Abstract
High biomass crops have recently attracted significant attention as an alternative platform for the renewable production of high energy storage lipids such as triacylglycerol (TAG). While TAG typically accumulates in seeds as storage compounds fuelling subsequent germination, levels in vegetative tissues are generally low. Here, we report the accumulation of more than 15% TAG (17.7% total lipids) by dry weight in Nicotiana tabacum (tobacco) leaves by the co-expression of three genes involved in different aspects of TAG production without severely impacting plant development. These yields far exceed the levels found in wild-type leaf tissue as well as previously reported engineered TAG yields in vegetative tissues of Arabidopsis thaliana and N. tabacum. When translated to a high biomass crop, the current levels would translate to an oil yield per hectare that exceeds those of most cultivated oilseed crops. Confocal fluorescence microscopy and mass spectrometry imaging confirmed the accumulation of TAG within leaf mesophyll cells. In addition, we explored the applicability of several existing oil-processing methods using fresh leaf tissue. Our results demonstrate the technical feasibility of a vegetative plant oil production platform and provide for a step change in the bioenergy landscape, opening new prospects for sustainable food, high energy forage, biofuel and biomaterial applications.
Keywords: triacylglycerol, Nicotiana tabacum, leaf, WRI1, DGAT1, oleosin
Introduction
Worldwide demand for vegetable oil is projected to double within the next thirty years due to increasing food, fuel and industrial requirements (FAO, 2003). Limitations on arable land mean that it will be difficult to meet this additional demand with current oilseed crops. High biomass crops engineered for increased lipid content in vegetative tissues such as leaves have been proposed as a novel platform for meeting global production needs for low-cost, energy-dense lipids such as triacylglycerol (TAG; Carlsson et al., 2011; Chapman et al., 2013; Dyer et al., 2012; Ohlrogge and Chapman, 2011; Troncoso-Ponce et al., 2013). The accumulation of TAG to levels above 10% on a dry weight basis in a dedicated energy crop has been postulated to increase the energy yield in such a crop by at least 30% (Ohlrogge and Chapman, 2011). Current metabolic engineering attempts, however, have only achieved low levels of storage lipid accumulation in nonseed plant tissues far below this benchmark. This can largely be attributed to the fact that single-gene strategies have been the focus of most studies thus far (Figure1a). Examples include the overexpression of DGAT or MGAT genes in leaf tissue (Andrianov et al., 2010; Bouvier-Nave et al., 2000; Petrie et al., 2012b; Sanjaya et al., 2013), reducing TAG and fatty acid turnover (James et al., 2010; Kelly et al., 2013; Slocombe et al., 2009), overexpression of the acetyl-CoA carboxylase gene (ACCase) in potato tubers (Klaus et al., 2004), ectopic expression of transcription factors (LEC1, LEC2, WRI1) that regulate seed development and maturation processes (Mu et al., 2008; Sanjaya et al., 2011; Santos Mendoza et al., 2005; Stone et al., 2008), blocking the lipid transfer between the endoplasmic reticulum and plastid (Xu et al., 2005) as well as down-regulating metabolic pathways that compete for available carbon (Sanjaya et al., 2011).
Figure 1.
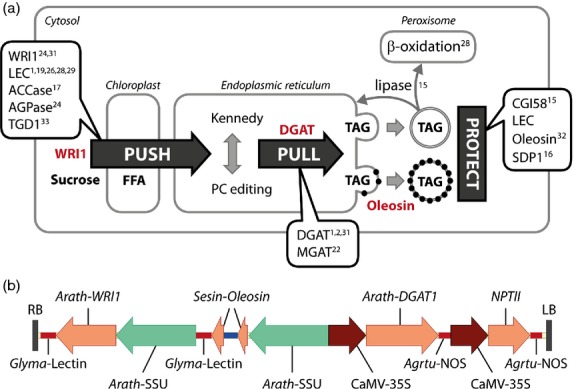
Increasing triacylglycerol (TAG) levels in leaf tissue by an integrated metabolic engineering approach. (a) Fatty acids are produced and exported from the chloroplast (Push) into the endoplasmic reticulum where they are assembled into TAG (Pull). TAG accumulates either as oil droplets or oleosin-coated oil bodies (Protect). Previously tested genes for each approach are listed and referenced. (b) Map of the binary T-DNA region used to co-express the Arabidopsis thaliana (Arath) WRI1, A. thaliana DGAT1 and Sesamum indicum (Sesin) OLEOSIN genes. Red lines show the terminator regions; promoter regions are represented by green and brown arrows; orange arrows indicate the coding regions.
It has become apparent from the above-mentioned studies that achieving industrially relevant levels of storage lipids in vegetative tissues likely requires optimizing the flux of carbon into TAG on multiple metabolic levels including fatty acid synthesis (‘Push’), TAG assembly (‘Pull’) and lipid turnover (‘Protect’). Consequently, some groups have attempted to further increase TAG accumulation in nonseed tissues by modifying the expression of gene pairs. One recent elegant example is the co-expression of a DGAT1 and an engineered oleosin oil-body protein in Arabidopsis thaliana (Winichayakul et al., 2013). Oleosins typically coat and stabilize oil bodies in oilseeds but are normally not expressed in leaf tissues. Interestingly, the authors not only reported improved stability of the lipid droplets but also noted a remarkable increase in biomass. An alternative approach consisting of diverting the flux of carbon from starch-to-lipid biosynthesis by silencing ADP-glucose-pyrophosphorylase (AGPase), while overexpressing the WRI1 transcription factor was reported by Sanjaya et al. (2011). In a third study, Slocombe et al. (2009) overexpressed the LEC2 transcription factor in the A. thaliana pxa1 β-oxidation mutant. While further encouraging increases in TAG yields were obtained in the first two cases, maximum TAG levels were still modest at best and corresponded to 2.1% on a leaf dry weight basis.
