = 5 WT, n = 5 Cre-Atg7−/−, n = 4 CD1d−/−, and n = 4 Jα18−/−. (B) Enhanced BrdU incorporation in CD4 Cre-Atg7−/− iNKT cells. Thymic iNKT cells (α-GalCer-CD1d tetramer+ TCR-β+ cells) were analyzed for BrdU incorporation and Ki-67 expression 24 h after injection of 2 mg BrdU. One representative experiment of two; n = 3. (C and D) Atg7-deficient T cells are more susceptible to apoptosis. Thymocytes (C) and splenocytes (D) of control (SJL:WT) and SJL:CD4 Cre-Atg7−/− chimeras were cultured 4 h in complete medium before staining with Annexin V antibodies to detect apoptotic cells. Depicted are the percentages (mean ± SD) of apoptotic CD45.1 (black bars) and CD45.2 cells (white bars) among CD4, CD8, and iNKT cells.
Proliferative Capacity of CD4 Cre Atg7−/− iNKT Cells.
We next determined whether the decreased iNKT cell numbers in Atg7-deficient mice could be due to decreased proliferation as immature iNKT cells undergo population expansion in the thymus. We observed increased BrdU incorporation and Ki-67 staining in CD4 Cre Atg7−/− iNKT cells compared with WT iNKT cells (Fig. 6B). This difference was not apparent at the immature NK1.1− CD44− CD24+ stage, but it was evident at the NK1.1− CD44+ CD24− stage, when the developmental block in CD4 Cre Atg7−/− iNKT cells occurs (Fig. S5 A and B). Although these results suggest that terminal maturation of Atg7-deficient iNTK cells is not due to defective proliferation, it remains possible that autophagy might regulate cell cycle progression and, ultimately, cell survival.
Defective iNKT Cell Survival in CD4 Cre-Atg7−/− Mice.
It has previously been shown that Atg7 and Atg5 play a role in survival of peripheral T cells (22, 23). In agreement with previously published data (23, 24), splenic T cells were reduced in percentages and absolute numbers (Fig. 2B and Fig. S3A), and survival of naive (CD62L+ CD44−) and central memory T cells (CM, CD62L+ CD44+) was particularly affected (Fig. S6 A and B). The phenotype was replicated in the bone marrow chimeric mice, where survival of Atg7-deficient T cells was compromised with a marked decrease in memory T cells (Fig. 5B and Fig. S7).
To investigate the contribution of Atg7 in iNKT cell survival, thymocytes and splenocytes from chimeric mice were cultured in complete medium for 4 h and the rate of apoptosis was measured by Annexin V staining. In the thymus, comparable survival was observed for CD4+ and CD8+ T, irrespective of the donor origin (CD45.1 or CD45.2). However, survival of Atg7-deficient iNKT cells (CD45.2) was markedly impaired (Fig. 6C). In the spleen of CD4 Cre-Atg7−/− chimeric mice, we observed increased apoptosis in both Atg7-deficient CD4+ and CD8+ cells (Fig. 6D), consistent with the impaired survival of Atg7-deficient mature T cells (23). Increased apoptosis of Atg7-deficient iNKT cells was also observed ex vivo (Fig. S5 C and D). Consistent with these results, we observed that upon in vitro culture with α-GalCer, CD4 Cre-Atg7−/− thymic iNKT cells underwent a threefold lower expansion compared with WT counterparts (Fig. S5E), despite evidence of cell proliferation and activation, as measured by carboxyfluorescein succinimidyl ester (CFSE) dilution and CD25 up-regulation (Fig. S5F).
Mitochondrial Content and Autophagy Are Developmentally Regulated in iNKT Cells.
