Abstract
Replication of the bacterial chromosome initiates at a single origin of replication that is called oriC. This occurs via the concerted action of numerous proteins, including DnaA, which acts as an initiator. The origin sequences vary across species, but all bacterial oriCs contain the information necessary to guide assembly of the DnaA protein complex at oriC, triggering the unwinding of DNA and the beginning of replication. The requisite information is encoded in the unique arrangement of specific sequences called DnaA boxes, which form a framework for DnaA binding and assembly. Other crucial sequences of bacterial origin include DNA unwinding element (DUE, which designates the site at which oriC melts under the influence of DnaA) and binding sites for additional proteins that positively or negatively regulate the initiation process. In this review, we summarize our current knowledge and understanding of the information encoded in bacterial origins of chromosomal replication, particularly in the context of replication initiation and its regulation. We show that oriC encoded instructions allow not only for initiation but also for precise regulation of replication initiation and coordination of chromosomal replication with the cell cycle (also in response to environmental signals). We focus on Escherichia coli, and then expand our discussion to include several other microorganisms in which additional regulatory proteins have been recently shown to be involved in coordinating replication initiation to other cellular processes (e.g., Bacillus, Caulobacter, Helicobacter, Mycobacterium, and Streptomyces). We discuss diversity of bacterial oriC regions with the main focus on roles of individual DNA recognition sequences at oriC in binding the initiator and regulatory proteins as well as the overall impact of these proteins on the formation of initiation complex.
Keywords: oriC, DnaA, initiation of chromosome replication, orisome, replication regulation, regulatory proteins, bacteria
Introduction
In contrast to the situation in Eukaryotes, chromosomal replication in bacteria begins at a single site on the chromosome: the origin of replication (oriC) (Leonard and Méchali, 2013). Using various in silico approaches (Mackiewicz et al., 2004; Gao et al., 2013), researchers have predicted the locations of the oriCs for more than 1500 bacterial chromosomes. However, in vivo replication activity has been confirmed for only a dozen such origins. Over the last 30 years, researchers have made considerable progress in understanding the mechanisms of replication initiation, particularly the organization and function of the oriC region in Escherichia coli (Figure 1), which is a model microorganism for the study of chromosomal replication (for reviews, see references, Fuller et al., 1984; Hwang and Kornberg, 1992; Messer, 2002; Kaguni, 2006, 2011; Leonard and Méchali, 2013). These studies have shown that replication is initiated through the cooperative binding of the initiator protein, DnaA, to multiple DnaA-recognition sites (boxes) within the oriC region. This triggers separation of the DNA strands at the AT-rich DNA unwinding element (DUE), providing an entry site for helicase and later on the other enzymes (e.g., primase and DNA Pol III) that are responsible for DNA synthesis.
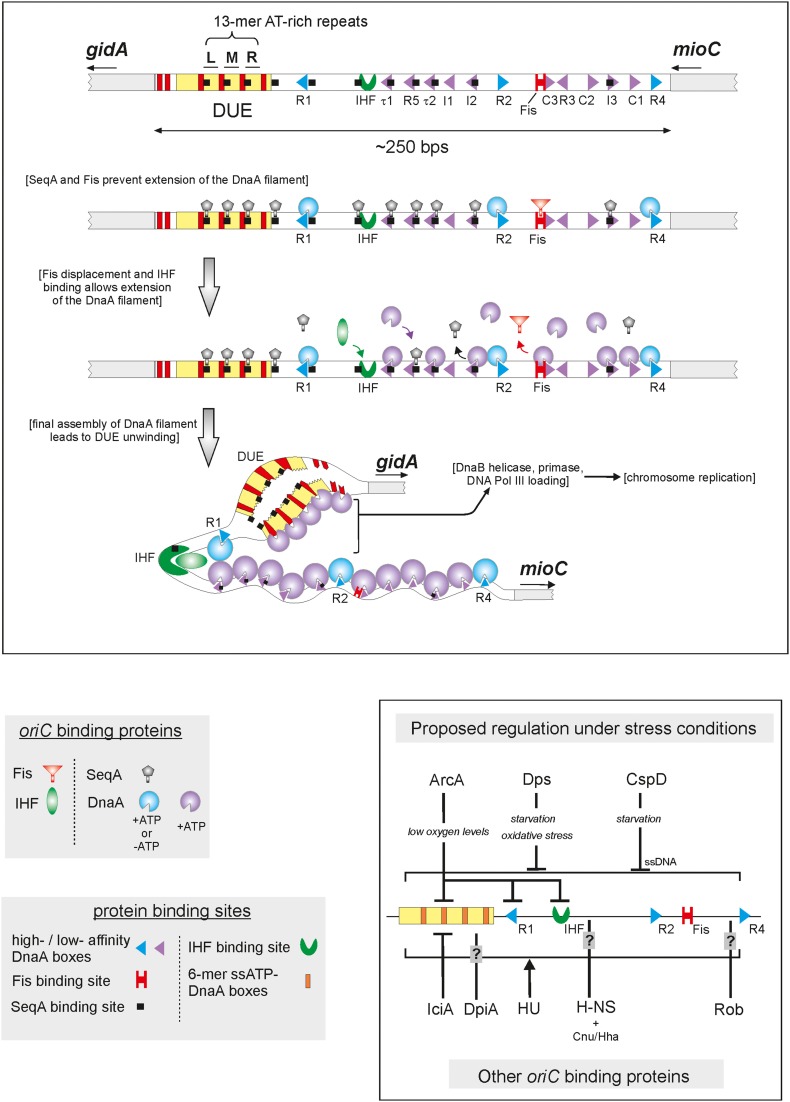
Figure 1.
A model of initiation replication and its regulation in E. coli by origin binding proteins (oriBPs). Large panel presents assumed sequence of events during the replication initiation and roles of particular oriBPs. The unwound DUE is accessible to the replication proteins complex (e.g., helicase DnaB, primase, and DNA Pol III). Small panel shows additional oriBPs divided in two subgroups, those involved in alternative scenarios that may occur under environmental stress conditions (upper part of the panel) and others, including those of unknown function (bottom part of the panel). Triangles' directions represent orientations of DnaA binding sites. Nucleotide bound status of DnaA is represented by blue and violet incomplete circles. Small arrows below gene names indicate gene orientations. In the small panel, different types of vertical lines represent type of action, activation (arrow), inhibition (bar-headed line) or unknown (question mark line). Horizontal lines indicate unspecific binding to oriC.
Comparative sequence analysis has demonstrated that the origin regions with confirmed in vivo functions differ in their sequences, organizations, and sizes, with only closely related organisms exhibiting fairly high overall similarities in their oriC sequences (Jakimowicz et al., 1998; Zawilak-Pawlik et al., 2005). In addition to a diverse repertoire of DnaA boxes, oriC regions also include various binding sites for accessory and regulatory proteins.
Chromosomal replication is mainly controlled at the initiation step (Mott and Berger, 2007; Zakrzewska-Czerwińska et al., 2007; Katayama et al., 2010; Leonard and Grimwade, 2010, 2011; Skarstad and Katayama, 2013). Therefore, the activities of the oriC region must be tightly regulated to guarantee that chromosomal DNA is entirely replicated only once per cell cycle. This is achieved by regulating the accessibility of oriC to DnaA, which occurs mainly via the binding of other proteins. Additionally, replication initiation is regulated by the modulation of DnaA protein activity.
The main goal of this review is to highlight the diversity of oriC regions in the context of replication regulation and the bacterial cell cycle. We focus on how oriC regions have adjusted to coordinate the regulation of chromosome replication and the progression of the cell cycle. We postulate that oriC regions encode species- and genus-specific instructions for the orderly binding of DnaA and other proteins responsible for forming the functional initiation complex (orisome) and/or regulating the assembly of this complex.
oriC chromosomal localisation and nucleotide sequence are not strictly conserved
The oriC regions are usually flanked by the dnaA gene and sometimes also the dnaN gene (Figure 2). These genes encode two pivotal proteins for initiating and continuing replication in the bacterial chromosome: DnaA, which is described above, and DnaN, which encodes a beta sliding clamp responsible for the processivity of DNA polymerase III. In linear chromosomes, such as those of Streptomyces coelicolor and (presumably) Borrelia burgdorferi, oriC is located in the center of the chromosome (Zakrzewska-Czerwińska and Schrempf, 1992; Mackiewicz et al., 2004). The region of gene synteny around oriC, which includes the highly preserved gene cluster of rnpA-rpmH-dnaA-dnaN-recF-gyrB-gyrA, is conserved in some (even distantly related) bacterial species. For a long time, the presence of these genes was assumed to mark the chromosomal localization of oriC (Ogasawara et al., 1991; Briggs et al., 2012). However, in many bacteria, including the model bacterium, E. coli, the oriC region is located in another gene context, indicating that this conserved gene cluster is not important for oriC function (Briggs et al., 2012). The localization of oriC is not random either as recent studies have indicated that the oriC-proximal gene context is conserved in certain group(s) of bacteria (e.g., genus or species). This is thought to enable a robust response to unfavorable conditions by allowing bacteria to increase the gene dosage in response to stress-induced initiation events (Moriya et al., 2009; Slager et al., 2014).
