Abstract
Human genome-wide association studies (GWASs) have identified numerous associations between single nucleotide polymorphisms (SNPs) and pulmonary function. Proving that there is a causal relationship between GWAS SNPs, many of which are noncoding and without known functional impact, and these traits has been elusive. Furthermore, noncoding GWAS-identified SNPs may exert trans-regulatory effects rather than impact the proximal gene. Noncoding variants in 5-hydroxytryptamine (serotonin) receptor 4 (HTR4) are associated with pulmonary function in human GWASs. To gain insight into whether this association is causal, we tested whether Htr4-null mice have altered pulmonary function. We found that HTR4-deficient mice have 12% higher baseline lung resistance and also increased methacholine-induced airway hyperresponsiveness (AHR) as measured by lung resistance (27%), tissue resistance (48%), and tissue elastance (30%). Furthermore, Htr4-null mice were more sensitive to serotonin-induced AHR. In models of exposure to bacterial lipopolysaccharide, bleomycin, and allergic airway inflammation induced by house dust mites, pulmonary function and cytokine profiles in Htr4-null mice differed little from their wild-type controls. The findings of altered baseline lung function and increased AHR in Htr4-null mice support a causal relationship between genetic variation in HTR4 and pulmonary function identified in human GWAS.—House, J. S., Li, H., DeGraff, L. M., Flake, G., Zeldin, D. C., London, S. J. Genetic variation in HTR4 and lung function: GWAS follow-up in mouse.
Keywords: pulmonary function, airway hyperresponsiveness, mouse models
Spirometry is used to assess clinically relevant pulmonary function parameters. Forced expiratory volume in the first second (FEV1) is an indicator of airway obstruction, forced vital capacity (FVC) is an index of lung size, and their ratio (FEV1/FVC) is a size-independent indicator of airflow obstruction (1). These pulmonary function measures are used clinically to diagnose lung diseases including chronic obstructive pulmonary disease (COPD), and to monitor the severity and control of asthma, COPD, and other respiratory conditions.
Heritability estimates support genetic contributions to pulmonary function (2, 3). Alpha-1-antitrypsin deficiency, the first identified genetic risk factor for COPD, is infrequent, and later candidate gene studies did not identify many other replicable associations (4, 5). Since 2009, genome-wide association studies (GWASs) have identified at least 33 loci for pulmonary function (6–10), including some also associated with the clinical phenotypes of airflow obstruction, COPD, or asthma (11–22). Although GWAS has been extremely successful in identifying novel loci, it is not well suited for identifying the specific causal variants within genes or even the causal gene within a multigene locus. Additional methods are needed for this purpose. Experimental studies to follow-up the myriad of GWAS hits for pulmonary function or other traits are labor intensive and costly and therefore scarce. Complicating GWAS follow-up is the fact that the vast majority of GWAS hits are located in noncoding sequences with unknown regulatory impact (23, 24). Noncoding GWAS hits may exert their effects by trans-regulatory actions, and intronic GWAS single nucleotide polymorphisms (SNPs) cannot be assumed to affect the residing or closest gene (25). Thus, experimental animal models in which the perturbation of a gene recapitulates associated phenotypes in human GWASs can provide powerful evidence that the gene implicated by GWAS variants is causally related to that associated trait.
A GWAS meta-analysis from our group identified intronic SNPs in 5-hydroxytryptamine (serotonin) (5-HT) receptor 4 (HTR4) related to pulmonary function in humans (8). These associations have been replicated by others (26), and the same HTR4 SNPs have since been associated with the clinical phenotypes of airflow obstruction and COPD (15, 27) and asthma (28). Given the human GWAS evidence for HTR4 and pulmonary function, we sought to follow up with genetically engineered mice. Unlike human studies, mice can be engineered with altered or removed genes and can be exposed to environmental agents in various injury models to perturb the system being studied.
The serotonin receptor family consists of 7 genes (HTR1–HTR7) and at least 17 different splice variants, all but 1 of which are GPCRs. On stimulation with their endogenous ligand serotonin (5-HT), HTRs activate intracellular secondary messenger cascades that result in both excitatory and inhibitory responses through the release of a variety of neurotransmitters and hormones (29).
HTR4 was first identified in localized brain neurons within the olfactory tubercle, basal ganglia, and hippocampus, and in the respiratory rhythm generating pre-Bötzinger complex (30, 31). Peripherally, HTR4 is broadly expressed in the gastrointestinal tract, vasculature, adrenal glands, lower urinary tract, and myocardium (32, 33) and has lower but detectable expression in whole lung, airway epithelial cells, and airway smooth muscle cells (34, 35). HTR4 has numerous splice variants (36, 37) with discrete isoform tissue and neuron distribution. Recent work has noted differential temporal expression of HTR4 in fetal human lung supporting a role for this gene in development (37).
We hypothesized that Htr4-null (Htr4−/−) mice would exhibit altered lung function. Htr4−/− mice have been developed and characterized with a variety of behavioral phenotypes including altered memory formation, eating, response to novel environments, and a propensity to seizures (38–41). To date, the role of HTR4 in pulmonary function has not been examined. Our study examines the role of HTR4 in baseline pulmonary function using Htr4−/− and wild-type (WT) mice. We also evaluated the response of these mice to well-established perturbation models using bacterial LPS to elicit an acute inflammatory response, bleomycin to elicit lung fibrosis, and house dust mite (HDM) to examine allergic airway inflammation.
MATERIALS AND METHODS
Animals
Htr4−/− mice (B6.129P2-Htr4tm1Dgen/J) were purchased from The Jackson Laboratory (005767; Bar Harbor, ME, USA) and maintained as Htr4+/− × Htr4+/− breeders to generate sibling Htr4+/+ (WT) and Htr4−/− mice. Male mice were between 9 and 12 wk of age for LPS and bleomycin studies at the time of exposure. Experiments with naïve mice included 9- to 12-wk-old male animals in addition to an aged population group (∼40 wk old). Males at 17 wk of age were used for body weight and lung volume assessment. For HDM experiments, 12- to 15-wk-old male mice were sensitized with 10 µg HDM (Dermatophagides pteronyssinus, Greer XPB82D3A2.5) in 50 µl PBS via oropharyngeal aspiration (OPA) on days 1 and 8, followed by challenge on days 15, 16, and 17 with 2 µg HDM, and collected on day 18. All animal work described in this study was conducted according to U.S. National Institutes of Health guidelines and approved by the National Institute of Environmental Health Sciences animal care and use committee.
Lung function assessment
Invasive lung function analysis was performed on either untreated (naïve) or treated mice with flexiVent (SCIREQ, Montreal, QC, Canada) according to the manufacturer’s instructions. Briefly, mice were anesthetized with urethane (1.5 g/kg) intraperitoneally (i.p.), and tracheostomized with a 19-guage stainless steel cannula. Mice were then given pancuronium bromide (0.8 mg/kg, i.p.) to paralyze the diaphragm and prevent autonomous breathing, placed on a 37°C heated pad, and connected to a computer-controlled ventilation device. Ventilation was set to 150 breaths/min, with a tidal volume of 7.5 ml/kg and a positive end expiratory pressure of 3.0 cm of H2O. Lung function metrics were calculated as described previously (42, 43).
After 3 baseline measurements of lung function using “snapshot” and “quickprime” perturbations, mice were given either increasing aerosolized doses of methacholine (MCH; 3.125, 6.25, 12.5, and 25 mg/ml) or 5-HT (4.25, 8.5, and 17 mg/ml) for 10 s without altering the ventilation pattern, followed by assessment with 8 separate “snapshot” and “quickprime” perturbations every 30 s. Between each dose of MCH or 5-HT, a total lung compliance loop was initiated to reset lung hysteresis. For each genotype/dose combination, responses with coefficient of determination >80% were kept, outliers >3 sd from the mean were excluded, and the 3 maximal [for compliance (C) minimal] values per mouse per dose were averaged and reported as means ± sem. Linear regression was used to interpolate values for provocative concentrations of broncho-constrictor (MCH or 5-HT) needed to elicit a 200% increase in baseline response for R, Rn, G, and H (PC200) or 50% decrease in C (PC50).
For lung inflammation studies, mice were anesthetized with isoflurane/oxygen and administered 50 µl of either endotoxin-free 0.9% saline (Sigma S8776 [certificate 7647-14-5 (<0.005 U/ml endotoxin)]; Sigma-Aldrich, St. Louis, MO, USA) or 50 µg LPS (Sigma L2630; Sigma-Aldrich) in 50 µl 0.9% saline via OPA. Mice were monitored hourly for 4 h and given supplemental heat as needed followed by lung function assessment as described above.
For lung fibrosis studies, mice were anesthetized with isoflurane/oxygen and administered 50 µl of either endotoxin-free 0.9% saline [Sigma S8776 (7647-14-5); Sigma-Aldrich] or 1.4 U/kg bleomycin (Sigma B5507; Sigma-Aldrich) via OPA. Mice were given supplemental heat and mash after treatment. To examine repair of fibrotic injury differed by genotype, lung function static compliance measurements were taken with flexiVent 20–21 d after exposure.
Cell and cytokine analyses
Bronchoalveolar lavage was performed with two × 1.0 ml of HBSS (Sigma H6648; Sigma-Aldrich) after lung function measurement via flexiVent; recovery was greater than 80% for each mouse. Bronchoalveolar lavage fluid (BALF) was pooled for each mouse and centrifuged (2000 rpm, 6 min, 4°C) to separate cells from supernatant. The cytokines in the cell-free supernatant were measured using the Bio-Plex mouse cytokine assay and Bio-Plex suspension array system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instruction. All cells were treated with 1.0 ml ammonium-chloride-potassium buffer for 1 min to lyse red blood cells, diluted in 5.0 ml HBSS, and gently pelleted by centrifugation, and the supernatant was removed. Cells were resuspended in 1.0 ml HBSS for quantification with a hemocytometer, and cytospins were prepared with 200 µl of suspended cells for cell differential analysis.
