Abstract
Ribonuclease P (RNase P) complexed with external guide sequence (EGS) represents a nucleic acid-based gene interference approach to knock-down gene expression. Unlike other strategies, such as antisense oligonucleotides, ribozymes, and RNA interference, the RNase P-based technology is unique because a custom-designed EGS molecule can bind to any complementary mRNA sequence and recruit intracellular RNase P for specific degradation of the target mRNA. In this study, we demonstrate that the RNase P-based strategy is effective in blocking gene expression and growth of Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), the causative agent of the leading AIDS-associated neoplasms, such as KS and primary-effusion lymphoma. We constructed 2′-O-methyl-modified EGS molecules that target the mRNA encoding KSHV immediate-early transcription activator Rta, and we administered them directly to human primary-effusion lymphoma cells infected with KSHV. A reduction of 90% in Rta expression and a reduction of ≈150-fold in viral growth were observed in cells treated with a functional EGS. In contrast, a reduction of <10% in the Rta expression and viral growth was found in cells that were either not treated with an EGS or that were treated with a disabled EGS containing mutations that preclude recognition by RNase P. Our study provides direct evidence that EGSs are highly effective in inhibiting KSHV gene expression and growth. Exogenous administration of chemically modified EGSs in inducing RNase P-mediated cleavage represents an approach for inhibiting specific gene expression and for treating human diseases, including KSHV-associated tumors.
Keywords: gene targeting, gene inactivation, antisense, cancer therapy
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) (also referred as human herpesvirus 8) (1) is believed to be the causative agent of KS as well as primary-effusion lymphoma (PEL) and multicentric Castleman's disease (2). These diseases, especially KS, are among the leading neoplasms of AIDS patients (2). Like all other herpesviruses, KSHV has the capability to engage both lytic and latent infections (2, 3). Although most of the tumor cells are latently infected, a spontaneous switch from latent to lytic replication is common in KS lesions, and expression of lytic gene products (including chemokines and proinflammatory cytokines encoded by KSHV) can be found (2). This spontaneous activation of the lytic cycle is believed to play a central role in KSHV pathogenesis and KS development (4, 5). Indeed, treatment of AIDS patients at risk for KS with anti-herpes compounds, including ganciclovir [a drug that blocks lytic but not latent KSHV replication (6)], reduced the incidence of KS development remarkably (7, 8). Thus, blocking viral lytic replication is central in the treatment of KSHV infection and for the inhibition and prevention of KS development.
Nucleic acid-based gene interference strategies, such as antisense oligonucleotides, ribozymes or DNAzymes, and RNA interference, are powerful research tools and promising therapeutic agents for human diseases (9-13). Each of these approaches has its own advantages and limitations in terms of targeting efficacy, sequence specificity, toxicity, and delivery efficiency in vivo (14). Improving these current technologies and developing nucleic acid-based strategies will provide exciting tools and reagents for basic research and clinical applications including therapeutic interventions.
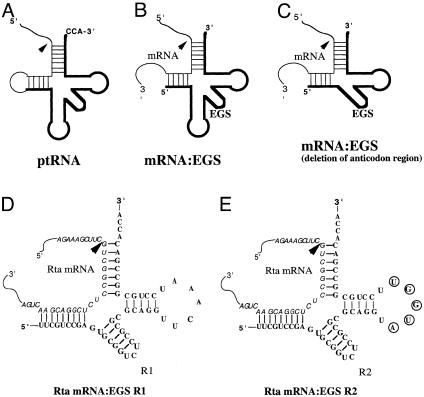
RNase P has been proposed as an RNA-based gene interference strategy for knocking down gene expression (15, 16). This enzyme, which can be found in all living organisms, catalyzes a hydrolysis reaction to remove the leader sequence of tRNA precursors by recognizing the common structure shared among all tRNAs (Fig. 1A) (16, 17). Altman and colleagues (18-20) proposed that RNase P can be recruited to cleave any mRNA by using a custom-designed external guide sequence (EGS) that hybridizes with the target mRNA to form a structure resembling a tRNA substrate (Fig. 1 B and C). Recent studies (21, 22) on substrate recognition and RNase P cleavage requirement provide significant insight into the construction of highly active EGS for gene-targeting applications. Compared with the RNA interference approach, which induces the cellular RNA-induced silencing complex RNase to cleave a target mRNA (14, 23), the EGS-based technology is unique in inducing endogenous RNase P for targeted cleavage. Moreover, the RNase P-mediated cleavage is highly specific and does not generate nonspecific “irrelevant cleavage” that is observed in RNase H-mediated cleavage induced by conventional antisense phosphothioate molecules (16, 22). Thus, EGSs represent a class of agents that may lead to highly effective and specific inhibition of gene expression (24-26).
Fig. 1.
Schematic representation of substrates for RNase P. (A) A natural substrate precursor tRNA for RNase P. (B and C) Hybridized complex of a target RNA (e.g., mRNA) and an EGS that resembles the structure of a tRNA and can be cleaved by RNase P. C results from B by deletion of the anticodon domain of the EGS, which is dispensable for EGS-targeting activity (21). Arrowheads indicate the site of cleavage by RNase P. (D and E) Complexes between the Rta mRNA sequence and EGS R1 and R2, respectively. The sequence of these EGSs equivalent to the tRNA sequence was derived from tRNAser and resembles the T-stem and loop and a variable region of the tRNA molecule.
To date, extensive studies exploiting the potential of antisense, ribozymes, and RNA interference molecules have been carried out, including several clinical trials using these technologies for treating certain human diseases (13, 14). However, there are only a few studies evaluating the usefulness of the RNase P-based technology to target disease-associated mRNAs in human cells. In this study, we provide direct evidence that exogenous administration of chemically synthesized EGSs with 2′-O-methyl modification is highly effective in treating KSHV-infected cells and abolishing KSHV replication.
