Abstract
Bacteria form communities, termed biofilms, in which cells adhere to each other within a matrix, typically comprised of polysaccharides, proteins, and extracellular DNA. Biofilm formation by the marine bacterium Vibrio fischeri requires the Syp polysaccharide, but the involvement of matrix proteins is as yet unknown. Here we identified three genes, termed bmpA, -B, and -C (biofilm maturation protein), with overlapping functions in biofilm maturation. A triple bmpABC mutant, but not single or double mutants, was defective in producing wrinkled colonies, a form of biofilm. Surprisingly, the triple mutant was competent to form pellicles, another biofilm phenotype, but they generally lacked a three-dimensional architecture. Transmission electron microscopy revealed that the extracellular matrix of the bmp mutant contained electron-dense, thread-like structures that were also present in the wild type but lacking in syp mutant strains. We hypothesized that the bmp mutant produces the Syp polysaccharide but fails to produce/export a distinct matrix component. Indeed, a mixture of the bmp and syp mutants produced a wrinkled colony. Finally, BmpA could be detected in cell-free supernatants from disrupted pellicles. Thus, this work identifies a new matrix protein necessary for biofilm maturation by V. fischeri and, based on the conservation of bmp, potentially other microbes.
INTRODUCTION
Bacteria can be found as planktonic cells or within sessile communities called biofilms, which are composed of bacterial cells encased in an extracellular matrix (1–3). The matrix is generally self-produced and comprised of polysaccharides, proteins, extracellular DNA, and other bacterial products, such as outer membrane vesicles (OMVs). These matrix components promote adherence of the bacteria to each other and to a variety of surfaces, including host tissues and medical devices, such as catheters (reviewed in reference 4). Furthermore, due at least in part to the protective nature of the matrix, resident cells exhibit increased resistance to predation (5), desiccation (6), host defenses (4), and antibiotics (7). While much is known about the process of biofilm formation and its regulation from the study of a variety of bacteria, we still know little about the constituents of the matrix, in particular the matrix proteins, and their contributions to biofilm formation.
Matrix proteins can be anchored to or associated with the cell surface or released into the extracellular space, where they appear to promote cell-matrix, cell-cell, matrix-matrix, and cell-surface interactions. For example, in Pseudomonas aeruginosa, the matrix protein CdrA is both cell associated and secreted and likely promotes cell-matrix and matrix-matrix interactions by binding to the Psl polysaccharide (8), which itself is cell associated and secreted (9). Additionally, the lectins LecA, specific for d-galactose (10), and LecB, specific for l-fucose (11), are thought to promote cell-cell interactions (12) during P. aeruginosa biofilm formation.
Perhaps the most is known about the role of matrix proteins during biofilm formation by Vibrio cholerae, due to two recent studies. In the first study, the V. cholerae matrix proteins Bap1 and RbmA were found to have overlapping functions yet distinct localizations within the mature biofilm (13); this was one of the first studies to show spatial and functional differentiation between biofilm matrix proteins within a biofilm. The second study examined the expression, localization, and interaction of the three matrix proteins (Bap1, RbmA, and RbmC) with the VPS polysaccharide during the initial stages of biofilm formation (14). Together, these studies provide important insights into the spatial and temporal distributions and interactions of these matrix proteins not only with themselves but also with the VPS polysaccharide.
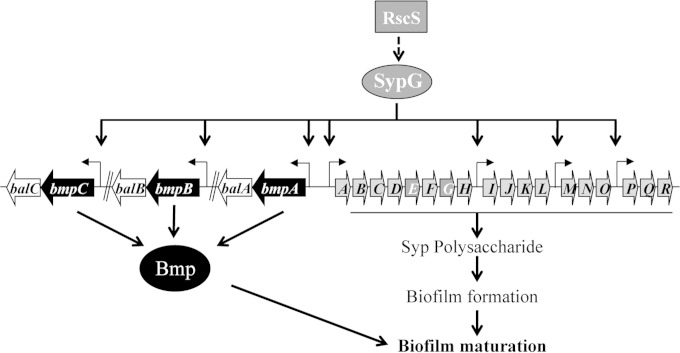
In contrast to the case of V. cholerae, relatively little is known about the matrix composition of the biofilm formed by the bioluminescent marine bacterium Vibrio fischeri. This organism is of interest because a biofilm is formed at a critical stage during colonization by V. fischeri of its symbiotic host squid (15–18). Biofilm formation by V. fischeri requires the symbiosis polysaccharide (syp) locus, an 18-gene locus that encodes proteins for the production and transport of a polysaccharide critical for biofilm formation and host colonization (17, 19, 20). The syp locus is regulated by a two-component signal transduction system that includes the sensor kinase RscS (17, 21) and the response regulator SypG (19, 22, 23). Upon sensing an unknown signal, RscS is predicted to initiate a phosphorelay that leads to the phosphorylation and activation of SypG (Fig. 1). In its phosphorylated form, SypG serves as the direct transcriptional activator of the syp locus by binding to the syp enhancer (SE), a conserved sequence located upstream of each of four operons within this locus, and promoting transcription in conjunction with RNA polymerase loaded with the alternative sigma factor σ54 (19, 24). The signal recognized by RscS is unknown, but overexpression of the rscS gene is sufficient to induce syp transcription and promote biofilm phenotypes, including the formation of wrinkled colonies (normally smooth) and pellicles (not normally formed) (17). Overexpression of sypG induces syp transcription to an even greater extent but fails to promote biofilm formation due to the presence of an additional RscS-responsive control mechanism that functions below syp transcription (18, 23, 25, 26).
FIG 1.
Control over biofilm formation by V. fischeri. RscS, a sensor kinase, is predicted to recognize a signal (currently unknown) and initiate a signal transduction cascade, which ultimately results in activation of the response regulator SypG and deactivation of a downstream inhibitor of Syp polysaccharide production, SypE (not depicted). Activated SypG promotes transcription of four syp operons as well as the three sets of bmp-bal genes. The Syp polysaccharide appears to be sufficient for biofilm formation, while the Bmp proteins appear to be necessary for biofilm maturation (i.e., 3D architecture, such as wrinkling). The coordinately produced Bal proteins play an unknown role in bioluminescence regulation (not shown). Arrows represent individual genes (not to scale).
Although much is known about the regulatory control over Syp polysaccharide production, important aspects of V. fischeri biofilm formation, including the identities of other matrix components, such as matrix proteins, remain unknown. Recently, however, we identified new putative members of the SypG regulon (24). Specifically, three two-gene operons, one of which was immediately adjacent to the syp locus, were identified via bioinformatics based on the presence in their upstream intergenic regions of an SE sequence, indicating a putative SypG binding site, and a predicted σ54 binding site (24). Intriguingly, the proteins encoded by the first gene in each operon are similar to each other (27, 28), suggesting that they may have overlapping functions. However, they contain no domains of known function, and thus the exact roles of these proteins are unknown. Due to their apparent regulation by SypG, a critical regulator of biofilm formation, we hypothesized that these genes function to control biofilm formation by V. fischeri. Here we demonstrate that these genes do in fact have overlapping roles in biofilm formation, specifically in biofilm maturation. Furthermore, we found that the protein product of at least one of them is secreted and is present in the supernatant fraction of disrupted pellicles. Thus, this study identifies the first known V. fischeri matrix protein and uncovers a role for it in biofilm maturation.