Breathing assay
Mice underwent surgery to place a jugular vein cannula 3 d prior to the experiment. On the day of the experiment, mice were acclimated to a whole body plethysmography chamber (BUXCO/Data Sciences International, Minneapolis, MN, USA) for 10 min prior to baseline assessment for 10 min. Mice were then administered 0.4 mg/kg fentanyl (0641-6030-01; West-Ward Pharmaceuticals, Cherry Hill, NJ, USA) via jugular vein cannula and monitored for 5 min. Five minutes after fentanyl administration, 4 mg/kg BIMU8 (4374; Tocris Bioscience, Bristol, United Kingdom), a 5-HT4 receptor selective agonist, was administered via jugular vein cannula, and mice were monitored for another 10 min.
RNA and quantitative PCR
The whole cardiac lobe from each mouse was homogenized and RNA was isolated and purified following the manufacturer’s instructions (RNeasy Kit 74104; Qiagen, Valencia, CA, USA). Following purification of RNA, cDNA was created with equal amounts of template RNA and subjected to real time RT-PCR analysis using TaqMan-based assays [Htr4 (Mm00434129_m1), Gapdh (Mm99999915_g1); Applied Biosystems/Life Technologies, Grand Island, NY, USA]. Relative expression was calculated with the ΔΔCt method using Gapdh as the housekeeping gene.
Inflammation and fibrosis scoring
Lungs from animals treated with LPS or bleomycin were removed, lavaged with 1 ml × 2 of HBSS, and fixed in 4.0% paraformaldehyde. They were then embedded in paraffin, and 5 μm sections were cut and stained with hematoxylin and eosin stain or Masson’s trichrome. Fibrosis was scored according to Ashcroft’s methodology (44) with blinded samples by a board-certified pathologist. Inflammation scoring for LPS- and saline-treated animals was done by summing the total of the following observations in left and right mouse lungs: margination of neutrophils, perivascular neutrophils, peribronchial neutrophils, and evidence of perivas/peribron hemorrhage. Each inflammation parameter was scored from 0 to 4 on the basis of both the local intensity and the extent of the lung involved, resulting in a total sum score ranging from 0 to 16. Scoring was based on infiltration of neutrophils only and not on the presence of mononuclear cells, because the latter are sometimes present in perivascular or peribronchial tissue of untreated mice. Isolated neutrophils adjacent to the endothelium of vessels were not considered evidence of margination; that is, only when several neutrophils appeared to be attached to the endothelium was margination considered to be present. For HDM scoring, the following inflammation parameters were scored: perivascular inflammation (PVI, 0–8), peribronchial inflammation (PBI, 0–8), and goblet cell hyperplasia (0–4). This resulted in a combined total score ranging from 0 to 20.
Collagen assay
Collagen content in each lung caudal lobe of mice treated with bleomycin or saline was measured using the Sircol Soluble Collagen Assay kit (# S1000; Biocolor, Carrickfergus, United Kingdom) following the manufacturer’s instructions. In brief, the entire caudal lobe of each mouse was snap frozen on dry ice and stored at −80°C until assay. The lobe was thawed on ice, gently blotted dry, and weighed. It was then minced in PBS to clear blood and centrifuged at 10,000 rpm at 4°C for 5 min, and the supernatant was removed. The minced tissue was incubated with 1 ml of cold 0.5 M acetic acid with 0.1 mg/ml of pepsin on a gentle shaker at 4°C overnight. The acid-pepsin extraction solution was centrifuged at 12,000 rpm for 10 min, and the supernatant was collected for use in the assay.
Lung volume
Mice were anesthetized with FatalPlus (Patterson, Devens, MA, USA) via i.p. injection until no longer responsive to toe pinch and weighed for whole body weight. Mouse whole lungs were isolated and removed with the heart intact and hung under 25 cm pressure of 4% neutral buffered formalin for 2 h. Lungs were tied off under pressure, the heart and esophagus were removed, and lung volume was calculated by displacement of fixative and adjusted for specific gravity.
Statistical analysis
For the breathing assay, the general linear model procedure (Proc GLM; SAS, Cary, NC, USA) was used to test differences by genotype for the respiratory rate at both 4 min after fentanyl administration and 10 min after BIMU8 administration, treating time as a repeated measure. For baseline lung function analyses, age-adjusted means and sems were calculated by genotype using SAS Proc GLM. For assessment of genotype differences in MCH- or 5-HT–induced airway hyperresponsiveness (AHR), Proc GLM was used to calculate age- and dose-adjusted means and sems. The same analysis was used to determine genotype differences in MCH-induced AHR 4 h after LPS. Two-tailed Student’s t test was used to test genotype differences in cytokines and cell counts, as well as genotype differences in static compliance/elastance after bleomycin treatment. All error bars in all figures represent sems. Values were reported as significant if P < 0.05.
RESULTS
Htr4−/− mouse characterization
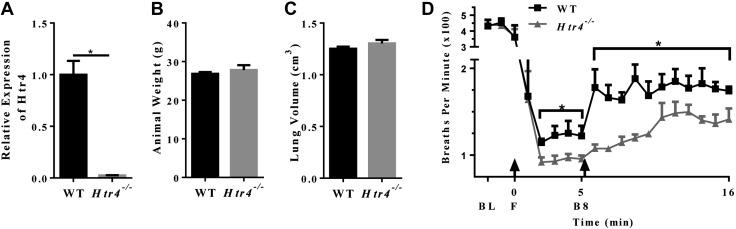
Attempts to confirm loss of HTR4 by immunohistochemistry or immunoblotting with multiple antibodies in WT and Htr4−/− mice yielded inconclusive results (data not shown). Antibody specificity with neurotransmitter receptors in rodents is a known problematic issue (45, 46). Therefore, quantitative real-time RT-PCR was used to ascertain expression of Htr4 in WT and Htr4−/− mice (Fig. 1A). The expression of Htr4 was first measured in a battery of tissues from WT mice. Consistent with previous studies (31, 47), expression was highest in brain and gastrointestinal tract with moderate expression in lung (Supplemental Fig. 1A). Real-time RT-PCR using primers spanning the exon3/exon4 (deletion cassette) junction showed detectable message in lungs from WT but not Htr4−/− mice (Fig. 1A). Histologic examination of lung architecture (hematoxylin and eosin stain) revealed no obvious differences between the genotypes (data not shown). Furthermore, we observed no differences between age-matched WT and Htr4−/− mice in either body weight or lung volume as measured by displacement (Fig. 1B, C).
Figure 1.
A) Relative expression of HTR4 in WT and Htr4−/− mouse whole lung (n = 7/genotype). B) Animal weights of age-matched WT and Htr4−/− mice (n = 5/genotype). C) Lung volume of age-matched WT and Htr4−/− mice (n = 5/genotype). D) Respiratory rate of WT and Htr4−/− mice. Baseline measurements (BL) were taken, followed by fentanyl bolus (F), followed by measurement for 5 min, followed by BIMU8 bolus (B8), and followed by measurement for 11 min. Baseline measurements are 5 min averages; all other points are 1 min averages (n = 4/genotype). Mean values per group with sem are plotted. *P < 0.05 vs. WT.
Real-time RT-PCR data also indicated that postdeletion cassette transcription (using primers spanning exon5/exon6 junction) occurred in Htr4−/− mice (data not shown). Work by Manzke et al. demonstrated opiate-induced respiratory depression could be mitigated with HTR4-specific agonists (48). Therefore, to prove we had a functionally HTR4-deficient mouse, we conducted a breathing assay using the opiate receptor agonist fentanyl. We observed that WT and Htr4−/− mice exhibited marked respiratory depression immediately after intravenous administration of fentanyl (Fig. 1D). In the 10 min prior to fentanyl administration, WT and Htr4−/− mice did not differ in respiratory rates. However, after fentanyl administration, Htr4−/− mice had a further reduction in respiratory rate compared with WT mice (Fig. 1D). Importantly, after fentanyl-induced respiratory depression, administration of the HTR4-specific agonist BIMU8 resulted in a marked increase in respiratory rate in WT mice, an effect that was not observed in Htr4−/− mice (Fig. 1D). This provides confirmation that the HTR4 receptor is nonfunctional in Htr4−/− mice.
Htr4−/− mice exhibit increased whole lung and airway resistance at baseline
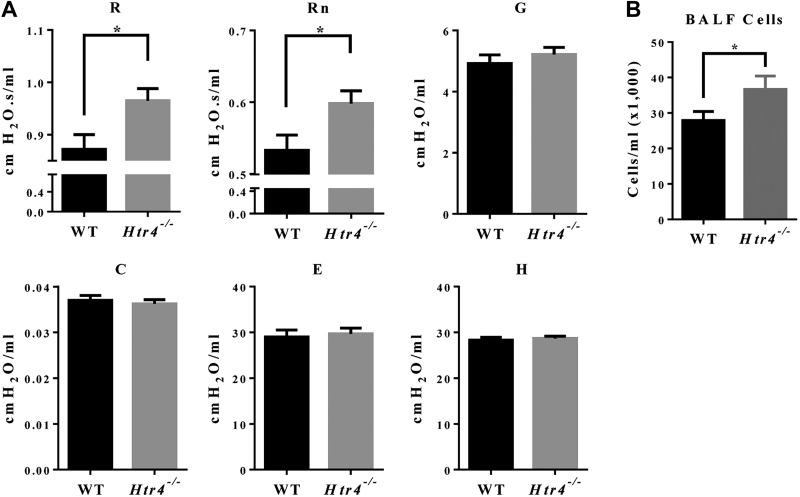
To assess whether Htr4−/− mice had altered lung function, we used flexiVent. At baseline, compared with WT mice, Htr4−/− mice had statistically significantly higher whole lung resistance (R) and Newtonian resistance (Rn), a measure of resistance in the conducting airways (Fig. 2A). In contrast, there were no differences in tissue resistance (G), compliance (C), elastance (E), and tissue elastance (H) between the genotypes (Fig. 2A).