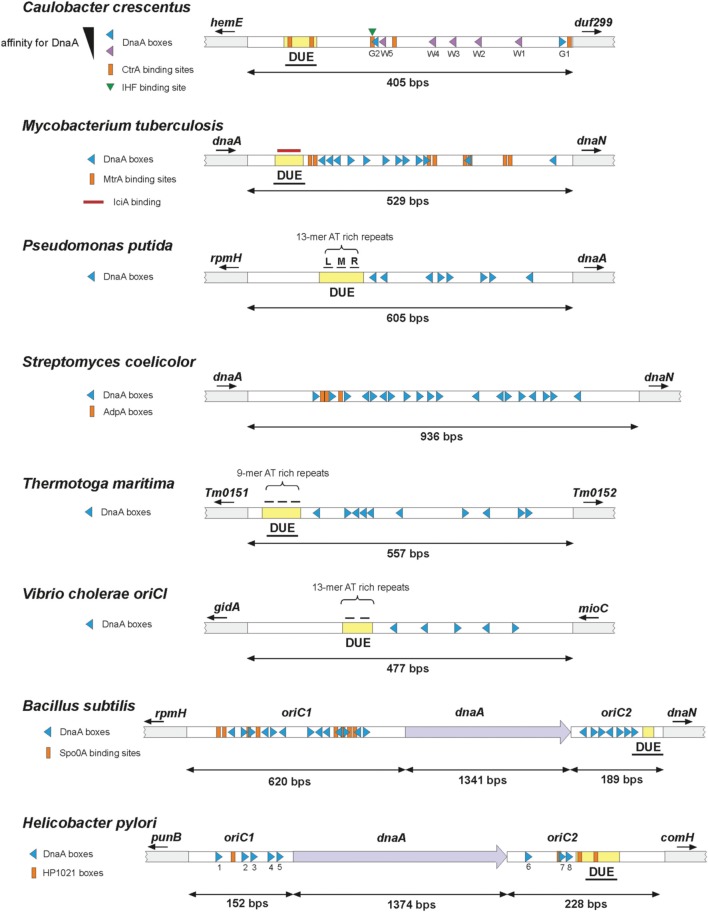
Figure 2.
The structures of selected bacterial origins. Continuous origin – upper part of the panel; bipartite origin – lower part of the panel. OriBP regulators' binding sites are presented if described in the literature. DUE – DNA unwinding element, underlined DUE indicates experimentally confirmed unwinding. Triangles' directions represent orientations of particular DnaA boxes. Small arrows below gene names indicate gene orientations.
Interestingly, a few obligate endosymbiotic bacteria, such as Wigglesworthia glossinidia, Blochmannia floridanus, and Candidatus Endolissoclinum faulkneri, lack the dnaA gene (Akman et al., 2002; Gil et al., 2003; Mackiewicz et al., 2004; Kwan and Schmidt, 2013). It is not known how these bacteria initiate chromosome replication and whether their oriCs resemble the bacterial origins characterized to date. Indeed, it has been postulated that the typical DnaA box cluster might not exist/be functional in these bacteria.
The nucleotide sequences of the oriC regions are highly diverse across unrelated species. Thus, they are not active in or interchangeable between unrelated bacteria (O'Neill and Bender, 1988), and sequence homology alignment of oriC is not used to identify unknown origins in bacterial genomes. In closely related bacteria, however, the sequence homology (and thus the organization) of the entire oriC region might be high enough to enable the oriC region from one species to autonomously replicate in another species (Harding et al., 1982; Takeda et al., 1982; Zyskind et al., 1983; Roggenkamp, 2007). It might also be possible to substitute the origin of one species with that of a related species, as was shown in the successful substitution of the E. coli oriC for the Vibrio cholerae origin of replication in chromosome I (Demarre and Chattoraj, 2010; Koch et al., 2010). It is important to note that when bacteria possess two or more chromosomes, only one undergoes replication initiated by the DnaA protein at the origin typical for bacterial chromosomes (e.g., the V. cholerae oriCI). The replication origin on the other chromosome is plasmid like, and it is activated by initiators that lack homology to DnaA (e.g., V. cholerae oriCII is initiated by a RctB protein) (Egan and Waldor, 2003; Duigou et al., 2006). Such diversification avoids the need for the chromosomes to compete for initiator proteins, and yields better control of their separate but coordinated replications (Duigou et al., 2006; Jha et al., 2012; Baek and Chattoraj, 2014).
The oriC region can be continuous or bipartite and consists of functional modules
A replication origin may be continuous or bipartite. Most of the bacteria studied to date contain a continuous oriC region that includes all of the functional modules within a single intergenic region (Figure 2). The divided origins, in contrast, are composed of two subregions, each of which contains a cluster of DnaA boxes, and one of which harbors the DUE region (Figure 2). They also differ in length: the continuous origins range from ~250 (E. coli) to ~950 bps (Streptomyces) (Zakrzewska-Czerwińska and Schrempf, 1992; Jakimowicz et al., 1998), while the bipartite origins are longer, up to ~2000 bps, because they contain a spacer gene (usually dnaA) between the oriC subregions (Figure 2; see below for mollicute origins). We do not yet know why some origins are split. Experimental data have shown that the spacer is important per se, although it may be altered to some extent without the loss of oriC activity. For example, a study showed that the spacer linking the oriC1 and oriC2 regions in Bacillus subtilis can be shortened (Moriya et al., 1992). Up until recently, the bipartite origin was assumed to be characteristic of a few Gram-positive bacteria (B. subtilis and Streptococcus pyogenes) and Mollicutes (Mycoplasma sp., Spiroplasma sp.) (Krause et al., 1997; Moriya et al., 1999; Suvorov and Ferretti, 2000; Lee et al., 2008; Briggs et al., 2012). However, bipartite origins were also recently identified in Gram-negative bacteria (e.g., Helicobacter pylori) (Donczew et al., 2012, 2014b), suggesting that bipartite origins might be more common than previously thought in diverse bacterial species. The origins of mollicutes were reported to have unusual properties, with interchangeability observed between species having divergent organizations of their oriCs (e.g., differences in the number, orientation and sequence of the DnaA boxes and/or the localization of the AT-rich regions) (Lartigue et al., 2003; Lee et al., 2008). The origins in mollicutes should be analyzed with caution, however, because they were identified solely by the minichromosome approach and no detailed characterization has yet been performed.
The study of the bacterial replication origin has progressed beyond the characterization of bacterial DnaA proteins, their assembly onto oriC, and the modes of initiation complex (orisome) formation in various bacterial species. However, it remains difficult to interpret the differences in bacterial oriCs in the context of their activities. As discussed above, the oriC regions characterized to date are quite diverse in terms of their chromosomal loci, genetic contexts, nucleotide sequences, lengths and continuities. However, they are all composed of three basic functional modules: a cluster (or multiple clusters) of DnaA boxes, the DUE region, and other sequences that are recognized by regulatory proteins. These modules constitute the central management system for orisome formation, but their numbers and relative localizations vary across organisms (Figure 2).
In each species, this organized information provides a perfect molecular scaffold for DnaA oligomerization, controls DNA opening, and regulates the initiation of chromosomal replication. These processes are detailed in the following sections.
The arrangements of the DnaA boxes and “DUE” are crucial for oriC activity
The role of particular oriC modules has been widely studied in E. coli, providing a comprehensive example of a replication initiation mechanism and its interplay with cellular regulatory circuits. The DnaA boxes constitute a framework for the binding of DnaA monomers, which interact with oriC to form a structure that is able to disturb the DNA double-helix. The unique layout of low- and high-affinity DnaA boxes in E. coli oriC regulates the formation of a specific DnaA oligomer, which (according to the current model) adopts the structure of a right-handed helical filament to directly stimulate DNA unwinding (Erzberger et al., 2006; Zorman et al., 2012). The particular DnaA molecules involved in the filament introduce a bend in the DNA helix, which is gradually wrapped around the filament's outer surface (Fujikawa et al., 2003; Erzberger et al., 2006). The final complex introduces a superhelical tension in the DNA helix; this is likely to be focused in the DUE region, and triggers the initial unwinding (Erzberger et al., 2006). In the subsequent steps, the ATP-DnaA oligomer is believed to bind the newly formed single-stranded DNA segments, stabilizing and stretching them to promote further extension of the initiation bubble (Duderstadt et al., 2011; Ozaki and Katayama, 2012). The formation of a similar helical DnaA oligomer was recently shown for B. subtilis in the presence of both single-stranded and double-stranded DNA (Scholefield et al., 2012). Scholefield et al. suggested that separate oligomers may be involved in the unwinding and subsequent stabilization of the single-stranded DUE in this case, but we do not yet understand the mechanism underlying oligomer formation in B. subtilis in terms of the bipartite structure of its origin or the subsequent steps of the initiation process. It also remains to be seen whether other bacterial initiation complexes involve the formation of similar higher-order structures. Nevertheless, it is plausible that the formation of a DnaA-containing oligomer is essential for the unwinding of DNA at the DUE region, and is thus a common feature of all bacterial origins.