MATERIALS AND METHODS
Bacterial strains and media.
V. fischeri strains utilized in this study are listed in Table 1 and in Table S1 in the supplemental material. All strains used in this study were derived from strain ES114, a bacterial isolate from Euprymna scolopes (29–31). V. fischeri strains were grown in a complex medium, either LBS (32) or SWTO (33). All derivatives of V. fischeri were generated via conjugation as previously described (34). Escherichia coli strains GT115 (Invivogen, San Diego, CA), TAM1 (Active Motif, Carlsbad, CA), and π3813 (35) were used for the purposes of cloning, plasmid maintenance, and conjugation. E. coli strains were grown in LB (36). Solid media were made by using agar at a final concentration of 1.5%. The following antibiotics were added to growth media as necessary, at the indicated final concentrations: chloramphenicol (Cm), 2.5 μg/ml (V. fischeri) or 12.5 or 25 μg/ml (E. coli); erythromycin (Em), 5 μg/ml (V. fischeri) or 150 μg/ml (E. coli); tetracycline (Tc), 5 μg/ml in LBS or 30 μg/ml in SWTO (V. fischeri) or 15 μg/ml (E. coli); and ampicillin (Ap), 100 μg/ml (E. coli). Along with any necessary antibiotics, thymidine was added to a final concentration of 0.3 mM for E. coli strain π3813.
TABLE 1.
V. fischeri strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| ES114 | Wild type | 29 |
| KV1787 | ΔsypG | 22 |
| KV5069 | ΔsypL | 20 |
| KV6475 | ΔsypG attTn7::sypG-FLAG | This study |
| KV6638 | ΔbmpB | This study |
| KV6712 | ΔbmpB ΔbmpC | This study |
| KV6787 | ΔbmpC | This study |
| KV6886 | ΔbmpA | This study |
| KV6897 | ΔbmpA ΔbmpB ΔbmpC | This study |
| KV7060 | ΔbmpA ΔbmpB ΔbmpC ΔsypL | This study |
| KV7062 | ΔbmpA ΔbmpB ΔbmpC attTn7::bmpA-balA | This study |
| KV7078 | ΔbmpA ΔbmpB | This study |
| KV7079 | ΔbmpA ΔbmpC | This study |
| KV7216 | attTn7::PbmpA -lacZ Emr | This study |
| KV7220 | attTn7::PbmpC -lacZ Emr | This study |
| KV7274 | ΔbmpA ΔbmpB ΔbmpC attTn7::bmpA-FLAG | This study |
Molecular techniques.
All plasmids were constructed using standard molecular biology techniques with restriction and modification enzymes obtained from Thermo Fisher Scientific (Pittsburgh, PA). Plasmids and primers used in this study are listed in Tables S2 and S3 in the supplemental material, respectively. In some cases where PCR was used to generate DNA fragments, the PCR cloning vector pJET1.2 (Fisher Scientific, Pittsburgh, PA) was used as an intermediate vector prior to cloning into the final vector. Gibson Assembly (New England BioLabs, Beverly, MA) was also used to construct plasmids in some instances. Unmarked deletions in V. fischeri were generated as previously described (35, 37). For complementation in single copy from the chromosome, the appropriate DNA fragment was cloned with its native promoter into the mini-Tn7 delivery vector pEVS107. Insertion at the Tn7 site of the chromosome was performed via tetraparental mating (38) with wild-type V. fischeri, E. coli carrying pEVS104 (39), E. coli carrying the pEVS107 derivative, and E. coli carrying the Tn7 transposase plasmid pUX-BF13 (40). All plasmids constructed in this study were sequenced at ACGT (Wheeling, IL) to ensure that they contained the desired sequences.
β-Galactosidase assay.
To assay β-galactosidase activity from the reporter fusions, strains containing either the vector control (VC) or sypG plasmid were grown in LBS containing Tc. Samples (50 μl) were collected at 24 h, and 50 μl of Pierce β-galactosidase assay reagent (Pierce Biotechnology, Rockford, IL) was added to each sample. Measurements were taken in a microtiter dish by using an ELx800 absorbance microplate reader (BioTek, Winooski, VT) with the appropriate settings. β-Galactosidase activity was determined as previously described (41). P values were calculated using Student's t test.
Wrinkled colony assay.
V. fischeri strains were cultured overnight at 28°C with shaking in LBS containing Tc and then subcultured 1:100 into fresh LBS containing Tc and grown under the same conditions for 2 to 4 h the next day. Subcultures were standardized to an optical density at 600 nm (OD600) of 0.2, and 10-μl aliquots were placed as spots on LBS agar plates containing Tc and incubated at 24°C. The spot cultures were then monitored until wrinkled colony development ceased or the appropriate data set was collected. Each set of strains for a particular experiment was examined on the same plate to account for any minor plate-to-plate variations. Each assay was performed at least 2 or 3 times.
Pellicle assay.
V. fischeri strains were cultured overnight at 28°C with shaking in LBS containing Tc and then subcultured 1:100 into fresh LBS containing Tc and grown under the same conditions for 2 to 4 h the next day. Subcultures were standardized to an OD600 of 0.2 in 2 ml in a 24-well plate. Plates were incubated statically at 24°C for up to 72 h.
Stickiness assay.
V. fischeri strains were grown as indicated above for the wrinkled colony assay. At the indicated time point, the spots were disturbed with a toothpick. A spot was considered sticky if it exhibited biofilm-like properties (i.e., the spot was pulled away intact from the agar surface).
Exogenous complementation assay.
V. fischeri strains were grown as indicated above for the wrinkled colony assay (in LBS containing Tc). For the mixing experiments, 10 μl of each subculture standardized to an OD600 of 0.2 was mixed, and then 10 μl of the mixture was applied as a spot. For spot-touching experiments, the standardized cultures were applied as spots close to each other and monitored over time as they grew close to or into each other.
Sample preparation for TEM.
Strains ES114, KV6897 (ΔbmpABC), and KV5069 (ΔsypL), all overexpressing rscS, were grown as described above for the wrinkled colony assay and sampled individually as spots. KV6897 and KV5069 were also grown and used for a spot-touching experiment. Samples were harvested after 72 h, as indicated below. For the individual spots, a sample from the edge of the spot was removed. For the touching spots, a sample at the interface of the touching spots was removed; note that since KV6897 overexpressing rscS is sticky, the sample mostly contained this strain. The samples were then fixed in the presence of ruthenium red. Briefly, 1 ml of solution containing 2.4% glutaraldehyde, 0.1 M sodium cacodylate, and 0.1% ruthenium red was added to the samples and incubated for 1 h at 37°C. The liquid was then removed, and the samples were washed with 1.4 ml of 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 at pH 7.4). Next, 1 ml of a 2% osmium tetroxide, 0.1 M sodium cacodylate, and 0.1% ruthenium red solution was added to each sample and incubated for 3 h at room temperature. The samples were then enrobed in agarose, dehydrated, embedded, sectioned, and analyzed by transmission electron microscopy (TEM) as described previously (42).