Figure 2.
A) Baseline lung function parameters [whole lung resistance (R), Newtonian resistance (Rn), tissue resistance (G), compliance (C), elastance (E), tissue elastance (H)] were examined on flexiVent in WT and Htr4−/−-naive animals. Age-adjusted least-squares means and sems are plotted. B) Total cells were counted from BALF of untreated mice (n ≥ 20 mice/genotype). Mean values per group with sem are plotted. *P < 0.05 vs. WT.
To examine whether Htr4−/− mice differed in steady-state immune cell readouts, we examined BALF from naïve mice. BALF cytokines were either undetectable or not different between WT and Htr4−/− mice (data not shown). However, we observed a 31% higher count of BALF cells in Htr4−/− mice compared with WT; as expected, nearly all cells were macrophages (>98%) (Fig. 2B).
Htr4−/− mice exhibit AHR to inhaled MCH and 5-HT
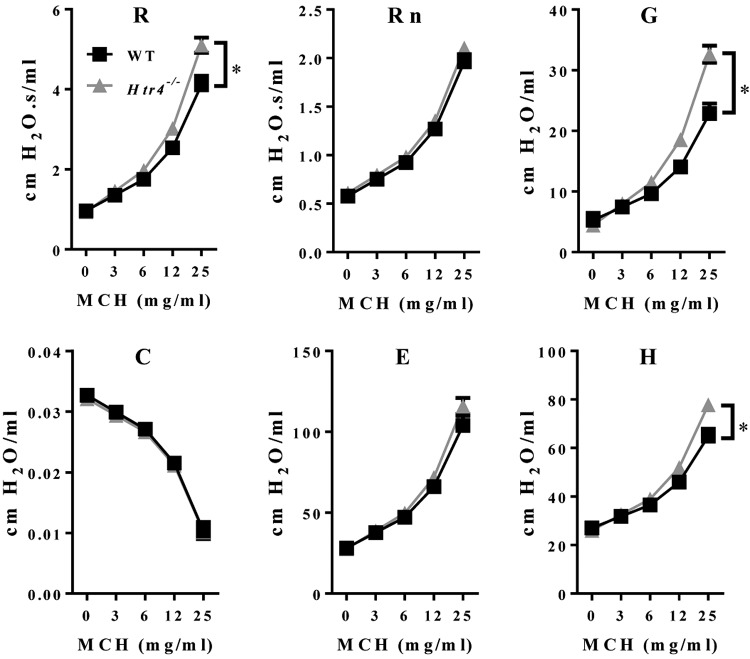
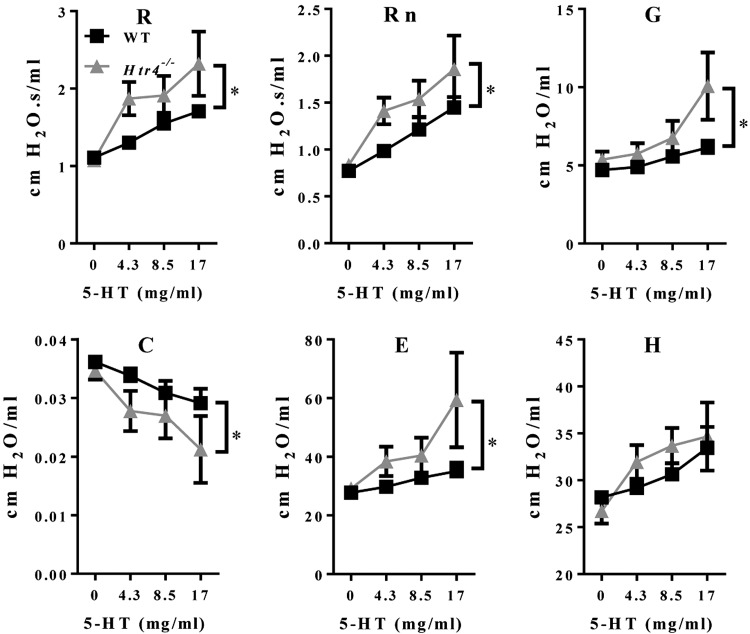
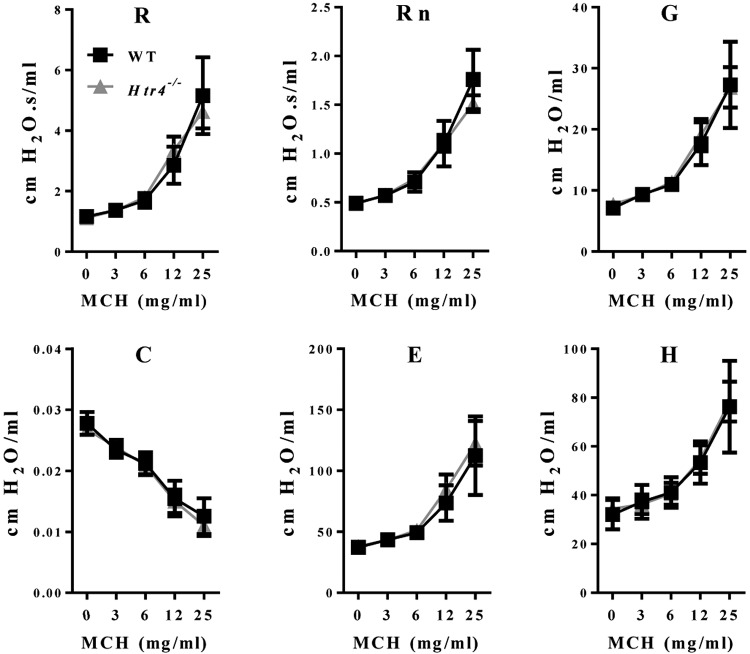
We used the broncho-constrictor MCH to assess AHR in naïve WT and Htr4−/− mice. After adjusting for age and dose of MCH, Htr4−/− mice were more responsive to MCH using the parameters of whole lung resistance (R), tissue resistance (G), and tissue elastance (H) (Fig. 3; Table 1). 5-HT is another broncho-constrictor with known mode of action pharmacologically linked to HTR1-3. To test whether 5-HT signaling also affects broncho-constriction through HTR4, we treated mice with increasing doses of 5-HT and assessed pulmonary function. Similar to MCH-induced AHR, Htr4−/− mice were more responsive than WT mice to 5-HT (28–72%) in all lung function parameters measured with the exception of tissue elastance (H) (Fig. 4; Table 1).
Figure 3.
Assessment of AHR to MCH in WT and Htr4−/− mice. Whole lung resistance (R), Newtonian resistance (Rn), tissue resistance (G), compliance (C), elastance (E), and tissue elastance (H) were measured with flexiVent. Resistance (R), tissue resistance (G), and tissue elastance (H) were higher in Htr4−/− animals relative to WT. Least-squares means and sems are plotted after adjusting for age (n ≥ 20/genotype). *P < 0.05 vs. WT.
TABLE 1.
Assessment of AHR in WT and Htr4−/− mice
| Parameter | Broncho-constrictor | PC200/PC50 mg/ml (WT) | PC200/PC50 mg/ml (Htr4−/−) | Percent change |
|---|---|---|---|---|
| Resistance (R) | MCH | 15.2 | 11.1 | −27% |
| Tissue resistance (G) | MCH | 14.8 | 7.74 | −48% |
| Tissue elastance (H) | MCH | 35.5 | 25.0 | −30% |
| Resistance (R) | 5-HT | 61.6 | 29.5 | −52% |
| Newtonian resistance (Rn) | 5-HT | 37.9 | 27.3 | −28% |
| Tissue resistance (G) | 5-HT | 107 | 39.3 | −63% |
| Compliance (C) | 5-HT | 42.4 | 21.7 | −49% |
| Elastance (E) | 5-HT | 31.0 | 8.54 | −72% |
PC200 or PC50 was calculated as the concentration needed to increase baseline parameters by 200% (for R, Rn, G, and H) or decrease baseline parameter by 50% (for C).
Figure 4.
Assessment of AHR to 5-HT in WT and Htr4−/− mice. Whole lung resistance (R), Newtonian resistance (Rn), tissue resistance (G), compliance (C), elastance (E), and tissue elastance (H) were measured with flexiVent. Resistance (R), Newtonian resistance (Rn), tissue resistance (G), and elastance (E) are higher and compliance (C) is lower in Htr4−/− mice relative to WT. Mean values per dose with sem are plotted (n ≥ 4/genotype). *P < 0.05 vs. WT.
AHR in Htr4−/− mice after LPS exposure
To examine acute inflammatory responses in Htr4−/− mice, mice were dosed with either saline or bacterial LPS and examined on flexiVent for genotype differences in MCH-induced AHR 4 h later. WT mice treated with LPS had increased lung resistance compared with saline-treated mice (data not shown). There were no significant differences in lung function parameters between genotypes 4 h after LPS challenge (Fig. 5).