The number of DnaA boxes in the studied origins ranges from five in Pseudomonas aeruginosa or V. cholerae to 19 in S. coelicolor (Figure 2). In most cases, these DnaA boxes are asymmetrical nine-nucleotide-long specific motifs (with the exception of the 12-nucleotide boxes found in Thermotoga maritima), whose exact sequences, numbers and layouts reflect the diversity of the various organisms. DnaA boxes from different bacteria do, however, share a common core sequence (Table 1). No analyzed origin contains boxes, which deviate by more than two mismatches from the so-called “perfect” box sequence (i.e., that which binds with the highest affinity) of E. coli (TTATCCACA), with the exception of that of T. maritima. In closely related organisms, the high-affinity box sequence is conserved, as seen in E. coli, V. cholerae, Pseudomonas putida and P. aeruginosa, which all belong to a branch of the γ-proteobacteria (Yee and Smith, 1990; Weigel et al., 1997; Egan and Waldor, 2003) (Table 1). Interestingly, even B. subtilis, which is evolutionarily distant from E. coli, shares the same conserved “perfect” box sequence (Fukuoka et al., 1990). As noted above, the “perfect” DnaA box in other species almost always differs from this E. coli DnaA box by only one or two nucleotides. In Actinomycetes (M. tuberculosis and S. coelicolor), which are considered high-GC organisms, the “perfect” DnaA box contains G or C at the third position: TT(G/C)TCCACA (Jakimowicz et al., 2000; Zawilak et al., 2004; Tsodikov and Biswas, 2011). Similarly, Caulobacter crescentus G-boxes (see Figure 2 and Table 1) differ from the E. coli consensus sequence by a single nucleotide (in this case in the second position; TGATCCACA) (Shaheen et al., 2009). Among the studied origins, the nine-nucleotide DnaA boxes most distant from that of E. coli are found in H. pylori, which contain two mismatches (at the second and fifth positions; TCATTCACA) with respect to the “perfect” E. coli sequence (Zawilak et al., 2001; Donczew et al., 2014b). An interesting exception from this general rule is the T. maritima origin, where ten 12-nucleotide DnaA boxes were identified (consensus DnaA box: AAACCTACCACC) (Ozaki et al., 2006). As T. maritima is one of the most ancient bacteria, it has been proposed that its DnaA boxes may resemble a sequence recognized by the initiator protein in a last common ancestor of the unicellular organisms (Ozaki et al., 2006). Indeed, the T. maritima DnaA box sequence shares some similarity to the ORC-binding sites in Saccharomyces cerevisiae (TAAACATAAAA) and the Orc1/Cdc6-binding sequences in Archaea (e.g., in Methanothermobacter thermoautotrophicus – TTACAGTTGAAA) (Ozaki et al., 2006). Thus, it is probable that the E. coli-like nine-mer DnaA box sequence has evolved from this original 12-nucleotide sequence, becoming shortened to nine nucleotides at some point. The last six nucleotides of the T. maritima consensus sequence (ACCACC) are similar to the corresponding part of the nine-nucleotide DnaA boxes. The importance of this six-nucleotide motif as an integral part of a bacterial DnaA box is further supported by the recent study of the C. crescentus oriC, where five six-nucleotide boxes (termed W-boxes) were identified in addition to the two known nine-nucleotide high-affinity G-boxes (Taylor et al., 2011). The W-box consensus sequence is TCCCCA, which deviates from the last six nucleotides of the E. coli-like box at the fourth position, and shows a very weak but detectable binding by the DnaA protein. Researchers have also identified atypical six-nucleotide-long DnaA boxes in E. coli; located directly in the DUE region and bound only in a single-stranded form, this consensus sequence (AGATCT) represents an alternative type of DnaA-recognized sequence (albeit so far exclusive for E. coli) (Speck and Messer, 2001).
Table 1.
Sequences of high-affinity DnaA boxes from various bacteria.
| Organism | High-affinity DnaA box sequence |
|---|---|
| Escherichia coli | TTATCCACA |
| Vibrio cholerae | TTATCCACA |
| Pseudomonas putida | TTATCCACA |
| Bacillus subtilis | TTATCCACA |
| Caulobacter crescentus | TGATCCACA |
| Helicobacter pylori | TCATTCACA |
| Mycobacterium tuberculosis | TTGTCCACA |
| Streptomyces coelicolor | TTGTCCACA |
| Thermotoga maritima | AAACCTACCACC |
The origins of closely related bacteria may be interchangeable to some degree between species, as shown in vivo (Koch et al., 2010) and in vitro (Jiang et al., 2006) for members of the γ-proteobacteria branch. Among evolutionarily distant bacteria, the situation is more complicated. Considering the similarity of DnaA boxes from different organisms, it is not surprising that the DnaA protein is often able to recognize DnaA boxes and/or whole oriC regions in heterologous systems in vitro, albeit often with a lower affinity and/or specificity. For example, S. coelicolor oriC (ScoriC) is bound efficiently by the M. tuberculosis and E. coli DnaA proteins, but neither protein was able to bend the ScoriC structure in the manner of the native DnaA protein (Jakimowicz et al., 2000; Zawilak-Pawlik et al., 2005). Furthermore, the origins of S. coelicolor, M. tuberculosis and H. pylori were not found to be active in E. coli cells, even though they are efficiently bound by the E. coli DnaA in vitro. Interestingly, the H. pylori DnaA interacts very poorly with the E. coli oriC, indicating that even in vitro DnaA/oriC systems from different species may be interchangeable in one setting (i.e., E. coli DnaA/H. pylori oriC) but not the other (i.e., H. pylori DnaA/E. coli oriC) (Zawilak-Pawlik et al., 2005). Furthermore, the DnaA proteins of E. coli and B. subtilis exhibit high affinities toward the same DnaA box sequence and were found to interact in heterologous systems in vitro, creating similar oligomeric structures as was visualized by EM (Krause et al., 1997). However, neither was found to trigger open-complex formation on the heterologous origin. This provides further evidence that even when there are apparent similarities, the DnaA-oriC systems of individual species are not easily interchangeable.
Such observations may reflect that the mode through which DnaA interacts with particular boxes can differ among bacterial organisms. For example, the E. coli DnaA protein interacts efficiently with single, double or multiple DnaA boxes (Weigel et al., 1997; Speck and Messer, 2001). In contrast, the DnaA proteins of some other organisms have been found to strongly prefer two or more boxes over a single box. For example, the M. tuberculosis DnaA does not interact with a single box, while the closely related S. coelicolor DnaA interacts only weakly with a single box (Zawilak-Pawlik et al., 2005). Similarly, the H. pylori DnaA protein has a higher affinity for two boxes vs. a single box (Zawilak et al., 2003). Such observations suggest that the joint action of multiple DnaA monomers may be required for efficient binding in many cases. This is especially true for longer origins, in which a greater number of boxes appears to correlate with an increased importance of cooperative interactions among multiple DnaA monomers, such as suggested for the origins of the Actinomycetes – M. tuberculosis and S. coelicolor (Zawilak-Pawlik et al., 2005). The affinity of individual DnaA boxes can also vary within and across bacterial origins, with low-, medium-, and high-affinity DnaA boxes present in the origins. Interestingly, the number of low-affinity boxes often exceeds the number of high-affinity sites, such as seen in the oriC regions of E. coli and C. crescentus (Figures 1, 2; Rozgaja et al., 2011). Studies in E. coli showed that the number and distribution of low- and high-affinity sites is crucial for the activity of the oriC region, including its ability to control the frequency of initiation (Grimwade et al., 2007; Leonard and Grimwade, 2011). Low-affinity DnaA boxes provide a scaffold for DnaA oligomerization, whereas the high-affinity boxes (R1, R2, and R4; Figure 2) are believed to provide nucleation sites for the DnaA molecules. Low affinity-sites in E. coli oriC are organized into two oppositely oriented arrays separated by box R2 and flanked by boxes R1 and R4, which act as nucleation sites for DnaA oligomers (Rozgaja et al., 2011). The two DnaA oligomers were proposed to be extended by sequential interactions of DnaA monomers with arrayed low-affinity sites to finally form a contiguous DnaA filament (Rozgaja et al., 2011). It was suggested that such mode of a DnaA oligomer formation is directly implicated in origin unwinding since the two arrays of low-affinity sites are not helically phased and connection of the two halves of the oligomer would require specific twisting of the DNA strand, which would create a torsional stress (Rozgaja et al., 2011). It is worth noting that at least some of the origins of other bacteria also exhibit particular orientations of clusters of boxes (e.g., all H. pylori boxes share the same orientation) (Figure 2), which might indicate sequential binding of DnaA molecules and organized formation of a DnaA oligomer, as in E. coli.
The E. coli high-affinity boxes (R1, R2, and R4; Figure 1) appear to be occupied for the majority of the cell cycle, regardless of the nucleotide state of DnaA. The low-affinity boxes, on the other hand, are preferentially bound by ATP-DnaA (Miller et al., 2009; Rozgaja et al., 2011). The nucleotide state of DnaA is subjected to complex regulation system by RIDA inactivation, DARS-reactivation and rejuvenation as well as de novo protein synthesis (for details see Katayama et al., 2010; Leonard and Grimwade, 2011; Kasho et al., 2014) The binding of DnaA to low-affinity sites is additionally facilitated by the DiaA protein, which has been shown to stimulate the assembly of specific ATP-DnaA-oriC complexes (Keyamura et al., 2007), as well as specific oriC-binding proteins like SeqA, IHF and Fis (see “oriC activity is regulated by specific origin-binding proteins”).