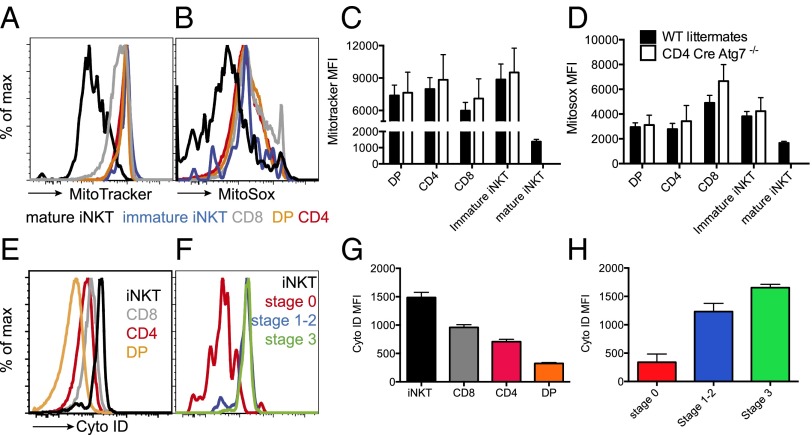
Autophagy-dependent changes in mitochondrial content mark the developmental transition from thymocytes to circulating peripheral T cells (23). We therefore asked whether the different impact of Atg7 in the thymic development of iNKT versus conventional T cells correlated with a distinct regulation of mitochondrial content and autophagy during development of the two populations. Toward this aim we stained cells with the lipophilic thiol-reactive dye MitoTracker Green that selectively labels the mitochondrial inner membrane and matrix (25) and that has previously been used to measure mitochondrial content in hematopoietic cells (14). We observed that within the thymus mature CD4+ and CD8+ T cells have a higher staining with MitoTracker Green compared with DP thymocytes and immature iNKT cells (Fig. 7 A and C). In addition, in iNKT cells MitoTracker Green staining is decreased during transition between immature CD24+ cells and mature CD44+ cells (Fig. 7 A and C). In parallel with the reduction in mitochondrial staining, mature iNKT cells also showed lower staining with the superoxide-specific probe MitoSox (Fig. 7 B and D). Although we observed only a minor increase in MitoTracker Green and MitoSox staining in the thymus of CD4 Cre-Atg7−/− mice, the difference reached statistical significance in chimeric mice and for conventional T cells in the spleen. Although a minor difference in MitoTracker Green staining was previously observed in lck Cre-Atg7−/− mice, this was not the case in Atg5 fetal liver chimeric mice, suggesting subtle differences in different model systems, possibly due to the timing of the deletion (13, 23).
Fig. 7.
Mitochondrial content and autophagy are developmentally regulated in iNKT cells. (A and B) Representative histograms of WT thymocytes stained with MitroTracker Green (A) and MitoSox (B). Black lines: mature iNKT cells; blue: immature iNKT cells (CD24+ CD44− CD3+ α-GalCer-CD1d tetramer+); gray: CD8; red: CD4; orange: double positive (DP). (C and D) Mean fluorescence intensity (MFI) (mean ± SD) of MitoTracker Green (C) and MitoSox (D) staining of WT (black bars) and CD4 Cre-Atg7−/− thymocytes. n = 4 mice/group, one experiment representative of four. (E) Representative histograms of WT thymocytes stained with Cyto ID. Black lines: iNKT cells (CD3+ α-GalCer-CD1d tetramer+); gray: CD8; red: CD4; orange: double positive (DP). (F) Representative histograms of WT iNKT cells (CD3+ α-GalCer-CD1d tetramer+) stained with Cyto ID. Red: stage 0 (CD24+ CD44−); blue: stages 1–2 (CD24− CD44−/+); green: stage 3 (CD44+ NK1.1+). (G and H) MFI (mean ± SD) of Cyto ID staining of WT thymocyte populations (G) and iNKT cells (H) color coded as in E and F, respectively. n = 4 mice/group, one experiment representative of four.
We next examined differences in autophagy levels in developing iNKT cells versus conventional T cells, using the Cyto ID green autophagy dye, which has been previously validated for use in flow cytometry (26). iNKT cells displayed the highest Cyto ID staining in the thymus, whereas SP CD4+ and CD8+ T cells had intermediate staining, and DP precursor the lowest (Fig. 7 E and G). Furthermore, Cyto ID staining increased with iNKT cell maturation from stage 0 (lowest) to stage 3 (highest) (Fig. 7 F and H).
One possible interpretation of these results is that, whereas conventional T cells control their mitochondrial content (as defined by MitoTracker Green staining) during the transition from the thymus to the periphery, iNKT cells need to have already done so in the thymus. This process is likely to require autophagy, which through mitophagy is known to regulate mitochondrial homeostasis.
Mitophagy Is Required for Memory T-Cell Development and Memory T-Cell Metabolic Requirements.