We recently investigated the effect of the combined transient overexpression of the A. thaliana WRI1 transcription factor and A. thaliana DGAT1 genes in Nicotiana benthamiana and found a significant synergistic effect on leaf TAG content (Vanhercke et al., 2013). We reasoned that the stable expression of this particular gene combination might result in even greater TAG accumulation due to a longer gene expression window. To reduce the likelihood of the newly synthesized TAG being degraded in a nonseed environment, we decided to also co-express a gene coding for the oleosin oil-body protein from Sesamum indicum (Scott et al., 2010). In a similar manner, Kelly et al. (2013) recently described transgenic A. thaliana plants overexpressing WRI1 and DGAT1 genes in a SUGAR-DEPENDENT1 (sdp1) mutant background. This approach resulted in up to 5% TAG by dry weight in leaf biomass of soil-grown events although a reduction in biomass was also reported. Here, we show that the combined expression of WRI1, DGAT1 and OLEOSIN genes in the broad leaf crop N. tabacum results in dramatically increased TAG yields in transgenic leaf tissue, while total leaf starch content is reduced. Importantly, the accumulation of more than 15% oil in senescing leaves did not have a detrimental effect on plant development and seed viability. Finally, the high levels of storage lipids obtained allowed us to explore the efficiency of several different extraction solvents on fresh leaf tissue.
Results
We designed a T-DNA construct consisting of the WRI1, DGAT1 and OLEOSIN genes placed under the control of the constitutive 35S and RuBisCO small subunit (SSU) promoters (Figure1b). The strong constitutive viral promoter CaMV-35S was selected for DGAT1 expression as this promoter had earlier been successfully used to express the DGAT1 gene resulting in a TAG increase without a significant negative phenotype (Bouvier-Nave et al., 2000; Vanhercke et al., 2013). The A. thaliana SSU promoter was used for the expression of the WRI1 and OLEOSIN genes as it is highly active in leaf, our target tissue. In addition, we hypothesized that, as the SSU promoter follows a circadian expression rhythm (Coruzzi et al., 1984), TAG biosynthesis might only be increased during part of the circadian cycle, thereby reducing possible unwanted metabolic impacts on leaf function and plant development that might be the result of a restriction in photosynthate supply.
Following transformation and antibiotic selection, mature fully expanded green leaves were sampled from primary transformants and wild-type plants at flowering stage. Total lipid extracts were analysed for the presence of TAG by thin-layer chromatography (TLC) for rapid visual screening. Several independent events exhibited more prominent TAG spots than the wild-type control (Figure S1). Subsequent quantification of TAG in senescing leaf tissue in selected lines revealed three transgenic events containing more than 10% leaf TAG on a dry weight basis including line #3, which was shown by Southern blot hybridization analysis to have a single-copy T-DNA insertion (Figure S2). In addition, the highest levels were detected in this particular line, which exhibited 17.1% TAG and 23.4% total lipids (on a dry weight basis) in senescing leaves (Table S1). This line was therefore maintained for progeny analysis. Seeds of this line were viable with oil content and fatty acid profile comparable with wild-type N. tabacum, albeit with a slight increase of 18:1 and decrease of 18:2 (Table S2). The next T1 generation was established directly in soil without screening.
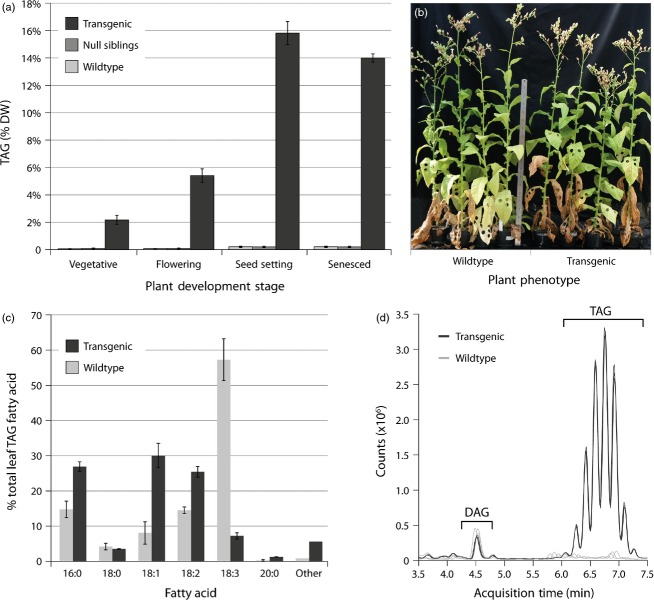
Initial TLC analysis of independent 6-week-old T1 plants identified numerous lines containing elevated TAG levels in young green leaves. To follow the accumulation of oil in the leaves during plant development, we selected three T1 plants with increased leaf TAG levels, three wild-type plants and three null segregant progeny plants exhibiting wild-type-like TAG levels. Samples were taken from fully expanded yellow-green leaves at three different stages during plant development: vegetative stage before flowering, during flowering and at four months after sowing when plants were producing dried seed capsules (seed setting). Both wild-type and null segregant control leaves contained very low TAG levels (<0.2% on a dry weight basis) at all three stages (Figure2a). In leaves of the transgenic plants, TAG contents were high and continued to increase with maturity throughout the three stages with highest levels detected during seed setting. Maximum TAG and total lipids detected in one of the three T1 plants at this stage averaged 15.8% and 17.7%, respectively, on a dry weight basis (Figure2a and Table S3). This corresponded to a 76-fold and 24-fold increase compared to wild-type TAG and total lipids levels, respectively, at a similar stage. Surprisingly, brown desiccated leaves from this particular line still contained 14% TAG on a leaf dry weight basis (Figure2a). Despite a delay in initial seedling establishment compared to the wild-type control, once established, T1 transgenic plants that accumulated high levels of TAG in their leaf tissue did not display any major negative phenotype or impairment throughout their development compared to wild-type and null segregant control plants (Figure2b). Expression of all transgenes in this particular plant was confirmed by RT-PCR and Northern blotting.
Figure 2.
Leaf triacylglycerol (TAG) content, fatty acid profile and phenotype of transgenic Nicotiana tabacum T1 plants. (a) TAG content on a dry weight (DW) in leaves of control (wild-type and null segregant) plants and the transgenic tobacco line exhibiting maximum TAG levels. ‘Vegetative’ stage was harvested 69 days after sowing (DAS), ‘Flowering’ at 104 DAS, ‘Seed Setting’ at 139 DAS and fully ‘Senesced’ leaves at 207 DAS. Error bars represent standard deviations of minimum three samples. (b) Seedling establishment was delayed, but no discernible negative phenotype was observed in mature transgenic T1 events compared to null segregants or wild-type plants. (c) Fatty acid profile of TAG from both transgenic and wild-type tobacco leaves at the onset of senescence. Error bars represent standard deviations (n = 3). (d) Total ion scan of TAG and DAG from triplicate samples, normalized on a dry weight basis, of transgenic and wild-type leaf total lipids by LC-MS. Detailed LC-MS analysis of different PC, DAG and TAG molecular species is shown in Figure S4.