The DUE region is a typically AT-rich stretch of nucleotides (comprehensively reviewed by Rajewska et al., 2012) that often includes characteristic repeated AT-rich sequences (e.g., that of E. coli comprises three 13-mer repeats) separated by short, non-AT-rich insertions. DUE regions are thermodynamically unstable compared to their neighboring sequences, rendering them susceptible to superhelical stress arising from the formation of the DnaA oligomer (Erzberger et al., 2006). The initially unwound region ranges from 20 to 60 bps in size, depending on the organism, which seems to provide sufficient space to accommodate a replicative helicase, DnaB (Figure 1) (Sutton et al., 1998; Abe et al., 2007; Mott et al., 2008; Keyamura et al., 2009). After the initial unwinding in E. coli, DnaA binds to single-stranded six-mer ATP-DnaA boxes (6-mer ssATP-DnaA boxes; Figure 1) located in the DUE (showing a strong preference for one of the strands), thereby stabilizing the initiation bubble prior to helicase loading (Speck and Messer, 2001). The bacterial DUE regions are always located upstream or downstream one or more DnaA box cluster(s), never in the midst of a cluster. It is important to note that the distance between the DUE and its proximal DnaA-box cluster is critical, as even slight changes were found to inhibit oriC unwinding (Hsu et al., 1994).
In sum, the existing evidence clearly shows that the cognate DnaA protein and DnaA boxes coevolved to achieve an optimal level of interaction. The orientation and spacing of DnaA boxes are both important for proper activity of the origin. For example, a change in the length of one helical turn between selected boxes does not affect initiation, but changes corresponding to part of a helical turn are highly detrimental (Woelker and Messer, 1993). At the level of an entire oriC region, the arrangement of individual boxes that differ in their affinities generates a specific order and assembly rate for the DnaA oligomer, which unwinds DNA in a precisely selected region called the DUE. From there, initiation events are further controlled by regulatory proteins that bind oriC at specific sites, as discussed below.
oriC activity is regulated by specific origin-binding proteins
Transmission of genetic material to nascent cells requires precise regulation of chromosome replication and its coordination with the cell cycle. Since chromosomal replication is mainly regulated at the initiation stage, the principal activity of the oriC region (i.e., unwinding DNA) is tightly controlled. The relevant protein regulators are primarily involved in controlling the initial assembly of the DnaA oligomer along the origin of replication. The formation of an active orisome depends on the presence of proteins that: (i) regulate DnaA protein activity (e.g., Hda, which regulates the nucleotide-bound state of DnaA); (ii) facilitate the interactions between DnaA monomers (e.g., DiaA, which facilitates the assembly of the DnaA oligomer); or (iii) bind oriC and modulate the interaction of the DnaA protein with the origin of replication (Katayama et al., 1998; Kato and Katayama, 2001; Keyamura et al., 2007). In this section, we focus on various sequences that are targeted by the origin binding proteins (oriBPs) (other than DnaA) (Table 2) regulating the cell-cycle timing of replication from the oriC region (called “oriBP regulators”).
Table 2.
OriBP (origin binding protein) regulators.
| OriBP | Binding sequence and features | Reference |
|---|---|---|
| ESCHERICHIA COLI | ||
| SeqA | 5′-GATC-3′ | Slater et al., 1995; Brendler et al., 1995; Taghbalout et al., 2000; Nievera et al., 2006 |
| (sequestration factor A) | Binds to oriC and inhibits DnaA binding at low affinity sites. | |
| Fis | 5′-GAACAACAGTTGTTC-3′ | Gille et al., 1991; Filutowicz et al., 1992; Ryan et al., 2004 |
| (factor for inversion stimulation) | Binds to single site in oriC and inhibits DnaA binding. | |
| IHF | 5′-GATCAACAACCTG-3′ Binds to single site in oriC, stimulates DnaA binding at low affinity sites. | Filutowicz and Roll, 1990; Polaczek, 1990; Grimwade et al., 2000 |
| (integration host factor) | ||
| HU | Binds non-specifically to oriC and influences: DnaA oligomer stability at oriC, IHF binding, and stability of ds DNA helix. | Bonnefoy and Rouvière-Yaniv, 1992; Hwang and Kornberg, 1992; Ryan et al., 2002 |
| (histone like U-factor) | Interacts with N-terminus of DnaA. | |
| Dps | Binds non-specifically to oriC and interacts with N-terminus of DnaA. | Chodavarapu et al., 2008b |
| (DNA-binding protein from starved cells) | ||
| ArcA | Binds to 13-mer AT-rich repeats, and to DnaA, IHF, IciA binding sites in oriC. | Lee et al., 2001 |
| (aerobic respiration control) | Influences DnaA interaction with AT-rich region. | |
| IciA | Binds to 13-mer AT-rich repeats in oriC and inhibits unwinding. | Hwang and Kornberg, 1990; Thöny et al., 1991; Hwang et al., 1992 |
| (inhibitor of chromosomal initiation) | ||
| CspD | No apparent target sequence, binds exclusively to ssDNA. Inhibits replication initiation. | Yamanaka et al., 2001 |
| (stationary phase-induced, stress response protein in the CspA family) | ||
| Rob | 5′-ATCGCACGATCTGTATACTT-3′ | Skarstad et al., 1993; Martin et al., 1999 |
| (right oriC-binding) | Binds to single site in oriC. No clear function in initiation regulation. | |
| H-NS | 5′-ATGATCGGTGATCCTG-3′ | Kim et al., 2005; Yun et al., 2012a,b |
| (heat-stable nucleoid structural protein) | Binds to single site in oriC. Probably requires both Cnu and/or Hha proteins to bind DNA. | |
| DpiA | Binds to 13-mer AT-rich repeats in oriC. No function in initiation regulation. | Miller et al., 2003 |
| (destabilizer of plasmid inheritance) | ||
| BACILLUS SUBTILIS | ||
| Spo0A | 5′-TG[TA]CGAA-3′ | Strauch et al., 1990; Castilla-Llorente et al., 2006 |
| (sporulation specific sigma factor) | Binds to oriC and inhibits DnaA binding. | |
| CAULOBACTER CRESCENTUS | ||
| CtrA | 5′-TTAA[Nx7]TTAA-3′ | Siam and Marczynski, 2000; Taylor et al., 2011 |
| (cell transcriptional regulator) | Binds to Cori and inhibits DnaA binding. | |
| IHF | 5′-TAACGCTCTGTT-3′ | Siam et al., 2003 |
| (integration host factor) | Binds to single site in Cori, displaces CtrA, which facilitates bending of DNA and promotes chromosome replication. | |
| HELICOBACTER PYLORI | ||
| HP1021 | 5′-TGTT[TA]C[TA]-3 | Donczew et al., 2014a |
| atypical response regulator | Binds to oriC1 and oriC2 and blocks DNA unwinding at the DUE (within the oriC2). | |
| STREPTOMYCES COELICOLOR | ||
| AdpA | 5′-TGGCSNGWWY-3′ | Wolański et al., 2012 |
| (A-factor dependent protein) | Binds to oriC and inhibits DnaA binding. | |
| MYCOBACTERIUM TUBERCULOSIS | ||
| IciA (Rv1985c) | Binds to AT-rich region within oriC and blocks DNA unwinding. | Kumar et al., 2009 |
| (inhibitor of chromosomal initiation) | ||
| MtrA | 5′-GTCACAGCG-3′ Mechanism not known. | Rajagopalan et al., 2010 |
| (Mycobacterium tuberculosis response regulator A) | ||
The oriBP regulators can be divided into three classes depending on their target sequences: (i) those that interact with DnaA boxes or in their close vicinity; (ii) those that interact with AT-rich sequences within the DUE; and (iii) those that interact with other sequences within oriC (Figure 1). The oriBPs can also be classified by their sequence specificity and/or function: they may specifically or non-specifically interact with oriC to positively or negatively influence the unwinding of the origin. They confer their direct effects by binding to DnaA (or other oriBPs) binding sites, and exert their indirect effects by changing the DNA structure of the origin to modulate the binding of additional oriBPs.
The proteins that regulate replication initiation have been best described for E. coli, in which ~11 oriC binding proteins have been identified (Table 2). However, we do not yet fully understand the roles played by all of these oriBPs in regulating replication. Here, we use the E. coli model to discuss the roles of particular oriBP regulators in the sequential events that are believed to occur following the initiation of replication. When possible, we also describe the roles of counterpart proteins in other bacteria and discuss alternative initiation regulators that are not found in E. coli.