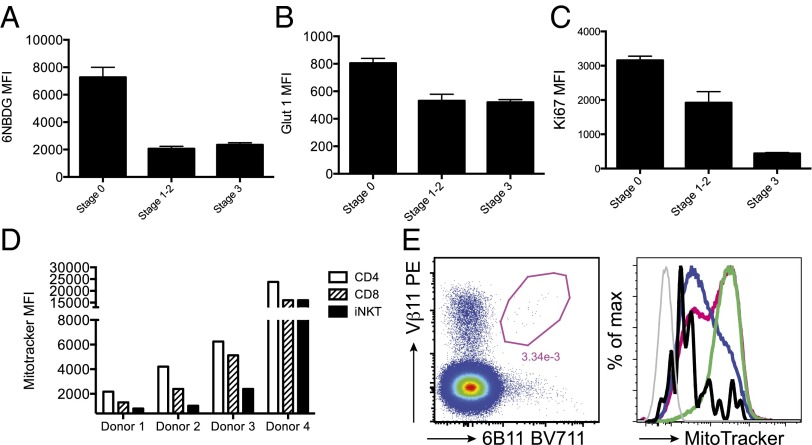
In splenic T cells, MitoTracker Green staining is lower in CD8+ than in CD4+ T cells, and among CD8+ T cells, CM and naive cells have lower levels of MitoTracker Green staining than effector memory and effectors (Fig. S6C). Interestingly, CD8+ CM and naive T cells are mostly affected by lack of functional Atg7 (Fig. S6A) (23). The observation that lack of Atg7 affects iNKT cells earlier than conventional T cells, in addition to the fact that iNKT cells differentiate into memory cells while in the thymus, suggests that mitochondrial clearance might be related to changes in cell metabolism during effector-memory differentiation. It is well known that naive T cells rely on oxidative phosphorylation for their metabolic demands but switch to aerobic glycolysis upon activation (27). This metabolic reprogramming is then reversed in memory T cells, which switch back from aerobic glycolysis to oxidative metabolism, allowing survival of memory T cells for extended periods in the absence of glucose or growth factors (28). Survival of T-cell precursors in the thymus depends on Notch and IL-7–dependent up-regulation of glucose uptake and glycolysis (29). The high metabolic demand of T-cell precursors correlates with extensive proliferation during thymic β-selection. Unlike conventional T cells, iNKT cells undergo a second proliferative wave after selection, which leads to expansion of iNKT cell numbers as development progresses from stage 0 to stage 3 (30). We therefore investigated glucose uptake in iNKT cells by incubating thymocytes with the fluorescent glucose analog 6-NBDG. When gating on iNKT cells, we observed higher uptake by immature stage 0 iNKT cells than by stage 3 mature iNKT cells (Fig. 8A). This was accompanied by a higher expression of the glucose transporter Glut1 (Fig. 8B) and correlated with higher proliferation of immature iNKT cells, as assessed by Ki67 staining (Fig. 8C). Following this population expansion phase, mature iNKT cells differentiate into memory cells. During this transition they enter a quiescent phase in which we speculate that their energy supplies may rely mainly on catabolic processes and autophagy (consistent with the Cyto ID staining data shown in Fig. 7H).
Fig. 8.
(A–C) Reduced glucose uptake in maturing iNKT cells. Thymocytes were incubated with the fluorescent glucose analog 6-NBDG, and uptake was assayed by flow cytometry. iNKT cells were gated as CD3+ α-GalCer-CD1d tetramer+ cells and staged as described in Fig. 7F. (A) MFI (mean ± SD) of 6-NBDG in iNKT cell subsets. (B) MFI (mean ± SD) of Glut-1 expression in gated iNKT cell subsets. (C) MFI (mean ± SD) of intracellular Ki67 expression in iNKT cell subsets. (D and E) Mitochondrial content is developmentally regulated in human thymuses. (D) Summary of MitoTracker Green MFI values in CD4, CD8, and iNKT cells in four human thymuses. (E) Representative FACS plots (donor 2) depicting iNKT cell identification with 6B11 and Vβ11 antibodies (Left) and overlay histograms for MitoTracker Green staining (Right). Black line: iNKT cells; blue line: CD8; pink line: CD4; green line: double-positive cells; gray line: negative control. Due to a limited number of iNKT cells it was not possible to further analyze different subsets.
All together, these results support the notion that Atg7-dependent autophagy is required for memory T-cell development and demonstrate that mitochondrial content (as measured by MitoTracker Green staining) is developmentally regulated in iNKT cells.
Human Thymic iNKT Cells Have Reduced MitoTracker Green Staining.
CD1d and iNKT cells are highly conserved in evolution and cross-species reactivity is common: indeed, human iNKT cells react with murine α-GalCer-CD1d complexes and vice versa (Fig. 4) (31). We therefore investigated whether human thymic iNKT cells also down-regulated their mitochondrial content during development. In four human thymus samples of different ages, we observed consistently lower MitoTracker Green staining in iNKT cells compared with mature CD4+ and CD8+ T cells (Fig. 8 D and E). In three of four donors, we also observed higher Cyto ID staining in iNKT cells compared with double-positive T-cell precursors, although there was no significant difference with mature CD4+ and CD8+ T cells (Fig. S8).
These results confirm and extend our previous findings on the developmental regulation of mitochondrial content in murine iNKT cells.