The fatty acid profile of lipids has important implications for the functioning of biological membranes as well as for postharvest applications. We therefore closely examined the TAG fatty acid profile of leaf samples from both wild-type and transgenic plants at the onset of senescence. Gas chromatography (GC) analysis revealed a shift from polyunsaturated towards monounsaturated and saturated fatty acids in the transgenic plants when compared with wild-type (Figure2c). We consistently observed increased proportions of palmitic acid (C16:0), oleic acid (C18:1△9) and linoleic acid (C18:2△9,12) in all high oil transgenic lines, while the proportion of α-linolenic acid (C18:3△9,12,15) was reduced dramatically compared to wild-type leaf tissue. Expression of the genes coding for the Δ15-fatty acid desaturases (FAD) in the plastid (FAD7) and endoplasmic reticulum (FAD3) were found to be down-regulated in one transgenic plant tested (Figure S3). More detailed lipid analysis by liquid-chromatography mass spectrometry (LC-MS) showed increased levels of almost all TAG molecular species in the analysed transgenic plant (Figure2d and Figure S4C). Further identification by MS/MS analysis (Q-TOF) confirmed GC results and revealed major increases in diacylglycerol (DAG) and TAG species composed of C16:0, C18:1 and/or C18:2 acyl chains (Figure S4B and C and Table S4). On the other hand, proportions of both DAG (36:6) and TAG (52:7) molecular species containing one or more C18:3 acyl chains were lower in the transgenic plant compared to the wild-type control, confirming the shift in fatty acid profile as found by GC analysis. Similarly, PC molecular species likely containing polyunsaturated acyl chains were reduced in the transgenic plant, while PC species with fewer double bonds were increased (Figure S4A).
To further investigate oil accumulation during leaf development, we compared TAG levels in young green and mature yellow-green leaf tissue originating from one transgenic line during seed setting. In addition, we quantified TAG in stem and roots to determine whether storage oils also accumulated in these vegetative tissues. TAG amounts were found to be maximal when plants were carrying seed pods and the leaves entered senescence (Figure3a). Both stem and roots exhibited increased TAG levels compared to respective wild-type control samples (Figure3b). TAG contents in both tissues, however, were much lower compared to leaf tissue from the same plant. Interestingly, the TAG fatty acid profile in roots exhibited a similar shift as observed in transgenic leaf tissue (polyunsaturated fatty acids tended to decrease while monounsaturated tended to increase, Table S5). Quantification of total starch in samples taken from young expanding leaves at the end of daylight hours when starch accumulation typically peaks showed a decreased starch content in transgenic leaf tissue (Figure3c). Analysis of surface lipid content and composition on wild-type and transgenic leaf surface also showed some noticeable differences (Figure S5 and Table S6). Total wax load on leaves of T1 transgenic plants was decreased by 35% compared to wild-type control plants. Among the different wax compounds, branched alkanes exhibited the largest reduction by almost 47%.
Figure 3.
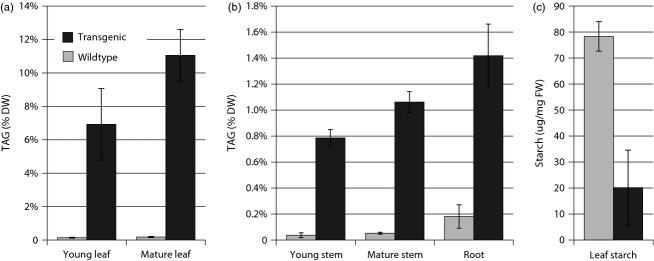
Triacylglycerol (TAG) and starch quantification in wild-type and T1 transgenic tissues. (a) TAG quantification on a dry weight (DW) basis in young green vs. mature leaf at the onset of senescence. (b) TAG quantification on a DW basis in young stem, mature stem and mature root tissues. (c) Leaf starch on a fresh weight (FW) basis in mature leaf tissue. Error bars represent standard deviations (n = 3).
TAG accumulation within transgenic leaf tissue was visualized by two independent methods. Confocal microscopy revealed a large number and increased size of lipid droplets accumulating within mesophyll cells, while wild-type leaves contained very few lipid droplets (Figure4a). MALDI-MS imaging (MSI) analysis showed that TAG content was elevated dramatically in transgenic leaf tissue when normalized to PC content (Figure4c). Total PC content was similar in both tissues (results not shown). Analysis of the spatial distribution of major TAG and PC molecular species by MSI confirmed their distribution throughout the entire leaf cross-section (Figure4d). We observed differences in the composition of the individual TAG and PC molecular species, likely as a result of differences in the age and cross-section positioning within the leaves sampled. Nevertheless, average molecular species compositions visualized in MALDI-MSI were in line with the Q-TOF LC-MS results (results not shown).
Figure 4.
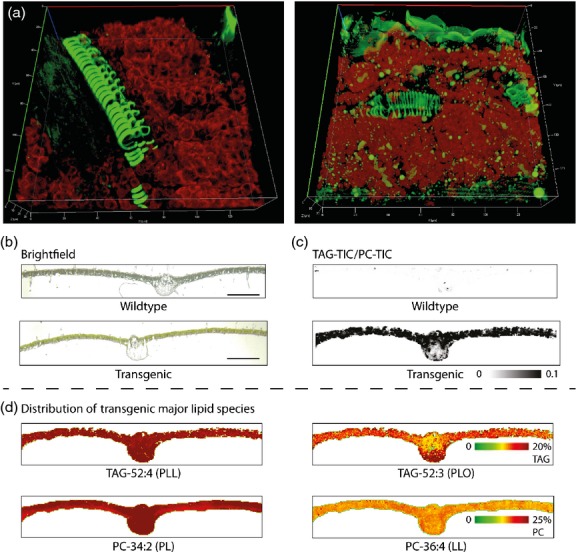
Accumulation of lipid droplets and spatial distribution of different triacylglycerol (TAG) molecular species within transgenic leaf tissue. (a) 3D reconstructed z-stacks of confocal images from wild-type (left) and transgenic (right) leaf tissue. Neutral lipids including TAG, stained with BODIPY, appear as green droplets within the leaf mesophyll cells that contain visible chloroplasts (red autofluorescence). Note that non-TAG features also fluorescing green include the vascular bundle and leaf epidermis. (b) Bright-field microscopy images of wild-type (upper) and transgenic (lower) leaf cross-sections subsequently used for MALDI-Orbitrap imaging. Scale bars correspond to 1000 microns. (c) Ratio of TAG total ion counts (TIC; all TAG molecular species summed at each analysed position) to PC-TIC as detected by MALDI-Orbitrap imaging in wild-type (upper) and transgenic (lower) leaf cross-sections. (d) Spatial distribution of selected dominant TAG and PC species across a transgenic leaf cross-section as observed by MALDI-Orbitrap.
