Abstract
Heme is an essential component of numerous hemoproteins with functions including oxygen transport, energy metabolism, and drug biotransformation. In nonerythropoietic cells, 5-aminolevulinate synthase (ALAS1) is the rate-limiting enzyme in heme biosynthesis. Upon exposure to drugs that induce cytochromes P450 and other drug-metabolizing enzymes, ALAS1 is transcriptionally up-regulated, increasing the rate of heme biosynthesis to provide heme for cytochrome P450 hemoproteins. We used a combined in silico-in vitro approach to identify sequences in the ALAS1 gene that mediate direct transcriptional response to xenobiotic challenge. We have characterized two enhancer elements, located 20 and 16 kb upstream of the transcriptional start site. Both elements respond to prototypic inducer drugs and interact with the human pregnane X receptor NR1I2 and the human constitutive androstane receptor NR1I3. Our results suggest that the fundamental mechanism of drug induction is the same for cytochromes P450 and ALAS1. Transcriptional activation of the ALAS1 gene is the first step in the coordinated up-regulation of apoprotein and heme synthesis in response to exogenous and endogenous signals controlling heme levels. Understanding the direct effects of drugs on heme synthesis is of clinical interest, particularly in patients with hepatic porphyrias.
Heme is indispensable for mammalian life. It is an essential component of numerous heme proteins, with functions including oxygen transport, energy metabolism, and drug bio-transformation. The two major sites of heme synthesis are the bone marrow, where hemoglobin is produced, and the liver, where various hemoproteins [in particular, microsomal cytochromes P450 (CYP)] rely on prosthetic heme to catalyze the oxidation of endogenous and exogenous compounds (1, 2). The rate of heme synthesis in the liver is controlled at the first committed step: the condensation of glycine and succinyl-CoA to 5-aminolevulinate, which is catalyzed by 5-aminolevulinic acid synthase (ALAS). The regulatory role of ALAS is underlined by the fact that ALAS mRNA is markedly increased under physiological conditions demanding more heme, whereas expression levels of the other enzymes in the pathway do not change significantly (1).
In mammals and birds, two isoforms of ALAS are known, which exhibit differential tissue distribution and regulation (3-6). The housekeeping isoform ALAS1 is expressed ubiquitously and is encoded by a gene located on human chromosome 3p21 (7). It provides heme for numerous cellular hemoproteins. In mitochondrial biogenesis, coordinated increase of heme synthesis and the synthesis of respiratory apocytochromes is a prerequisite for functional energy metabolism (8).
Intracellular free heme levels are extremely low because increased levels of free heme are cytotoxic, and heme biosyn-thesis, therefore, is tightly regulated (1). Negative feedback by heme controls the pathway at various steps. Most importantly, a regulatory pool of heme negatively influences ALAS1 activity by multiple mechanisms, such as (i) preventing the transfer of ALAS1 precursor to the mitochondrial inner membrane (9), (ii) reducing ALAS1 mRNA stability (1), and (iii) repressing ALAS1 mRNA transcription in mammals but not in birds (10).
Also, high concentrations of free heme induce heme oxygenase and, therefore, stimulate heme catabolism (11).
The rate of heme biosynthesis must also be responsive to increased demands, for instance, during induction of drug-metabolizing CYPs. In this situation, the levels of ALAS1 mRNA are swiftly up-regulated to provide sufficient heme to nascent apocytochromes. Recent studies in our laboratory and other laboratories (12) have demonstrated that this increase is not a consequence of heme feedback regulation but a direct activation of ALAS1 transcription by the drug.
Accordingly, we found drug-responsive sequences within the chicken and murine ALAS1 genes, and we tested them in regard to their interaction with nuclear receptors (NRs) (13, 14). The central role of orphan NRs in drug-induced expression of CYPs is well established (see refs. 15-19 and references therein). With few exceptions, NRs bind as homodimers or heterodimers to cognate response elements (REs) consisting of two hexameric half-sites. The sequence and orientation of half sites, as well as the distance between them, account for the specificity of a given RE to a particular NRs dimer (20). Within drug-responsive enhancer sequences of CYP2B, CYP2H, and CYP3A genes, NR REs were shown to be essential for drug-mediated activation (21-26), and heterodimers of the 9-cis retinoic acid X receptor (RXR) with the orphan NRs constitutive androstane receptor (CAR), pregnane X receptor (PXR), and chicken xenobiotic-sensing receptor (CXR) have been shown to bind to these elements and to be activated by drugs directly or indirectly (23, 27, 28).