The target is the mRNA encoding KSHV ORF50 [also known as Rta (replication and transcription activator)]. Rta is an immediate-early gene that encodes a transcription factor activating the expression of most of viral early and late genes (27-30). Moreover, Rta may play an important role in the progression of KS and KSHV-associated tumors because this protein can induce the expression of cellular cytokines, including IL-6, which acts through autocrine and paracrine mechanisms to stimulate proliferation of B cells and angiogenesis and is critical for the development of KS (31-33). Thus, this gene may represent an ideal target for anti-KSHV drug development. However, whether direct blocking of Rta transcription and/or translation can achieve a significant reduction of viral growth has not been demonstrated because of the difficulty of constructing mutants with deletions of viral essential genes and generating complementing cell lines expressing the essential functions. Our results provide direct evidence that the chemically synthesized 2′-O-methyl EGS oligonucleotides are highly effective in inhibiting KSHV gene expression and growth in human cell culture, and furthermore, they demonstrate the feasibility of using the EGSs for studying viral infection and for treating KSHV-associated diseases.
Materials and Methods
Viruses, Cells, and Antibodies. Human PEL BCBL-1 cells containing KSHV were propagated as described (3). KSHV lytic replication was induced by treating BCBL-1 cells with tetradecanoyl phorbol acetate (TPA) (20 ng/ml) (Sigma). Antibodies against KSHV Rta, ORF65, ORF59, and K8.1 were provided by Ren Sun (University of California, Los Angeles) or purchased from Advanced Biotechnologies (Columbia, MD) (34, 35). The mAb against human actin was purchased from Sigma.
EGS and Rta mRNA Substrate. Plasmid pRTA contains the entire coding sequence of Rta. Construct prta-S contains the sequence encoding the RNA substrate rta-S, which includes the target Rta mRNA sequence. To generate construct prta-S, PCR products were first amplified by using pRTA as the template and the following primers: RTAEGSPRIME, 5′-GGGAAGCTTGGCCTTCAGTTCGTCCGAGAGGCCGACGAAGCTTTCTGGATCCGCG-3′; and SUBPRIMER2, 5′-CGCGGATCCA-3′. They were then digested with BamHI and HindIII and inserted between the BamHI and HindIII sites of plasmid pT7, which was derived from pUC19 and contained the promoter sequence of T7 RNA polymerase at the SmaI site. RNA substrate rta-S was synthesized in vitro from prta-S by using T7 RNA polymerase. EGS R1 (5′-UUCGUCCGAGUGCGGUCUCCGCGCGCAGGUUCAAAUCCUGCGGCCGACACCA-3′) and R2 (5′-UUCGUCCGAGUGCGGUCUCCGCGCGCAGGUAUGGUUCCUGCGGCCGACACCA-3′) were chemically synthesized by using a DNA synthesizer (Dharmacon, Lafayette, CO). The 2′-hydroxyl groups in these EGS molecules were replaced with an O-methyl group. A DNA-based EGS, TK1 (5′-TACGTCGGTGCGGTCTCCGCGCGCAGGTTCAAATCCTGCCGCAGACACCA-3′), which targets the mRNA coding for the thymidine kinase of herpes simplex virus 1 (HSV-1) and also contains a 5′-FITC label, was synthesized by using a DNA synthesizer, as described (36).
In Vitro Binding and Cleavage of Rta mRNA. Human RNase P was prepared from HeLa cellular extracts as described (20). The EGSs and 32P-labeled rta-S were incubated with human RNase P at 37°C in buffer A (50 mM Tris, pH 7.4/100 mM NH4Cl/10 mM MgCl2) (20). Cleavage products were separated in denaturing gels and analyzed with a STORM 840 PhosphorImager (Molecular Dynamics). The procedures to measure the Kd of the EGS-rta-S complexes were carried out as described (21, 37). HeLa nuclear extracts and rabbit reticulocyte lysates were purchased from GIBCO or Promega.
EGS Internalization and Treatment of Human Cells and Fluorescence-Activated Cell Sorting (FACS) Analysis. Lipofectamine 2000 reagent (GIBCO) was diluted in medium with the EGS to give a final concentration of 10 μg/ml lipid/100 nM EGS. The transfection experiments were carried out by using a final concentration of 80 nM R1 or R2 and 20 nM FITC-labeled TK1. The EGS-lipid mixtures were prepared according to the manufacturer's recommendation (GIBCO), incubated with cells for 7 h, and then removed. The transfection efficiency was evaluated by detecting the FITC staining of the transfected cells by using fluorescence microscopy. Under these settings, we consistently achieved an optimal transfection efficiency of 10%. At 7 h after transfection, cells were subjected to FACS by using a FACSVantage SE sorter (Becton Dickinson), and a population of the transfected cells (usually 105 cells with a positive fluorescence of >99%) was isolated.
Assays of KSHV Gene Expression and Growth in EGS-Treated Cells. We first treated 2 × 106 cells with liposome complexes containing TK1 and in the absence and presence of R1 or R2, and we then subjected them to FACS analysis at7haftertransfection. Cells containing the EGSs were isolated and incubated in culture media either in the absence or presence of 20 ng/ml TPA for 4-48 h. At 12-72 h after induction, total cellular RNAs and proteins were isolated from cells, as described in ref. 30. For Northern blot analyses, RNA fractions were separated in 0.8-2.5% agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, hybridized with the 32P-radiolabeled DNA probes that contained the KSHV DNA sequences, and analyzed with a STORM 840 PhosphorImager. For Western blot analyses, the denatured, solubilized polypeptides were separated on 7.5% or 9% (vol/vol) SDS/PAGE cross-linked with N,N′′-methylenebisacrylamide, transferred electrically to nitrocellulose membranes, and reacted to the antibodies against human β-actin, KSHV Rta, ORF59, ORF65, and K8.1. The membranes were subsequently stained with a chemiluminescent substrate with the aid of a Western chemiluminescent substrate kit (Amersham Biosciences) and quantitated with a STORM 840 PhosphorImager (36).