Cell-free supernatant collection and TCA precipitation.
Cell-free supernatant was collected via centrifugation (5,000 rpm for 15 min) of V. fischeri cultures (25 ml) grown in LBS overnight at 24°C with vigorous shaking; the pellet was saved for Western blot analysis (see below). The supernatant was subjected to trichloroacetic acid (TCA) precipitation. Briefly, TCA was added to the supernatant at a final concentration of 14%, followed by vortexing and a 30-min incubation on ice. The samples were centrifuged at 15,000 rpm for 30 min, and the pellet (precipitated proteins) was washed with acetone and centrifuged again. The sample was then air dried and utilized for Western blot analysis as described below.
Cell-free supernatant collection from pellicles.
To search for the presence of BmpA in the matrix, strains were inoculated into 24-well plates containing 2 ml LBS broth with Tc. After pellicle formation, six pellicles were collected and rinsed briefly in PBS, and then the samples were homogenized by vortexing with glass beads in test tubes. One milliliter of each sample was removed to a microcentrifuge tube, and samples were centrifuged for 1 min. After centrifugation, three layers were observed. A cell pellet was present at the bottom of the tube (pellet fraction), a clear liquid fraction was present at the top (supernatant fraction), and in between was a sticky layer that appeared to contain extracellular polymeric substance (EPS) and/or biofilm matrix. The fractions were separated into fresh tubes, and 0.5 ml of PBS was added to the matrix layer, which was vortexed again and then centrifuged. The same three layers were observed, indicating that the matrix layer still contained cells. The first pellet and both supernatant fractions were separated by SDS-PAGE and subjected to Western blot analysis (see below).
Western blot analysis.
TCA-precipitated samples and the pellets collected as described in the previous section were resuspended in 2× SDS loading buffer (4% SDS, 10% 2-mercaptoethanol, 0.005% bromophenol blue, 20% glycerol, 0.1 M Tris, pH 7), boiled for 5 min, and then loaded onto a 12% SDS-polyacrylamide gel. After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and probed with anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO). Protein bands were visualized using a horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence (ECL) reagents (SuperSignal West Pico chemiluminescence substrate; Pierce Biotechnology, Rockford, IL).
RESULTS
bmp and bal constitute part of the SypG regulon.
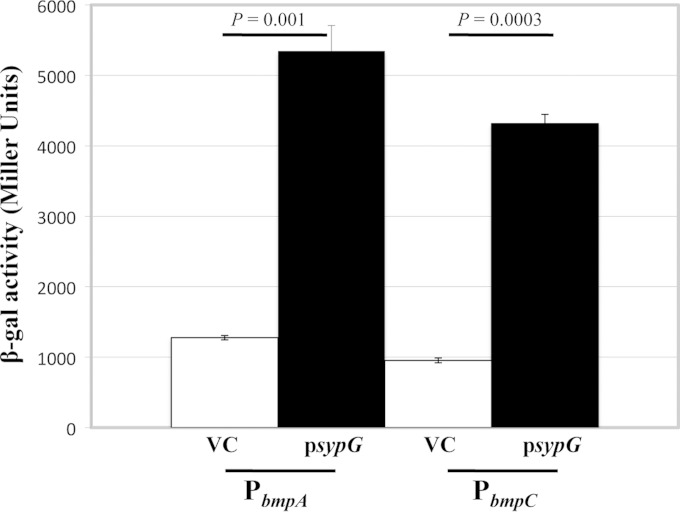
We previously identified three unlinked two-gene sets (pairs of genes), namely, VF_A1019-VF_A1018, VF_A0120-VF_A0121, and VF_A0550-VF_A0549, as putative members of the SypG regulon (24). Our subsequent investigation of these genes, as described below, prompted us to rename them bmp, for biofilm maturation protein (bmpA [VF_A1019], bmpB [VF_A0120], and bmpC [VF_A0550]), and bal, for biofilm-associated lipoprotein (balA [VF_A1018], balB [VF_A0121], and balC [VF_A0449], respectively) (Fig. 1). As part of our original work, we demonstrated that SypG could bind to the bmpB promoter region (24). To confirm that SypG could also regulate bmpA and bmpC, we generated transcriptional reporter constructs that fuse the promoter regions of these genes to lacZ and introduced the fusions, in single copy, into the chromosome of wild-type V. fischeri at a benign site (the Tn7 site). Since transcription of other SypG-regulated genes (e.g., the syp genes) is minimal in the absence of sypG (or rscS) overexpression, we predicted that the same would be true for the bmp genes. Therefore, we introduced either a multicopy SypG expression plasmid or the vector control (VC) into these strains. Finally, we measured the β-galactosidase activity of each reporter to determine relative promoter activities. As predicted, expression of SypG in these strains led to a substantial increase in β-galactosidase activity relative to that in the VC-containing strains (Fig. 2), indicating that bmpA and bmpC are indeed regulated by SypG and thus comprise part of the SypG regulon.
FIG 2.
SypG induces transcription from bmp promoters. The ability of SypG to induce transcription from the bmpA and bmpC promoters was assessed using reporter fusions to a promoterless lacZ gene. β-Galactosidase activity was assessed as a measure of promoter activity from sypG-overexpressing (psypG; pEAH73) and vector control (VC; pKV69) derivatives of KV7216 (PbmpA-lacZ) and KV7220 (PbmpC-lacZ), grown as described in Materials and Methods. β-Galactosidase activities are shown in Miller units. Error bars represent standard deviations, and P values refer to the variations between the samples indicated by the lines. These data are representative of at least two independent experiments.
BmpA, BmpB, and BmpC are conserved proteins, while BalA, BalB, and BalC are conserved lipoproteins.
Bioinformatic analyses of the bmp and bal gene sets indicated that bmpA and balA, which are located immediately adjacent to the syp locus, clearly form an operon, as the two genes overlap by 38 nucleotides, while bmpB-balB and bmpC-balC likely do so as well, as these genes are separated by only 11 and 12 nucleotides, respectively. Importantly, the predicted V. fischeri proteins are similar to each other: the three Bmp proteins are 37% identical and 53% similar to each other, while the three Bal proteins are 26% identical and 50% similar to each other (see Fig. S1 in the supplemental material) (27, 28). The Bmp proteins contain no domains of known or unknown function, but they each contain a putative Sec-dependent signal sequence (see Fig. S2A) (43), suggesting that these proteins are exported to the periplasmic space or out of the cell. The Bal proteins appear to be lipoproteins: in addition to a predicted lipoprotein signal sequence, the Bal proteins contain a lipobox sequence (44) that includes an invariant cysteine predicted to become acylated following transport across the inner membrane (45) (see Fig. S2B). The apparent absence of inner membrane retention signals suggests that the Bal proteins may be sorted to the outer membrane (46). Finally, while the syp locus is primarily conserved only in Vibrio spp., the bmp-bal gene sets are present not only in Vibrio spp. but also in numerous other bacteria, primarily marine bacteria (see Fig. S3). This conservation suggests that the bmp and bal genes play similar roles in the physiology and/or ecology of various other microbes.
The bmp genes contribute to wrinkled colony formation.