In this article, we describe two sequence elements in the distal 5′-flanking region of the human ALAS1 gene that mediate direct transcriptional activation in response to drugs. For both elements, we show binding and activation in response to drugs by human CAR and PXR. Understanding the direct effect of drugs on heme synthesis is of clinical interest. In patients with hepatic porphyrias (inherited diseases caused by partial deficiencies of enzymes of heme synthesis), the administration of drugs that induce hepatic CYP and ALAS levels can precipitate acute porphyric attacks.
Methods
Reagents and Plasmids. Cell culture reagents were obtained from Invitrogen. Propylisopropylacetamide (PIA) was a gift from P. Sinclair (Veterans Administration Hospital, White River Junction, VT). Glutethimide, metyrapone (2-methyl-1,2-di-3-pyridylpropadone), rifampicin (Rif), and phenobarbital (PB) sodium salt were purchased from Fluka. All other reagents and supplies were obtained from standard commercial sources.
The pGL3tk luciferase reporter gene vector was constructed by excising the thymidine kinase promoter from the pBLCAT5 reporter vector (29) with BamHI and BglII and inserting it into the BglII site of the promoterless pGL3-basic reporter vector (Promega). The pRSV-βGal β-galactosidase expression vector was a gift from A. Kralli (The Scripps Research Institute, La Jolla, CA). Expression plasmids for chicken xenobiotic-sensing receptor (CXR) and human CAR are described in refs. 27 and 30. The expression plasmid for human PXR was a gift from S. A. Kliewer (University of Texas Southwestern Medical Center, Dallas). Human RXR-α expression plasmid was provided by P. Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France).
Isolation of a Genomic Clone Containing the ALAS1 Gene. We isolated a clone harboring the ALAS1 gene from a P1 artificial chromosome (PAC) human genomic library (Incyte Pharmaceuticals, Palo Alto, CA). As probe for hybridization, a 546-bp 5′-fragment of human ALAS1 cDNA was amplified by using the following primers: 5′-CAGCGCAGTCTTTCCACAG-3′ and 5′-TACTACAGGACCTTCAAGAA-3′.
Analysis of Drug-Responsive Fragments Within the ALAS1 Gene. From GenBank, the sequence for a P1 artificial chromosome (PAC) clone containing the human ALAS1 gene was retrieved (Gen-Bank accession no. AC006252), containing 29,120 bp of the 5′-flanking region of this gene. The sequence was subjected to analysis for predicted NR binding sites by using the NUBIScan algorithm (24). Hypothesizing that the NRs CAR and PXR may be involved in drug-mediated induction of ALAS1 transcription, we screened this sequence for the presence of NR REs that had been described to interact with these receptors. Specifically, we screened for matches to DR3-, DR4-, DR5-, ER6-, ER7-, and ER8-type REs. From the resulting predictions, the best-ranking of each were selected, all of which had a z score >6.4. Subsequently, fragments harboring the predicted REs were selected such that (i), whenever possible, several predicted sites were grouped together and (ii), to preserve context around the binding sites, at least 200 bp were added in front of the most 5′ site of the group and after the most 3′ site. Selected fragments were amplified by PCR, cloned into the pGL3tk reporter vector, and verified by sequencing.
Mutations in putative NR binding sites were introduced by PCR using standard overlap techniques, as described in ref. 24. All mutated constructs were verified by sequencing. Gel mobility-shift assays with human RXR-α, PXR, and CAR were performed, as described in ref. 24.
Cell Culture and Transient Transfection. Leghorn male hepatoma (LMH) cells were used in reporter gene studies, as described in ref. 24. After 24 h of drug induction, cells were lysed and assayed luciferase activity was normalized against β-galactosidase activity to compensate for varying transfection efficiency.
Transcriptional activation assays were performed in HepG2 cells. Cells were kept in DMEM/F12 medium without phenol red, supplemented with 10% charcoal-stripped FBS. Cells were plated at a density of 625,000 cells per well in six-well cell culture dishes. Cells in each well were transfected with 150 ng of PXR or CAR expression vector, 150 ng of RXR-α expression vector, 100 ng of reporter vector, 500 ng of pRSV-β-gal, and 1.1 μg of carrier with FuGene 6 reagent. At 24 h after transfection, cells were exposed to inducer compounds or vehicle for another 24 h before cell extracts were prepared and analyzed as described for LMH cells.
Results
Isolation of a Human ALAS1 Genomic Clone. To test regions of the ALAS1 gene for the presence of regulatory elements conveying drug inducibility, we isolated a clone containing the human ALAS1 gene from a PAC human genomic library. The library was screened with a probe derived from the 5′ part of the ALAS1 mRNA. By restriction digestion, the insert size of the clone was estimated to be >80 kb. It contains at least 29.1 kb of ALAS1 5′-flanking region, as determined by PCR analysis with primers based on the sequence used (GenBank accession no. AC006252), which spans the ALAS1 gene (data not shown).