To determine the level of inhibition of viral growth, 2 × 106 BCBL-1 cells were first incubated with liposome complexes containing TK1 and in the absence and presence of EGS R1 or R2. Those transfected ones (≈2 × 105 cells) were isolated by using FACS analysis and induced in the presence of TPA (20 ng/ml) for 24-48 h. Supernatants were isolated from the cultured cell media harvested at 1, 2, 3, 4, 6, 8, and 10 days after induction and then treated with DNase I (GIBCO). The DNase-resistant viral DNAs were isolated by using phenol extraction, followed by ethanol precipitation (30). The purified viral DNAs were subsequently denatured for 10 min at room temperature by adding 0.2 N NaOH and diluting with blotting buffer 1× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7), transferred to a Zeta-Probe nylon membrane by using a dot-blot apparatus (Bio-Rad). The bound DNA on the filter was then hybridized with a 32P-labeled probe containing KSHV ORF65 sequence, and the signals were quantitated by using a STORM 840 PhosphorImager. The background signal from supernatants collected in the absence of TPA induction was <1% of the values detected from the equivalent amount of supernatant in the presence of TPA induction and in the absence of EGSs. We determined the values that represent the folds of increase in KSHV virion DNA level in samples, as compared with the level of virion DNA found in cells that were treated with liposome complexes in the absence of R1 or R2 and were incubated in the absence of TPA. The results were derived from three independent experiments.
Results
To achieve efficient targeting, it is critical to choose a target region that is accessible to binding of EGSs because most mRNA species inside cells are usually either present in folded conformations or associated with proteins. To identify the accessible regions, human PEL BCBL-1 cells that are latently infected with KSHV were first incubated with TPA for 12 h to induce the Rta expression and then placed in medium containing dimethyl sulfate (DMS), which can pass the cell membrane and modify the exposed regions of cellular mRNAs (38, 39). The modified regions were mapped by primer-extension analysis of the mRNAs isolated from the DMS-treated cells. By using this in vivo mapping method, we mapped the region of Rta mRNA and chose a position (37 nucleotides downstream from the 5′ terminus of Rta exon 2) (40), as the cleavage site for human RNase P. This site appears to be one of the most accessible regions to DMS modification (data not shown) and would presumably be accessible also to EGS binding.
Two EGSs, with substitution of the 2′-hydroxyl group with 2′-O-methyl residues, were constructed (Fig. 1 D and E): R1, which resembles a part of the tRNAser structure, contains a T-loop and stem, and a variable region but not the anticodon region that is dispensable for EGS activity (Fig. 1 C and D) (21); and R2, which was derived from R1 by introducing base-substitution mutations in five positions of the T-loop. The nucleotides in these five positions are highly conserved among tRNA molecules and are important for folding of the tRNA molecules and their recognition by RNase P (16, 17). The 2′-O-methyl-modified EGSs were chemically synthesized in vitro, and the 2′-hydroxyl group of each nucleotide was replaced with an O-methyl residue. RNA analogs with the 2′-O-methyl modification have been shown to be stable against various endonucleases and exonucleases (41). More importantly, 2′-O-methyl-modified oliogonucleotides, when complexed with target mRNAs, can form a RNA-like duplex (42), of which the conformation is important for RNase P recognition (16). However, the duplex between the target mRNA and 2′-O-methyl-modified oligonucleotides, unlike that of a DNA-RNA duplex, is not a substrate for RNase H (41, 42). Thus, these modified EGSs can be used to assess the efficacy of the RNase P-mediated cleavage in antiviral application in the absence of the context of the RNase H-mediated antisense effect.