The large quantities of storage oil accumulating in senescing leaves of the transgenic T1 plants prompted us to investigate extraction efficiency using a variety of organic solvents of different polarity. We examined fresh, rather than freeze-dried, leaf tissue given that the highest TAG levels were observed before the leaves had fully senesced. Green leaves were harvested at seed-setting stage, followed by overnight extraction and quantification of different lipid classes by TLC-flame ionization detection (TLC-FID). After the initial extraction, leaf samples were subjected to Bligh and Dyer extraction to quantify residual lipid and calculate overall TAG recovery from fresh leaf tissue. Extraction with hexane resulted in lowest extraction efficiency from fresh leaf due to less polar nature of this solvent and the presence of water in the fresh leaf samples (Figure5). Consequently, a considerable proportion of total leaf lipids could still be extracted following a Bligh and Dyer extraction. Up to 89% of all extractable lipids could be recovered from fresh green leaf tissue in a single extraction round with acetone. Intermediate results were obtained with methanol as the extraction solvent.
Figure 5.
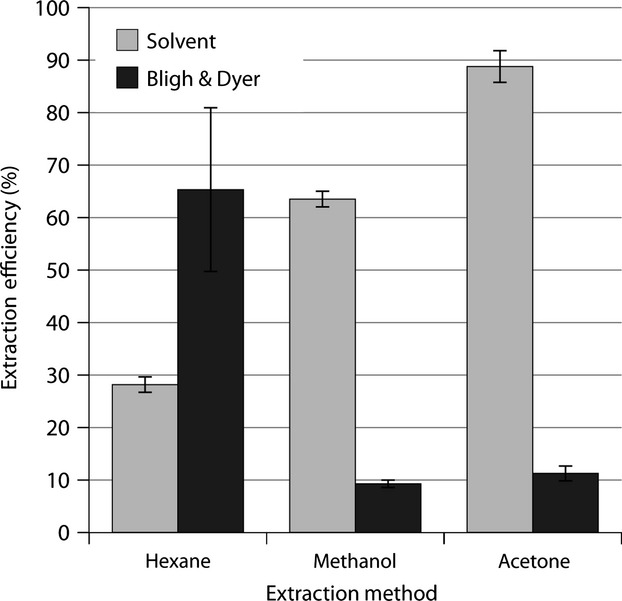
Triacylglycerol (TAG) extraction efficiencies from fresh transgenic leaf tissue obtained with solvents of different polarity. Subsequent Bligh and Dyer extractions were performed on the fresh leaf samples to recover any residual lipid and to determine overall extraction efficiency. Extraction efficiencies are calculated using the formula 100 × % lipid extracted/combined yield following extraction with acetone and subsequent BD. Error bars represent standard deviations (n = 3).
Discussion
Most biotechnological approaches directed at increasing the storage lipid content in seed and vegetative tissues have focused on single genes involved either directly or indirectly in TAG biosynthesis or degradation. In this work, we report the combined overexpression of three genes that play critical roles in fatty acid biosynthesis, neutral lipid assembly and lipid droplet biogenesis. This integrated metabolic engineering strategy resulted in dramatically elevated levels of the storage lipid TAG in N. tabacum leaf tissue. The concomitant increase in total leaf lipids points to a net synthesis of TAG, rather than a mere redistribution between storage and membrane or surface lipid pools. Remarkably, TAG was found to accumulate during plant development with maximum levels detected in mature leaves during seed setting.
Our results differ in two aspects from many earlier findings in A. thaliana. Surprisingly, constitutive overexpression of identical DGAT1 and Oleosin genes in A. thaliana only slightly increased total lipid content in leaf tissue (Winichayakul et al., 2013). However, we previously uncovered a synergistic effect on TAG levels in N. benthamiana infiltrated leaves transiently expressing WRI1 and DGAT1 genes (Vanhercke et al., 2013). It is therefore likely that the expression of both WRI1 and DGAT1 genes is crucial in achieving the current engineered TAG yields in N. tabacum. Secondly, TAG yields in A. thaliana plants expressing a DGAT2 acyltransferase gene were found to be reduced in soil-grown plants when compared to young seedlings (Sanjaya et al., 2013). The combined co-expression of an oil-body protein in our study might explain the accumulation of TAG throughout the plant development as well as the substantial amount of storage lipids detected in completely senesced leaves. Interestingly, increased TAG levels as well as similar changes in fatty acid profile were also detected in roots. These observations are likely to be the result of the 35S-driven constitutive overexpression of the A. thaliana DGAT1. However, currently, we cannot rule out phloem-mediated transport of fatty acids or their ester derivatives from leaf tissue to root organs (Guelette et al., 2012).
To determine whether the accumulation of high levels of storage lipids in leaf tissues has an indirect effect on other major carbon pools, we quantified starch levels in wild-type and transgenic N. tabacum leaf tissue. Transitory starch accumulates in leaf chloroplasts during the day and represents a dominant pool of carbon fixed by photosynthesis. During the night, this photosynthate reserve is converted to sucrose to ensure the supply of reduced carbon for plant development. The accumulation of TAG in transgenic N. tabacum leaf tissue was found to come at the expense of total leaf starch levels that were reduced compared to the wild-type control. This altered partitioning suggests a shift in carbon metabolism resulting in additional carbon being used for fatty acid biosynthesis. Unlike N. tabacum, A. thaliana transgenic events engineered for increased TAG content in vegetative tissues did not display reduced total leaf starch yields. Silencing of the A. thaliana AGPase small subunit in combination with WRI1 overexpression did not affect starch levels further in whole seedling tissue compared to the AGPase silenced control line, while TAG levels were increased (Sanjaya et al., 2011). In addition, overexpression of the DGAT2 yielded increased TAG and starch levels in leaves (Sanjaya et al., 2013). The reduction in starch levels observed in this study may be due to the different gene(s) targeted, our choice of the SSU promoter and of tobacco as a model system. The delay that was observed in seedling establishment was interesting although little difference was noticed on the eventual phenotype of mature plants (Figure2). Further work is needed to fully uncover the molecular, biochemical and physiological mechanisms underlying the observed changes in carbon allocation.