In E. coli, shortly after chromosomal replication the SeqA protein binds to several sites within the oriC region to strictly prevent the initiation of new rounds of replication via a mechanism called “sequestration.” SeqA specifically binds the short palindromic sequence, GATC, which is overrepresented within oriC compared to the rest of the bacterial chromosome. Newly replicated origins are hemimethylated for about 1/3 of the E. coli cell cycle, and SeqA preferentially binds hemimethylated GATC sequences over the fully methylated sequences. Thus, SeqA sequesters the oriC region until the GATC sites are fully methylated by the Dam methylase (Campbell and Kleckner, 1990; Lu et al., 1994; Brendler et al., 1995; Slater et al., 1995). SeqA predominantly inhibits replication initiation by blocking DnaA from binding to the R5, I2, I3, τ1, and τ2 sites, which overlap with the GATC sequences (Taghbalout et al., 2000; Nievera et al., 2006). This prevents the DnaA filament from being elongated from the high-affinity DnaA boxes, R1, R2, and R4, although it does not alter their occupation by DnaA (Samitt et al., 1989; Cassler et al., 1995; Nievera et al., 2006). This sequestration mechanism appears to be exclusive to a few DamMT-specifying proteobacteria, as homologs of the seqA gene have been identified only in this subset of Gram-negative bacteria (Brézellec et al., 2006).
Another negative regulator of initiation in E. coli, the Fis protein, associates with oriC throughout most of the cell cycle; similar to SeqA, Fis negatively influences replication initiation by regulating the occupation of DnaA on low-affinity sites (Cassler et al., 1995; Ryan et al., 2004). Fis specifically binds to a single site that is located between R2 and R3, and overlaps with the C3 DnaA binding site (Figure 1) (Gille et al., 1991; Filutowicz et al., 1992). Fis binding is thought to competitively inhibit the interaction of DnaA with this region (Ryan et al., 2004), and Fis exhibits a DNA-bending activity that plays a yet-unknown role (Finkel and Johnson, 1992; Ryan et al., 2004).
In addition to competing with DnaA for binding to oriC, both Fis and SeqA also negatively regulate the interaction of another oriBP, IHF, with the origin. In contrast to the former two proteins, IHF positively regulates replication initiation (Hwang and Kornberg, 1992; Grimwade et al., 2000; Ryan et al., 2002). As the time of initiation draws near, increasing levels of DnaA trigger the displacement of Fis and the full methylation of DNA weakens SeqA binding, ending the repressive activities of these proteins (Slater et al., 1995; Ryan et al., 2004). The release of SeqA reveals the IHF binding site; displacement of Fis promotes IHF binding; and IHF binding leads to bending of the DNA (Polaczek, 1990; Cassler et al., 1995; Rice et al., 1996; Weisberg et al., 1996; Swinger and Rice, 2004). IHF then stimulates the binding of DnaA-ATP to low-affinity sites (thus redistributing the DnaA protein) and induces the unwinding of oriC (Grimwade et al., 2000). Notably, the transcription of the dnaA gene is also subject to regulation by the SeqA protein (Campbell and Kleckner, 1990; Theisen et al., 1993; Bogan and Helmstetter, 1997). Thus, the increased DnaA concentrations that trigger the displacement of Fis displacement presumably reflect the earlier release of the dnaA promoter from inhibition by SeqA. In C. crescentus, the protein that corresponds to IHF also binds to a single site within the oriC of this species (Cori). Here, the recognition sequence for IHF overlaps the C-binding site for CtrA, which negatively regulates chromosomal replication in C. crescentus (for more on CtrA, see below). In this system, IHF binding leads to the displacement of CtrA from Cori, allowing the DNA to bend and promoting replication (Siam et al., 2003).
In E. coli, HU is a second positive regulator of initiation. Although this histone-like protein was believed to non-specifically bind DNA, some evidence has suggested that it may interact with oriC in a specific manner (Bonnefoy and Rouvière-Yaniv, 1992; Ryan et al., 2002). HU enhances the DnaA-dependent unwinding of oriC. This presumably occurs through its ability to bend and destabilize DNA (Hwang and Kornberg, 1992; Ryan et al., 2002). However, HU was further shown to interact with the N-terminal part of DnaA to stabilize the DnaA oligomer assembled at oriC (Chodavarapu et al., 2008a), suggesting that oriC unwinding may also be stimulated through this additional mechanism. Interestingly, HU was also shown to reduce the binding of DnaA at the DnaA-I3 site (Ryan et al., 2002), and modulate the binding of IHF to oriC in a manner dependent on the relative concentrations of IHF and HU (Bonnefoy and Rouvière-Yaniv, 1992).
The oriBPs, Dps, and ArcA, negatively regulate replication initiation in response to oxidative stress and oxygen depletion, respectively (Almirón et al., 1992; Lee et al., 2001; Chodavarapu et al., 2008b). Dps non-specifically binds DNA and interacts with the N-terminus of the DnaA protein to inhibit DNA unwinding (Almirón et al., 1992; Chodavarapu et al., 2008b). It has been suggested that Dps may act as a checkpoint during oxidative stress, delaying initiation until the oxidative DNA damage has been repaired (Chodavarapu et al., 2008b). Under anaerobic conditions, in contrast, ArcA is phosphorylated by a cognate kinase of the two-component system. ArcA-P transcriptionally regulates the genes required to maintain anaerobic growth (Lee et al., 2001), and it is also thought to regulate the activity of oriC. In vitro, ArcA-P binds a region that contains AT-rich 13-mers and the binding sites for IHF and DnaA (R1 box). It prevents the formation of the open complex without displacing IHF or DnaA from the DNA (Lee et al., 2001), suggesting that ArcA-P may disrupt the interaction between the DnaA protein and the AT-rich region.
Interestingly, ArcA-P is capable of displacing another oriBP, IciA, which specifically binds to the 13-mer AT-rich region and inhibits the unwinding of oriC (Hwang and Kornberg, 1990, 1992; Thöny et al., 1991). Interestingly, IciA is also capable of transcriptionally regulating genes known to be involved in DNA replication (e.g., dnaA) and amino acid metabolism (Lee et al., 1996; Nandineni and Gowrishankar, 2004; Bouvier et al., 2008). A study of the IciA counterpart in Mycobacterium tuberculosis showed that this protein also binds to the AT-rich region of the oriC and in vitro blocks DnaA-dependent helix opening, and may play a role in maintaining mycobacterial latency (during which DNA replication is arrested) (Kumar et al., 2009).
Regarding additional oriBPs in E. coli, the CspD protein reportedly inhibits both the initiation and elongation of chromosomal replication in vitro (Yamanaka et al., 2001). Finally, additional proteins capable of specifically binding oriC have been identified and described (e.g., Rob, H-NS, and DpiA), but their specific roles and contributions to the replication initiation process are not yet known (Skarstad et al., 1993; Martin et al., 1999; Miller et al., 2003; Kim et al., 2005; Yun et al., 2012a,b).
In bacteria that undergo a complex life cycle (e.g., Bacillus, Caulobacter, and Streptomyces), the regulation of replication initiation must also be adjusted to the developmental stage to ensure that each nascent cell receives a single copy of the chromosome (Wolański et al., 2014). Recently, master transcription factors known to regulate the expression levels of hundreds of genes involved in cell cycle progression and cell differentiation were demonstrated to be also involved in controlling frequency of chromosomal replication initiation events. Examples of these are Spo0A, CtrA, and AdpA proteins, which temporally and spatially coordinate chromosome replication with developmental program in B. subtilis, C. crescentus, and S. coelicolor, respectively (Laub et al., 2000, 2002; Molle et al., 2003; Fujita and Losick, 2005; Fujita et al., 2005; Ohnishi et al., 2005; Wolański et al., 2011). They bind specifically to relevant recognition sequences within the origin of replication and inhibit the binding of DnaA, thereby disrupting assembly of the DnaA oligomer and inhibiting replication initiation (Siam and Marczynski, 2000; Castilla-Llorente et al., 2006; Taylor et al., 2011; Wolański et al., 2012; Boonstra et al., 2013; reviewed in Wolański et al., 2014). In all three cases, the binding sites for these regulators overlap with one or more DnaA binding sites, setting up a competition between the regulator and initiator for binding to oriC (Figure 2). Interestingly, the activities of Spo0A and CtrA are regulated by phosphorylation, which enhances their binding to DNA. Increasing the levels of these active proteins inhibits chromosomal replication and stimulates the expression levels of various genes responsible for differentiation.
In the pathogenic Mycobacterium, M. tuberculosis, in addition to IciA, the MtrA protein has been shown to bind the oriC region and regulate chromosomal replication (Rajagopalan et al., 2010). MtrA binds specifically to four MtrA boxes that are dispersed throughout the oriC, between the DnaA boxes (Figure 2). Each MtrA box consists of two direct repeats of GTCACAgcg-like sequences. Mutations in the MtrA binding sequences were found to compromise the replication of the minichromosome (an oriC containing plasmid), whereas increased levels of MtrA appear to be associated with deficient autonomous replication of the minichromosome (Rajagopalan et al., 2010). Thus, MtrA may play both positive and negative roles in the initiation of replication. The exact action mechanism of MtrA at oriC is not yet known, but it has been suggested that this protein may facilitate or hinder the ability of DnaA to oligomerize at oriC, rather than interfering with the direct binding of the initiator protein. MtrA has been identified as a response regulator component of the signal transduction system, MtrAB, which suggests that its role in replication initiation might depend on its phosphorylation status (Via et al., 1996; Fol et al., 2006; Rajagopalan et al., 2010). Interestingly, it has been recently shown that in other pathogenic bacterium, H. pylori, the orisome assembly is controlled by HP1021 protein – the orphan response regulator, which was previously shown to affect expression of nearly 80 genes (Pflock et al., 2007). HP1021 competes with DnaA for the binding sites at oriC and inhibits DNA unwinding at the DUE site (Donczew et al., 2014a). It suggests that HP1021 controls initiation of H. pylori chromosome replication in response to yet unknown stimuli. It is very likely that in numerous bacteria chromosome replication is regulated by signal transduction systems in response to cellular or external stimuli affecting bacterial growth.