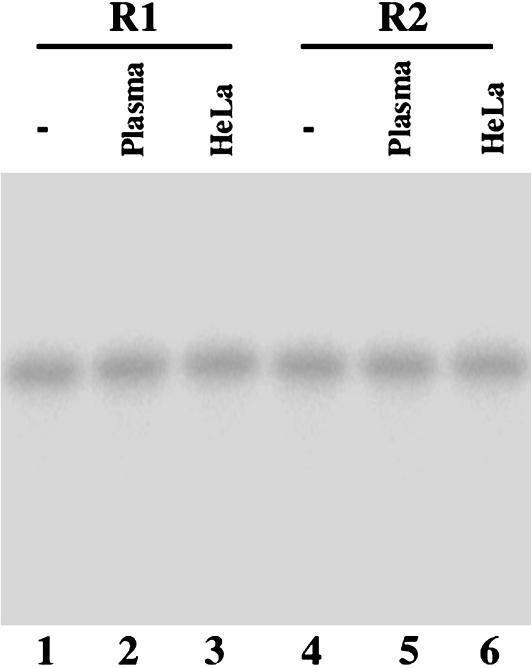
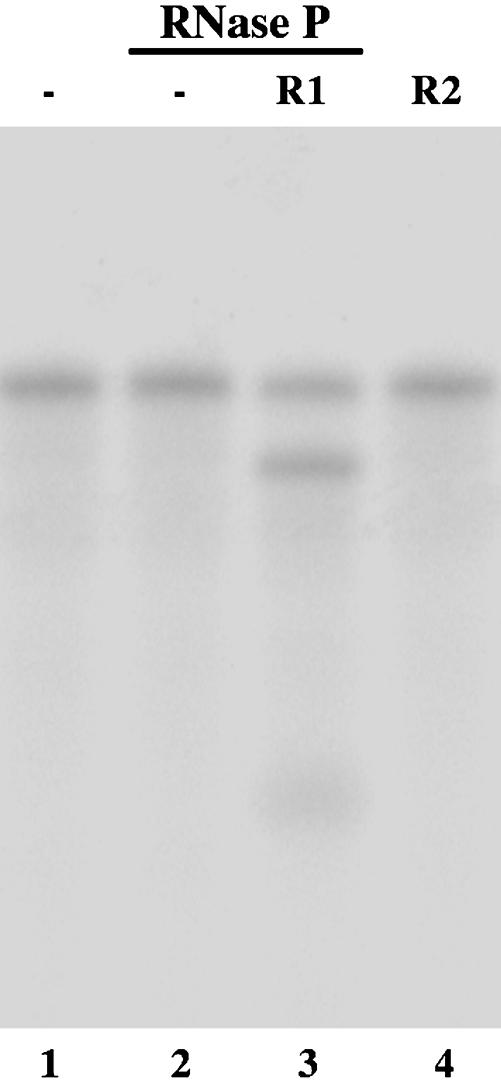
Indeed, most of the 2′-O-methyl modified R1 and R2 are stable during a 24-h incubation at 37°C in the presence of human plasma and culture media (Fig. 2). Moreover, most of R1 and R2 remained intact at 37°C in the presence of rabbit reticulocyte lysates and HeLa nuclear extracts for 24 h (Fig. 2, and data not shown), suggesting that the modified EGSs are also stable in cellular environment. The EGSs were subsequently incubated with human RNase P and substrate rta-S, which contains the targeted sequence of Rta mRNA. Apparent cleavage of rta-S by RNase P was observed in the presence of R1 (Fig. 3, lane 3). However, cleavage was barely detected in the presence of R2 (Fig. 3, lane 4). To determine whether the differential cleavage efficiencies observed with R1 and R2 were possibly due to their different binding affinities to the Rta mRNA sequence, the binding between the EGSs and rta-S was assayed in the absence of human RNase P, and the Kd values were determined. These results indicate that the binding affinity of R2 to rta-S (Kd = 130 ± 25 nM) is similar to that of R1 (Kd = 147 ± 31 nM). Meanwhile, cleavage products were barely detected in the presence of R2, even under high concentrations of RNase P and a prolonged incubation period. These observations indicate that the T-loop mutations do not significantly affect the binding affinity between R2 and Rta mRNA sequence but disrupt the recognition of EGS-Rta mRNA complex by RNase P. Thus, R2 may be used as a control for the antisense effect in our experiments in cultured cells (see below).
Fig. 2.
Stability of 2′-O-methyl-modified EGS R1 and R2 in the presence of human plasma and HeLa nuclear extracts. EGSs were either loaded directly on polyacrylamide denaturing gels (-, lanes 1 and 4) or incubated in either DMEM containing 10% human plasma (Plasma, lanes 2 and 5) or in HeLa nuclear extracts (Promega) (HeLa, lanes 3 and 6) at 37°C for 24 h before loading on the gels.
Fig. 3.
Cleavage of 32P-labeled substrate rta-S by human RNase P in the presence of different EGSs. No RNase P was added to the reaction mixture in lane 1. We incubated 5 nM of Rta-S substrate alone (lanes 1-2) or in the presence of 5 nM R1 (lane 3) or R2 (lane 4) in the presence of 2 units of RNase P. Cleavage reactions were carried out in buffer A (50 mM Tris, pH 7.0/100 mM NH4Cl/10 mM MgCl2) at 37°C for 1 h.
The EGS molecules were complexed with Lipofectamine 2000 liposomes (GIBCO) and delivered into BCBL-1 cells. Studies (27, 28) have suggested that the BCBL-1 cells are not highly permissible to transfection. To assess and normalize the transfection efficiency, a DNA-based EGS, TK1, which targets the mRNA coding for thymidne kinase (TK) of herpes simplex virus 1 (HSV-1) and contains a conjugated-fluorescein (FITC) at its 5′ terminus (36), was included in the transfection mixture along with either R1 or R2 and was used as the internal control for monitoring the transfection efficiency. The transfection efficiency was determined by using FACS analysis, which detects the presence of TK1 in the FITC-staining cells. Treatment of the cells with the EGS-liposome complexes by using our transfection protocol consistently yielded a transfection efficiency of ≈10%. No cleavage of rta-S by RNase P was found in vitro in the presence of TK1 (data not shown).
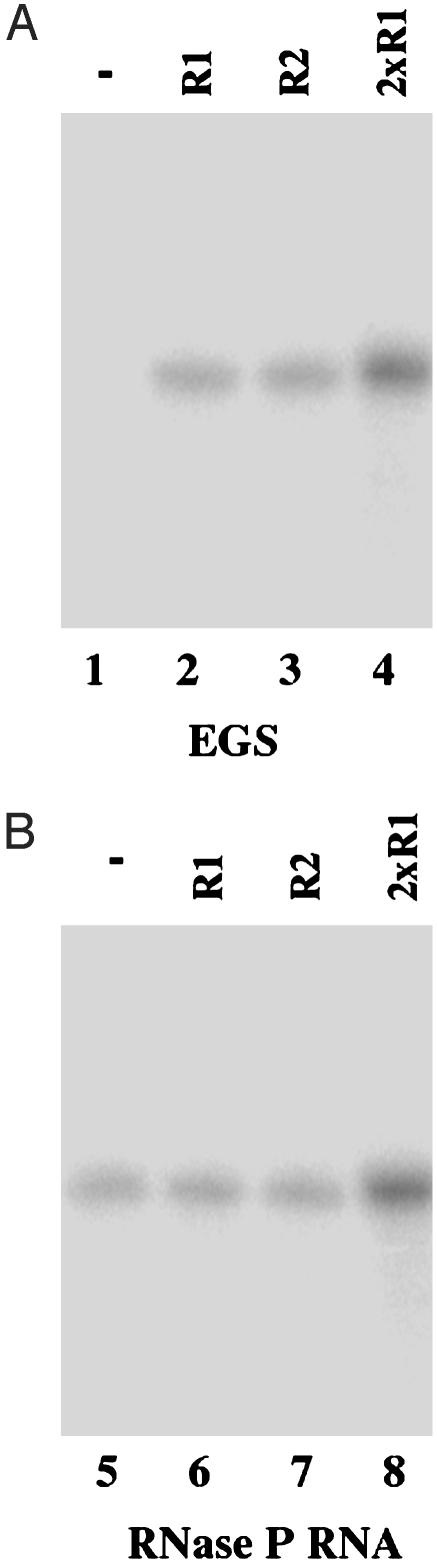
To investigate the distribution of the internalized EGS in the transfected cells, cells were isolated by using FACS analysis at 7 h after transfection. Cytoplasmic and nuclear RNAs were isolated from these cells, and the presence of the internalized EGS in these samples was detected by Northern blot analysis. A substantial amount of intact EGSs was found in the nuclear RNA fractions (Fig. 4, lanes 1-4) but not in the cytoplamsic fractions (data not shown). Thus, the internalized R1 and R2 appear to be in the nuclei, where RNase P is exclusively localized.