Figure 5.
Assessment of MCH-induced AHR 4 h after treatment with LPS (via OPA) in WT and Htr4−/− mice. Whole lung resistance (R), Newtonian resistance (Rn), tissue resistance (G), compliance (C), elastance (E), and tissue elastance (H) were measured with flexiVent. Mean values per group with sem are plotted (n ≥ 7/genotype). *P < 0.05 vs. WT.
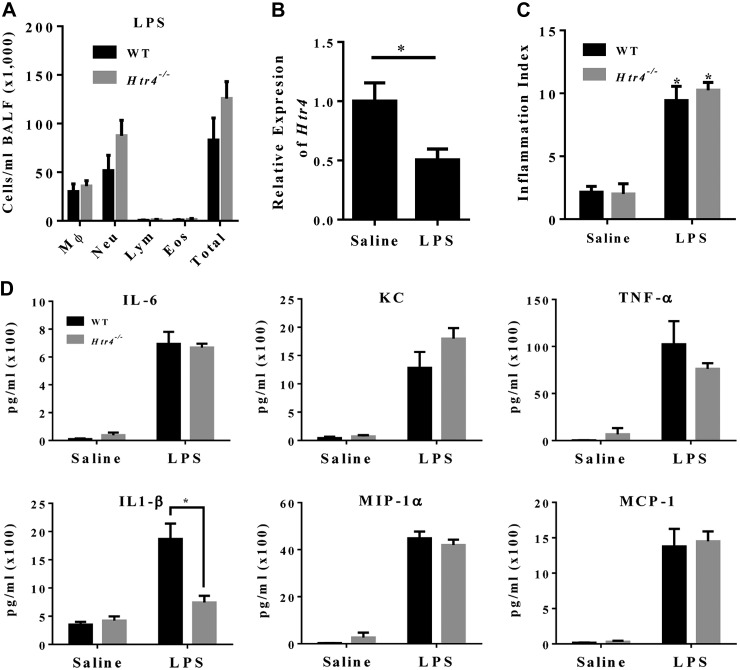
Examination of BALF collected from LPS-treated mice revealed no differences between the genotypes in total cells or in cell differential populations (Fig. 6A). Expression of Htr4 mRNA was decreased in LPS-treated animals (Fig. 6B). Lung sections were scored by a pathologist blinded to genotype to assess degree of inflammation, and there were no genotype differences observed (Fig. 6C). We also examined cytokines involved in LPS signaling (IL-1β, IL-6, keratinocyte-derived protein, macrophage inflammatory protein -1α, TNF-α, and monocyte chemoattractant protein-1). Most of the cytokines exhibited no differences in LPS response between the genotypes (Fig. 6D). The exception was IL-1β, for which the expected LPS induction confirmed in WT mice was mitigated in Htr4−/− mice (Fig. 6D). Non–LPS-responsive cytokines (IL-4, IL-5, IL-10, and IL-12) were below the limits of detection (data not shown). Examination of serum cytokines from mice treated with either saline or LPS revealed no genotype differences (Supplemental Fig. 1B).
Figure 6.
A) Cell differentials by genotype in LPS-treated WT and Htr4−/− mice (n ≥ 7/genotype). B) Expression of HTR4 in saline and LPS-treated WT mice (n = 7/group). *P < 0.05 vs. saline. C) Inflammatory index of LPS- and saline-treated mice 4 h after LPS treatment (n ≥ 7/group). *P < 0.05 vs. saline. D) Cytokine concentrations in BALF from LPS- and saline-treated WT and Htr4−/− mice (n ≥ 5/group in saline; n ≥ 9/group in LPS). Mean values per group with sem are plotted. *P < 0.05 vs. WT.
Responses to bleomycin exposure in Htr4−/− mice
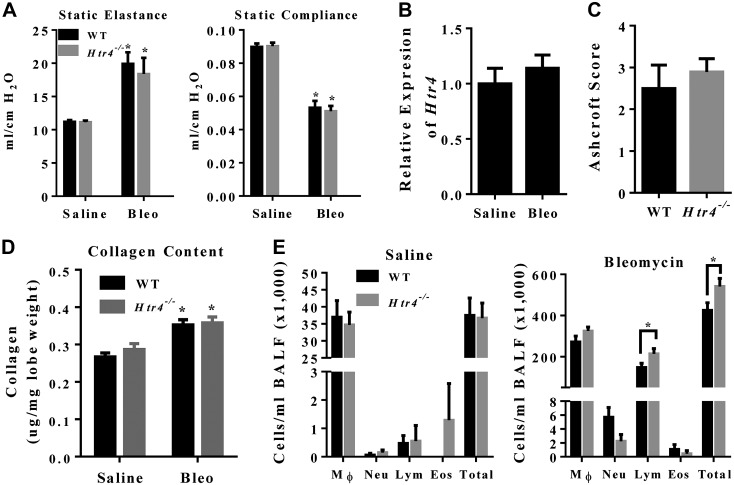
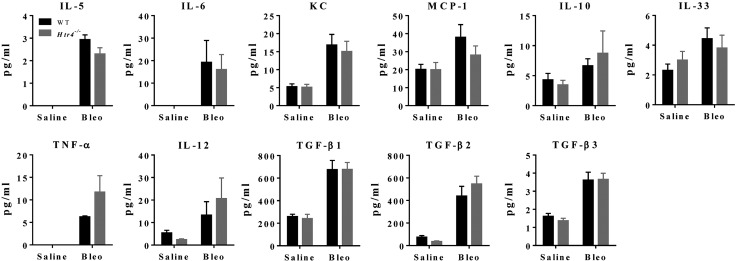
To determine whether mice deficient in HTR4 exhibit differences in bleomycin-induced fibrosis, we examined mice 20—21 d after exposure to bleomycin or saline. Mice treated with bleomycin had increased static elastance and decreased static compliance, but with no difference between genotypes (Fig. 7A). Expression of Htr4 was unchanged by bleomycin exposure (Fig. 7B). No fibrosis was observed in saline-treated animals (data not shown). Fibrotic scoring and collagen content did not differ between Htr4−/− mice and WT mice treated with bleomycin (Fig. 7C, D). Total cells in BALF increased dramatically in bleomycin treated mice (Fig. 7E, right). After bleomycin treatment, there were more total cells and more lymphocytes in Htr4−/− mice than in WT mice (Fig. 7E, right). There were no significant differences in BALF cytokines between the genotypes (Fig. 8). Similar to LPS results, we observed no differences between WT and Htr4−/− in levels of serum cytokines (Supplemental Fig. 2).
Figure 7.
A) flexiVent measurements of static elastance and compliance 21 d after saline or bleomycin (1.75 mg/kg) by genotype (n ≥ 10/group). *P < 0.05 vs. saline. B) Expression of Htr4 21 d after saline or bleomycin treatment in WT nice (n = 5/group). C) Fibrosis scoring of bleomycin-treated animals (n ≥ 9/group). D) Collagen content of whole caudal lobe 21 d after saline or bleomycin treatment (*P < 0.05 different from controls; n ≥ 8/group). E) Cell differentials from BALF 21 d after saline or bleomycin (1.75 mg/kg) by genotype (n ≥ 5/group in saline; n ≥ 10/group in bleomycin). Mean values per group with sem are plotted. *P < 0.05 vs. WT.
Figure 8.
Cytokine concentrations in BALF collected from WT and Htr4−/− mice 21 d after treatment with saline or bleomycin (1.75 mg/kg; n ≥ 6/group in saline; n ≥ 8/group in bleomycin).
Allergic airway inflammation in Htr4−/− mice
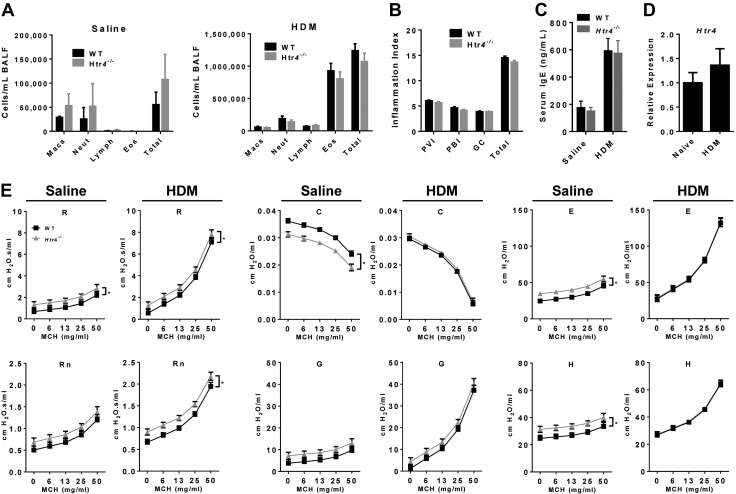
We examined the responses of Htr4−/− and WT mice to acute allergen challenge with the environmentally relevant allergen HDM. As expected, HDM treatment dramatically induced eosinophilia in BALF compared with saline-treated controls (Fig. 9A), indicating a robust type II response. However, there were no genotype differences, suggesting HTR4 does not play a significant role in this model of adaptive immunity. Similarly, there were no genotype differences on pathologic examination of inflammation (Fig. 9B) or in total serum IgE levels (Fig. 9C). We also found no evidence of subepithelial fibrosis after HDM in either genotype (data not shown). The expression of Htr4 was unchanged in HDM-treated WT mice (Fig. 9D). We assessed the levels of multiple cytokines involved in the modulation of allergic airway inflammation. HDM treatment induced changes in levels of type I and type II cytokines but without variation by genotype (Supplemental Figs. 3 and 4).