To probe the functions of the Bmp and Bal proteins, we generated in-frame deletion mutants defective for each bmp or bal gene. Because the similarity of the proteins to each other suggested that they might have overlapping or redundant functions, we also generated double and triple bmp and bal mutants. We first evaluated the impacts of the mutations on motility, luminescence, and growth. None of the mutations substantially affected these phenotypes, with the exception that the balC mutant exhibited an increase in bioluminescence and the balB mutant, in some genetic contexts, exhibited a modest but reproducible decrease in bioluminescence (see Fig. S4 in the supplemental material; also data not shown); a greater understanding of the role of these genes in luminescence awaits further investigation.
Next, because of their coordinate regulation with the syp locus, which is critical for biofilm formation by V. fischeri (19, 20, 26), we asked if the bmp and/or bal genes might also play a role in biofilm formation. We induced biofilm formation by overexpressing the sensor kinase gene rscS and then evaluated wrinkled colony formation over time. As we observed previously (17), the control strain produced a wrinkled colony under these conditions (Fig. 3; see also Fig. S5 in the supplemental material). The bal genes played little to no role in biofilm formation: a strain deleted for all three bal genes exhibited wrinkled colony formation similar to that of the control (see Fig. S5). The biggest impact was exerted by balB, which when deleted alone or in the context of a balA deletion caused a minor (∼5 h) delay in wrinkled colony formation (see Fig. S5). However, at a later time point (70 h), there was no difference in wrinkled colony formation for the balB or balAB mutant compared to the control (see Fig. S5). Taken together, these data suggest that the bal genes play a relatively minor role in biofilm formation, with balB exerting the largest impact.
FIG 3.
Impact of bmp mutations on biofilm formation. To assess the impact of the bmp genes on biofilm formation, we applied cultures of various bmp mutants to LBS medium containing Tc and incubated them at room temperature. All strains overexpressed rscS (pKG11). Images were collected for up to 70 h for the following strains: wild-type control (ES114) and ΔbmpA (KV6886), ΔbmpB (KV6638), ΔbmpC (KV6787), ΔbmpAB (KV7078), ΔbmpAC (KV7079), ΔbmpBC (KV6712), and ΔbmpABC (KV6897) mutants. The images are representative of at least two independent experiments.
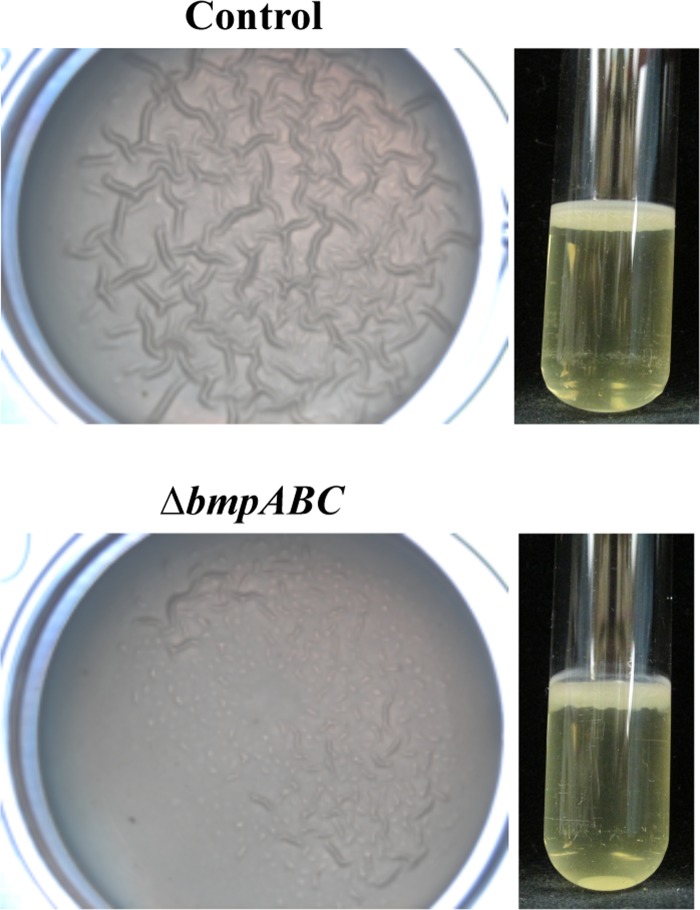
In contrast, the triple bmp mutant exhibited a severe defect in wrinkled colony formation, largely failing to wrinkle (Fig. 3). This defect could not be attributed to a single bmp gene, as deletion of individual bmp genes had no substantial impact on wrinkled colony formation relative to that of the control (Fig. 3). However, deletions of combinations of two bmp genes revealed that bmpA and bmpB are the more important genes: only the bmpAB double mutant was severely defective for wrinkled colony formation, though this mutant did exhibit some wrinkling at later time points (Fig. 3). These data indicate that the presence of either bmpA or bmpB is sufficient to promote wrinkled colony formation. Furthermore, they suggest that there is likely some overlap in function of the Bmp proteins. Indeed, complementation of the triple mutant with the bmpA-balA operon alone permitted robust wrinkled colony formation similar to that of the control (see Fig. S6 in the supplemental material). Because the triple bmp mutant exhibited the most severe defect in wrinkled colony formation, the remainder of our work focused on the phenotypes associated with this triple mutant, henceforth termed the bmp mutant for simplicity.
The bmp mutant retains the ability to form a pellicle.
We next examined the ability of the bmp mutant to produce a pellicle, a biofilm that forms at the air-liquid interface of a static liquid culture. This phenotype, like wrinkled colonies, is induced by rscS overexpression and depends upon the syp locus (17, 23). Although we previously observed a strong correlation between the formation of wrinkled colonies and the formation of pellicles (17, 20, 23, 25, 26), this was not the case for the bmp mutant: despite being unable to form a wrinkled colony, the bmp mutant was competent to form a pellicle (Fig. 4). We noted, however, that while pellicles formed by the control exhibited a wrinkled phenotype, those formed by the bmp mutant consistently exhibited reduced or no wrinkling (Fig. 4). These data suggested that while the bmp genes are not required for pellicle formation per se, they may be involved in the maturation of the V. fischeri biofilm by building and/or maintaining the three-dimensional (3D) architecture of the biofilm. Additionally, because the syp locus is necessary for pellicle formation (20, 23, 25), these data suggested that the bmp mutant may still produce the Syp polysaccharide, a possibility that we address below.
FIG 4.
Pellicle formation by the bmp mutant. To evaluate the role of bmp genes in pellicle formation, we grew rscS (pKG11)-containing wild-type (control; ES114) and bmp mutant (ΔbmpABC; KV6897) strains statically in LBS containing Tc for 72 h. Pellicle production can be observed as a 3D architecture visible on the surface or in a side view of a static culture. The images are representative of at least two independent experiments.
Because of the difference in pellicle architecture of the bmp mutant, we also examined the pellicle formation and architecture of the triple bal mutant expressing rscS. However, the triple bal mutant was proficient at forming a pellicle that was similar to that of the control (data not shown). These data further indicate that even though the individual bal genes are likely part of operons with their associated bmp genes, their loss does not substantially diminish biofilm formation.
The bmp mutant retains the ability to produce the Syp polysaccharide.