The Proximal Promoter Is Not Sufficient to Convey ALAS1 Drug Inducibility. In rat, sequences close to the transcriptional start site have been proposed to confer drug-mediated activation of ALAS1 (31). We tested the promoter of human ALAS1 for elements mediating drug induction. Two promoter fragments, spanning -234 to -1 bp or -1,249 to -1 bp, were inserted into the pGL3-basic reporter vector, and reporter gene activity was assayed in LMH cells. These chicken hepatoma cells maintain PB-type drug induction completely, despite being a continuously dividing cell line (30, 32). Although both fragments exhibited basal promoter activity, neither fragment caused increased transcription in response to 400 μM PB, 10 μM Rif, or 250 μM PIA, which are prototypic inducers of ALAS1 (see Fig. 6, which is published as supporting information on the PNAS web site). We concluded that human ALAS1 drug response is conveyed by elements outside of the proximal promoter.
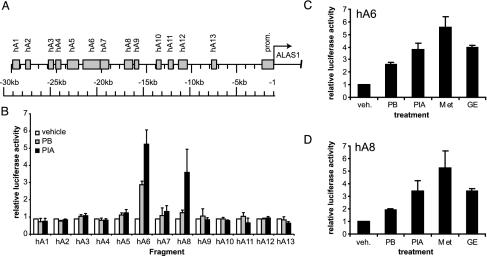
Screening of Regions of Interest Containing Likely REs. A large segment of the 5′-flanking region of the ALAS1 gene was screened for the presence of putative NR binding sites by using a bioinformatic approach (24). Of particular interest to us were recognition sites for the xenosensors PXR and CAR. Our search included NR REs that have been described as binding sites for PXR or CAR. The best-scoring predictions were grouped into 13 fragments (Fig. 1, and see Table 1, which is published as supporting information on the PNAS web site), which were amplified subsequently by PCR and cloned into the pGL3tk reporter vector.
To verify our in silico predictions, we tested these fragments for drug induction in the LMH cell line (Fig. 1B). Reporter vectors containing the putative response regions were transfected into cells that were then exposed to 400 μM PB, 250 μM PIA, or vehicle (0.1% DMSO). Of all fragments tested, only two fragments, hA6 and hA8, showed considerable activation by PB (3.2- and 1.4-fold, respectively) and PIA (5.9- and 4.3-fold, respectively).
Fig. 1.
Combined in silico-in vitro screening of the 5′-flanking region of the human ALAS1 gene. (A) Graphical representation of NUBIScan prediction clusters in the 5′-flanking region of the ALAS1 gene. Shaded boxes designate fragments, each containing one or more possible xenosensor binding sites. (B) Reporter gene assays in LMH cells. Reporter vectors carrying the fragments were tested in LMH cells. After 24 h of induction with 400 μM PB, 250 μM PIA, or vehicle (0.1% DMSO), reporter activities were assayed, and they are expressed relative to that of vehicle-treated cells. (C and D) Activation of the hA6 and hA8 fragment by a panel of xenobiotics. hA6 or hA8 reporter constructs were tested in LMH cells subjected to vehicle (veh.; 0.1% DMSO) or the following drugs: PB (400 μM), PIA (250 μM), metyrapone (MET, 400 μM), and glutethimide (GE, 500 μM). Data represent the mean of at least three independent experiments plus one SD.
To characterize the spectrum of drugs to which the hA6 and hA8 fragments respond, we assayed their response to a collection of known CYP inducers. In addition to PB (400 μM) and PIA (250 μM), we chose metyrapone (400 μM) and glutethimide (500 μM), which are classical CYP3A inducers (Fig. 1 C and D). The fragments exhibited similar strong responses to all four of the inducers.
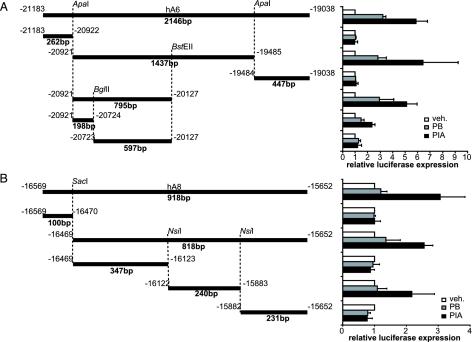
Definition of ALAS1 Drug-Responsive Enhancer Sequences (ADRESs) Within the Responsive Fragments. To determine the exact location of enhancer elements mediating drug induction, both responsive fragments were dissected further. The hA6 fragment was cut with ApaI, yielding three fragments of 262, 1,437, and 447 bp, respectively, which were inserted into the pGL3tk vector and tested in LMH cells. Activation was observed solely with the 1,437-bp fragment. Drug responsiveness could further be localized to a 795-bp ApaI/BstEII fragment, which was subsequently split into 198- and 597-bp fragments by cleavage with BglII. Of these two, only the 198-bp ADRES element showed inducibility (Fig. 2A).