High levels of monounsaturated fatty acids are desirable for both food and biodiesel applications due to improved oxidative stability, ignition quality and cold-temperature flow properties (Rogalski and Carrer, 2011). Detailed GC and LC-MS analysis of the fatty acid composition of transgenic N. tabacum leaf tissue revealed a shift towards acyl chains containing fewer double bonds. This is in line with earlier observations (Andrianov et al., 2010; Bouvier-Nave et al., 2000) upon the stable expression of the A. thaliana DGAT1 in tobacco leaves and could be due in part to the substrate selectivity of DGAT1. Whether the extensive reduction in α-linolenic acid levels in transgenic leaf tissue is due to altered expression of the fatty acid desaturases such as FAD3 and FAD7 and/or a change in the flux of acyl chains between the Kennedy pathway and membrane phospholipids will be the subject for further detailed biochemical studies.
The current TAG levels achieved support the industrial feasibility for harvesting energy-dense oil from plant vegetative biomass at projected yields per hectare that exceed conventional oilseed crops. High biomass yields have been reported for N. tabacum (Andrianov et al., 2010), and the stable production of up to 16% TAG under these conditions would result in oil yields that compare favourably with current oilseed crops. Further improvements in oil yield could be envisaged by combining TAG accumulation in both seed and nonseed tissues of the same plant. One example could be the expression of current leaf oil technology reported here in an elite tobacco cultivar germplasm that has been selected for increased seed oil yields (Fogher, 2008; Fogher et al., 2011). Such a ‘dual purpose’ crop strategy has the potential to result in oil yields that are nearing those of oil palm, currently the highest yielding oil crop. It was encouraging that some existing oil extraction methods were effective and applicable for even fresh leaf material. For instance, acetone which yielded the highest lipid recovery using fresh N. tabacum leaves is a standard method used to extract krill oil which is high in relatively unstable polyunsaturated fatty acids on an industrial scale from biomass with high water content (Gigliotti et al., 2011 and references therein). Extraction of oil from dried tissue (e.g. the senesced leaf in Figure2) would be significantly easier.
This study provides a basis for further optimization of the ‘Push’, ‘Pull’ and ‘Protect’ integrated concept of TAG accumulation by extending the number of target genes (Chapman et al., 2013) and engineering other vegetative tissues such as stems, tubers or roots to serve as novel TAG stores. Up to 8% TAG has recently been demonstrated in A. thaliana root tissue without sucrose supplementation by a similar integrated ‘Push’, ‘Pull’ and ‘Protect’ integrated approach (Kelly et al., 2013). It will be worthwhile to determine whether high TAG levels can be achieved simultaneously in all vegetative tissues, thereby optimally using the entire vegetative biomass for renewable production of storage lipids. The demonstrated engineered high levels of storage oils in vegetative tissues should also enable and motivate further studies of TAG biosynthesis and turnover in nonseed tissues and of the intricate relationship with other carbon pools such as starch. Due to the myriad of applications of a high vegetative oil production platform, including biofuels, food and oleochemical feedstocks, we are confident that the global plant lipid research community will rapidly increase the level of TAG in several application species with fatty acid profiles tailored according to a variety of specific needs.
Experimental procedures
Construct design and N. tabacum transformation
An intron-interrupted S. indicum Oleosin gene was obtained from pRSh1-PSP1, kindly provided by Dr. Nick Roberts (AgResearch, Palmerston North, New Zealand). The coding region for this gene, flanked by NotI, was inserted into a pORE04-based binary expression vector, which contained a double enhancer-region 35S promoter expressing the NPTII kanamycin resistance gene. A codon-optimized DNA fragment encoding the A. thaliana WRI1 gene was subsequently cloned as an EcoRI fragment into the binary expression vector. Finally, the wild-type A. thaliana DGAT1 gene was inserted into the AsiSI site generating pJP3502.
Agrobacterium tumefaciens-mediated transformation of N. tabacum cultivar Wisconsin 38 with the binary vector pJP3502 was essentially carried out as described by Horsch et al. (1985). Shoots were developed on selective ½ MS agar supplemented with 1% sucrose to reduce any abnormal phenotypic effects from the expression of the WRI1 gene (Cernac and Benning, 2004). Plants were grown in the glasshouse without artificial light.
Lipid analyses
Total lipids were isolated from freeze-dried N. tabacum leaf, stem and root material or 1 mg seed as described (Vanhercke et al., 2013). Total extracted lipids were separated by TLC using a hexane/diethyl ether/acetic acid (70:30:1, v:v:v) solvent system. TAG fractions were visualized with primuline, transmethylated into fatty acid methyl esters (FAME) and analysed by GC as described (Vanhercke et al., 2013). For quantification, either 10–15 μg (wild type and null segregants) or 70–100 μg C17:0 TAG was added as internal standard.
For LC-MS analysis, total leaf lipids were extracted as described (Petrie et al., 2012b) and resuspended in 5 µL chloroform per mg dry leaf weight. Lipids from 3 mg dry weight were dried under nitrogen and dissolved in 0.1 mL of 10 mm butylated hydroxytoluene in butanol/methanol (1:1, v:v) and analysed by LC-MS essentially as described (Petrie et al., 2012a). Ammonium adducts of PC, DAG and TAG species with fatty acids C16 to C18 were analysed by selected ion monitoring (SIM). These species were quantified by calculating the ratio of abundance of the individual species to the abundance of a 50 µm tristearin external standard. To identify the fatty acid composition, product ion spectra of selected TAG species were collected by Q-TOF LC-MS. The auto-MS/MS scan function was used and a preferred MS/MS list created containing major TAG species. Product ion spectra were collected when the preferred ion intensity was >20 000 counts using collision energy of 25 V.