Because the bmp mutant was unable to form a wrinkled colony yet remained competent to form a pellicle, we explored the colony phenotype further. Specifically, we wondered whether the lack of wrinkled colony formation was caused by a defect in the ability to build/maintain 3D architecture, similar to what we observed for pellicle formation. We therefore compared the colony morphology of the bmp mutant with those of the biofilm-competent positive control and a representative syp mutant, the ΔsypL strain; the sypL mutant is defective for the formation of both wrinkled colonies and pellicles (20). At the indicated time point, the positive control exhibited a wrinkled phenotype with “sticky” properties: when perturbed with a flat toothpick, the whole colony was readily pulled away intact from the agar surface (Fig. 5A). In contrast, the toothpick slid through the sypL mutant colony, resulting in a clear path, with the rest of the colony remaining unperturbed (Fig. 5B). When we assessed the morphology of the bmp mutant, we found that this mutant exhibited “sticky” properties not unlike those of the positive control: despite the lack of wrinkling, the colony was pulled away intact from the agar surface by the toothpick (Fig. 5C). These data suggested that, similar to what we observed for pellicle formation, the bmp mutant is capable of forming a biofilm but unable to promote biofilm maturation. Thus, these data further support our hypothesis that the bmp genes are involved in building and/or maintaining the 3D architecture of the mature V. fischeri biofilm.
FIG 5.
The bmp mutant colony is sticky. To assess the “stickiness” of various strains, we applied cultures as spots on LBS medium containing Tc and incubated them at room temperature for 48 h. All strains overexpressed rscS (pKG11). Images were collected for each spot before (left) and after (right) disruption (with a toothpick) for the following strains: wild type (ES114) (A), ΔsypL mutant (KV5069) (B), ΔbmpABC mutant (KV6897) (C), and ΔsypL ΔbmpABC mutant (KV7060) (D). When a sticky colony is perturbed with a toothpick, the whole colony is readily pulled away intact from the agar surface (regions of the colony distal to the toothpick are dislodged). In contrast, perturbation of nonsticky colonies dislodges only cells within the path of the toothpick. The images are representative of at least two independent experiments.
Because the bmp mutant exhibited “sticky” properties that were lacking in the sypL mutant, we next questioned whether the stickiness of the bmp mutant depended upon an intact syp locus. We therefore generated a bmp sypL mutant and expressed rscS to examine the resulting colony morphology. Similar to the sypL single mutant (Fig. 5B), the bmp sypL mutant was not “sticky” (Fig. 5D), indicating that an intact syp locus is necessary for the “stickiness” of the bmp mutant. Thus, the bmp mutant retains the ability to produce the Syp polysaccharide. Furthermore, these results suggest that while the bmp genes are regulated coordinately with the syp locus, the bmp gene products function in a pathway distinct from that of Syp polysaccharide production (Fig. 1).
The wrinkled colony defect of the bmp mutant can be complemented exogenously.
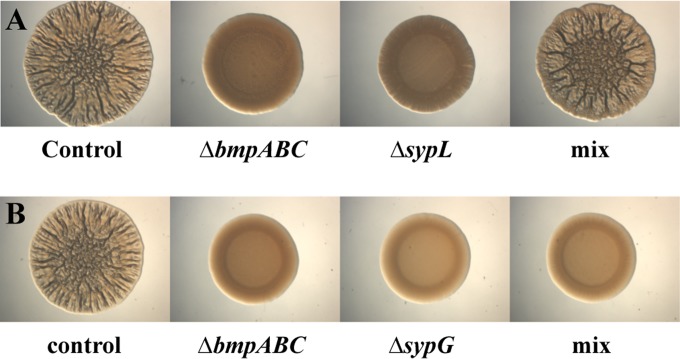
Because bmp and syp appear to function in distinct pathways to control biofilm formation, we wondered whether a mixture of the rscS-overexpressing bmp and sypL mutants could produce wrinkled colonies (i.e., could one mutant exogenously complement the other?). Indeed, whereas neither mutant alone could produce wrinkled colonies, a mixture of the two strains resulted in wrinkled colony formation (Fig. 6A). This result was not limited to mixtures of the bmp and sypL mutants, as mixtures of the bmp mutant with any of the syp structural mutants behaved in the same manner (see Fig. S7 in the supplemental material). Moreover, neither a mixture of the bmp and bmp sypL mutants nor a mixture of the sypL and bmp sypL mutants resulted in exogenous complementation (data not shown). These data suggest that both bmp and syp are necessary for exogenous complementation and, ultimately, wrinkled colony formation. Finally, in contrast to the case with the syp structural mutants, mixing the bmp mutant with the sypG mutant, which cannot activate expression of syp or bmp, did not result in wrinkled colony formation (Fig. 6B). Thus, not surprisingly, exogenous complementation requires a SypG-dependent product(s). We predict that either the bmp mutant provides the Syp polysaccharide to the syp mutants or, alternatively, the syp mutant provides Bmp or a Bmp-dependent product to the bmp mutant; we address these possibilities below. Regardless of the directionality, it is clear that both Bmp and Syp are required for wrinkled colony (and wrinkled pellicle) formation, further supporting the idea that Bmp and Syp comprise separate but necessary pathways leading to the production of a mature biofilm.
FIG 6.
A mixture of biofilm-defective strains permits wrinkled colony formation. (A) We assessed the ability of the biofilm-defective syp and bmp mutants to complement each other for wrinkled colony formation by applying a mixture (mix) of the two strains (pKG11-containing ΔsypL mutant [KV5069] and pKG11-containing ΔbmpABC mutant [KV6897]) to LBS medium containing Tc. As controls, we applied the two strains separately, as well as the biofilm-proficient pKG11-containing wild-type strain (ES114). In the experiment shown, cultures were applied as spots to LBS plates containing Tc and incubated at room temperature for 70 h. (B) To assess the requirement for sypG in exogenous complementation of the bmp mutant, we applied a mixture (mix) of the pKG11-containing ΔbmpABC mutant and the pKG11-containing ΔsypG mutant (KV1787). As controls, we applied the two strains separately, as well as the biofilm-proficient pKG11-containing wild-type strain (ES114). In the experiment shown, cultures were applied to plates as spots and incubated at room temperature for 53 h. The images are representative of at least two independent experiments.
Next, because mixtures of the bmp and syp mutants resulted in wrinkled colony formation, we wondered whether exogenous complementation could occur if the mutant strains (bmp and ΔsypL strains) were applied as spots adjacent to each other. We found that if the colonies did not touch, no wrinkling occurred (Fig. 7A). However, when the two strains were placed sufficiently close that they grew into each other, wrinkling occurred at the interface of the two spots (Fig. 7B). Moreover, the wrinkling appeared only on the bmp mutant side of the interface: while the bmp mutant sometimes exhibited weak wrinkling, this wrinkling never occurred at the edges of the colony unless it was in contact with the sypL mutant. We then used epifluorescence microscopy to examine touching spots of bmp and sypL mutant cells expressing green fluorescent protein (GFP) and red fluorescent protein (RFP), respectively (see Fig. S8 in the supplemental material). With those strains, we similarly observed wrinkling only at the interface. Although some RFP-labeled sypL mutants had clearly penetrated into the bmp mutant spot, wrinkling could be observed beyond the sites of penetration (see Fig. S8). From these data, we conclude that the bmp mutant receives some secreted (or cell-associated) factor, either Bmp itself or a molecule whose synthesis or secretion is Bmp dependent, from the syp mutant.