Figure 9.
A) Cell differentials of saline- and HDM-treated WT and Htr4−/− mice collected from BALF (n = 17/group). B) Histopathological scoring of HDM-treated WT and Htr4−/− mice for PVI (0–8), PBI (0–8), goblet cell hyperplasia (GC, 0–4), and the combined total (0–20; n = 17/group). C) Total serum IgE levels in saline- (n = 6/genotype) or HDM-treated WT and Htr4−/− mice (n = 18/genotype). D) Expression of Htr4−/− in whole lungs of WT mice after treatment with HDM (n = 6/12 for naïve/HDM). E) Assessment of MCH-induced AHR in saline- vs. HDM-treated WT and Htr4−/− mice. Whole lung resistance (R), Newtonian resistance (Rn), tissue resistance (G), compliance (C), elastance (E), and tissue elastance (H) were measured with flexiVent. Mean values per group with sem are plotted (n = 6/genotype for saline and n = 17/genotype for HDM, *P < 0.05).
We examined whether HTR4-deficient mice respond to HDM-induced allergic airway inflammation differently than their WT counterparts. Treatment with HDM induced expected increases in AHR (Fig. 9E). After HDM treatment, Htr4−/− mice had higher AHR in whole lung resistance (R) and Newtonian resistance (Rn) parameters, but this is similar to the baseline phenotype (Figs. 2 and 3) and that seen in the saline-treated HDM control mice (Fig. 9E).
DISCUSSION
HTR4 is a member of the 5-HT receptor family and is expressed in the lung (26). Prior research on HTR4 focused on its expression and roles in the brain and gastrointestinal tract. Because of recent, well-replicated human GWAS findings that variants in HTR4 are related to both baseline lung function and airflow obstruction, we conducted follow-up research in mouse models. None of the HTR4 human GWASs of lung function-associated SNPs have clear functional relevance, and thus experimental mouse models can shed light on the causality of the observed associations. We therefore measured lung function in WT and Htr4−/− mice at baseline, after treatment with 2 broncho-constrictors (MCH and 5-HT), and after exposure to bacterial LPS and bleomycin.
When we examined expression of the Htr4 transcripts, we confirmed that the targeted region was absent in Htr4−/− mice; however, we also found in-frame downstream message in Htr4−/− mice. Therefore, to verify that the Htr4−/− mice were truly deficient in functional HTR4, we conducted a breathing assay. On the basis of work done by Manzke et al. that opiate-induced respiratory depression in rats could be rescued with BIMU8, an HTR4-specific agonist (48), we hypothesized that Htr4−/− mice would fail to respond to BIMU8 after opiate administration. We showed that not only do Htr4−/− mice fail to respond to selective HTR4 agonism, but they also respond with greater opiate-induced respiratory depression than their WT counterparts. Thus, we confirmed that the B6.129P2-Htr4tm1Dgen/J mice that we studied are functionally HTR4 deficient.
Our discovery phenotype in human GWAS was baseline lung function. After ensuring that Htr4−/− mice were functionally deficient in HTR4, we tested our hypothesis that baseline lung function would be altered in Htr4−/− mice relative to WT. We found this to indeed be the case, providing confirmation for the human GWAS findings on baseline lung function. In human GWASs, the sentinel SNP (rs11168048) in HTR4 was negatively associated with baseline lung function (8, 26). In mice, deficiency of HTR4 was related to impaired baseline lung function as exhibited by increased whole lung and conducting airway resistance (R and Rn, respectively).
It is well known that 5-HT is a broncho-constrictor in humans and mice. Pharmacological experiments in rodents suggest that HTR4 does not play a role in 5-HT–induced broncho-constriction (49, 50). Given we had an HTR4-deficient mouse, we were able to confirm that HTR4 was not responsible for the 5-HT–induced broncho-constriction response. Indeed, we found that Htr4−/− mice exhibited greater sensitivity to broncho-constriction induced by both 5-HT and MCH. These data suggest that Htr4−/− mice are primed for AHR to exogenous and endogenous broncho-constrictors. It is possible that increased AHR over time could contribute to decreased lung function, consistent with the finding from human longitudinal studies that AHR is an independent risk factor for accelerated decline in pulmonary function (51) and development of respiratory symptoms (52, 53).
Given recent work by Hodge et al. (37) that suggests HTR4 may play a developmental role in the lung, combined with our findings of baseline pulmonary function differences in Htr4−/− mice, we examined lung size and architecture in age-matched male Htr4−/− and WT mice. There were no discernible differences between untreated Htr4−/− and WT mice with regard to lung volume (an indicator of size), polarized gas imaging, or histopathological examination (data not shown). It is not surprising we were unable to identify structural differences between genotype in our mice; in humans, modest variation in pulmonary function parameters within the normal range is not related to discernible structural differences in the lung or airways.
Work by Bayer et al. demonstrated that activation of HTR4 in human airway epithelial cell lines regulates release of IL-6 (34). We did not find that Htr4−/− mice had altered IL-6 in either serum or BALF at baseline or after treatment with either LPS or bleomycin. This may be due to compensatory effects during development or off-target pharmacological effects.
Exposing humans to injurious agents is difficult; however, in mice, airway perturbation models can be used to examine changes in responses to agents involved in lung injury and fibrosis. Endotoxin or LPS, a component of gram-negative bacterial cell membranes, can elicit an acute immune response in mammals. Given that untreated Htr4−/− mice had increased macrophages in BALF, we investigated whether Htr4−/− and WT mice would exhibit differences in acute inflammatory responses to LPS. With the exception of mitigation of IL-1β induction after LPS, we did not observe any genotype differences in response to LPS with respect to other cytokines or immune cell responses. This suggests that the immune responses to LPS were largely unaffected by HTR4 disruption. Furthermore, we did not observe any differences in lung function in LPS treated Htr4−/− mice. Collectively, these findings suggest that HTR4 does not play a significant role in response to LPS.
To examine whether WT and Htr4−/− mice respond differently to fibrotic injury, we treated mice with bleomycin. Bleomycin is a chemotherapeutic agent that causes pulmonary fibrosis in humans and mice. In mice, bleomycin administration, regardless of route, results in acute pulmonary epithelial injury. Repair of this injury results in collagen deposition and fibrosis. Unlike in humans, where fibrosis is irreversible, mice can repair the fibrotic lesions. Similar to LPS, we found little difference between WT and Htr4−/− mice in their ability to resolve fibrotic injury. Htr4−/− mice did have slightly higher levels of lymphocytes and total cells from lavage, but we found no differences in bleomycin injury-related cytokines. Thus, the biologic relevance of the differences in cell responses is not clear. These data suggest that repair of fibrotic injury is not dependent on HTR4.
Serotonergic receptors have been associated with asthma in human airway epithelial cells, and Bayer et al. demonstrated that activation of HTR4 in human lung cell lines could modulate the release of IL-6 (34). There is only 1 study relating HTR4 SNPs with asthma (28), and HTR4 has not been related to asthma in GWASs. We did not observe any differences by genotype in release of IL-6 or other proinflammatory cytokines after HDM treatment. The genotype differences we observed in whole lung resistance (R) and conducting airway resistance (Rn) after treatment with HDM are very similar to the differences by genotype in naive and saline-treated HDM control mice. The fact that we saw no substantive differences by genotype in 22 different cytokines examined in BALF and serum after HDM-induced allergic airway inflammation further suggests that differences in lung function at baseline and after HDM are not likely due to immune system modulation.
HTR4 is heavily expressed in neurons of the pre-Bötzinger complex, a region in the brainstem responsible for rhythmogenesis of inspiration and expiration in mammals. Our functional breathing assay replicated work done in rat (48) showing HTR4 agonism could overcome opiate-induced respiratory depression. We surmised that differences in baseline lung function might result from persistent perturbations in control of breathing due to the absence of functional HTR4 in the pre-Bötzinger complex. To test whether HTR4 was related to control of breathing, we conducted whole body plethysmography on WT and Htr4−/− mice; however, we did not find any differences by genotype in baseline control of breathing parameters (data not shown).
In summary, we found Htr4−/− mice have altered baseline lung function (increased baseline lung resistance) and exhibit increased AHR to both endogenous and exogenous broncho-constrictors. These data recapitulate human findings from GWASs, lending evidence for a causal relationship between lung function and HTR4. This is important because, although human GWASs established an association of HTR4 SNPs with lung function, the HTR4 SNPs most strongly related to lung function were not clearly functional. We did not find that HTR4 deficiency leads to altered responses to either bacterial LPS or bleomycin. Combined with published work showing no difference in response to cigarette smoke between WT and Htr4−/− mice (54), the evidence suggests that the differences in baseline lung function do not reflect differential susceptibility to these common environmental exposures. Our work showing that WT and Htr4−/− mice differ in baseline lung function, combined with recent work showing HTR4 is temporally expressed in the developing human lung (37), suggest a possible role for HTR4 in lung development. In conclusion, we present here strong evidence for a causal relationship between HTR4 and baseline lung function in mice that bolsters human GWAS findings.
Supplementary Material
Acknowledgments
The authors thank Spencer Lacy, J. Alyce Bradbury, Grace Kissling, Artiom Gruzdev, Rohan Virgincar, Matthew Freeman, and the Center for In Vivo Microscopy (National Institute of Biomedical Imaging and Bioengineering; P41 EB015897) for help and assistance. This work was supported with funds from the Intramural Research Program of the U.S. National Institutes of Health, National Institutes of Environmental Health Sciences (Grants Z01 025043, Z01 ES025045, and Z01 ES043012). The authors declare no conflicts of interest.