Discussion
Autophagy plays an essential role during development, immunity, and cell death (1). As it is known that autophagy is important in MHC class II antigen presentation (8) and in shaping the T-cell repertoire (32), we investigated whether it also regulates CD1d-dependent lipid antigen presentation. We have shown that Atg7-dependent autophagy does not influence presentation of exogenous or self-lipid antigens to iNKT cells; however, it has a T-cell–intrinsic role in regulating iNKT cell development, which is not seen during thymic development of conventional T cells.
Although we found normal presentation of soluble synthetic iNKT cell agonists in the absence of Atg7, it remains to be determined whether presentation of particulated antigens, such as lipids complexed in bacterial cell walls, might depend on a functional autophagy pathway. Indeed, autophagy is known to be required for bacterial handling (33). Furthermore, although we did not detect altered autoreactivity of iNKT cells toward CD4 Cre-Atg7−/− thymocytes, in our bone marrow chimeras the reconstitution of type II NKT cells by WT and CD4 Cre-Atg7−/− precursors was almost comparable (unlike that of iNKT cells, Fig. S4 D and E), raising the possibility that CD1d-dependent presentation of other classes of lipids may not be autophagy-independent. However, due to technical challenges in tracking type II NKT cells, we did not investigate this question further.
We observed that mice with a deletion of the essential autophagy gene Atg7 in the T-cell compartment (CD4 Cre-Atg7−/−) have a severe block in iNKT cell development, with the majority of thymic iNKT cells arresting at stage 0/1 and mature iNKT cells absent from peripheral organs. This early developmental block is specific to iNKT and noninvariant TCRβ+ NK1.1+ T cells, whereas conventional CD4+ and CD8+ T cells are reduced in numbers and percentages only in peripheral organs. The reason for the observed increase in thymic γδ T cells (consistent with their thymic development branching off at the double-negative stage, and hence not affected by the CD4-Cre transgene) warrants further investigation.
Recent findings have challenged the linear maturation process of iNKT cell development, suggesting that iNKT cells might instead differentiate into three subsets characterized by specific effector functions and tissue homing and hallmarked by the expression of distinct transcription factors: T-bet (iNKT1), PLZF (iNKT2), and RORγt (iNKT17) (34). This classification roughly correlates with the conventional staging system in B6 mice, as NKT1 cells were shown to be predominantly stage 3 and NKT2 cells stages 1 and 2, although NKT17 cells cannot be distinguished in this way (34). In CD4 Cre-Atg7−/− mice, we observed a marked reduction in T-bet+ NKT1 cells with comparable ratios between NKT2 (Tbet− RORγt−) and NKT17 (Tbet− RORγt+) cells (Fig. S9). Furthermore, we also observed an overall increase in ICOS+ NK1.1− iNKT cells (Fig. S2), which in Tsc1−/− mice correlated with NKT17 function (35). However, upon stimulation of thymocytes with α-GalCer, we observed reduced secretion of IFN-γ and IL-17 with no change in IL-4 secretion (Fig. S9). These results are consistent with a functional impairment of both NKT1 and NKT17 iNKT cell subsets.
We have observed normal rearrangement of Vα14-Jα18 segments, thus excluding altered survival of DP cells as a possible cause for iNKT cell development. Furthermore, we have observed normal proliferative capacity of stage 0 iNKT cells, but enhanced incorporation of BrdU and Ki-67 staining in stage 1 iNKT cells. However, upon α-GalCer stimulation we observed reduced expansion of CD4 Cre-Atg7−/− iNKT cells. Although these results suggest normal proliferative signals during early stages of Atg7-deficient iNKT cell development, they also highlight a defect in their survival, which could be due to a combination of increased apoptosis (see below) and/or to autophagy-dependent regulation of cell cycle. Loss of autophagy might result in the abnormal persistence of cell cycle regulators (e.g., cyclin-dependent kinase inhibitors) and aberrant cell cycle entry, although the molecular mechanisms of this effect remain ill-defined (36).
We have shown that iNKT cells’ mitochondrial content (indirectly measured with the MitoTracker Green Probe) is developmentally regulated in human and mice thymuses and that in the absence of mitophagy murine cells accumulate superoxide species (indirectly measured with the MitoSox Probe) and die of apoptosis. A similar mechanism has been proposed to explain the decline of conventional T-cell numbers in the periphery of several autophagy-deficient mouse models (22–24, 37). The observation that conventional T cells are affected during critical metabolic checkpoints in the transition phase between effector and memory cells is consistent with the fact that iNKT cells have already developed as memory cells in the thymus and prompted us to investigate the link between metabolism and autophagy in developing iNKT cells. Indeed, metabolic reprogramming has emerged as a critical modulator of T-cell fate and function (38), and it is well known that the mTOR pathway controls the link between metabolism and immunity, in addition to inhibiting autophagy (39, 40). We have shown that immature iNKT cells have higher uptake of a fluorescent glucose analog. Upon maturation, reduction of glucose uptake by iNKT cells is accompanied by lower proliferation and increased autophagy levels. These results suggest that mature iNKT cells, like memory T cells, are more quiescent, and we speculate that they may also need to revert to a metabolism mainly dependent on oxidative phosphorylation.