Surface lipid analysis
Surface lipids were extracted by immersing fresh leaves for 30 s in chloroform and concentrated under nitrogen. Wax molecular species were separated by TLC using hexane/diethyl ether/acetic acid (90:7.5:1, v:v:v) as a resolving solvent and visualized under UV light after spraying with primuline. Bands corresponding to each class of compound were recovered from TLC plates, extracted twice with 5 mL chloroform and dried under nitrogen. Fatty acids were converted to FAME and fatty alcohols to trimethylsilyl (TMS) derivatives. Samples were analysed by GC and compounds identified using MS or quantified using FID and suitable internal molecular standards.
Microscopy analyses
Freshly harvested tobacco leaves were fixed in 4% paraformaldehyde for 30 min. Following a thorough rinse in 50 mm PIPES buffer (pH 7.2), the fixed leaf tissues were embedded in CRYO-GEL™ embedding medium (Instrumedics, St. Louis, MO). The mounted samples were then frozen at −80 °C for 20 min and kept at −20 °C for 48 h prior to sectioning using a microtome. Sections of 30 μm thickness were laid on a glass slide for microscopy examination.
Glass microscope slides containing N. tabacum leaf sections were incubated with 1 μg/mL solution of BODIPY FL (Life Technologies, Carlsbad, CA) for 1 min. Following rinsing off the excess stain, the BODIPY FL-stained lipid droplets were imaged using a Zeiss LSM 710 confocal scanning microscope. The excitation wavelength was 488 nm. Lipid droplets were imaged at 63× magnification, with the gain set to 766 and 670 for BODIPY stain and chlorophyll, respectively, and analysed using Zen blue software (Zeiss, Sydney, NSW, Australia).
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) was used to analyse spatial distribution of lipid accumulation in N. tabacum leaves following methods described by Horn et al. (2012) for seed tissues. Essentially, the leaf cross-sections were adhered to glass slides and coated with the matrix 2,5-dihydroxybenzoic acid (Sigma-Aldrich, St. Louis, MO). Sections were scanned at 25 micron step-size in a MALDI linear ion-trap-Orbitrap hybrid mass spectrometer (MALDI LTQ Orbitrap-XL; Thermo Fisher Scientific, Bremen, Germany). MALDI-MS images were reconstructed from mass spectral data using software Metabolite Imager (Horn et al., 2012) and converted into a coloured or grey-scale colour-scheme according to overall sum or relative mol% of lipid class.
TAG extraction from green leaf tissue
Tobacco leaves were harvested and immediately prepared by first removing the main vein and then cutting into small pieces. Between 3 and 6 g leaf was weighed in triplicate, dipped into liquid nitrogen to aid disruption of tissue and immediately placed in 100 mL of each solvent in separatory funnels. Solvents tested included hexane, methanol and acetone. Samples were shaken several times and left overnight at room temperature. The next day the lipid extracts were drained, followed by a second rinse using 100 mL of the same solvent while shaking. The two lipid extracts were combined and concentrated using a rotary evaporator. A subsequent Bligh and Dyer extraction using the solvent system chloroform/methanol/water (1:2:0.8, v:v:v) was included to determine the amount of residual lipid left after each initial extraction step. As acetone extraction followed by Bligh and Dyer extraction resulted in the highest extraction efficiency, this yield was set as 100% recovery in the calculation for the extraction efficiencies of the other solvents used.
The lipid extracts obtained by methanol and acetone extraction were each redissolved in 100 mL of the Bligh and Dyer solvent mixture. Solvent compositions were adjusted to chloroform/methanol/water (1:1:0.9, v:v:v) by the addition of chloroform and water to partition and remove any nonlipid components extracted using these solvents such as sugars and amino acids. The lower chloroform lipid layer was subsequently recovered and concentrated by rotary evaporation. The lipid extract was transferred to a preweighed GC vial and all solvent evaporated under a stream of nitrogen gas.
Lipid classes were analysed and quantified by TLC-FID (Iatroscan Mark V; Iatron Laboratories, Tokyo, Japan) using hexane/diethyl ether/glacial acetic acid (70:10:0.1, v:v:v) as the developing solvent system in combination with Chromarod S-III silica on quartz rods and suitable calibration curves of representative standards obtained from Nu-Chek Prep, Inc. (Elysian, MN). Data were processed using SIC-480II software (SISC version: 7.0-E, System Instruments Co., Hachioji, Japan).
Quantitative real-time PCR (qRT-PCR)
One microgram of total RNA extracted from N. tabacum leaf tissue was used to prepare cDNA using First-Strand cDNA synthesis mix (Origene, Rockville, MD). The prepared cDNA was diluted five times and used as template for qRT-PCR using CFX real-time system (Bio-Rad, Gladesville, NSW, Australia). Primer validation was performed on five dilutions of each target gene along with control genes Nicotiana benthamiana ACTIN and N. benthamiana GAPDH. Reactions included initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 10 s, 58 °C for 30 s and 68 °C for 30 s. Single amplicons were confirmed by melt-curve analysis (a temperature increase from 60 °C to 95 °C was applied at 0.5 °C per 5 s) and agarose gel electrophoresis. Relative fold differences were determined by ΔΔCT method (Livak and Schmittgen, 2001).
Southern blotting
Total DNA was extracted from wild-type and transgenic N. tabacum leaf tissue using the caesium chloride gradient method. Genomic DNA (2 µg) was digested using HindIII or BamHI, and digestion products were separated on a 0.8% agarose gel. DNA was subsequently transferred onto Hybond N+ membranes (GE Healthcare Life Sciences, Rydalmere, NSW, Australia) in 0.4 m NaOH. A 652-bp fragment of nptII region of the binary vector construct was PCR-amplified using primers NPTIIF (5′-CAGGTTCTCCGGCCGCTTGG-3′) and NPTIIR (5′-AAGCGGTCAGCCCATTCGCC-3′). Following gel purification, the PCR product was 32P-labelled using the Megaprime DNA labelling system (GE Healthcare Life Sciences) and used as probe.