Fig. 2.
Dissection of the drug-responsive fragments leads to definition of ADRES elements. (A) The 2,146-bp hA6 fragment was cut with ApaI to yield three smaller fragment of 262, 1,437, and 447 bp, respectively. Fragments were introduced into the pGL3tk vector and tested in LMH cells for induction by vehicle, PB, or PIA, as described. The responsive 1,437-bp fragment was further digested with BglII and BstEII, yielding fragments of 795, 198, and 597 bp. The 1,089-bp fragment spanning from the BstEII site to the 3′ end of the hA6 fragment showed no response to drugs (data not shown). Data represent the mean of at least three independent experiments plus one SD. (B) The hA8 fragment was cut at SacI and NsiI restriction sites, and the resulting fragments of 100 and 818 bp, as well as subfragments of the 818-bp piece (347, 240, and 231 bp), were cloned into the pGL3tk reporter vector. The ability of these fragments to mediate induction in LMH cells was tested as described. Data represent the mean of at least three independent experiments plus one SD.
Analogously, digestion of the 918-bp hA8 fragment by SacI digestion yielded two subfragments of 100 and 818 bp. Inducibility was observed with the 818-bp fragment, which was further cut with NsiI into three smaller units 347, 240, and 231 bp in size, which were tested for drug-responsiveness. Only the 240-bp fragment showed inducibility and, therefore, was defined as a second ADRES element (Fig. 2B).
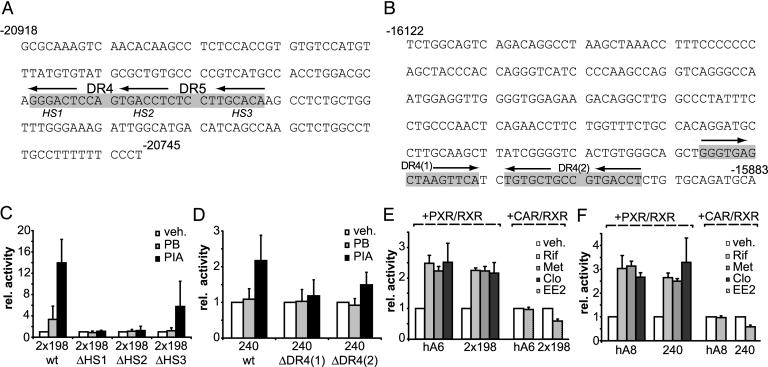
DR4 Type REs Are Essential for Drug Induction in both ADRES Elements. Detailed computational analysis of the 198- and 240-bp ADRES elements revealed the presence of overlapping DR4 and DR5 motifs formed by three half-sites within the 198-bp ADRES (Fig. 3A) and two potential DR4 elements within the 240-bp element (Fig. 3B). We, thus, performed mutagenesis studies to clarify the functional role of the DR4 and DR5 motifs within the ADRES elements.
Fig. 3.
Drug induction of the two ADRES elements is mediated by NR REs. (A) Sequence of the 198-bp core ADRES element contained in hA6. The two contiguous potential NR REs (DR4 and DR5) are shown in gray. Arrows indicate the position of each of the three half-sites HS1, HS2, and HS3. Sequence numbers are relative to the ALAS1 transcriptional start site. (B) Sequence of the 240-bp core ADRES element of hA8. Numbering is relative to the ALAS1 transcriptional start site. Gray boxes show the predicted NR DR4 (1) and DR4 (2) REs. Arrows indicate the individual half-sites. (C) Reporter gene assays in LMH cells with two copies of wild-type and mutant (ΔHS1 to ΔHS3) 198-bp ADRES fragments. Cells were treated with vehicle, 400 μM PB, or 250 μM PIA. (D) LMH cell reporter gene assays with wild-type and mutant [DR4 (1) or DR4 (2)] 240-bp ADRES fragments. Treatment was analogous to treatment described for C.(E) Transactivation assays with hA6 fragment or two copies of the 198-bp ADRES. Reporter constructs were cotransfected into HepG2 cells with expression plasmids for human RXR-α and either human PXR or human CAR. To assess PXR-mediated activation, cells were treated with vehicle (0.1% DMSO), 10 μM Rif, or 400 μM metyrapone (Met). For human CAR, cells were treated with vehicle (0.1% DMSO) or 10 μM EE2. (F) Transactivation assays with hA8 fragment or the 240-bp ADRES contained within the hA8 fragment. Cells were treated as described for E. Data represent the mean of three independent experiments plus one SD.