Conclusion and outlook
In sum, the bacterial origins differ across organisms in the organization of their DNA modules, but all origins encode comprehensive instructions for the assembly and disassembly of the orisome-forming proteins, enabling the timely regulation of this first and crucial step in chromosomal replication. The instructions direct the sequential binding of DnaA molecules to the available array of high- and low-affinity DnaA boxes to form a nucleoprotein complex that triggers the unwinding of DNA within the AT-rich region of the oriC. The oriC-encoded instructions also guide a number of other oriC-binding proteins that directly or indirectly respond to environmental signals and induce or repress formation of the DnaA-oriC complex, thereby modulating replication initiation. Tight regulation of the initiation process is achieved in all bacteria, albeit via different strategies involving various oriC binding proteins, many of which play additional roles in cell-cycle regulation. In pathogens, the functions of some initiation regulators may also depend on interactions with the host cell cycle; however, such interactions have not yet been thoroughly elucidated. It is important to remember that origins do not contain universal instructions. Only origins from very closely related organisms exhibit similar organizations, and the repertoire of regulatory proteins is unique for each species or group of related organisms. That enables a bacterium to perfectly adjust its replication to the cell cycle and coordinate its growth with external stimuli. As reviewed herein, we know a great deal about origins and their structures. To continue progressing in this field, we need detailed analyses of orisome formation, as has already been done for E. coli and (to a lesser extent) a limited number of other organisms (e.g., B. subtilis or M. tuberculosis). Future studies should examine how differences in origin structure are translated to the species-specific characteristics of DnaA oligomerization and its control by regulatory proteins. In addition, many important aspects of the replication initiation process remain to be discovered, particularly in pathogens, including the answers to questions, such as:
How is replication initiation coordinated with the cell cycles of different pathogens upon host infection?
How do host signals modulate or influence replication initiation in pathogenic bacteria?
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dagmara Jakimowicz for providing helpful comments on the manuscript. This work was supported by the National Science Centre, Poland (Maestro, Grant 2012/04/A/NZ1/00057) and by Wroclaw Research Centre EIT+ under the project “Biotechnologies and advanced medical technologies” (BioMed; POIG.01.01.02-02-003/08), which is financed through the European Regional Development Fund (Operational Programme Innovative Economy, 1.1.2). The cost of publication was financed by the Wroclaw Centre of Biotechnology, programme the Leading National Research Centre (KNOW) for years 2014–2018.
References
- Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. (2007). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. 10.1074/jbc.M701841200 [DOI] [PubMed] [Google Scholar]
- Akman L., Yamashita A., Watanabe H., Oshima K., Shiba T., Hattori M., et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. 10.1038/ng986 [DOI] [PubMed] [Google Scholar]
- Almirón M., Link A. J., Furlong D., Kolter R. (1992). A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6, 2646–2654. 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- Baek J. H., Chattoraj D. K. (2014). Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 10:e1004184. 10.1371/journal.pgen.1004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan J. A., Helmstetter C. E. (1997). DNA sequestration and transcription in the oriC region of Escherichia coli. Mol. Microbiol. 26, 889–896. 10.1046/j.1365-2958.1997.6221989.x [DOI] [PubMed] [Google Scholar]
- Bonnefoy E., Rouvière-Yaniv J. (1992). HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 11, 4489–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra M., de Jong I. G., Scholefield G., Murray H., Kuipers O. P., Veening J.-W. (2013). Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol. Microbiol. 87, 925–938. 10.1111/mmi.12141 [DOI] [PubMed] [Google Scholar]
- Bouvier J., Stragier P., Morales V., Rémy E., Gutierrez C. (2008). Lysine represses transcription of the Escherichia coli dapB gene by preventing its activation by the ArgP activator. J. Bacteriol. 190, 5224–5229. 10.1128/JB.01782-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T., Abeles A., Austin S. (1995). A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 14, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brézellec P., Hoebeke M., Hiet M.-S., Pasek S., Ferat J.-L. (2006). DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics 22, 1935–1941. 10.1093/bioinformatics/btl336 [DOI] [PubMed] [Google Scholar]
- Briggs G. S., Smits W. K., Soultanas P. (2012). Chromosomal replication initiation machinery of low-G+C-content Firmicutes. J. Bacteriol. 194, 5162–5170. 10.1128/JB.00865-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. (1990). E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979. 10.1016/0092-8674(90)90271-F [DOI] [PubMed] [Google Scholar]
- Cassler M. R., Grimwade J. E., Leonard A. C. (1995). Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 14, 5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla-Llorente V., Munoz-Espin D., Villar L., Salas M., Meijer W. J. J. (2006). Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J. 25, 3890–3899. 10.1038/sj.emboj.7601266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu S., Felczak M. M., Yaniv J. R., Kaguni J. M. (2008a). Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 67, 781–792. 10.1111/j.1365-2958.2007.06094.x [DOI] [PubMed] [Google Scholar]
- Chodavarapu S., Gomez R., Vicente M., Kaguni J. M. (2008b). Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67, 1331–1346. 10.1111/j.1365-2958.2008.06127.x [DOI] [PubMed] [Google Scholar]
- Demarre G., Chattoraj D. K. (2010). DNA Adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 6:e1000939. 10.1371/journal.pgen.1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew R., Lurz R., Zakrzewska-Czerwinska J., Zawilak-Pawlik A. (2012). Helicobacter pylori oriC–the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 40, 9647–9660. 10.1093/nar/gks742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew R., Makowski L., Jaworski P., Bezulska M., Nowaczyk M., Zakrzewska-Czerwińska J., et al. (2014a). The atypical response regulator HP1021 controls formation of the Helicobacter pylori replication initiation complex. Mol. Microbiol. 10.1111/mmi.12866 [DOI] [PubMed] [Google Scholar]
- Donczew R., Mielke T., Jaworski P., Zakrzewska-Czerwińska J., Zawilak-Pawlik A. (2014b). Assembly of Helicobacter pylori initiation complex is determined by sequence-specific and topology-sensitive DnaA-oriC interactions. J. Mol. Biol. 10.1016/j.jmb.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Duderstadt K. E., Chuang K., Berger J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. 10.1038/nature10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duigou S., Knudsen K. G., Skovgaard O., Egan E. S., Løbner-Olesen A., Waldor M. K. (2006). Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J. Bacteriol. 188, 6419–6424. 10.1128/JB.00565-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan E. S., Waldor M. K. (2003). Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114, 521–530. 10.1016/S0092-8674(03)00611-1 [DOI] [PubMed] [Google Scholar]
- Erzberger J. P., Mott M. L., Berger J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. 10.1038/nsmb1115 [DOI] [PubMed] [Google Scholar]
- Filutowicz M., Roll J. (1990). The requirement of IHF protein for extrachromosomal replication of the Escherichia coli oriC in a mutant deficient in DNA polymerase I activity. New Biol. 2, 818–827. [PubMed] [Google Scholar]
- Filutowicz M., Ross W., Wild J., Gourse R. L. (1992). Involvement of Fis protein in replication of the Escherichia coli chromosome. J. Bacteriol. 174, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel S. E., Johnson R. C. (1992). The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 6, 3257–3265. 10.1111/j.1365-2958.1992.tb02193.x [DOI] [PubMed] [Google Scholar]
- Fol M., Chauhan A., Nair N. K., Maloney E., Moomey M., Jagannath C., et al. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657. 10.1111/j.1365-2958.2006.05137.x [DOI] [PubMed] [Google Scholar]
- Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. 10.1093/nar/gkg309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., González-Pastor J. E., Losick R. (2005). High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187, 1357–1368. 10.1128/JB.187.4.1357-1368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Losick R. (2005). Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19, 2236–2244. 10.1101/gad.1335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka T., Moriya S., Yoshikawa H., Ogasawara N. (1990). Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 107, 732–739. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. (1984). The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38, 889–900. 10.1016/0092-8674(84)90284-8 [DOI] [PubMed] [Google Scholar]
- Gao F., Luo H., Zhang C.-T. (2013). DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41, D90–93. 10.1093/nar/gks990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R., Silva F. J., Zientz E., Delmotte F., González-Candelas F., Latorre A., et al. (2003). The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. U.S.A. 100, 9388–9393. 10.1073/pnas.1533499100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Egan J. B., Roth A., Messer W. (1991). The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 19, 4167–4172. 10.1093/nar/19.15.4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade J. E., Ryan V. T., Leonard A. C. (2000). IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35, 835–844. 10.1046/j.1365-2958.2000.01755.x [DOI] [PubMed] [Google Scholar]