It is known that stage 0 iNKT cells undergo massive population expansion, controlled by the transcription factor c-myc. Ablation of c-myc at the double-positive stage specifically abrogates iNKT cell, but not conventional T-cell, development (30). c-myc is also a crucial transcription factor for the metabolic switch to glucose metabolism that occurs upon naive T-cell activation, in addition to controlling key cell cycle regulators (41). It is therefore possible that the increased glucose uptake that we observed in immature iNKT cells is driven by c-myc, as it occurs in a highly proliferative stage of iNKT cell development.
Similarly, the nuclear orphan receptor Nur77, which is up-regulated in developing iNKT cells following strong TCR signaling (42), is capable of regulating the expression of genes linked to glucose utilization (43). Strong TCR engagement in conventional T cells has been shown to induce Nur77 translocation to the mitochondria, association with Bcl2, and exposure of the Bcl2 proapoptotic BH3 domain. In developing iNKT cells, however, concomitant signaling by the SLAM family of coreceptors delivers a survival signal that rescues iNKT cells from apoptotic death (44). Sustained TCR signaling following recognition of CD1d self-lipids by developing iNKT cells at stage 0 leads to Egr1/Egr2-mediated up-regulation of PLZF (17), the transcription factor that dictates acquisition of an iNKT cell innate-like phenotype (16, 45). In stem cells, PLZF opposes mTORC1 activity by inducing expression of the mTORC1 inhibitor Redd1 (46). Redd1 inhibits mTORC1 activity through regulation of Tsc1/Tsc2 (47). In hematopoietic stem cells, the Tsc-mTOR axis is important to maintaining quiescence by repressing mitochondrial biogenesis and reactive oxygen species (48), consistent with the ability of Redd1 to localize to the mitochondria and regulate mitochondrial metabolism, reducing ROS production (49). By inhibiting mTORC1, Tsc1 also induces a quiescence program in naive T cells, in the absence of which T cells undergo apoptosis (50). The phenotype of CD4 Cre Atg7−/− mice is reminiscent of that of CD4 Cre Tsc1−/−, which have fewer T cells in peripheral lymphoid organs, an altered ratio of CD4+-to-CD8+ T cells, and expansion of CD44hi CD62Llow cells, with a corresponding reduction of naive cells and a defect in iNKT cell development. In addition, CD4 Cre Tsc1−/− mice exhibit a block in iNKT differentiation at stage 2, with reduced IFN-γ and enhanced IL-17 secreting iNKT cells; however, the developmental block does not seem as severe as that observed in CD4 Cre Atg7−/− mice, and discrete α-GalCer-CD1d tetramer+ cells can be detected in the spleen and liver of CD4 Cre Tsc1−/− mice (35). Furthermore, the IFN-γ/IL-17 imbalance in iNKT cell subsets and the developmental block can be substantially corrected by rapamycin injection, suggesting that activation of the autophagy pathway downstream of the Tsc-mTOR axis might be critical for iNKT cell development. The observation that Tsc1 deficiency affects preferentially IFN-γ–secreting iNKT cells is interesting, as it is has been shown that conventional Th-17 cell differentiation is driven by an mTOR and HIF-1α–dependent glycolytic program (51). Conversely, memory T-cell differentiation requires a lower metabolic activity (28, 52).
Recently, two other metabolic regulators have been shown to regulate iNKT cell development. The serine/threonine kinase LKB1 is essential for the development of iNKT cells and other innate cells such as intraepithelial T cells, but not of conventional T cells (53). LKB1 plays an important role during the proliferative burst of iNKT cells that follows positive selection, when such rapid proliferation imposes high metabolic demands. One possible target of LKB1 is the NUAK1 kinase, which induces cell cycle arrest in tumor cells exposed to energy stress, protecting them from apoptosis. Thus, NUAK1 may play an important role to return positively selected T lymphocytes to a quiescent state. The metabolic regulator Folliculin-interacting protein 1 (Fnip1) is also crucial for iNKT and noninvariant NKT cell development (54). Residual Fnip1−/− iNKT cells are more sensitive to apoptosis, which has been suggested to be secondary to altered mitochondrial homeostasis and metabolic stress. Fnip1 interacts with the serine/threonine kinase AMPK (adenosine monophosphate-activated protein kinase) that regulates energy consumption inhibiting mTOR via Tsc2 and Raptor phosphorylation and, in doing so, is required for the development of CD8+ memory cells (55). Furthermore, in response to low energy, AMPK stimulates ATP and nutrient production, stimulating autophagy by activating the autophagy protein Vps34. Consistent with this, both excessive mTOR activation following conditional disruption of Tsc1 (50) and inhibition of autophagy by conditional disruption of Vps34 (24) have been shown to cause an early block in iNKT cell development. However, conditional T-cell deletion of AMPKα does not affect percentages or absolute numbers of thymic iNKT cells, whereas their differentiation into subsets and functional capacity remains to be investigated (53).