Starch quantification
Starch was first extracted by grinding frozen plant tissue samples in 10 mL extraction buffer (40 mm Tris–HCl, pH 8.0; 10 mm EDTA) using a mortar and pestle, followed by filtering through two layers of miracloth. After a first centrifugation at 3000 g for 10 min to remove cell debris, starch granules were isolated using a 15 mL sedimentation gradient of 90% Percol solution centrifuged at 3000 g for 30 min. Starch pellets were washed twice with 80% ethanol solution and stored at 4 °C. Total starch was quantified using the total starch quantification kit from Megazyme (Wicklow, Ireland).
Acknowledgments
We would like to thank Nathalie Niesner, Anne Mackenzie, Dawar Hussain, Anna Mechanicos, Cheryl Blundell, Luch Hac, Alex Miller, Lina Ma, Lijun Tian and Jeni Pritchard for technical assistance. We also thank Dr Damien Callahan at Metabolomics Australia, University of Melbourne and Dr. Matt Taylor at CSIRO Ecosystem Sciences for assistance with LC-MS, Dr. Rosemary White for help with microscopy and Dr. Allan Green for helpful discussions and advice. This work was funded by the CSIRO Food Futures National Research Flagship, CSIRO Plant Industry and the CSIRO Office of the Chief Executive Postdoctoral Fellowship Scheme. LC-MS analysis was supported by Bioplatforms Australia Services. Research in the UNT Center for Plant Research on oil accumulation in leaves is supported by a grant from the U.S. Department of Energy, BER Division, DE-FG02-09ER64812. Acquisition of the imaging instruments at UNT was made possible, in part, by grants from the Hoblitzelle Foundation and the US National Science Foundation (MRI#112605).
Supporting Information
Additional Supporting information may be found in the online version of this article
TLC screening of leaf tissue of Nicotiana tabacum T0 events at flowering stage as described in materials and methods. Lanes 42–44 are wild-type controls, which have relatively low TAG compared with the T0 transgenic event in lane 3 (described in this study).
Figure S2 Southern blot of transgenic Nicotiana tabacum leaf DNA with NPTII probe demonstrating that the T0 line displaying maximum TAG levels was a single-copy event. This was corroborated by the segregation pattern of subsequent generations.
Figure S3 Expression analysis of fad3 and fad7 fatty acid desaturase genes in wild-type and transgenic T1 Nicotiana tabacum leaf tissue as quantified by qRT-PCR. Error bars denote standard deviations (n = 3).
Figure S4 LC-MS analysis of different phosphatidylcholine (A), diacylglycerol (B) and triacylglycerol (C) molecular species in wild-type and transgenic T1 Nicotiana tabacum leaf tissue. Error bars denote standard deviations (n = 3).
Figure S5 GC analysis of cuticular wax composition on wild-type and T1 transgenic leaves. Alk, alkanes; Br Alk, branched alkanes; WE, wax esters; FA, fatty acids; Alc, fatty alcohols. Error bars denote standard deviations (n = 3).
Table S1 Representative triacylglycerol (TAG) and total fatty acid (TFA) levels (% dry weight) and fatty acid profiles in leaves of Nicotiana tabacum primary transformant #3 at the onset of senescence. A TLC chromatogram of this line is depicted in lane 3 of Figure S1.
Table S2 Triacylglycerol content (% seed weight) and fatty acid profile in wild-type (WT) and pooled T1 seed from Nicotiana tabacum primary transformant #3.
Table S3 Total fatty acid levels (% dry weight) and fatty acid profile in wild-type (WT) and T1 transgenic Nicotiana tabacum senescing leaf tissue at the onset of senescence. Error bars represent standard deviations (n ≥ 3).
Table S4 MS/MS (Q-TOF) identification of major triacylglycerol (TAG) and diacylglycerol (DAG) molecular species in wild-type and T1 transgenic Nicotiana tabacum leaf tissue.
Table S5 Triacylglycerol (TAG) levels (% dry weight) and TAG fatty acid profile in stem and root tissues of wild-type (WT) and T1 transgenic Nicotiana tabacum line. Error bars represent standard deviations (n = 2, wild type; N = 3, transgenic).
Table S6 Cuticular wax content (µg/cm2) and composition (% each molecular class) on the surface of wild-type (WT) and T1 transgenic Nicotiana tabacum leaves. Error bars represent standard deviations (n = 3).
References
- Andrianov V, Borisjuk N, Pogrebnyak N, Brinker A, Dixon J, Spitsin S, Flynn J, Matyszczuk P, Andryszak K, Laurelli M, Golovkin M, Koprowski H. Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 2010;8:277–287. doi: 10.1111/j.1467-7652.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H. Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferases. Eur. J. Biochem. 2000;267:85–96. doi: 10.1046/j.1432-1327.2000.00961.x. [DOI] [PubMed] [Google Scholar]
- Carlsson AS, Lindberg Yilmaz J, Green AG, Stymne S, Hofvander P. Replacing fossil oil with fresh oil – with what and for what? Eur. J. Lipid Sci. Technol. 2011;113:812–831. doi: 10.1002/ejlt.201100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004;40:575–585. doi: 10.1111/j.1365-313X.2004.02235.x. [DOI] [PubMed] [Google Scholar]
- Chapman K, Dyer JM, Mullen RT. Why don't plant leaves get fat? Plant Sci. 2013;207:128–134. doi: 10.1016/j.plantsci.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Broglie R, Edwards C, Chua NH. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-biphosphate carboxylase. EMBO J. 1984;3:1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer J, Mullen R, Chapman K. Oil in biomass: a step-change for bioenergy production? Inform. 2012;23:193–272. [Google Scholar]
- FAO. World Agriculture: Towards 2015/2030: An FAO Perspective. 2003. http://www.fao.org/docrep/005/y4252e/y4252e05c.htm.
- Fogher C. 2008. Mutagenized tobacco plant as seed culture for production of oil for energetic, industrial and alimentary uses. Patent application WO/2008/110876.