- Grimwade J. E., Torgue J. J.-C., McGarry K. C., Rozgaja T., Enloe S. T., Leonard A. C. (2007). Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol. Microbiol. 66, 428–439. 10.1111/j.1365-2958.2007.05930.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding N. E., Cleary J. M., Smith D. W., Michon J. J., Brusilow W. S., Zyskind J. W. (1982). Chromosomal replication origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae are functional in Escherichia coli. J. Bacteriol. 152, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., Bramhill D., Thompson C. M. (1994). Open complex formation by DnaA initiation protein at the Escherichia coli chromosomal origin requires the 13-mers precisely spaced relative to the 9-mers. Mol. Microbiol. 11, 903–911. 10.1111/j.1365-2958.1994.tb00369.x [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kornberg A. (1990). A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell 63, 325–331. 10.1016/0092-8674(90)90165-B [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kornberg A. (1992). Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267, 23083–23086. [PubMed] [Google Scholar]
- Hwang D. S., Thöny B., Kornberg A. (1992). IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J. Biol. Chem. 267, 2209–2213. [PubMed] [Google Scholar]
- Jakimowicz D., Majkadagger J., Konopa G., Wegrzyn G., Messer W., Schrempf H., et al. (2000). Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298, 351–364. 10.1006/jmbi.2000.3686 [DOI] [PubMed] [Google Scholar]
- Jakimowicz D., Majka J., Messer W., Speck C., Fernandez M., Martin M. C., et al. (1998). Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology 144(Pt 5), 1281–1290. 10.1099/00221287-144-5-1281 [DOI] [PubMed] [Google Scholar]
- Jha J. K., Baek J., Venkova-Canova T., Chattoraj D. K. (2012). Chromosome dynamics in multichromosome bacteria. Biochim. Biophys. Acta 1819, 826–829. 10.1016/j.bbagrm.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Yao S., Helinski D., Toukdarian A. (2006). Functional analysis of two putative chromosomal replication origins from Pseudomonas aeruginosa. Plasmid 55, 194–200. 10.1016/j.plasmid.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Kaguni J. M. (2006). DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60, 351–375. 10.1146/annurev.micro.60.080805.142111 [DOI] [PubMed] [Google Scholar]
- Kaguni J. M. (2011). Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613. 10.1016/j.cbpa.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. 10.1093/nar/gku1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. (1998). The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94, 61–71. 10.1016/S0092-8674(00)81222-2 [DOI] [PubMed] [Google Scholar]
- Katayama T., Ozaki S., Keyamura K., Fujimitsu K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. 10.1038/nrmicro2314 [DOI] [PubMed] [Google Scholar]
- Kato J., Katayama T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. 10.1093/emboj/20.15.4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. 10.1074/jbc.M109.002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., et al. (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099. 10.1101/gad.1561207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Bae S.-H., Yun S. H., Lee H. J., Ji S. C., Lee J. H., et al. (2005). Cnu, a novel oriC-binding protein of Escherichia coli. J. Bacteriol. 187, 6998–7008. 10.1128/JB.187.20.6998-7008.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B., Ma X., Løbner-Olesen A. (2010). Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation. J. Bacteriol. 192, 3903–3914. 10.1128/JB.00311-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Rückert B., Lurz R., Messer W. (1997). Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274, 365–380. 10.1006/jmbi.1997.1404 [DOI] [PubMed] [Google Scholar]
- Kumar S., Farhana A., Hasnain S. E. (2009). In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PloS ONE 4:e4139. 10.1371/journal.pone.0004139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan J. C., Schmidt E. W. (2013). Bacterial endosymbiosis in a chordate host: long-term co-evolution and conservation of secondary metabolism. PloS ONE 8:e80822. 10.1371/journal.pone.0080822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue C., Blanchard A., Renaudin J., Thiaucourt F., Sirand-Pugnet P. (2003). Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res. 31, 6610–6618. 10.1093/nar/gkg848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M. T., Chen S. L., Shapiro L., McAdams H. H. (2002). Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U.S.A. 99, 4632–4637. 10.1073/pnas.062065699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M. T., McAdams H. H., Feldblyum T., Fraser C. M., Shapiro L. (2000). Global analysis of the genetic network controlling a bacterial cell cycle. Science 290, 2144–2148. 10.1126/science.290.5499.2144 [DOI] [PubMed] [Google Scholar]
- Lee S.-W., Browning G. F., Markham P. F. (2008). Development of a replicable oriC plasmid for Mycoplasma gallisepticum and Mycoplasma imitans, and gene disruption through homologous recombination in M. gallisepticum. Microbiology 154, 2571–2580. 10.1099/mic.0.2008/019208-0 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Han J. S., Jeon Y., Hwang D. S. (2001). The arc two-component signal transduction system inhibits in vitro Escherichia coli chromosomal initiation. J. Biol. Chem. 276, 9917–9923. 10.1074/jbc.M008629200 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Kim H., Hwang D. S. (1996). Transcriptional activation of the dnaA gene encoding the initiator for oriC replication by IciA protein, an inhibitor of in vitro oriC replication in Escherichia coli. Mol. Microbiol. 19, 389–396. 10.1046/j.1365-2958.1996.485902.x [DOI] [PubMed] [Google Scholar]
- Leonard A. C., Grimwade J. E. (2010). Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 13, 766–772. 10.1016/j.mib.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Grimwade J. E. (2011). Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65, 19–35. 10.1146/annurev-micro-090110-102934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Méchali M. (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5:a010116. 10.1101/cshperspect.a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Campbell J. L., Boye E., Kleckner N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. 10.1016/0092-8674(94)90156-2 [DOI] [PubMed] [Google Scholar]
- Mackiewicz P., Zakrzewska-Czerwinska J., Zawilak A., Dudek M. R., Cebrat S. (2004). Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32, 3781–3791. 10.1093/nar/gkh699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. G., Gillette W. K., Rhee S., Rosner J. L. (1999). Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34, 431–441. 10.1046/j.1365-2958.1999.01599.x [DOI] [PubMed] [Google Scholar]
- Messer W. (2002). The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374. 10.1111/j.1574-6976.2002.tb00620.x [DOI] [PubMed] [Google Scholar]
- Miller C., Ingmer H., Thomsen L. E., Skarstad K., Cohen S. N. (2003). DpiA binding to the replication origin of Escherichia coli plasmids and chromosomes destabilizes plasmid inheritance and induces the bacterial SOS response. J. Bacteriol. 185, 6025–6031. 10.1128/JB.185.20.6025-6031.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. T., Grimwade J. E., Betteridge T., Rozgaja T., Torgue J. J.-C., Leonard A. C. (2009). Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U.S.A. 106, 18479–18484. 10.1073/pnas.0909472106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V., Fujita M., Jensen S. T., Eichenberger P., González-Pastor J. E., Liu J. S., et al. (2003). The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50, 1683–1701. 10.1046/j.1365-2958.2003.03818.x [DOI] [PubMed] [Google Scholar]
- Moriya S., Atlung T., Hansen F. G., Yoshikawa H., Ogasawara N. (1992). Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 6, 309–315. 10.1111/j.1365-2958.1992.tb01473.x [DOI] [PubMed] [Google Scholar]
- Moriya S., Imai Y., Hassan A. K., Ogasawara N. (1999). Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid 41, 17–29. 10.1006/plas.1998.1381 [DOI] [PubMed] [Google Scholar]
- Moriya S., Kawai Y., Kaji S., Smith A., Harry E. J., Errington J. (2009). Effects of oriC relocation on control of replication initiation in Bacillus subtilis. Microbiology 155, 3070–3082. 10.1099/mic.0.030080-0 [DOI] [PubMed] [Google Scholar]
- Mott M. L., Berger J. M. (2007). DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354. 10.1038/nrmicro1640 [DOI] [PubMed] [Google Scholar]
- Mott M. L., Erzberger J. P., Coons M. M., Berger J. M. (2008). Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135, 623–634. 10.1016/j.cell.2008.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandineni M. R., Gowrishankar J. (2004). Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J. Bacteriol. 186, 3539–3546. 10.1128/JB.186.11.3539-3546.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievera C., Torgue J. J.-C., Grimwade J. E., Leonard A. C. (2006). SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24, 581–592. 10.1016/j.molcel.2006.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. A., Bender R. A. (1988). Klebsiella pneumoniae origin of replication (oriC) is not active in Caulobacter crescentus, Pseudomonas putida, and Rhodobacter sphaeroides. J. Bacteriol. 170, 3774–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. (1991). Initiation of chromosome replication: structure and function of oriC and DnaA protein in eubacteria. Res. Microbiol. 142, 851–859. 10.1016/0923-2508(91)90065-I [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Yamazaki H., Kato J.-Y., Tomono A., Horinouchi S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439. 10.1271/bbb.69.431 [DOI] [PubMed] [Google Scholar]
- Ozaki S., Fujimitsu K., Kurumizaka H., Katayama T. (2006). The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells 11, 425–438. 10.1111/j.1365-2443.2006.00950.x [DOI] [PubMed] [Google Scholar]