Within a reporter construct carrying two copies of the 198-bp ADRES, we mutated each of the three half-sites in both ADRES copies. Half-site 1 (HS1) was replaced by a EcoRV site, half-site 2 (HS2) was replaced by a XbaI site, or half-site 3 (HS3) was replaced by a SacII restriction site (Fig. 3A). We then assayed drug-induction mediated by wild-type or mutant fragments in LMH cells (Fig. 3C). Whereas mutations of HS1 or HS2 abolished induction, mutation of the HS3 site led only to minor reduction, which may indicate some activity of the DR5 element but may also be due to unspecific changes in the immediate environment of the active DR4 element.
To investigate the role of the predicted DR4 elements in the 240-bp ADRES, we disrupted these sequences by replacing the hexameric half-sites with restriction sites. The DR4 (1) element was replaced by SmaI and EcoRV sites, and the DR4 (2) was replaced by XbaI and XhoI sites. Although ablation of the DR4 (1) half-sites abolished induction, the lack of an effect of mutational disruption of DR4 (2) on induction suggests no function of DR4 (2) in drug induction (Fig. 3D).
We next assessed the ability of the human drug-sensing receptors PXR and CAR to transactivate the ADRES elements. To this end, HepG2 human hepatoma cells were transfected with expression plasmids for human RXR-α, as well as expression plasmids for either human PXR or human CAR. Also, reporter plasmids harboring either two copies of the 198-bp ADRES or a single copy of the hA 240-bp ADRES were cotransfected. PXR-transfected cells were subsequently exposed to known activators of human PXR, and cells transfected with CAR were treated with 17α-ethynyl-3,17β-estradiol (EE2), a compound shown to repress basal CAR activity (33). Both ADRES elements, as well as the longer elements hA6 and hA8, can be activated by human PXR in response to a number of well characterized PXR activators, essentially reflecting our observations in a priori responsive LMH cells (Fig. 3 E and F).
In CAR cotransfections, the 198-bp ADRES, as well as the 240-bp ADRES, responded to EE2. Surprisingly, this ≈2-fold repression was not observed when EE2 was tested on constructs harboring the larger hA6 or hA8 fragments. This finding indicates that elements beyond the core ADRES may influence the binding capacity of CAR.
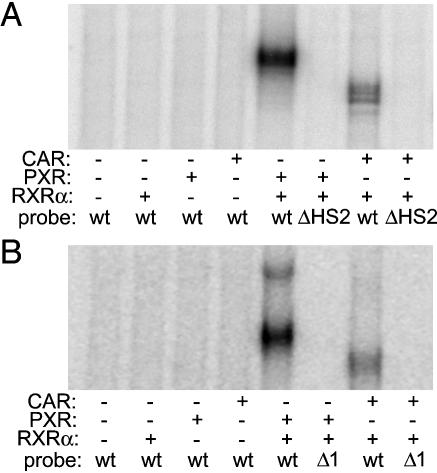
Human PXR and Human CAR Bind to NR REs in both ADRESs. To examine direct interaction of the xenosensing receptors PXR and CAR with the two enhancer elements, we performed gel mobility-shift assays with both fragments (Fig. 4). Both CAR and PXR bound to radiolabeled wild-type 198-bp ADRES. Observed binding depended on the presence of human RXR-α. Neither receptor bound to the radiolabeled 198-bp ADRES that carried the ΔHS2 mutation described above. When in vitro binding of proteins to the 240-bp ADRES was examined, results were analogous (Fig. 4B). Both CAR and PXR bind to the 240-bp ADRES, and mutation of the DR4 (1) RE abolishes binding.
Fig. 4.
Human PXR and CAR bind to the ALAS1 drug-responsive enhancers in vitro. (A) Gel mobility-shift assay with wild-type (wt) and ΔHS2 mutant 198-bp ADRES as probes. The first four lanes show wild-type 198-bp ADRES radiolabeled and incubated with no receptor or in vitro transcribed and translated human RXR-α, PXR, or CAR. The fifth and seventh lanes show wild-type 198-bp ADRES incubated with RXR-α and PXR or RXR-α and CAR. The sixth and eighth lanes show the same conditions as described for the fifth and seventh lanes, except instead of wild-type probe, radiolabeled ΔHS2 mutant was used. (B) Gel mobility-shift assays with wild-type and ΔDR4 (1) mutant 240-bp ADRES. The first four lanes show radiolabeled wild-type 240-bp ADRES incubated with no receptor or in vitro transcribed and translated human RXR-α, PXR, or CAR. The fifth and seventh lanes show wild-type 240-bp ADRES incubated with RXR-α and PXR or RXR-α and CAR. The sixth and eighth lanes show the same conditions as described for the fifth and seventh lanes, except that radiolabeled ΔDR4 (1) mutant was used instead of wild-type probe.