It has been suggested that environmental factors such as higher oxygen tension in the blood and secondary lymphoid organs compared with the thymus may cause stress to T lymphocytes if their mitochondrial content is not reduced and that CD8+ T cells somehow have less tolerance for superfluous mitochondria than CD4+ T cells (23). In conjunction with strong TCR signaling, the relatively hypoxic thymic microenvironment might itself contribute to driving T-cell quiescence via HIF-1α–dependent up-regulation of the Redd1-TCS pathway (49). Thus, although in activated CD8+ T cells HIF1-α can sustain the glycolytic response downstream of mTOR (56, 57), it remains to be established whether, in addition to c-myc, it also plays an important role in regulating iNKT cell development, perhaps by sustaining the glycolytic switch during population expansion.
HIF1-α also activates a negative feedback control mechanism through transcriptional regulation of Redd1 expression and the Redd1/Tsc axis.
Altogether the above results suggest that a fine balance between the autophagy and mTOR pathways is required during a developmental phase, resulting in massive cell expansion and metabolic stress. Indeed, ablation of mTORC1 signaling also causes a severe early defect in iNKT cell development (58, 59). We speculate that iNKT cells undergo an early glycolytic shift driven by c-myc, but at the end of their expansion phase they have to be able to enter a quiescent, less metabolically active phase that relies on fatty acids, amino acids, and glucose as energy sources (60). The reduced staining with mitochondrial probes, here used as surrogate for mitochondrial content, ultimately reflects quiescence of mature iNKT cells and naive/memory conventional T cells, which become more dependent on autophagy to provide the T-cell with sufficient metabolic fuel (61). It remains to be determined whether in these cells autophagy also regulates the cell-intrinsic lipolysis recently shown to support the metabolic programming necessary for CD8+ memory T-cell development (62).
It has recently been reported that the metabolic checkpoint kinase mTOR is important for the development and activation of natural killer cells (63). It remains to be determined whether other innate-like cells marked by PLZF expression (64) might have similar metabolic requirements for the acquisition of effector function and/or peripheral survival.
Materials and Methods
Mice.
Mice were bred and housed in the Department of Biomedical Services, University of Oxford, in individually ventilated cases. Atg7flox/flox mice (15) were crossed to CD4-Cre mice (65) (from Adeline Hajjar, University of Washington, Seattle) to obtain CD4 Cre-Atg7−/− on a C57BL/6 background. Genotyping was performed on ear genomic DNA as described (15). Male and female mice were used equally in all experiments. CD4 Creneg-Atg7flox/flox were used as littermate controls. C57BL/6 WT, Jα18-deficient mice (a gift from Paolo Dellabona and Giulia Casorati, Opsedale San Raffaele, Milan) and CD1d-deficient mice (purchased from Jackson Laboratories) were used as tetramer staining controls. All animal experiments were approved by the University of Oxford ethical review committee and performed under a Home Office license (PPL 40/3636). Mice 7–9 wk of age were used for experiments.
Medium and Reagents.
A detailed list of medium and reagents, including working conditions for probes and tetramers, is provided in SI Materials and Methods.
Generation of iNKT Cells.
Human iNKT cells were isolated and maintained as described (66). The murine hybridoma DN32 was kindly provided by A. Bendelac (University of Chicago, Chicago).
Bone Marrow DC Stimulation Assays.
BMDCs were differentiated from mouse bone marrow in complete medium in the presence of 20 ng/mL GM-CSF (Peprotech) for 7 d. Fresh complete medium and GM-CSF were added every 2 d. BMDC phenotype was confirmed by flow cytometry (CD11b, CD11c, and CD1d, all from eBiosciences). BMDCs were collected, washed, and plated at 100,000/well in U-bottom 96-well plates and used to stimulate iNKT cells (30,000–50,000/well in duplicate or triplicate) in the presence of lipids at the indicated concentration.
iNKT cell activation was assessed by ELISA (human IFN-γ and mouse IL-2 from BD Pharmingen) on supernatants harvested after 36 h.
Mixed Bone Marrow Chimera.