FIG 7.
The syp mutant complements the bmp mutant. As an initial test of the nature and direction of the complementation that occurs between the syp and bmp mutants, we applied the mutants as spots separately, both adjacent but not touching (A) and touching (B). In the experiment shown, pKG11-containing ΔsypL (KV5069) and ΔbmpABC (KV6897) strains were used. Cultures were applied to plates and incubated at room temperature for 92 h (A) or 66 h (B). Wrinkling can be observed on the bmp mutant side of the interface of the touching colonies. The images are representative of at least two independent experiments.
TEM analysis reveals differences in the extracellular matrixes of the wild type and the bmp and sypL mutants.
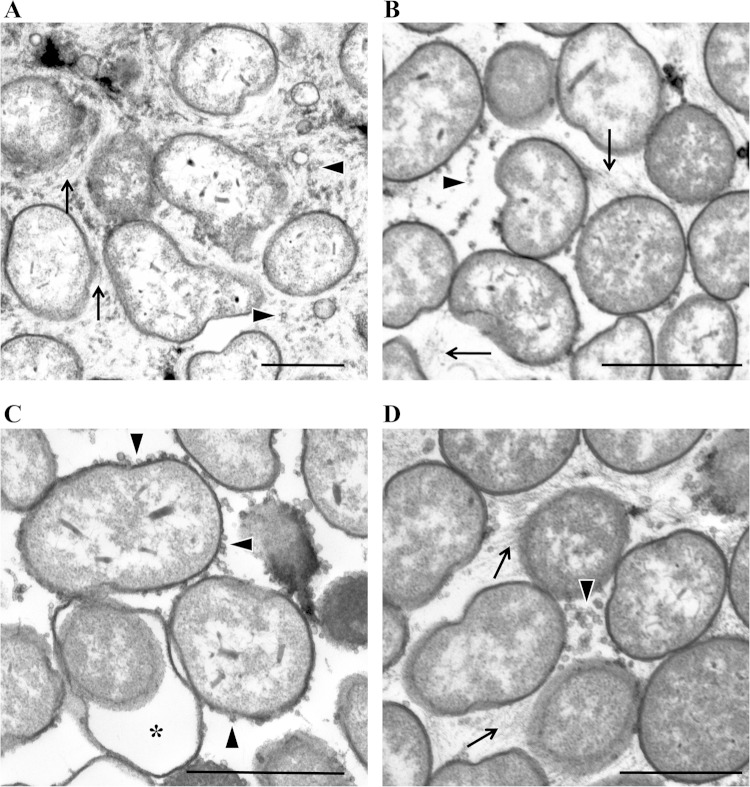
Since neither the bmp nor sypL mutant could promote wrinkled colony formation separately but the mutant strains could do so when mixed or applied as spots so that they would grow into each other, we asked whether we could observe differences in the extracellular matrixes of these mutants by microscopy. Therefore, we used TEM to analyze ultrathin sections of biofilm colonies produced by rscS-overexpressing strains, including the biofilm-competent wild-type control, the bmp mutant, and the sypL mutant, as well as the interface of the bmp and sypL mutants obtained from the spot-touching experiments. We used the stain ruthenium red to enhance staining of polysaccharide. As expected, cells in close association were present in all samples (Fig. 8). An electron-dense, thread-like material was readily detected in the extracellular matrixes of the control, bmp mutant, and interface samples but not in the sypL mutant sample (Fig. 8). The thread-like material appeared to be more abundant in the control strain colonies than in the bmp mutant colonies (Fig. 8A and B). We speculate that the thread-like material may be the Syp polysaccharide, since it was absent in the sypL mutant (Fig. 8C). We also observed numerous outer membrane vesicles in all the samples. Intriguingly, those seen in the sypL sample, but not those in the other samples, appeared to be largely cell associated. Thus, SypL specifically or Syp polysaccharide, in general, may promote release of outer membrane vesicles. Note that some cells of the sypL mutant exhibited a “swollen cell” phenotype, which was previously observed for other syp mutants (20); in addition, the cells of this mutant did not appear to be as tightly packed as cells within the samples of the other strains. Taken together, these data suggest that there are distinct differences between the control and the bmp and sypL mutants. While these experiments do not reveal the exact function of the Bmp proteins, they do provide insights into what they do not do, as the absence of the Bmp proteins did not prevent production of either outer membrane vesicles or the thread-like material.
FIG 8.
TEM analysis of biofilm mutants. We used TEM to visualize wrinkled and smooth colonies formed by applying the following pKG11-containing strains to LBS medium containing Tc: wild type (ES114) (A), bmpABC mutant (KV6897) (B), and sypL mutant (KV5069) (C). (D) We also collected samples from the interface of touching cultures of the pKG11-containing bmpABC and sypL mutant strains. Arrows indicate the thread-like material (likely a polymer or polysaccharide), arrowheads indicate outer membrane vesicles, and a star indicates the “swollen cell” phenotype (present only in the sypL mutant strain). Bars, 100 nm.
BmpA is secreted into the biofilm matrix.
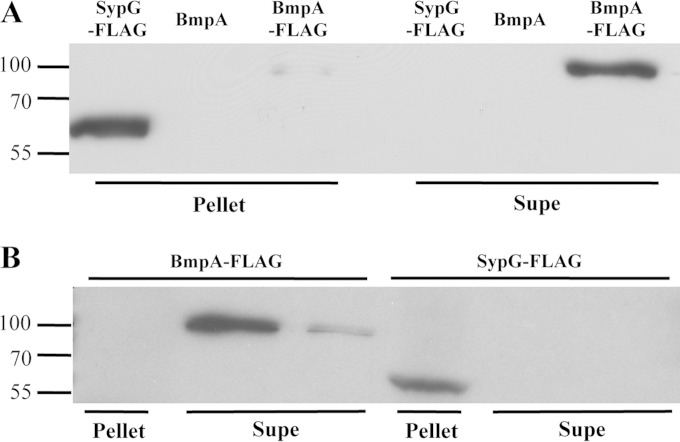
Our cell-mixing experiments suggested that Bmp or a Bmp-dependent product present in the cell matrix promotes biofilm maturation. Because the Bmp proteins contain a putative signal sequence, we hypothesized that these proteins may themselves be secreted into the matrix. To assess this possibility, we selected BmpA as a representative Bmp protein and generated a construct that encodes an epitope-tagged protein (BmpA-FLAG). We introduced this fusion construct into the chromosome of the Δbmp mutant and then induced biofilm formation with RscS. Expression of the BmpA-FLAG protein complemented the bmp mutant for wrinkled colony formation (see Fig. S6 in the supplemental material). We then examined the presence of BmpA-FLAG in cell-free supernatants of the bmp mutant via Western blot analysis with an anti-FLAG antibody. We observed a band at ∼100 kDa, which is slightly larger than the predicted size of BmpA (∼75 kDa) (Fig. 9A); we address the apparent size discrepancy in Discussion. This band was absent in a strain that expressed untagged BmpA, suggesting that, despite the apparent molecular mass difference, the antibody detected BmpA in the supernatant fraction (Fig. 9A). BmpA could also be observed in the cell pellet, though to a considerably lesser extent (Fig. 9A). Finally, as a control, we collected and examined cell pellets and supernatants from a strain that expresses SypG-FLAG; as a transcription factor (19, 24), SypG is expected to be located in the cytoplasm. Indeed, SypG could be found in the cell pellet but not in the supernatant fraction (Fig. 9A). These data indicate that BmpA is located outside the cell, where it may be positioned to directly affect biofilm architecture.