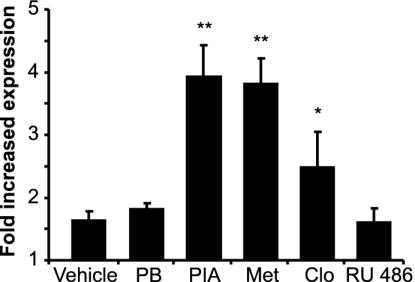
Effects of the ADRES Elements in Genomic Context. To approximate the genomic situation, we generated a minigene in which the role of the two upstream enhancers in drug-mediated regulation of ALAS1 could be investigated. The 795-bp element encompassing the 198-bp ADRES (see Fig. 2 A), followed by the 818-bp element containing the 240-bp ADRES (see Fig. 2B), as well as the 234-bp human ALAS1 promoter, were inserted into a reporter construct. Reporter levels obtained with this construct in LMH cells were compared with levels obtained with a vector harboring only the 234-bp promoter (Fig. 5). Expression was increased 1.6-fold in the absence of drugs, presumably by means of basal activation mediated by unliganded NR. Moreover, expression was significantly up-regulated by 250 μM PIA (3.93-fold), 400 μM metyrapone (3.80-fold), and 10 μM clotrimazole (2.48-fold). Notably, 400 μM PB and 10 μM mifepristone (RU486) had only nonsignificant effects on this test system.
Fig. 5.
The ADRES elements mediate drug induction in a larger genomic context. A reporter construct harboring the 795- and 818-bp drug-responsive elements (see Fig. 2 A and B) directly upstream of the 234-bp hALAS1 promoter was tested for induction in LMH cells with vehicle (0.1% DMSO), 400 μM PB, 250 μM PIA, 400 μM metyrapone (Met), 10 μM clotrimazole (Clo), or 10 μM mifepristone (RU486). By comparing reporter activities with activities obtained with a reporter construct containing only the 234-bp hALAS1 promoter, the fold increase of expression caused by the two ADRES is shown. Data represent the mean of four independent experiments plus one SD. *, P < 0.05; **, P < 0.01.
Discussion
In the present study, we describe the discovery of two distal regulatory elements in the human ALAS1 gene that mediate drug response. Both elements respond to drugs with a relative potency that reflects in vivo observations. Human CAR and PXR can bind to DR4-type hexamer repeats found in both elements, and exposure to agonists of PXR or inverse agonists of CAR affect reporter gene expression.
The two REs exhibit remarkable similarity in their response to drugs, even though no extensive similarity is found at the sequence level. The presence of several regions cooperatively mediating drug induction has been observed in the regulation of CYP2B, CYP2H, CYP3A genes. When analyzed in their natural context and coupled to the natural ALAS1 promoter, these elements elicit increased basal expression as well as a transcriptional response to inducer drugs. This fact strongly suggests the in vivo functionality of these REs.
Despite the demonstrated role of the REs described here, the existence of further drug REs within the ALAS1 gene is not excluded. Our studies covered only 58% of the 30-kb 5′-flanking sequence of the ALAS1 coding region, and further REs may exist in untested upstream regions, introns, or the 3′ flanking region. However, the fact that induction observed with the ADRES fragments reflects in vivo data to a high degree strongly suggests their key role in ALAS1 regulation.
What are the roles of PXR and CAR in ALAS1 drug induction? Our studies of human, chicken (13), and mouse (14) ALAS1 regulation, combined with recent studies in transgenic mice and human hepatocytes (34-36), reveal a complex picture. Loss-of-function experiments in mice lacking either PXR or CAR clearly showed involvement of PXR in hepatic ALAS1 induction, which was decreased in PXR-deficient mice. Remaining inducibility must be caused by a PXR-independent mechanism. Interestingly, ablation of PXR abolished induction in small intestine, a tissue in which CAR signaling is deficient (35). Mice lacking CAR exhibit decreased basal levels of ALAS1 mRNA, but induction is retained, suggesting redundancy between CAR and PXR signaling pathways. Specific agonists point out the contribution of CAR to ALAS1 regulation. A recent study in human hepatocytes has revealed induction of ALAS1 mRNA by a selective human CAR agonist (36).
Our findings show interaction between the ADRES elements and PXR, as well as CAR, and they support a role for both receptors in ALAS1 regulation, as evidenced by transactivation studies and gel mobility-shift assays. Double knock-out studies in mice lacking both CAR and PXR would clarify the importance of further pathways in ALAS1 drug induction. To our knowledge, such experiments have not yet been published.
In conclusion, our data define enhancer sequences causing direct transcriptional response of human ALAS1 to drugs and, furthermore, demonstrate that induction of ALAS1 is mediated by the same mechanisms controlling drug-mediated expression of CYPs. This coordinated induction of CYP apoprotein and heme biosynthesis makes physiological sense. In a second step, negative feedback by free heme can then adjust ALAS1 expression at the mRNA (9) and protein transport (1) levels. This scenario is consistent with experiments showing a strong initial increase of ALAS1 mRNA within few hours after exposure to drug (37, 38). At this time point, CYP mRNA and apoprotein levels are not yet markedly increased.