- Fogher C, Di Norscia N, Tommasini S. Developing tobacco's potential as a novel, high-yielding, renewable energy plant. Inform. 2011;22:631–634. [Google Scholar]
- Gigliotti JC, Davenport MP, Beamer SK, Tou JC, Jaczynski J. Extraction and characterization of lipids from Antarctic krill (Euphausia superb. Food Chem. 2011;125:1028–1036. [Google Scholar]
- Guelette BS, Benning UF, Hoffmann-Benning S. Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 2012;63:3603–3616. doi: 10.1093/jxb/ers028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee YJ, Chapman KD. Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell. 2012;24:622–636. doi: 10.1105/tpc.111.094581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- James CN, Horn PJ, Case CR, Gidda SK, Zhang D, Mullen RT, Dyer JM, Anderson RG, Chapman KD. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc. Natl Acad. Sci. USA. 2010;107:17833–17838. doi: 10.1073/pnas.0911359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ. The SUGAR-DEPENDENT1 lipase limits triacylglycerols accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 2013;162:1282–1289. doi: 10.1104/pp.113.219840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus D, Ohlrogge JB, Neuhaus HE, Dormann P. Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase. Planta. 2004;219:389–396. doi: 10.1007/s00425-004-1236-3. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–428. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, Zuo J. LEAFY COTYLEDON 1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008;148:1042–1054. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Chapman K. The seeds of green energy. Biochemist. 2011;33:34–38. [Google Scholar]
- Petrie J, Shrestha P, Zhou XR, Mansour MP, Liu Q, Belide S, Nichols PD, Singh SP. Metabolic engineering plant seeds with fish oil-like levels of DHA. PLoS ONE. 2012a;7:e49165. doi: 10.1371/journal.pone.0049165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie JR, Vanhercke T, Shrestha P, El Tahchy A, White A, Zhou XR, Liu Q, Mansour MP, Nichols PD, Singh SP. Recruiting a new substrate for triacylglycerol synthesis in plants: the monoacylglycerol acyltransferase pathway. PLoS ONE. 2012b;7:e35214. doi: 10.1371/journal.pone.0035214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Carrer H. Engineering plastid fatty acid biosynthesis to improve food quality and biofuel production in higher plants. Plant Biotechnol. J. 2011;9:554–564. doi: 10.1111/j.1467-7652.2011.00621.x. [DOI] [PubMed] [Google Scholar]
- Sanjaya, Durrett TP, Weise SE, Benning C. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 2011;9:874–883. doi: 10.1111/j.1467-7652.2011.00599.x. [DOI] [PubMed] [Google Scholar]
- Sanjaya, Miller R, Durrett TP, Kosma DK, Lydic TA, Muthan B, Koo AJ, Bukhman YV, Reid GE, Howe GA, Ohlrogge J, Benning C. Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell. 2013;25:677–693. doi: 10.1105/tpc.112.104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005;579:4666–4670. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Scott RW, Winichayakul S, Roldan M, Cookson R, Willingham M, Castle M, Pueschel R, Peng CC, Tzen JT, Roberts NJ. Elevation of oil body integrity and emulsion stability by polyoleosins, multiple oleosin units joined in tandem head-to-tail fusions. Plant Biotechnol. J. 2010;8:912–927. doi: 10.1111/j.1467-7652.2010.00522.x. [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Cornah J, Pinfield-Wells H, Soady K, Zhang Q, Gilday A, Dyer JM, Graham IA. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 2009;7:694–703. doi: 10.1111/j.1467-7652.2009.00435.x. [DOI] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl Acad. Sci. USA. 2008;105:3151–3156. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, Cao X, Yang Z, Ohlrogge JB. Lipid turnover during senescence. Plant Sci. 2013;205–206:13–19. doi: 10.1016/j.plantsci.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Shrestha P, Zhou XR, Singh SP, Petrie JR. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013;587:364–369. doi: 10.1016/j.febslet.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Winichayakul S, Scott RW, Roldan M, Hatier JH, Livingston S, Cookson R, Curran AC, Roberts NJ. In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol. 2013;162:626–639. doi: 10.1104/pp.113.216820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich JE, Awai K, Benning C. Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell. 2005;17:3094–3110. doi: 10.1105/tpc.105.035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TLC screening of leaf tissue of Nicotiana tabacum T0 events at flowering stage as described in materials and methods. Lanes 42–44 are wild-type controls, which have relatively low TAG compared with the T0 transgenic event in lane 3 (described in this study).
Figure S2 Southern blot of transgenic Nicotiana tabacum leaf DNA with NPTII probe demonstrating that the T0 line displaying maximum TAG levels was a single-copy event. This was corroborated by the segregation pattern of subsequent generations.
Figure S3 Expression analysis of fad3 and fad7 fatty acid desaturase genes in wild-type and transgenic T1 Nicotiana tabacum leaf tissue as quantified by qRT-PCR. Error bars denote standard deviations (n = 3).
Figure S4 LC-MS analysis of different phosphatidylcholine (A), diacylglycerol (B) and triacylglycerol (C) molecular species in wild-type and transgenic T1 Nicotiana tabacum leaf tissue. Error bars denote standard deviations (n = 3).
Figure S5 GC analysis of cuticular wax composition on wild-type and T1 transgenic leaves. Alk, alkanes; Br Alk, branched alkanes; WE, wax esters; FA, fatty acids; Alc, fatty alcohols. Error bars denote standard deviations (n = 3).
Table S1 Representative triacylglycerol (TAG) and total fatty acid (TFA) levels (% dry weight) and fatty acid profiles in leaves of Nicotiana tabacum primary transformant #3 at the onset of senescence. A TLC chromatogram of this line is depicted in lane 3 of Figure S1.
Table S2 Triacylglycerol content (% seed weight) and fatty acid profile in wild-type (WT) and pooled T1 seed from Nicotiana tabacum primary transformant #3.
Table S3 Total fatty acid levels (% dry weight) and fatty acid profile in wild-type (WT) and T1 transgenic Nicotiana tabacum senescing leaf tissue at the onset of senescence. Error bars represent standard deviations (n ≥ 3).
Table S4 MS/MS (Q-TOF) identification of major triacylglycerol (TAG) and diacylglycerol (DAG) molecular species in wild-type and T1 transgenic Nicotiana tabacum leaf tissue.
Table S5 Triacylglycerol (TAG) levels (% dry weight) and TAG fatty acid profile in stem and root tissues of wild-type (WT) and T1 transgenic Nicotiana tabacum line. Error bars represent standard deviations (n = 2, wild type; N = 3, transgenic).
Table S6 Cuticular wax content (µg/cm2) and composition (% each molecular class) on the surface of wild-type (WT) and T1 transgenic Nicotiana tabacum leaves. Error bars represent standard deviations (n = 3).