Fig. 4.
Internalization of EGSs in human cells. We complexed 20 nM 5′-fluorescein-labeled TK1 with 10 μg/ml Lipofectamine 2000 either in the absence or presence of R1 or R2 (80 nM), and it was then transfected into BCBL-1 cells. The transfected cells were isolated by using FACS analysis at 7 h after infection, and nuclear and cytoplasmic RNA fractions were purified. Northern blot analyses were carried out by using nuclear RNA fractions isolated from parental BCBL-1 cells (-, lanes 1 and 5) and cells that were treated with R1 (lanes 2, 4, 6, and 8) and R2 (lanes 3 and 7). We separated 30-μg (lanes 1-3, 5-7) and 60-μg RNA samples (2×, lanes 4 and 8) on 0.8% (B) and 2.5% (A) agarose gels that contained formaldehyde, and they were then transferred to a nitrocellulose membrane and hybridized to a 32P-radiolabeled probe that contained the DNA sequence coding for R1 (A) or RNase P H1 RNA (B). The RNase P RNA sequence was used as the internal control.
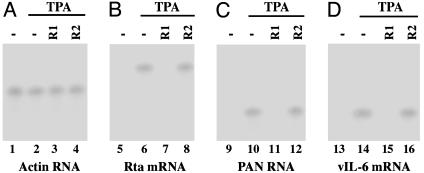
To determine whether the internalized EGSs inhibit viral Rta expression, the transfected cells treated with EGSs were isolated by using FACS analysis and then incubated with 20 ng/ml TPA to induce KSHV lytic cycle. The levels of viral Rta mRNA were determined by Northern blot analyses using the levels of human actin mRNA as an internal control (Fig. 5 A and B). Cells that were treated with R1 exhibited a reduction of 93 ± 6% in the level of Rta mRNA expression, whereas the R2-treated cells exhibited only a reduction of <10% (Table 1). No reduction in the expression level of Rta mRNA was observed in cells that were treated with liposome complexes in the absence of EGSs or in the presence of TK1. These results suggest that the significant reduction of Rta mRNA expression in the R1-treated cells was due to targeted cleavage by RNase P. The low level of inhibition observed in the R2-treated cells presumably was due to the antisense effects of the EGS. The expression level of Rta protein was also determined by Western blot analyses using expression of cellular β-actin as the internal control (Fig. 6 A and B). A reduction of 90 ± 5% in the expression level of Rta protein was observed in cells treated with R1, whereas a reduction of <10% was found in the R2-treated cells (Table 1).
Fig. 5.
Expression of KSHV mRNAs in EGS-treated cells. We first treated 2 × 105 BCBL-1 cells with liposome complexes containing EGSs and isolated them by using FACS analysis, and then we incubated them either in the absence (lanes 1, 5, 9, and 13) or presence (lanes 2-4, 6-8, 10-12, and 14-16) of 20 ng/ml TPA and finally harvested them at 24 h (A and B)or72h(C and D) after TPA induction. RNA samples were isolated from cells treated with liposome complexes in the absence of EGSs (-, lanes 1-2, 5-6, 9-10, and 13-14) or in the presence of R1 (lanes 3, 7, 11, and 15) and R2 (lanes 4, 8, 12, and 16). Equal amounts of each RNA sample (30 μg) were separated on agarose gels, transferred to a nitrocellulose membrane, and hybridized to a 32P-radiolabeled probe that contained the cDNA sequence of the actin mRNA (lanes 1-4), KSHV Rta (lanes 5-8), polyadenylated nuclear (PAN; lanes 9-12), and vIL6 transcripts (lanes 13-16).
Table 1. Levels of mRNA and protein expression inhibition of different viral genes in cells treated with liposome complexes in the presence of TK1 alone or plus R1 or R2 as compared with levels in cells treated with liposome complexes in the absence of EGSs (BCBL-1).
| KSHV gene | Viral gene class | BCBL-1, % | R1, % | R2, % | TK1, % |
|---|---|---|---|---|---|
| Rta mRNA | Immediate-early | 0 | 93 ± 6 | 9 | 1 |
| Polyadenylated nuclear RNA | Early | 0 | 83 ± 4 | 3 | 1 |
| vIL-6 mRNA | Early/late | 0 | 85 ± 5 | 2 | 0 |
| Rta protein | Immediate-early | 0 | 90 ± 5 | 5 | 2 |
| ORF59 protein | Early | 0 | 8 ± 5 | 3 | 1 |
| ORF65 mRNA | Late | 0 | 80 ± 5 | 2 | 2 |
| K8.1 protein | Late | 0 | 80 ± 6 | 3 | 1 |
The values shown are the means from triplicate experiments. SD values <5% are not shown.