In more practical terms, our study enhances the understanding of the mechanisms underlying drug-induction of heme biosyn-thesis in humans. The build-up of intermediates of the heme pathway is the biochemical hallmark of acute porphyrias. However, the exact relationship between disturbed heme synthesis and the neuropsychiatric symptoms of porphyria remains elusive (39, 40). Understanding the mechanisms underlying this induction is the first and necessary step in preventing drug-elicited porphyric attacks. Our findings can lead to screening tests for human ALAS1 induction, which promise to discover potential inducers of acute porphyric attacks. Moreover, because ALAS1 induction is an early sign of CYP induction, it may provide a sensitive general screen for induction of drug metabolism. Such assays can be deployed efficiently in the early stages of drug development, and they contribute to increased drug safety and prevent avoidable severe adverse drug reactions in susceptible patients.
Supplementary Material
Acknowledgments
We thank Dr. C. Gnerre for preparing the pGL3tk reporter gene vector; Drs. A. Kralli, S. A. Kliewer, and P. Chambon for kindly providing reporter plasmids and expression plasmids for NRs; and Dr. P. Sinclair for his generous gift of PIA. This work was supported by a grant from the Swiss National Science Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADRES, ALAS1 drug-responsive enhancer sequence; ALAS, 5-aminolevulinic acid synthase; CAR, constitutive androstane receptor; CYP, cytochrome P450; EE2, 17α-ethynyl-3,17β-estradiol; HSn, half-site n; LMH, leghorn male hepatoma; NR, nuclear receptor; PB, phenobarbital; PIA, propylisopropylacetamide; PXR, pregnane X receptor; RE, response element; Rif, rifampicin; RXR, 9-cis retinoic acid X receptor.
References
- 1.May, B. K., Dogra, S. C., Sadlon, T. J., Bhasker, C. R., Cox, T. C. & Bottomley, S. S. (1995) Prog. Nucleic Acid Res. Mol. Biol. 51, 1-51. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano, P. R. (1995) Cytochrome P-450: Structure, Mechanism, and Biochemistry (Plenum, New York).
- 3.Schoenhaut, D. S. & Curtis, P. J. (1989) Nucleic Acids Res. 17, 7013-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munakata, H., Yamagami, T., Nagai, T., Yamamoto, M. & Hayashi, N. (1993) J. Biochem. (Tokyo) 114, 103-111. [DOI] [PubMed] [Google Scholar]
- 5.Yomogida, K., Yamamoto, M., Yamagami, T., Fujita, H. & Hayashi, N. (1993) J. Biochem. (Tokyo) 113, 364-371. [DOI] [PubMed] [Google Scholar]
- 6.Riddle, R. D., Yamamoto, M. & Engel, J. D. (1989) Proc. Natl. Acad. Sci. USA 86, 792-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, D. F., Henderson, A. S. & Astrin, K. H. (1990) Genomics 7, 207-214. [DOI] [PubMed] [Google Scholar]
- 8.Li, B., Holloszy, J. O. & Semenkovich, C. F. (1999) J. Biol. Chem. 274, 17534-17540. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi, N., Watanabe, N. & Kikuchi, G. (1983) Biochem. Biophys. Res. Commun. 115, 700-706. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava, G., Borthwick, I. A., Maguire, D. J., Elferink, C. J., Bawden, M. J., Mercer, J. F. & May, B. K. (1988) J. Biol. Chem. 263, 5202-5209. [PubMed] [Google Scholar]
- 11.Cable, E. E., Miller, T. G. & Isom, H. C. (2000) Arch. Biochem. Biophys. 384, 280-295. [DOI] [PubMed] [Google Scholar]
- 12.Jover, R., Hoffmann, K. & Meyer, U. A. (1996) Biochem. Biophys. Res. Commun. 226, 152-157. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, D. J., Podvinec, M., Kaufmann, M. R. & Meyer, U. A. (2002) J. Biol. Chem. 277, 34717-34726. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, D. J., Zumsteg, A. & Meyer, U. A. (2003) J. Biol. Chem. 278, 39392-39401. [DOI] [PubMed] [Google Scholar]
- 15.Honkakoski, P. & Negishi, M. (2000) Biochem. J. 347, 321-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, S. A., Moore, L. B., Shenk, J. L., Wisely, G. B., Hamilton, G. A., McKee, D. D., Tomkinson, N. C., LeCluyse, E. L., Lambert, M. H., Willson, T. M., et al. (2000) Mol. Endocrinol. 14, 27-39. [DOI] [PubMed] [Google Scholar]
- 17.Sueyoshi, T. & Negishi, M. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 123-143. [DOI] [PubMed] [Google Scholar]
- 18.Wei, P., Zhang, J., Egan-Hafley, M., Liang, S. & Moore, D. D. (2000) Nature 407, 920-923. [DOI] [PubMed] [Google Scholar]
- 19.Waxman, D. J. (1999) Arch. Biochem. Biophys. 369, 11-23. [DOI] [PubMed] [Google Scholar]
- 20.Laudet, V. & Gronemeyer, H. (2002) The Nuclear Receptor Facts Book (Academic, London).