- Ozaki S., Katayama T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflock M., Bathon M., Schär J., Müller S., Mollenkopf H., Meyer T. F., et al. (2007). The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J. Bacteriol. 189, 2339–2349. 10.1128/JB.01827-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaczek P. (1990). Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 2, 265–271. [PubMed] [Google Scholar]
- Rajagopalan M., Dziedzic R., Al Zayer M., Stankowska D., Ouimet M.-C., Bastedo D. P., et al. (2010). Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285, 15816–15827. 10.1074/jbc.M109.040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewska M., Wegrzyn K., Konieczny I. (2012). AT-rich region and repeated sequences – the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 36, 408–434. 10.1111/j.1574-6976.2011.00300.x [DOI] [PubMed] [Google Scholar]
- Rice P. A., Yang S., Mizuuchi K., Nash H. A. (1996). Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87, 1295–1306. 10.1016/S0092-8674(00)81824-3 [DOI] [PubMed] [Google Scholar]
- Roggenkamp A. (2007). Phylogenetic analysis of enteric species of the family Enterobacteriaceae using the oriC-locus. Syst. Appl. Microbiol. 30, 180–188. 10.1016/j.syapm.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Rozgaja T. A., Grimwade J. E., Iqbal M., Czerwonka C., Vora M., Leonard A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. 10.1111/j.1365-2958.2011.07827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan V. T., Grimwade J. E., Camara J. E., Crooke E., Leonard A. C. (2004). Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 51, 1347–1359. 10.1046/j.1365-2958.2003.03906.x [DOI] [PubMed] [Google Scholar]
- Ryan V. T., Grimwade J. E., Nievera C. J., Leonard A. C. (2002). IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46, 113–124. 10.1046/j.1365-2958.2002.03129.x [DOI] [PubMed] [Google Scholar]
- Samitt C. E., Hansen F. G., Miller J. F., Schaechter M. (1989). In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 8, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G., Errington J., Murray H. (2012). Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31, 1542–1555. 10.1038/emboj.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen S. M., Ouimet M.-C., Marczynski G. T. (2009). Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155, 1215–1225. 10.1099/mic.0.025528-0 [DOI] [PubMed] [Google Scholar]
- Siam R., Brassinga A. K. C., Marczynski G. T. (2003). A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. J. Bacteriol. 185, 5563–5572. 10.1128/JB.185.18.5563-5572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R., Marczynski G. T. (2000). Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19, 1138–1147. 10.1093/emboj/19.5.1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Katayama T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Thöny B., Hwang D. S., Kornberg A. (1993). A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268, 5365–5370. [PubMed] [Google Scholar]
- Slager J., Kjos M., Attaiech L., Veening J.-W. (2014). Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157, 395–406. 10.1016/j.cell.2014.01.068 [DOI] [PubMed] [Google Scholar]
- Slater S., Wold S., Lu M., Boye E., Skarstad K., Kleckner N. (1995). E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82, 927–936. 10.1016/0092-8674(95)90272-4 [DOI] [PubMed] [Google Scholar]
- Speck C., Messer W. (2001). Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20, 1469–1476. 10.1093/emboj/20.6.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M., Webb V., Spiegelman G., Hoch J. A. (1990). The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1801–1805. 10.1073/pnas.87.5.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M. D., Carr K. M., Vicente M., Kaguni J. M. (1998). Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262. 10.1074/jbc.273.51.34255 [DOI] [PubMed] [Google Scholar]
- Suvorov A. N., Ferretti J. J. (2000). Replication origin of Streptococcus pyogenes, organization and cloning in heterologous systems. FEMS Microbiol. Lett. 189, 293–297. 10.1111/j.1574-6968.2000.tb09246.x [DOI] [PubMed] [Google Scholar]
- Swinger K. K., Rice P. A. (2004). IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14, 28–35. 10.1016/j.sbi.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Taghbalout A., Landoulsi A., Kern R., Yamazoe M., Hiraga S., Holland B., et al. (2000). Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro. Genes Cells 5, 873–884. 10.1046/j.1365-2443.2000.00380.x [DOI] [PubMed] [Google Scholar]
- Takeda Y., Harding N. E., Smith D. W., Zyskind J. W. (1982). The chromosomal origin of replication (oriC) of Erwinia carotovora. Nucleic Acids Res. 10, 2639–2650. 10.1093/nar/10.8.2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. A., Ouimet M.-C., Wargachuk R., Marczynski G. T. (2011). The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol. Microbiol. 82, 312–326. 10.1111/j.1365-2958.2011.07785.x [DOI] [PubMed] [Google Scholar]
- Theisen P. W., Grimwade J. E., Leonard A. C., Bogan J. A., Helmstetter C. E. (1993). Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10, 575–584. 10.1111/j.1365-2958.1993.tb00929.x [DOI] [PubMed] [Google Scholar]
- Thöny B., Hwang D. S., Fradkin L., Kornberg A. (1991). iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 88, 4066–4070. 10.1073/pnas.88.10.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov O. V., Biswas T. (2011). Structural and thermodynamic signatures of DNA recognition by Mycobacterium tuberculosis DnaA. J. Mol. Biol. 410, 461–476. 10.1016/j.jmb.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Via L. E., Curcic R., Mudd M. H., Dhandayuthapani S., Ulmer R. J., Deretic V. (1996). Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178, 3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C., Schmidt A., Rückert B., Lurz R., Messer W. (1997). DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 16, 6574–6583. 10.1093/emboj/16.21.6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg R. A., Freundlich M., Friedman D., Gardner J., Goosen N., Nash H., et al. (1996). Nomenclature of the genes encoding IHF. Mol. Microbiol. 19, 642. 10.1046/j.1365-2958.1996.t01-2-442924.x [DOI] [PubMed] [Google Scholar]
- Woelker B., Messer W. (1993). The structure of the initiation complex at the replication origin, oriC, of Escherichia coli. Nucleic Acids Res. 21, 5025–5033. 10.1093/nar/21.22.5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolański M., Donczew R., Kois-Ostrowska A., Masiewicz P., Jakimowicz D., Zakrzewska-Czerwinska J. (2011). The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 193, 6358–6365. 10.1128/JB.05734-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolański M., Jakimowicz D., Zakrzewska-Czerwińska J. (2012). AdpA, key regulator for morphological differentiation regulates bacterial chromosome replication. Open Biol. 2, 120097. 10.1098/rsob.120097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolański M., Jakimowicz D., Zakrzewska-Czerwińska J. (2014). Fifty years after the replicon hypothesis: cell-specific master regulators as new players in chromosome replication control. J. Bacteriol. 196, 2901–2911. 10.1128/JB.01706-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Zheng W., Crooke E., Wang Y. H., Inouye M. (2001). CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39, 1572–1584. 10.1046/j.1365-2958.2001.02345.x [DOI] [PubMed] [Google Scholar]
- Yee T. W., Smith D. W. (1990). Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl. Acad. Sci. U.S.A. 87, 1278–1282. 10.1073/pnas.87.4.1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. H., Ji S. C., Jeon H. J., Wang X., Kim S. W., Bak G., et al. (2012a). The CnuK9E H-NS complex antagonizes DNA binding of DicA and leads to temperature-dependent filamentous growth in E. coli. PloS ONE 7:e45236. 10.1371/journal.pone.0045236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. H., Ji S. C., Jeon H. J., Wang X., Lee Y., Choi B.-S., et al. (2012b). A mutational study of Cnu reveals attractive forces between Cnu and H-NS. Mol. Cells 33, 211–216. 10.1007/s10059-012-0006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Czerwińska J., Jakimowicz D., Zawilak-Pawlik A., Messer W. (2007). Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31, 378–387. 10.1111/j.1574-6976.2007.00070.x [DOI] [PubMed] [Google Scholar]
- Zakrzewska-Czerwińska J., Schrempf H. (1992). Characterization of an autonomously replicating region from the Streptomyces lividans chromosome. J. Bacteriol. 174, 2688–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilak A., Cebrat S., Mackiewicz P., Król-Hulewicz A., Jakimowicz D., Messer W., et al. (2001). Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 29, 2251–2259. 10.1093/nar/29.11.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilak A., Durrant M. C., Jakimowicz P., Backert S., Zakrzewska-Czerwińska J. (2003). DNA binding specificity of the replication initiator protein, DnaA from Helicobacter pylori. J. Mol. Biol. 334, 933–947. 10.1016/j.jmb.2003.10.018 [DOI] [PubMed] [Google Scholar]
- Zawilak A., Kois A., Konopa G., Smulczyk-Krawczyszyn A., Zakrzewska-Czerwińska J. (2004). Mycobacterium tuberculosis DnaA initiator protein: purification and DNA-binding requirements. Biochem. J. 382, 247–252. 10.1042/BJ20040338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilak-Pawlik A., Kois A., Majka J., Jakimowicz D., Smulczyk-Krawczyszyn A., Messer W., et al. (2005). Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 389, 471–481. 10.1042/BJ20050143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorman S., Seitz H., Sclavi B., Strick T. R. (2012). Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 40, 7375–7383. 10.1093/nar/gks371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. (1983). Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc. Natl. Acad. Sci. U.S.A. 80, 1164–1168. 10.1073/pnas.80.5.1164 [DOI] [PMC free article] [PubMed] [Google Scholar]