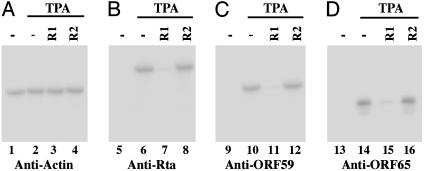
Fig. 6.
Expression of KSHV proteins in EGS-treated cells. We first treated 1 × 105 BCBL-1 cells with liposome complexes containing EGSs, isolated them by using FACS analysis, and then incubated them either in the absence (lanes 1, 5, 9, and 13) or the presence (lanes 2-4, 6-8, 10-12, and 14-16) of 20 ng/ml TPA, and we finally harvested them at either 24 (A and B)or72h(C and D) after TPA induction. Protein samples were isolated from cells treated with liposome complexes in the absence of EGSs (-, lanes 1-2, 5-6, 9-10, and 13-14) or in the presence of R1 (lanes 3, 7, 11, and 15) and R2 (lanes 4, 8, 12, and 16). The samples were separated by SDS/PAGE, transferred to membranes, and reacted with antibodies against human actin (A), KSHV Rta (B), ORF59 (C), and ORF65 proteins (D).
It has been shown that ectopic expression of Rta induces global gene expression and lytic replication of KSHV (28-30). Moreover, expression of a dominant-negative Rta mutant significantly inhibits the activation of the KSHV lytic replication program upon induction by TPA (27). Thus, reduction of Rta expression in the presence of EGS R1 is expected to lead to an inhibition of KSHV gene expression and growth. The expression levels of the polyadenylated nuclear RNA (a viral lytic transcript) and the mRNA coding for vIL6 (a viral early gene) in cells treated with R1 were examined by using Northern blot analysis (2, 43) (Fig. 5 C and D). The protein expression levels of ORF59 (a viral early protein) and ORF65 (a viral late protein) were also determined (29, 35) (Fig. 6 C and D). The expression levels of cellular actin mRNA and protein were used as the internal controls. A reduction of 80-85% in the expression of these genes was found in cells treated with R1, whereas no significant reduction was observed in cells that were either not treated with an EGS or treated with the control EGS, R2 (Table 1). Similar results were found also in the protein expression of K8.1 (a viral late gene) (Table 1).
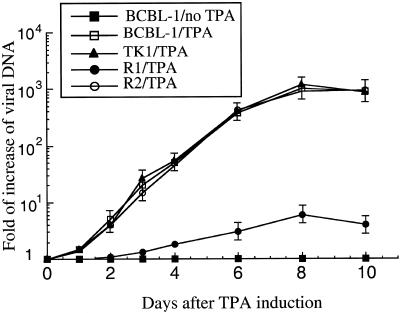
We next investigated whether the inhibition of Rta expression abolishes viral growth and whether Rta can be an effective antiviral target. Because there is currently no efficient de novo infection system that is permissible to KSHV, it is technically difficult to perform KSHV plaque assays and measure plaque-forming units. To assess the effects of EGS on viral production, we chose to determine the amount of viral DNA released into the media after TPA induction and treatment with EGSs. Cells were treated with liposome complexes either in the presence of 5′-FITC-labeled TK1 alone or in the presence of TK1 and R1 or R2. At 7 h after transfection, the cells were isolated by using FACS analysis based on their FITC staining and then incubated in the presence of TPA. Supernatants were collected at different time points after the TPA treatment and then treated with DNase I. The level of DNase-resistant viral DNA in the supernatants, which represents the extracellular virion DNA, was determined by using dot-blot analysis of the purified DNAs with a probe complementary to the KSHV ORF65 sequence. A substantial amount of DNase-resistant viral DNA was detected in the cells that were either treated with TK1 alone or with TK1 as well as R2 (Fig. 7). In contrast, DNase-resistant viral DNA was barely detected in the supernatants from cells treated with R1. Meanwhile, dot-blot analyses showed that similar amounts of intracellular viral DNA were found in these cells before TPA treatment (data not shown), indicating that the cells carry similar copy numbers of latent KSHV genome. These results suggest that the reduced DNase-resistant viral DNA level found in the R1-treated cells is due to the treatment of R1 but not due to the lower amount of viral latent genome DNA in these cells. Indeed, at 8 days after infection, a reduction of ≈150-fold in viral yield was observed in the R1-treated cells, whereas no significant reduction was found in cells that were treated with R2 (Fig. 7).
Fig. 7.
The level of extracellular KSHV DNAs isolated from supernatants of the culture of cells that were first treated with liposome complexes containing TK1 and in the presence of R1 or R2, isolated by FACS analysis, and then either incubated in the absence or presence of TPA. Supernatants were collected from cells at days 1-4, 6, 8, and 10 after TPA induction, and the levels of KSHV DNA were determined by dot-blot analysis. The values, which are the means from triplicate experiments, represent the folds of increase in KSHV virion DNA level in samples, as compared with the level of virion DNA found in cells that were treated with liposome complexes in the absence of R1 or R2 and were incubated in the absence of TPA (BCBL-1/no TPA). Bars indicate SD.
Discussion
RNase P is one of the most ubiquitous and essential enzymes found in nature because it is responsible for processing of all tRNA molecules, which accounts for 2% of all RNA species within a single cell (16, 17). The EGS-based technology represents an attractive approach for gene inactivation because it utilizes endogenous RNase P to generate highly efficient and specific cleavage of the target RNA. Moreover, RNase P-mediated cleavage directed by EGSs is specific and does not generate “irrelevant cleavage,” which is observed usually with RNase H-mediated cleavage induced by conventional antisense phosphothioate oligonucleotides (18, 20, 22). Our study uses exogenous administration of 2′-O-methyl-modified EGSs for antiviral applications. Moreover, our results demonstrate that RNase P-mediated targeting directed by the modified EGS is highly effective in inhibiting KSHV gene expression and growth in cultured cells.