- 21.Stoltz, C. & Anderson, A. (1999) Biochem. Pharmacol. 57, 1073-1076. [DOI] [PubMed] [Google Scholar]
- 22.Honkakoski, P., Moore, R., Washburn, K. A. & Negishi, M. (1998) Mol. Pharmacol. 53, 597-601. [DOI] [PubMed] [Google Scholar]
- 23.Sueyoshi, T., Kawamoto, T., Zelko, I., Honkakoski, P. & Negishi, M. (1999) J. Biol. Chem. 274, 6043-6046. [DOI] [PubMed] [Google Scholar]
- 24.Podvinec, M., Kaufmann, M. R., Handschin, C. & Meyer, U. A. (2002) Mol. Endocrinol. 16, 1269-1279. [DOI] [PubMed] [Google Scholar]
- 25.Handschin, C. & Meyer, U. A. (2000) J. Biol. Chem. 275, 13362-13369. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin, B., Hodgson, E. & Liddle, C. (1999) Mol. Pharmacol. 56, 1329-1339. [DOI] [PubMed] [Google Scholar]
- 27.Handschin, C., Podvinec, M. & Meyer, U. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10769-10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann, J. M., McKee, D. D., Watson, M. A., Willson, T. M., Moore, J. T. & Kliewer, S. A. (1998) J. Clin. Invest. 102, 1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boshart, M., Kluppel, M., Schmidt, A., Schutz, G. & Luckow, B. (1992) Gene 110, 129-130. [DOI] [PubMed] [Google Scholar]
- 30.Handschin, C., Podvinec, M., Stockli, J., Hoffmann, K. & Meyer, U. A. (2001) Mol. Endocrinol. 15, 1571-1585. [DOI] [PubMed] [Google Scholar]
- 31.Scassa, M. E., Varone, C. L., Montero, L. & Canepa, E. T. (1998) Exp. Cell Res. 244, 460-469. [DOI] [PubMed] [Google Scholar]
- 32.Ourlin, J. C., Baader, M., Fraser, D., Halpert, J. R. & Meyer, U. A. (2000) Arch. Biochem. Biophys. 373, 375-384. [DOI] [PubMed] [Google Scholar]
- 33.Makinen, J., Frank, C., Jyrkkarinne, J., Gynther, J., Carlberg, C. & Honkakoski, P. (2002) Mol. Pharmacol. 62, 366-378. [DOI] [PubMed] [Google Scholar]
- 34.Ueda, A., Hamadeh, H. K., Webb, H. K., Yamamoto, Y., Sueyoshi, T., Afshari, C. A., Lehmann, J. M. & Negishi, M. (2002) Mol. Pharmacol. 61, 1-6. [DOI] [PubMed] [Google Scholar]
- 35.Maglich, J. M., Stoltz, C. M., Goodwin, B., Hawkins-Brown, D., Moore, J. T. & Kliewer, S. A. (2002) Mol. Pharmacol. 62, 638-646. [DOI] [PubMed] [Google Scholar]
- 36.Maglich, J. M., Parks, D. J., Moore, L. B., Collins, J. L., Goodwin, B., Billin, A. N., Stoltz, C. A., Kliewer, S. A., Lambert, M. H., Willson, T. M. & Moore, J. T. (2003) J. Biol. Chem. 278, 17277-17283. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, G. & Ades, I. Z. (1989) Int. J. Biochem. 21, 1025-1031. [DOI] [PubMed] [Google Scholar]
- 38.Hamilton, J. W., Bement, W. J., Sinclair, P. R., Sinclair, J. F. & Wetterhahn, K. E. (1988) Biochem. J. 255, 267-275. [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer, U. A., Schuurmans, M. M. & Lindberg, R. L. (1998) Semin. Liver Dis. 18, 43-52. [DOI] [PubMed] [Google Scholar]
- 40.Lindberg, R. L., Martini, R., Baumgartner, M., Erne, B., Borg, J., Zielasek, J., Ricker, K., Steck, A., Toyka, K. V. & Meyer, U. A. (1999) J. Clin. Invest. 103, 1127-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.