Recipient B6.SJL (CD45.1) mice were γ-irradiated twice with 450 rad and then reconstituted with a mixture of bone marrow derived from CD4 Cre Atg7−/− (CD45.2) or C57BL/6 (CD45.2) and B6.SJL (totally 5 × 106 cells) to achieve a 50:50 chimera bone marrow. Eight weeks after reconstitution, mice were tested for chimerism, and at 10 wk chimeras were killed for analysis.
Flow Cytometry.
Thymus, spleen, and liver were harvested, and a single-cell suspension was generated (67). Cells were counted, and 10 million cells were stained for 15 min at room temperature with α-GalCer-CD1d tetramers [PE, PE CF594, or APC-labeled (68)] and live dead stain (Invitrogen, Aqua). Cells were washed and stained on ice. A detailed list of clones used is provided in SI Materials and Methods.
BrdU Analysis.
To study cell turnover in vivo, 6-wk-old mice were injected i.p. daily with 1 mg BrdU (Pharmingen) on 2 consecutive days. Twenty-four hours after the second injection, thymocytes were harvested, and the single-cell suspension was stained for surface markers followed by staining with anti-BrdU (Pharmingen) and Ki67 antibodies (eBioscience), following the manufacturer’s instructions.
In Vitro Culture.
Single-cell suspension of thymocytes (5 × 106) were CSFE-labeled (Invitrogen) and plated in complete medium supplemented with human IL-2 (1,000 U/mL) and murine IL-7 (20 ng/mL, Peprotech) in the presence or absence of α-GalCer (200 ng/mL). Cultures were stained at day 4 and supernatants used for IL-4, IFN-γ, and IL-17 ELISAs (eBioscience).
Human Thymus Tissue.
Thymus samples were obtained after informed consent and ethics committee approval from the University of Oxford and Great Ormond Street Hospital, London. The samples were from (i) one infant (aged 1 y ) undergoing corrective cardiac surgery; (ii) two patients with generalized myasthenia gravis (MG) but no detectable acetyl-choline receptor auto-antibodies, whose thymuses appeared normal (69), and one of whom was male, aged 13.9, and the other female, aged 23.5, and neither of whom had been given corticosteroids or other immunosuppressants; and (iii) one 12-y-old MG female with an anti-AChR titer of 4 nM, whose thymus showed lympho-follicular hyperplasia. Fresh thymic tissue was dispersed mechanically, and single-cell suspensions were washed and cryo-stored as described by Willcox et al. (70). Upon thawing in complete medium, cells were immediately stained with MitoTracker, cyto ID, live and dead staining, and the following antibodies (from Biolegend except when specified): CD3 (clone UCHT1, BD Pharmingen), 6B11 (anti-invariant NKT cells, BD Pharmingen), Vβ11 (clone C21, Beckman Coulter), CD4 (clone OKT4), CD8 (clone RPA-T8), CD161 (clone 3C10), CD45RO (clone UCHL1), and CD24 (clone ML5).
Statistical Analysis.
To compare differences between WT and KO values, a two-tailed Student t test was performed using GraphPad PRISM analysis software.
Supplementary Material
Acknowledgments
We thank Prof. Nick Willcox (University of Oxford) and Dr. Graham Davies (Great Ormond Street Hospital) for provision of human thymus samples, Drs. Giorgio Napolitani and Nicolas Prevot for helpful discussion and critical reading of the manuscript, and Dr. Uzi Gileadi and Thomas Riffelmacher for help with the bone marrow chimeras. This work was supported by Cancer Research UK [Programme Grant C399/A2291 (to V.C.)], the Medical Research Council, and The Harry Mahon Cancer Research Trust UK and the Wellcome Trust [84923 (to V.C.)]. D.J.P. is funded by the Medical Research Council Human Immunology Unit, The Allan & Nesta Ferguson Charitable Trust, and St. Catherine’s College. A.K.S. acknowledges funding from the Wellcome Trust (New Investigator Award WT103830MA and Project Grant 088098/Z/08/Z). G.S.B. acknowledges support in the form of a Personal Research Chair from Mr. James Bardrick and the Medical Research Council (MR/K012118/1). E.A. and G.A.H. acknowledge support from the National Institute for Health Research Biomedical Research Centre (Oxford).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413935112/-/DCSupplemental.