Compared with an all-RNA EGS, an EGS with 2′-O-methyl modification possesses several attractive features. First, 2′-O-methyl-modified oligonucleotides can be easily synthesized chemically by using standard protocols for DNA oligomers (44). Second, the modified EGSs are extremely resistant to degradation by various exonucleases and endonucleases (41, 42). These unique properties of the modified EGSs, along with their ability to bind to target mRNAs and form duplexes in a manner similar to an RNA EGS molecule, strongly support the notion that EGSs with 2′-O-methyl residues may represent a class of antiviral compounds that can be administered directly for gene-targeting applications. In this study, we show that these EGS molecules directed human RNase P to cleave the Rta mRNA sequence efficiently in vitro. Moreover, these EGSs were readily delivered in human cell culture. A reduction of 90-93% in the Rta expression was achieved with a functional EGS, R1, whereas a reduction of <10% was observed in cells that were treated with R2. R2 bound to Rta mRNA substrate rta-S in vitro as well as R1 but contained nucleotide mutations that disrupted RNase P recognition. These results suggest that the overall observed inhibition with R1 was primarily due to targeted cleavage by RNase P, as opposed to the antisense effect or other nonspecific effects of the EGSs.
RNase P-mediated targeting directed by the 2′-O-methyl-modified EGS appears to be specific. The EGSs did not exhibit significant cytotoxicity because cells treated with EGSs are indistinguishable from the cells treated with lipid complexes alone in the absence of the EGSs, in terms of cell growth and viability for 10 days (data not shown). Moreover, the antiviral effect of the EGS treatment (inhibition of viral growth) appears to be due to the reduction of the Rta expression. The expression of all of the viral early and late genes examined, including vIL6, polyadenylated nuclear RNA, ORF59, ORF65, and K8.1, was found to be reduced significantly in cells that were treated with R1 but not in those treated with R2 or TK1 (Table 1). The extent of the observed inhibition of the expression of most of these viral early and late genes correlated with that of the inhibition of the Rta expression. Meanwhile, no significant reduction in the level of K3 mRNA, a viral immediate-early transcript that is not regulated by Rta, was found in the cells treated with R1 (data not shown). Thus, the EGS is specific in inhibiting the expression of its target mRNA.
Delivery of the EGS into the nuclear compartment is essential to the success of the EGS technology because RNase P is localized exclusively in the nuclei (16). Our data also provide direct evidence that exogenous EGSs can be delivered into the nuclei. The efficient delivery and proper localization of the EGS may be mediated by cellular tRNA-binding proteins, which may interact with the tRNA-like domains of the EGS and target the EGS to the nuclear compartment.
The EGS-based strategy describe in this article can be generally applicable to define the function of KSHV genes by knocking down the expression of individual viral genes. KSHV is a member of the human herpesvirus family (2), which includes seven other different viruses, such as herpes simplex virus and Epstein-Barr virus (45, 46). The Rta is conserved among γ-herpesviruses and is among the first viral proteins expressed during viral lytic replication (28, 30). These properties, along with our results that blocking Rta expression can inhibit KSHV gene expression and growth effectively, strongly suggest that Rta represents an ideal target for antiviral therapy. To develop EGSs as a conventional drug that can be used as an exogenous agent for intracellular delivery in antiviral therapy, studies are needed to determine whether the chemically modified EGSs, when directly administered to the skin and blood, are effective in treating and preventing KSHV-associated diseases including KS and the PEL. Also, further studies on in vitro genetic engineering and different designs of EGSs for increasing their targeting activity, as well as on developing means for improving their delivery (21, 22), are needed to increase the efficacy of the EGSs in vivo. These studies would further facilitate the development of the EGS-based technology for gene-targeting applications in both basic research and clinical therapy.
Acknowledgments
We thank Dr. Ren Sun for invaluable suggestions and reagents, including anti-KSHV antibodies and KSHV DNA plasmid constructs. We also thank Hua Zou, Manfred Lee, and Ada Tam for technical assistance. P.T. is a recipient of a predoctoral fellowship from the American Heart Association (Western Affiliate). K.K. is supported by a predoctoral Block grant from University of California, Berkeley. F.L. is a Scholar of the Leukemia and Lymphoma Society and an Established Investigator of the American Heart Association. This research was supported by the National Institutes of Health.
Abbreviations: EGS, external guide sequence; FACS, fluorescence-activated cell sorting; KS, Kaposi's sarcoma; KSHV, KS-associated herpesvirus; PEL, primary-effusion lymphoma; TK, thymidine kinase; TPA, tetradecanoyl phorbol acetate.
References
- 1.Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M. & Moore, P. S. (1994) Science 266, 1865-1869. [DOI] [PubMed] [Google Scholar]
- 2.Moore, P. S. & Chang, Y. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 2803-2834.
- 3.Renne, R., Zhong, W., Herndier, B., McGrath, M., Abbey, N., Kedes, D. & Ganem, D. (1996) Nat. Med. 2, 342-346. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff, C., Endo, Y., Collins, P. D., Takeuchi, Y., Reeves, J. D., Schweickart, V. L., Siani, M. A., Sasaki, T., Williams, T. J., Gray, P. W., et al. (1997) Science 278, 290-294. [DOI] [PubMed] [Google Scholar]
- 5.Arvanitakis, L., Geras-Raaka, E., Varma, A., Gershengorn, M. C. & Cesarman, E. (1997) Nature 385, 347-350. [DOI] [PubMed] [Google Scholar]
- 6.Kedes, D. H. & Ganem, D. (1997) J. Clin. Invest. 99, 2082-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin, D. F., Kuppermann, B. D., Wolitz, R. A., Palestine, A. G., Li, H. & Robinson, C. A. (1999) N. Engl. J. Med. 340, 1063-1070. [DOI] [PubMed] [Google Scholar]
- 8.Glesby, M. J., Hoover, D. R., Weng, S., Graham, N. M. H., Phair, J. P., Detels, R., Ho, M. & Saah, A. J. (1996) J. Infect. Dis. 173, 1477-1480. [DOI] [PubMed] [Google Scholar]
- 9.Santoro, S. W. & Joyce, G. F. (1997) Proc. Natl. Acad. Sci. USA 94, 4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein, C. A. & Cheng, Y. C. (1993) Science 261, 1004-1012. [DOI] [PubMed] [Google Scholar]
- 11.Rossi, J. J. (1999) Chem. Biol. 6, R33-R37. [DOI] [PubMed] [Google Scholar]
- 12.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457-467. [DOI] [PubMed] [Google Scholar]
- 13.Wong-Staal, F., Poeschla, E. M. & Looney, D. J. (1998) Hum. Gene Ther. 9, 2407-2425. [DOI] [PubMed] [Google Scholar]
- 14.Scherer, L. & Rossi, J. (2003) Nat. Biotechnol. 21, 1457-1465. [DOI] [PubMed] [Google Scholar]
- 15.Altman, S. (1995) Biotechnology 13, 327-329. [DOI] [PubMed] [Google Scholar]
- 16.Altman, S. & Kirsebom, L. A. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 351-380.