FIG 9.
BmpA is a secreted protein that is found in the supernatant fraction of disrupted pellicles. (A) To determine whether BmpA is secreted from V. fischeri cells, we collected the cell pellet and TCA-concentrated cell supernatant (Supe) for the following rscS-overexpressing strains: ΔsypG attTn7::sypG-FLAG (KV6475), ΔbmpABC attTn7::bmpA-balA (KV7062), and ΔbmpABC attTn7::bmpA-FLAG (KV7274) strains. (B) To assess the presence of BmpA within pellicles, we disrupted and fractionated pellicles formed by the same strains. For both panels, the resulting cell pellet and supernatant (Supe) samples were resolved by SDS-PAGE (12%), and ultimately, the presence of SypG-FLAG and BmpA-FLAG was assessed by Western immunoblotting with an anti-FLAG antibody. Sizes of the marker proteins (kDa) are indicated on the left.
To determine more directly if BmpA is localized to the matrix within V. fischeri biofilms, we collected pellicles formed by the BmpA-FLAG-expressing strain grown under static conditions. We then disrupted the pellicles by vortexing and separated fractions (cells and supernatant) by centrifugation. As a control, we performed the same experiment with the SypG-FLAG-expressing strain. Western blot analysis of these samples showed the presence of SypG-FLAG in the cell pellet but not the supernatant, while BmpA was present predominantly in the supernatant fraction (Fig. 9B). Together, these data support the location of BmpA within the biofilm and strongly suggest that BmpA serves as a biofilm matrix protein.
DISCUSSION
The work described here identifies the first V. fischeri matrix protein, BmpA, a novel protein unlike other characterized matrix proteins, and uncovers a role for it in biofilm maturation. BmpA represents the first member of a family of similar but uncharacterized proteins that includes V. fischeri BmpB and BmpC as well as proteins encoded by a variety of other bacteria. The bmpA, bmpB, and bmpC genes were previously identified as putative members of the SypG regulon (24), and we demonstrated here via reporter assays that SypG indeed regulates transcription of the bmp genes. The coordinate regulation of bmp with the syp polysaccharide locus provided the first indication that bmp might function in biofilm formation.
Given the similarity of the Bmp proteins, it is perhaps not surprising that they appear to have overlapping functions with respect to biofilm formation: deletion of one or a combination of two of the bmp genes did not significantly affect wrinkled colony formation, while deletion of all three genes caused a severe defect (Fig. 3). The exception to this was the case of the bmpAB double mutant, which exhibited a significant defect in this phenotype (Fig. 3). Interestingly, BmpA and BmpB are more similar to each other than they are to BmpC, suggesting that their function may be more conserved. Furthermore, while deletion of bmpA alone did not substantially affect biofilm formation, complementation of the triple bmp mutant with just bmpA permitted nearly normal biofilm formation (see Fig. S6 in the supplemental material). These results are analogous to those found with V. cholerae for matrix proteins Bap1 and RbmC, which have sequence similarity and can partially complement each other (13, 14, 47). We hypothesize that like BmpA, BmpB and BmpC are secreted into the matrix, where they can serve similar functions in controlling biofilm architecture. The Bmp proteins are not similar to the V. cholerae matrix proteins or to other known matrix proteins. The function in biofilm formation that we uncovered here and the presence of Bmp-encoding genes in numerous bacterial genomes make these novel matrix proteins of interest for further investigation.
The bmp genes are located upstream of the bal lipoprotein genes, which we similarly predicted would be involved in biofilm formation. However, only loss of balB exerted a negative impact on biofilm formation, which was modest at best and did not occur at all when balC was also missing (see Fig. S5 in the supplemental material); it is currently unclear why this was the case. Instead, we obtained some evidence that balC and, to a lesser extent, balB may contribute to control over cellular bioluminescence (see Fig. S4B). While previous links have been established between bioluminescence and biofilm formation in V. fischeri (48, 49), the fact that the bmp and bal genes appear to comprise an operon yet affect diverse cellular functions was surprising. Furthermore, the impact of the bal genes on luminescence was observed in the absence of sypG (or rscS) overexpression, suggesting that either the level of basal transcription from the SypG-dependent bmp promoter is sufficient for bal expression or there is a second promoter that drives bal expression. If the latter explanation is correct, then the promoter must be located outside the bmp genes, since the triple bmp mutant did not exhibit a defect in light production (see Fig. S4A). Additional work is necessary to understand the contribution of the Bal proteins to cellular bioluminescence. Overall, these findings represent the third connection between biofilm formation and bioluminescence in V. fischeri and, to our knowledge, the first time that lipoproteins have been implicated in controlling bioluminescence in vibrios.
Our work investigating the contribution of Bmp to biofilm formation has permitted a deeper understanding of the role played by the Syp polysaccharide in biofilm formation. Because our studies of the syp genes had revealed a tight correlation between wrinkled colony formation and pellicle formation (20), we were initially surprised to find that the triple bmp mutant, which formed smooth colonies (Fig. 3), retained the ability to form a pellicle (Fig. 4). However, our work has shown that Bmp promotes the development of the 3D architecture observed for both colonies and pellicles, while Syp is responsible for the “stickiness” associated with biofilm formation. Our subsequent cell-mixing experiments indicated that these two distinct SypG-dependent processes function together (Fig. 6) and that the Syp polysaccharide may not be a communal (i.e., shared) product, as we did not observe wrinkling within the syp mutant spot for the cell-touching experiments (Fig. 7). Thus, the Syp polysaccharide may be cell associated or poorly diffusible. In this regard, the V. fischeri biofilm is similar to the VPS-dependent biofilm of Vibrio cholerae, as the VPS polysaccharide also does not appear to be a communal product (13). Finally, our TEM analyses also revealed a possible new role for sypL and/or the Syp polysaccharide in the release of OMVs; most of the OMVs for this mutant appeared to be surface associated, with few free-floating vesicles. These experiments have thus provided one or perhaps two new phenotypes (“stickiness” and OMV release) and a new approach (mutant mixing) to probing the pathways that lead to biofilm formation, which we will utilize in the future to characterize the communal nature of matrix components and to categorize additional biofilm-defective mutants.