References
- 1.Klionsky DJ. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanida I, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10(5):1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 8.Münz C. Antigen processing for MHC class II presentation via autophagy. Front Immunol. 2012;3:9. doi: 10.3389/fimmu.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadola SD, et al. Impaired selection of invariant natural killer T cells in diverse mouse models of glycosphingolipid lysosomal storage diseases. J Exp Med. 2006;203(10):2293–2303. doi: 10.1084/jem.20060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prigozy TI, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291(5504):664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 11.Schümann J, et al. Differential alteration of lipid antigen presentation to NKT cells due to imbalances in lipid metabolism. Eur J Immunol. 2007;37(6):1431–1441. doi: 10.1002/eji.200737160. [DOI] [PubMed] [Google Scholar]
- 12.Engel I, Kronenberg M. Making memory at birth: Understanding the differentiation of natural killer T cells. Curr Opin Immunol. 2012;24(2):184–190. doi: 10.1016/j.coi.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortensen M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107(2):832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortensen M, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208(3):455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13(3):264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salio M, et al. Saposins modulate human invariant Natural Killer T cells self-reactivity and facilitate lipid exchange with CD1d molecules during antigen presentation. Proc Natl Acad Sci USA. 2013;110(49):E4753–E4761. doi: 10.1073/pnas.1310050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303(5657):523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arase N, Arase H, Good RA, Onoé K. Contribution of host radioresistant T cells to the clonal elimination of minor lymphocyte stimulatory-1a reactive T cells in mouse bone marrow chimeras. Cell Immunol. 1994;156(1):13–23. doi: 10.1006/cimm.1994.1149. [DOI] [PubMed] [Google Scholar]
- 21.Hager E, Hawwari A, Matsuda JL, Krangel MS, Gapin L. Multiple constraints at the level of TCRalpha rearrangement impact Valpha14i NKT cell development. J Immunol. 2007;179(4):2228–2234. doi: 10.4049/jimmunol.179.4.2228. [DOI] [PubMed] [Google Scholar]
- 22.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182(7):4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 24.Parekh VV, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013;190(10):5086–5101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;793(1):141–150. doi: 10.1016/s1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- 26.Chan LL, et al. A novel image-based cytometry method for autophagy detection in living cells. Autophagy. 2012;8(9):1371–1382. doi: 10.4161/auto.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciolek JA, Pasternak JA, Wilson HL. Metabolism of activated T lymphocytes. Curr Opin Immunol. 2014;27:60–74. doi: 10.1016/j.coi.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6(9):881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 30.Dose M, et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci USA. 2009;106(21):8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brossay L, et al. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188(8):1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455(7211):396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol. 2014;12(2):101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14(11):1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J Clin Invest. 2014;124(4):1685–1698. doi: 10.1172/JCI69780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang RC, Levine B. Autophagy in cellular growth control. FEBS Lett. 2010;584(7):1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci USA. 2012;109(22):8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, Ye L, Araki K, Ahmed R. mTOR, linking metabolism and immunity. Semin Immunol. 2012;24(6):429–435. doi: 10.1016/j.smim.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208(6):1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao LC, et al. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21(9):2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27(5):751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142(3):468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205(10):2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horak P, et al. Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc Natl Acad Sci USA. 2010;107(10):4675–4680. doi: 10.1073/pnas.0907705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12(9):888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarrouk M, Rolf J, Cantrell DA. LKB1 mediates the development of conventional and innate T cells via AMP-dependent kinase autonomous pathways. PLoS ONE. 2013;8(3):e60217. doi: 10.1371/journal.pone.0060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park H, Tsang M, Iritani BM, Bevan MJ. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc Natl Acad Sci USA. 2014;111(19):7066–7071. doi: 10.1073/pnas.1406473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rolf J, et al. AMPKα1: A glucose sensor that controls CD8 T-cell memory. Eur J Immunol. 2013;43(4):889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209(13):2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin J, et al. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc Natl Acad Sci USA. 2014;111(8):E776–E783. doi: 10.1073/pnas.1315435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J Immunol. 2014;193(4):1759–1765. doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 60.Jones RG, Thompson CB. Revving the engine: Signal transduction fuels T cell activation. Immunity. 2007;27(2):173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 61.Hubbard VM, et al. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185(12):7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Sullivan D, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41(1):75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marçais A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. 2014;15(8):749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 66.Gadola SD, Dulphy N, Salio M, Cerundolo V. Valpha24-JalphaQ-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4(+) and CD8alphabeta(+) T lymphocytes. J Immunol. 2002;168(11):5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- 67.Crowe NY, et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med. 2005;202(9):1279–1288. doi: 10.1084/jem.20050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karadimitris A, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci USA. 2001;98(6):3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willcox N, Schluep M, Ritter MA, Newsom-Davis J. The thymus in seronegative myasthenia gravis patients. J Neurol. 1991;238(5):256–261. doi: 10.1007/BF00319736. [DOI] [PubMed] [Google Scholar]
- 70.Willcox HN, Newsom-Davis J, Calder LR. Greatly increased autoantibody production in myasthenia gravis by thymocyte suspensions prepared with proteolytic enzymes. Clin Exp Immunol. 1983;54(2):378–386. [PMC free article] [PubMed] [Google Scholar]
- 71.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202(4):485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.