- 17.Frank, D. N. & Pace, N. R. (1998) Annu. Rev. Biochem. 67, 153-180. [DOI] [PubMed] [Google Scholar]
- 18.Forster, A. C. & Altman, S. (1990) Science 249, 783-786. [DOI] [PubMed] [Google Scholar]
- 19.Guerrier-Takada, C., Li, Y. & Altman, S. (1995) Proc. Natl. Acad. Sci. USA 92, 11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan, Y., Hwang, E. & Altman, S. (1992) Proc. Natl. Acad. Sci. USA 89, 8006-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan, Y. & Altman, S. (1994) Science 263, 1269-1273. [DOI] [PubMed] [Google Scholar]
- 22.Ma, M., Benimetskaya, L., Lebedeva, I., Dignam, J., Takle, G. & Stein, C. A. (2000) Nat. Biotechnol. 18, 58-61. [DOI] [PubMed] [Google Scholar]
- 23.Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. J. (2000) Nature 404. [DOI] [PubMed]
- 24.McKinney, J., Guerrier-Takada, C., Wesolowski, D. & Altman, S. (2001) Proc. Natl. Acad. Sci. USA 98, 6605-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraus, G., Geffin, R., Spruill, G., Young, A. K., Seivright, R., Cardona, D., Burzawa, J. & Hnatyszyn, H. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plehn-Dujowich, D. & Altman, S. (1998) Proc. Natl. Acad. Sci. USA 95, 7327-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukac, D. M., Kirshner, J. R. & Ganem, D. (1999) J. Virol. 73, 9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukac, D. M., Renne, R., Kirshner, J. R. & Ganem, D. (1998) Virology 252, 304-312. [DOI] [PubMed] [Google Scholar]
- 29.Gradoville, L., Gerlach, J., Grogan, E., Shedd, D., Nikiforow, S., Metroka, C. & Miller, G. (2000) J. Virol. 74, 6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, R., Lin, S. F., Gradoville, L., Yuan, Y., Zhu, F. & Miller, G. (1998) Proc. Natl. Acad. Sci. USA 95, 10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ensoli, B., Barillari, G. & Gallo, R. C. (1992) Immunol. Rev. 127, 147-155. [DOI] [PubMed] [Google Scholar]
- 32.Deng, H., Chu, J. T., Rettig, M. B., Martinez-Maza, O. & Sun, R. (2002) Blood 100, 1919-1921. [DOI] [PubMed] [Google Scholar]
- 33.Taga, T. & Kishimoto, T. (1997) Annu. Rev. Immunol. 15, 797-819. [DOI] [PubMed] [Google Scholar]
- 34.Wu, L., Renne, R., Ganem, D. & Forghani, B. (2000) J. Clin. Virol. 17, 127-136. [DOI] [PubMed] [Google Scholar]
- 35.Chan, S. R., Bloomer, C. & Chandran, B. (1998) Virology 240, 118-126. [DOI] [PubMed] [Google Scholar]
- 36.Dunn, W., Trang, P. & Liu, F. (2001) Proc. Natl. Acad. Sci. USA 99, 12332-12337. [Google Scholar]
- 37.Pyle, A. M., McSwiggen, J. A. & Cech, T. R. (1990) Proc. Natl. Acad. Sci. USA 87, 8187-8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, F. & Altman, S. (1995) Genes Dev. 9, 471-480. [DOI] [PubMed] [Google Scholar]
- 39.Zaug, A. J. & Cech, T. R. (1995) RNA 1, 363-374. [PMC free article] [PubMed] [Google Scholar]
- 40.Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., Parry, J. P., Peruzzi, D., Edelman, I. S., Chang, Y. & Moore, P. S. (1996) Proc. Natl. Acad. Sci. USA 93, 14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesnik, E. A., Guinosso, C. J., Kawasaki, A. M., Sasmor, H., Zounes, M., Cummins, L. L., Ecker, D. J., Cook, P. D. & Freier, S. M. (1993) Biochemistry 32, 7832-7838. [DOI] [PubMed] [Google Scholar]
- 42.Cummins, L. L., Owens, S. R., Risen, L. M., Lesnik, E. A., Freier, S. M., McGee, D., Guinosso, C. J. & Cook, P. D. (1995) Nucleic Acids Res. 23, 2019-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, R., Lin, S. F., Staskus, K., Gradoville, L., Grogan, E., Haase, A. & Miller, G. (1999) J. Virol. 73, 2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma, S. & Eckstein, F. (1998) Annu. Rev. Biochem. 67, 99-134. [DOI] [PubMed] [Google Scholar]
- 45.Kieff, E. & Rickinson, A. B. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2511-2573. [Google Scholar]
- 46.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2399-2460. [Google Scholar]