Because our spot-touching experiments suggested that the bmp mutant was supplied with a missing factor from the syp mutant, and because bioinformatic analyses of the Bmp proteins revealed the presence of a Sec-dependent signal sequence at their N termini (43) (see Fig. S2A in the supplemental material), we predicted that Bmp might be a secreted factor. Indeed, we found BmpA in cell-free supernatants under biofilm-inducing conditions, as well as in the cell-free supernatant fraction collected from disrupted pellicles (Fig. 9). Why BmpA migrates on SDS-PAGE to a position that is slightly higher than expected is unclear. Perhaps BmpA is modified during export. Given the presence of a putative Sec-dependent signal sequence (see Fig. S2), it is likely that the Sec pathway is responsible for secretion of BmpA (and presumably the other Bmp proteins) into the periplasm. However, it is unclear how BmpA crosses the barrier of the outer membrane. It is unlikely to be dependent on the Syp proteins for export, as all of the structural syp mutants retained the ability to complement the bmp mutant in mixing experiments (see Fig. S7). In V. cholerae, type II secretion appears to be involved in secretion of at least one matrix protein (50). Alternatively, it is possible that BmpA is exported via OMVs. For V. cholerae, a recent proteomic analysis of OMVs indicated that all three matrix proteins (RbmA, RbmC, and Bap1) are associated with OMVs (51); whether the OMVs serve to shuttle/deliver these proteins during biofilm formation is unclear. If BmpA were shuttled via OMVs, then the deficiency of the sypL mutant in vesicle release could account for the limited diffusion of Bmp in the spot-touching experiments.
The relatively limited degree of exogenous complementation in the spot-touching experiments may instead have been due to the need for Bmp to be positioned properly to exert its effect(s). This possibility is supported by the results of experiments (unpublished data) designed to exogenously complement the bmp mutant with Bmp protein extracted from pellicles: wrinkled colonies did not form upon the addition of Bmp-containing extracts, indicating that the mere presence of Bmp protein is insufficient for complementation. Thus, evaluating how and where Bmp is localized will be an intriguing future direction for this work.
How do the Bmp proteins function to promote biofilm maturation? The answer to this question remains unclear. TEM analysis revealed the presence of a thread-like material between cells in the wild type (Fig. 8A) and, to a lesser extent, the bmp mutant (Fig. 8B). Since the thread-like material was absent in the sypL mutant colonies (Fig. 8C), we predict that this substance may be the Syp polysaccharide. Whether Bmp directly interacts with the Syp polysaccharide and whether it helps to organize the Syp polysaccharide or retain it on the cell surface or on an abiotic surface, or plays some other role, remain to be determined.
Due to the conservation of the Bmp proteins in other Vibrio spp. and marine bacteria, their function may be relevant to marine environments. However, our preliminary experiments did not reveal a role for bmp in initiation of symbiotic colonization (unpublished results). This is perhaps not surprising given that the biofilm formed by V. fischeri during symbiotic colonization is transient in nature, a process that may be independent of the development of biofilms with substantial 3D architecture. Because syp mutants exhibit severe colonization defects, it is likely that the “stickiness” contributed by the Syp polysaccharide is a key event in proficient colonization. It will be interesting to determine if/when during symbiotic colonization the Bmp protein can be found outside V. fischeri cells.
In summary, our study identifies the first biofilm matrix protein for V. fischeri, provides a new set of tools (cell mixing/touching and “stickiness”) to aid in our understanding of this process, and reveals that biofilm formation is a stage distinct from biofilm maturation for this organism. Because the Bmp proteins are unrelated to other known biofilm matrix proteins (27, 28), they represent a novel class of matrix proteins. Thus, understanding the exact function of the Bmp proteins in promoting the 3D architecture of biofilm formation and how their function compares to that of other characterized matrix proteins is an important area of future study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Allison Norsworthy for providing strain KV6475, Krystal Thomas-White, Cecilia Thompson, and Dörte Lehmann for experimental assistance, Alan Wolfe and Sean Crosson for helpful discussions, and members of the Visick lab for reviewing the manuscript.
This work was supported by funding from the National Institutes of Health (grant R01 GM59690) to K.L.V. and by a Schmitt fellowship awarded to V.A.R.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02292-14.
REFERENCES
- 1.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 2.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 4.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci U S A 102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang WS, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol 189:8290–8299. doi: 10.1128/JB.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mah TF. 2012. Biofilm-specific antibiotic resistance. Future Microbiol 7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 8.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilboa-Garber N, Mizrahi L, Garber N. 1972. Purification of the galactose-binding hemagglutinin of Pseudomonas aeruginosa by affinity column chromatography using Sepharose. FEBS Lett 28:93–95. doi: 10.1016/0014-5793(72)80685-9. [DOI] [PubMed] [Google Scholar]
- 11.Garber N, Guempel U, Gilboa-Garber N, Doyle R. 1987. Specificity of the fucose-binding lectin of Pseudomonas aeruginosa. FEMS Microbiol Lett 48:331–334. doi: 10.1111/j.1574-6968.1987.tb02619.x. [DOI] [Google Scholar]
- 12.Tielker D, Hacker S, Loris R, Strathmann M, Wingender J, Wilhelm S, Rosenau F, Jaeger KE. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- 13.Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog 7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 17.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris AR, Darnell CL, Visick KL. 2011. Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol Microbiol 82:114–130. doi: 10.1111/j.1365-2958.2011.07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 20.Shibata S, Yip ES, Quirke KP, Ondrey JM, Visick KL. 2012. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 194:6736–6747. doi: 10.1128/JB.00707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visick KL, Skoufos LM. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussa EA, O'Shea TM, Darnell CL, Ruby EG, Visick KL. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J Bacteriol 189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussa EA, Darnell CL, Visick KL. 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol 190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray VA, Eddy JL, Hussa EA, Misale M, Visick KL. 2013. The syp enhancer sequence plays a key role in transcriptional activation by the sigma54-dependent response regulator SypG and in biofilm formation and host colonization by Vibrio fischeri. J Bacteriol 195:5402–5412. doi: 10.1128/JB.00689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris AR, Visick KL. 2013. The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri. Mol Microbiol 87:509–525. doi: 10.1111/mmi.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris AR, Visick KL. 2013. Inhibition of SypG-induced biofilms and host colonization by the negative regulator SypE in Vibrio fischeri. PLoS One 8:e60076. doi: 10.1371/journal.pone.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A 102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandel MJ, Stabb EV, Ruby EG. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol 65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 34.DeLoney CR, Bartley TM, Visick KL. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol 184:5121–5129. doi: 10.1128/JB.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Shibata S, Visick KL. 2012. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol 194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCann J, Stabb EV, Millikan DS, Ruby EG. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol 69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/S0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 40.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]
- 41.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol 20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188:2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 46.Tokuda H, Matsuyama S. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta 1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Fong JC, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol 189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray VA, Visick KL. 2012. LuxU connects quorum sensing to biofilm formation in Vibrio fischeri. Mol Microbiol 86:954–970. doi: 10.1111/mmi.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyashiro T, Oehlert D, Ray VA, Visick K, Ruby EG. 25 September 2014. The putative oligosaccharide translocase SypK connects biofilm formation with quorum signaling in Vibrio fischeri. Microbiologyopen doi: 10.1002/mbo3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altindis E, Fu Y, Mekalanos JJ. 2014. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc Natl Acad Sci U S A 111:E1548–E1556. doi: 10.1073/pnas.1403683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.