Glossary
- 5-HT
5-hydroxytryptamine (serotonin)
- AHR
airway hyperresponsiveness
- BALF
bronchoalveolar lavage fluid
- COPD
chronic obstructive pulmonary disease
- FEV1
forced expiratory volume (in first second)
- FVC
forced vital capacity
- GWAS
genome-wide association study
- HDM
house dust mite
- HTR4
serotonin receptor 4
- MCH
methacholine
- OPA
oropharyngeal aspiration
- PBI
peribronchal inflammation
- PC200
provocative concentration 200
- PC50
provocative concentration 50
- PVI
perivascular inflammation
- SNP
single nucleotide polymorphism
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
References
- 1.Rybicki B. A., Beaty T. H., Cohen B. H. (1990) Major genetic mechanisms in pulmonary function. J. Clin. Epidemiol. 43, 667–675 [DOI] [PubMed] [Google Scholar]
- 2.Wilk J. B., Djousse L., Arnett D. K., Rich S. S., Province M. A., Hunt S. C., Crapo R. O., Higgins M., Myers R. H. (2000) Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet. Epidemiol. 19, 81–94 [DOI] [PubMed] [Google Scholar]
- 3.DeMeo D. L., Silverman E. K. (2003) Genetics of chronic obstructive pulmonary disease. Semin. Respir. Crit. Care Med. 24, 151–160 [DOI] [PubMed] [Google Scholar]
- 4.Cooper D. M., Hoeppner V., Cox D., Zamel N., Bryan A. C., Levison H. (1974) Lung function in alpha1-antitrypsin heterozygotes (Pi type MZ). Am. Rev. Respir. Dis. 110, 708–715 [DOI] [PubMed] [Google Scholar]
- 5.Silverman E. K., Sandhaus R. A. (2009) Clinical practice. Alpha1-antitrypsin deficiency. N. Engl. J. Med. 360, 2749–2757 [DOI] [PubMed] [Google Scholar]
- 6.Wilk J. B., Chen T. H., Gottlieb D. J., Walter R. E., Nagle M. W., Brandler B. J., Myers R. H., Borecki I. B., Silverman E. K., Weiss S. T., O’Connor G. T. (2009) A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 5, e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repapi E., Sayers I., Wain L. V., Burton P. R., Johnson T., Obeidat M., Zhao J. H., Ramasamy A., Zhai G., Vitart V., Huffman J. E., Igl W., Albrecht E., Deloukas P., Henderson J., Granell R., McArdle W. L., Rudnicka A. R., Barroso I., Loos R. J., Wareham N. J., Mustelin L., Rantanen T., Surakka I., Imboden M., Wichmann H. E., Grkovic I., Jankovic S., Zgaga L., Hartikainen A. L., Peltonen L., Gyllensten U., Johansson A., Zaboli G., Campbell H., Wild S. H., Wilson J. F., Gläser S., Homuth G., Völzke H., Mangino M., Soranzo N., Spector T. D., Polasek O., Rudan I., Wright A. F., Heliövaara M., Ripatti S., Pouta A., Naluai A. T., Olin A. C., Torén K., Cooper M. N., James A. L., Palmer L. J., Hingorani A. D., Wannamethee S. G., Whincup P. H., Smith G. D., Ebrahim S., McKeever T. M., Pavord I. D., MacLeod A. K., Morris A. D., Porteous D. J., Cooper C., Dennison E., Shaheen S., Karrasch S., Schnabel E., Schulz H., Grallert H., Bouatia-Naji N., Delplanque J., Froguel P., Blakey J. D., Holloway J. W., Lawlor D. A., Hui J., Nyberg F., Jarvelin M. R., Jackson C., Kähönen M., Kaprio J., Probst-Hensch N. M., Koch B., Hayward C., Evans D. M., Elliott P., Strachan D. P., Hall I. P., Tobin M. D.. Wellcome Trust Case Control Consortium, NSHD Respiratory Study Team. (2010) Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock D. B., Eijgelsheim M., Wilk J. B., Gharib S. A., Loehr L. R., Marciante K. D., Franceschini N., van Durme Y. M., Chen T. H., Barr R. G., Schabath M. B., Couper D. J., Brusselle G. G., Psaty B. M., van Duijn C. M., Rotter J. I., Uitterlinden A. G., Hofman A., Punjabi N. M., Rivadeneira F., Morrison A. C., Enright P. L., North K. E., Heckbert S. R., Lumley T., Stricker B. H., O'Connor G. T., London S. J. (2010) Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imboden M., Bouzigon E., Curjuric I., Ramasamy A., Kumar A., Hancock D. B., Wilk J. B., Vonk J. M., Thun G. A., Siroux V., Nadif R., Monier F., Gonzalez J. R., Wjst M., Heinrich J., Loehr L. R., Franceschini N., North K. E., Altmüller J., Koppelman G. H., Guerra S., Kronenberg F., Lathrop M., Moffatt M. F., O'Connor G. T., Strachan D. P., Postma D. S., London S. J., Schindler C., Kogevinas M., Kauffmann F., Jarvis D. L., Demenais F., Probst-Hensch N. M. (2012) Genome-wide association study of lung function decline in adults with and without asthma. J. Allergy Clin. Immunol. 129, 1218–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loth D. W., Artigas M. S., Gharib S. A., Wain L. V., Franceschini N., Koch B., Pottinger T. D., Smith A. V., Duan Q., Oldmeadow C., Lee M. K., Strachan D. P., James A. L., Huffman J. E., Vitart V., Ramasamy A., Wareham N. J., Kaprio J., Wang X. Q., Trochet H., Kähönen M., Flexeder C., Albrecht E., Lopez L. M., de Jong K., Thyagarajan B., Alves A. C., Enroth S., Omenaas E., Joshi P. K., Fall T., Viñuela A., Launer L. J., Loehr L. R., Fornage M., Li G., Wilk J. B., Tang W., Manichaikul A., Lahousse L., Harris T. B., North K. E., Rudnicka A. R., Hui J., Gu X., Lumley T., Wright A. F., Hastie N. D., Campbell S., Kumar R., Pin I., Scott R. A., Pietiläinen K. H., Surakka I., Liu Y., Holliday E. G., Schulz H., Heinrich J., Davies G., Vonk J. M., Wojczynski M., Pouta A., Johansson A., Wild S. H., Ingelsson E., Rivadeneira F., Völzke H., Hysi P. G., Eiriksdottir G., Morrison A. C., Rotter J. I., Gao W., Postma D. S., White W. B., Rich S. S., Hofman A., Aspelund T., Couper D., Smith L. J., Psaty B. M., Lohman K., Burchard E. G., Uitterlinden A. G., Garcia M., Joubert B. R., McArdle W. L., Musk A. B., Hansel N., Heckbert S. R., Zgaga L., van Meurs J. B., Navarro P., Rudan I., Oh Y. M., Redline S., Jarvis D. L., Zhao J. H., Rantanen T., O'Connor G. T., Ripatti S., Scott R. J., Karrasch S., Grallert H., Gaddis N. C., Starr J. M., Wijmenga C., Minster R. L., Lederer D. J., Pekkanen J., Gyllensten U., Campbell H., Morris A. P., Gläser S., Hammond C. J., Burkart K. M., Beilby J., Kritchevsky S. B., Gudnason V., Hancock D. B., Williams O. D., Polasek O., Zemunik T., Kolcic I., Petrini M. F., Wjst M., Kim W. J., Porteous D. J., Scotland G., Smith B. H., Viljanen A., Heliövaara M., Attia J. R., Sayers I., Hampel R., Gieger C., Deary I. J., Boezen H. M., Newman A., Jarvelin M. R., Wilson J. F., Lind L., Stricker B. H., Teumer A., Spector T. D., Melén E., Peters M. J., Lange L. A., Barr R. G., Bracke K. R., Verhamme F. M., Sung J., Hiemstra P. S., Cassano P. A., Sood A., Hayward C., Dupuis J., Hall I. P., Brusselle G. G., Tobin M. D., London S. J. (2014) Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat. Genet. 46, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M. H., Boutaoui N., Klanderman B. J., Sylvia J. S., Ziniti J. P., Hersh C. P., DeMeo D. L., Hunninghake G. M., Litonjua A. A., Sparrow D., Lange C., Won S., Murphy J. R., Beaty T. H., Regan E. A., Make B. J., Hokanson J. E., Crapo J. D., Kong X., Anderson W. H., Tal-Singer R., Lomas D. A., Bakke P., Gulsvik A., Pillai S. G., Silverman E. K. (2010) Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M. H., Castaldi P. J., Wan E. S., Siedlinski M., Hersh C. P., Demeo D. L., Himes B. E., Sylvia J. S., Klanderman B. J., Ziniti J. P., Lange C., Litonjua A. A., Sparrow D., Regan E. A., Make B. J., Hokanson J. E., Murray T., Hetmanski J. B., Pillai S. G., Kong X., Anderson W. H., Tal-Singer R., Lomas D. A., Coxson H. O., Edwards L. D., MacNee W., Vestbo J., Yates J. C., Agusti A., Calverley P. M., Celli B., Crim C., Rennard S., Wouters E., Bakke P., Gulsvik A., Crapo J. D., Beaty T. H., Silverman E. K., ICGN Investigators, ECLIPSE Investigators, COPDGene Investigators. (2012) A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 21, 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D. K., Cho M. H., Hersh C. P., Lomas D. A., Miller B. E., Kong X., Bakke P., Gulsvik A., Agustí A., Wouters E., Celli B., Coxson H., Vestbo J., MacNee W., Yates J. C., Rennard S., Litonjua A., Qiu W., Beaty T. H., Crapo J. D., Riley J. H., Tal-Singer R., Silverman E. K., ECLIPSE, ICGN, and COPDGene Investigators. (2012) Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 186, 1238–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillai S. G., Ge D., Zhu G., Kong X., Shianna K. V., Need A. C., Feng S., Hersh C. P., Bakke P., Gulsvik A., Ruppert A., Lødrup Carlsen K. C., Roses A., Anderson W., Rennard S. I., Lomas D. A., Silverman E. K., Goldstein D. B., ICGN Investigators. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 5, e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soler Artigas M., Wain L. V., Repapi E., Obeidat M., Sayers I., Burton P. R., Johnson T., Zhao J. H., Albrecht E., Dominiczak A. F., Kerr S. M., Smith B. H., Cadby G., Hui J., Palmer L. J., Hingorani A. D., Wannamethee S.G., Whincup P. H., Ebrahim S., Smith G. D., Barroso I., Loos R. J., Wareham N. J., Cooper C., Dennison E., Shaheen S. O., Liu J. Z., Marchini J., Dahgam S., Naluai A. T., Olin A. C., Karrasch S., Heinrich J., Schulz H., McKeever T. M., Pavord I. D., Heliövaara M., Ripatti S., Surakka I., Blakey J. D., Kähönen M., Britton J. R., Nyberg F., Holloway J. W., Lawlor D. A., Morris R. W., James A. L., Jackson C. M., Hall I. P., Tobin M. D., Medical Research Council National Survey of Health and Development (NSHD) Respiratory Study Team, SpiroMeta Consortium. (2011) Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am. J. Respir. Crit. Care Med. 184, 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Y. I., Shrine N. R., Soler Artigas M., Wain L. V., Blakey J. D., Moffatt M. F., Bush A., Chung K. F., Cookson W. O., Strachan D. P., Heaney L., Al-Momani B. A., Mansur A. H., Manney S., Thomson N. C., Chaudhuri R., Brightling C. E., Bafadhel M., Singapuri A., Niven R., Simpson A., Holloway J. W., Howarth P. H., Hui J., Musk A. W., James A. L., Brown M. A., Baltic S., Ferreira M. A., Thompson P. J., Tobin M. D., Sayers I., Hall I. P., Australian Asthma Genetics Consortium. (2012) Genome-wide association study to identify genetic determinants of severe asthma. Thorax 67, 762–768 [DOI] [PubMed] [Google Scholar]
- 17.Torgerson D. G., Ampleford E. J., Chiu G. Y., Gauderman W. J., Gignoux C. R., Graves P. E., Himes B. E., Levin A. M., Mathias R. A., Hancock D. B., Baurley J. W., Eng C., Stern D. A., Celedón J. C., Rafaels N., Capurso D., Conti D. V., Roth L. A., Soto-Quiros M., Togias A., Li, X, Myers R. A., Romieu I., Van Den Berg D. J., Hu D., Hansel N. N., Hernandez R. D., Israel E., Salam M. T., Galanter J., Avila P. C., Avila L., Rodriquez-Santana J. R., Chapela R., Rodriguez-Cintron W., Diette G. B., Adkinson N. F., Abel R. A., Ross K. D., Shi M., Faruque M. U., Dunston G. M., Watson H. R., Mantese V. J., Ezurum S. C., Liang L., Ruczinski I., Ford J. G., Huntsman S., Chung K. F., Vora H., Li X., Calhoun W. J., Castro M., Sienra-Monge J. J., del Rio-Navarro B., Deichmann K. A., Heinzmann A., Wenzel S. E., Busse W. W., Gern J. E., Lemanske R. F. Jr., Beaty T. H., Bleecker E. R., Raby B. A., Meyers D. A., London S. J., Gilliland F. D., Martinez F. D., Weiss S. T., Williams L. K., , Barnes K. C., Ober C., Nicolae D. L., Mexico City Childhood Asthma Study (MCAAS), Children’s Health Study (CHS) and HARBORS study Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE), Childhood Asthma Research and Education (CARE) Network, Childhood Asthma Management Program (CAMP), Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE), Genetic Research on Asthma in African Diaspora (GRAAD) Study. (2011) Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat. Genet. 43, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffatt M. F., Kabesch M., Liang L., Dixon A. L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., Heinzmann A., Simma B., Frischer T., Willis-Owen S. A., Wong K. C., Illig T., Vogelberg C., Weiland S. K., von Mutius E., Abecasis G. R., Farrall M., Gut I. G., Lathrop G. M., Cookson W. O. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 [DOI] [PubMed] [Google Scholar]
- 19.Li X., Hawkins G. A., Ampleford E. J., Moore W. C., Li H., Hastie A. T., Howard T. D., Boushey H. A., Busse W. W., Calhoun W. J., Castro M., Erzurum S. C., Israel E., Lemanske R. F. Jr., Szefler S. J., Wasserman S. I., Wenzel S. E., Peters S. P., Meyers D. A., Bleecker E. R. (2013) Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J. Allergy Clin. Immunol. 132, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes B. E., Hunninghake G. M., Baurley J. W., Rafaels N. M., Sleiman P., Strachan D. P., Wilk J. B., Willis-Owen S. A., Klanderman B., Lasky-Su J., Lazarus R., Murphy A. J., Soto-Quiros M. E., Avila L., Beaty T., Mathias R. A., Ruczinski I., Barnes K. C., Celedón J. C., Cookson W. O., Gauderman W. J., Gilliland F. D., Hakonarson H., Lange C., Moffatt M. F., O'Connor G. T., Raby B. A., Silverman E. K., Weiss S. T. (2009) Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 84, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffatt M. F., Gut I. G., Demenais F., Strachan D. P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W. O., Consortium G.; GABRIEL Consortium (2010) A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirota T., Takahashi A., Kubo M., Tsunoda T., Tomita K., Doi S., Fujita K., Miyatake A., Enomoto T., Miyagawa T., Adachi M., Tanaka H., Niimi A., Matsumoto H., Ito I., Masuko H., Sakamoto T., Hizawa N., Taniguchi M., Lima J. J., Irvin C. G., Peters S. P., Himes B. E., Litonjua A. A., Tantisira K. G., Weiss S. T., Kamatani N., Nakamura Y., Tamari M. (2011) Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 43, 893–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., Shafer A., Neri F., Lee K., Kutyavin T., Stehling-Sun S., Johnson A. K., Canfield T. K., Giste E., Diegel M., Bates D., Hansen R. S., Neph S., Sabo P. J., Heimfeld S., Raubitschek A., Ziegler S., Cotsapas C., Sotoodehnia N., Glass I., Sunyaev S. R., Kaul R., Stamatoyannopoulos J. A. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards S. L., Beesley J., French J. D., Dunning A. M. (2013) Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X., Fuller C. K., Song Y., Meng Q., Zhang B., Yang X., Li H. (2013) Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am. J. Hum. Genet. 92, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soler Artigas M., Loth D. W., Wain L. V., Gharib S. A., Obeidat M., Tang W., Zhai G., Zhao J. H., Smith A. V., Huffman J. E., Albrecht E., Jackson C. M., Evans D. M., Cadby G., Fornage M., Manichaikul A., Lopez L. M., Johnson T., Aldrich M. C., Aspelund T., Barroso I., Campbell H., Cassano P. A., Couper D. J., Eiriksdottir G., Franceschini N., Garcia M., Gieger C., Gislason G. K., Grkovic I., Hammond C. J., Hancock D. B., Harris T. B., Ramasamy A., Heckbert S. R., Heliovaara M., Homuth G., Hysi P. G., James A. L., Jankovic S., Joubert B. R., Karrasch S., Klopp N., Koch B., Kritchevsky S. B., Launer L. J., Liu Y., Loehr L. R., Lohman K., Loos R. J., Lumley T., Al Balushi K. A., Ang W. Q., Barr R. G., Beilby J., Blakey J. D., Boban M., Boraska V., Brisman J., Britton J. R., Brusselle G. G., Cooper C., Curjuric I., Dahgam S., Deary I. J., Ebrahim S., Eijgelsheim M., Francks C., Gaysina D., Granell R., Gu X., Hankinson J. L., Hardy R., Harris S. E., Henderson J., Henry A., Hingorani A. D., Hofman A., Holt P. G., Hui J., Hunter M. L., Imboden M., Jameson K. A., Kerr S. M., Kolcic I., Kronenberg F., Liu J. Z., Marchini J., McKeever T., Morris A. D., Olin A. C., Porteous D. J., Postma D. S., Rich S. S., Ring S. M., Rivadeneira F., Rochat T., Sayer A. A., Sayers I., Sly P. D., Smith G. D., Sood A., Starr J. M., Uitterlinden A. G., Vonk J. M., Wannamethee S. G., Whincup P. H., Wijmenga C., Williams O. D., Wong A., Mangino M., Marciante K. D., McArdle W. L., Meibohm B., Morrison A. C., North K. E., Omenaas E., Palmer L. J., Pietilainen K. H., Pin I., Pola Sbreve Ek O., Pouta A., Psaty B. M., Hartikainen A. L., Rantanen T., Ripatti S., Rotter J. I., Rudan I., Rudnicka A. R., Schulz H., Shin S. Y., Spector T. D., Surakka I., Vitart V., Volzke H., Wareham N. J., Warrington N. M., Wichmann H. E., Wild S. H., Wilk J. B., Wjst M., Wright A. F., Zgaga L., Zemunik T., Pennell C. E., Nyberg F., Kuh D., Holloway J. W., Boezen H. M., Lawlor D. A., Morris R. W., Probst-Hensch N., International Lung Cancer C., consortium G., Kaprio J., Wilson J. F., Hayward C., Kahonen M., Heinrich J., Musk A. W., Jarvis D. L., Glaser S., Jarvelin M. R., Ch Stricker B. H., Elliott P., O'Connor G. T., Strachan D. P., London S. J., Hall I. P., Gudnason V., Tobin M. D. (2011) Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 43, 1082–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilk J. B., Shrine N. R., Loehr L. R., Zhao J. H., Manichaikul A., Lopez L. M., Smith A. V., Heckbert S. R., Smolonska J., Tang W., Loth D. W., Curjuric I., Hui J., Cho M. H., Latourelle J. C., Henry A. P., Aldrich M., Bakke P., Beaty T. H., Bentley A. R., Borecki I. B., Brusselle G. G., Burkart K. M., Chen T. H., Couper D., Crapo J. D., Davies G., Dupuis J., Franceschini N., Gulsvik A., Hancock D. B., Harris T. B., Hofman A., Imboden M., James A. L., Khaw K. T., Lahousse L., Launer L. J., Litonjua A., Liu Y., Lohman K. K., Lomas D. A., Lumley T., Marciante K. D., McArdle W. L., Meibohm B., Morrison A. C., Musk A. W., Myers R. H., North K. E., Postma D. S., Psaty B. M., Rich S. S., Rivadeneira F., Rochat T., Rotter J. I., Artigas M. S., Starr J. M., Uitterlinden A. G., Wareham N. J., Wijmenga C., Zanen P., Province M. A., Silverman E. K., Deary I. J., Palmer L. J., Cassano P. A., Gudnason V., Barr R. G., Loos R. J., Strachan D. P., London S. J., Boezen H. M., Probst-Hensch N., Gharib S. A., Hall I. P., O'Connor G. T., Tobin M. D., Stricker B. H. (2012) Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am. J. Respir. Crit. Care Med. 186, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim T. H., An S. H., Cha J. Y., Shin E. K., Lee J. Y., Yoon S. H., Lee Y. M., Uh S. T., Park S. W., Park J. S., Kim Y. H., Choi J. S., Lee S. O., Park B. L., Shin H. D., Park C. S. (2011) Association of 5-hydroxytryptamine (serotonin) receptor 4 (5-HTR4) gene polymorphisms with asthma. Respirology 16, 630–638 [DOI] [PubMed] [Google Scholar]
- 29.Nichols D. E., Nichols C. D. (2008) Serotonin receptors. Chem. Rev. 108, 1614–1641 [DOI] [PubMed] [Google Scholar]
- 30.Reynolds G. P., Mason S. L., Meldrum A., De Keczer S., Parnes H., Eglen R. M., Wong E. H. (1995) 5-Hydroxytryptamine (5-HT)4 receptors in post mortem human brain tissue: distribution, pharmacology and effects of neurodegenerative diseases. Br. J. Pharmacol. 114, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compan V., Daszuta A., Salin P., Sebben M., Bockaert J., Dumuis A. (1996) Lesion study of the distribution of serotonin 5-HT4 receptors in rat basal ganglia and hippocampus. Eur. J. Neurosci. 8, 2591–2598 [DOI] [PubMed] [Google Scholar]
- 32.Hegde S. S., Eglen R. M. (1996) Peripheral 5-HT4 receptors. FASEB J. 10, 1398–1407 [DOI] [PubMed] [Google Scholar]
- 33.Berger M., Gray J. A., Roth B. L. (2009) The expanded biology of serotonin. Annu. Rev. Med. 60, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayer H., Müller T., Myrtek D., Sorichter S., Ziegenhagen M., Norgauer J., Zissel G., Idzko M. (2007) Serotoninergic receptors on human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 36, 85–93 [DOI] [PubMed] [Google Scholar]
- 35.Einstein R., Jordan H., Zhou W., Brenner M., Moses E. G., Liggett S. B. (2008) Alternative splicing of the G protein-coupled receptor superfamily in human airway smooth muscle diversifies the complement of receptors. Proc. Natl. Acad. Sci. USA 105, 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azim S., Banday A. R., Tabish M. (2012) Identification of alternatively spliced multiple transcripts of 5-hydroxytryptamine receptor in mouse. Brain Res. Bull. 87, 250–258 [DOI] [PubMed] [Google Scholar]
- 37.Hodge E., Nelson C. P., Miller S., Billington C. K., Stewart C. E., Swan C., Malarstig A., Henry A. P., Gowland C., Melén E., Hall I. P., Sayers I. (2013) HTR4 gene structure and altered expression in the developing lung. Respir. Res. 14, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segu L., Lecomte M. J., Wolff M., Santamaria J., Hen R., Dumuis A., Berrard S., Bockaert J., Buhot M. C., Compan V. (2010) Hyperfunction of muscarinic receptor maintains long-term memory in 5-HT4 receptor knock-out mice. PLoS ONE 5, e9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jean A., Conductier G., Manrique C., Bouras C., Berta P., Hen R., Charnay Y., Bockaert J., Compan V. (2007) Anorexia induced by activation of serotonin 5-HT4 receptors is mediated by increases in CART in the nucleus accumbens. Proc. Natl. Acad. Sci. USA 104, 16335–16340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conductier G., Dusticier N., Lucas G., Côté F., Debonnel G., Daszuta A., Dumuis A., Nieoullon A., Hen R., Bockaert J., Compan V. (2006) Adaptive changes in serotonin neurons of the raphe nuclei in 5-HT(4) receptor knock-out mouse. Eur. J. Neurosci. 24, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 41.Compan V., Charnay Y., Dusticier N., Daszuta A., Hen R., Bockaert J. (2004) [Feeding disorders in 5-HT4 receptor knockout mice]. J. Soc. Biol. 198, 37–49 [PubMed] [Google Scholar]
- 42.Card J. W., Carey M. A., Bradbury J. A., DeGraff L. M., Morgan D. L., Moorman M. P., Flake G. P., Zeldin D. C. (2006) Gender differences in murine airway responsiveness and lipopolysaccharide-induced inflammation. J. Immunol. 177, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dackor R. T., Cheng J., Voltz J. W., Card J. W., Ferguson C. D., Garrett R. C., Bradbury J. A., DeGraff L. M., Lih F. B., Tomer K. B., Flake G. P., Travlos G. S., Ramsey R. W. Jr., Edin M. L., Morgan D. L., Zeldin D. C. (2011) Prostaglandin E₂ protects murine lungs from bleomycin-induced pulmonary fibrosis and lung dysfunction. Am. J. Physiol. Lung Cell. Mol. Physiol. 301, L645–L655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashcroft T., Simpson J. M., Timbrell V. (1988) Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 41, 467–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones I. W., Wonnacott S. (2005) Why doesn’t nicotinic ACh receptor immunoreactivity knock out? Trends Neurosci. 28, 343–345 [DOI] [PubMed] [Google Scholar]
- 46.Abramowski D., Staufenbiel M. (1995) Identification of the 5-hydroxytryptamine2C receptor as a 60-kDa N-glycosylated protein in choroid plexus and hippocampus. J. Neurochem. 65, 782–790 [DOI] [PubMed] [Google Scholar]
- 47.Bockaert J., Claeysen S., Compan V., Dumuis A. (2004) 5-HT4 receptors. Curr. Drug Targets CNS Neurol. Disord. 3, 39–51 [DOI] [PubMed] [Google Scholar]
- 48.Manzke T., Guenther U., Ponimaskin E. G., Haller M., Dutschmann M., Schwarzacher S., Richter D. W. (2003) 5-HT4(a) receptors avert opioid-induced breathing depression without loss of analgesia. Science 301, 226–229 [DOI] [PubMed] [Google Scholar]
- 49.Buckner C. K., Dea D., Liberati N., Krell R. D. (1991) A pharmacologic examination of receptors mediating serotonin-induced bronchoconstriction in the anesthetized guinea pig. J. Pharmacol. Exp. Ther. 257, 26–34 [PubMed] [Google Scholar]
- 50.Martin T. R., Cohen M. L., Drazen J. M. (1994) Serotonin-induced pulmonary responses are mediated by the 5-HT2 receptor in the mouse. J. Pharmacol. Exp. Ther. 268, 104–109 [PubMed] [Google Scholar]
- 51.Rijcken B., Schouten J. P., Xu X., Rosner B., Weiss S. T. (1995) Airway hyperresponsiveness to histamine associated with accelerated decline in FEV1. Am. J. Respir. Crit. Care Med. 151, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 52.Vestbo J., Prescott E. (1998) Update on the “Dutch hypothesis” for chronic respiratory disease. Thorax 53(Suppl 2), S15–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hospers J. J., Postma D. S., Rijcken B., Weiss S. T., Schouten J. P. (2000) Histamine airway hyper-responsiveness and mortality from chronic obstructive pulmonary disease: a cohort study. Lancet 356, 1313–1317 [DOI] [PubMed] [Google Scholar]
- 54.Dupont L. L., Bracke K. R., De Maeyer J. H., Compan V., Joos G. F., Lefebvre R. A., Brusselle G. G. (2014) Investigation of 5-HT receptors in bronchial hyperresponsiveness in cigarette smoke-exposed mice. Pulmonary Pharmacol. Therap. 28, 60–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.