Abstract
Human milk oligosaccharides (HMOs) play a key role in shaping and maintaining a healthy infant gut microbiota. This article demonstrates the potential of combining recent advances in glycomics and genomics to correlate abundances of fecal microbes and fecal HMOs. Serial fecal specimens from two healthy breast-fed infants were analyzed by bacterial DNA sequencing to characterize the microbiota and by mass spectrometry to determine abundances of specific HMOs that passed through the intestinal tract without being consumed by the luminal bacteria. In both infants, the fecal bacterial population shifted from non-HMO-consuming microbes to HMO-consuming bacteria during the first few weeks of life. An initial rise in fecal HMOs corresponded with bacterial populations composed primarily of non-HMO-consuming Enterobacteriaceae and Staphylococcaeae. This was followed by decreases in fecal HMOs as the proportion of HMO-consuming Bacteroidaceae and Bifidobacteriaceae increased. Analysis of HMO structures with isomer differentiation revealed that HMO consumption is highly structure-specific, with unique isomers being consumed and others passing through the gut unaltered. These results represent a proof-of-concept and are consistent with the highly selective, prebiotic effect of HMOs in shaping the gut microbiota in the first weeks of life. The analysis of selective fecal bacterial substrates as a measure of alterations in the gut microbiota may be a potential marker of dysbiosis.
Keywords: Term infants, human milk, oligosaccharides, prebiotic, microbiota, mass spectrometry, HMOs
Introduction
Breast milk, the sole source of nourishment for newborns, has been under intense selective pressure over millions of years of evolution to meet the infant’s needs to grow and survive.1,2 Interestingly, the third most abundant chemical component class in human milk after lactose and lipids, human milk oligosaccharides (HMOs), is both non-nutritive and non-digestible.3 HMOs play a key role in creating and maintaining a healthy infant gut microbiota through two established mechanisms. First, HMOs have prebiotic effects (promoting growth of beneficial bacteria such as Bifidobacterium and Lactobacillus).4 Recent metagenomic studies have confirmed that bifidobacteria are enriched in the intestines of healthy breast fed infants.5 In vitro studies provide a reason for the prevalence of bifidobacterial and even bacteroides species: they selectively consume HMOs.6,7 A number of studies have characterized the enzymes in bifidobacteria that specifically degrade HMOs (reviewed in ref (8)).
Second, HMOs compete for specific pathogen binding with sites in the infant gut.9 Free HMOs and conjugated cell surface glycoforms are synthesized by similar glycosyltransferases and thus have common epitopes. Ingested HMOs can thus interact with enterocyte surface molecules, limiting binding to these sites by viruses such as HIV10 and rotavirus.11 In addition, because HMOs are also unbound, they can serve as free analogues of pathogen host receptors. Instead of binding to cell surface glycoproteins or glycolipids, pathogens bind to HMOs. HMOs therefore act as decoys and protect infants from infectious diseases.9 Anti-adhesive activity of free HMOs has been described for Vibrio cholerae,12Salmonella fyris,12 enteropathogenic12 and enterotoxigenic13Escherichia coli, Streptococcus pneumoniae,14 and caliciviruses.13 The large diversity of HMO structures suggests multiple functions.15 Separated HMO fractions have been shown to have differing activities. For instance, fucosylated HMOs inhibit the binding of Campylobactor jejuni to intestinal cells,16 whereas sialylated HMOs block the adhesion of E. coli to human erythrocytes.17
The functions of HMOs are related to their specific structures; however, identifying and quantifying those structures has been a major challenge in HMO analysis. HMOs can have diverse and complicated structures even though they commonly share a lactose core consisting of glucose and galactose linked via a β-1,4-linkage.18 Functional studies to date have involved either a limited number of structures or little or no structural information.
The benefits of breast feeding include the long-term health implications of a well-established, specific, and protective microbiota. Recent advances in glycomic analysis and genomic sequencing have allowed us to observe the correlation between HMO consumption and gut microbiota populations. In this study, we demonstrate two examples of direct correlation between the abundances of HMOs in feces and the composition of the infant fecal microbiota as proof-of-concept. HMOs from fecal samples of breast-fed infants were analyzed using advanced separation and mass spectrometry methods that allow quantitation of specific structures. Microbial populations were characterized by next-generation sequencing of 16S rDNA amplicons, specific qPCR, and Bifidobacterium species-specific terminal restriction fragment length polymorphism (Bif-TRFLP). The observed shift in the gut microbiota of these two healthy infants from a non-HMO-consuming population into a more saccharolytic microbiota with a corresponding decrease in fecal HMO intensities supports more extensive analyses of larger populations.
Methods
Sample Collection and Handling
The study was approved by the Institutional Review Board at UC Davis (protocol no. 200715509-4), and informed consent was obtained from the parents of infants prior to participation. The infants included were born vaginally at term and were not treated with antibiotics during the study period. Infant A received exclusive breast milk feeding. Infant B had formula supplementation for 4 days right after birth, from days 2 to 6, and then was solely breast fed. Details of the mothers’ diet and health status were not collected as part of this study.
Feces samples were collected from two full-term infants as follows: twice a week for the first month, twice a month in the second month, and once or twice a month thereafter. The samples were stored at −80 °C prior to analysis. Samples were thawed, reconstituted with water, and homogenized prior to analysis. Homogenate was left in the shaker at 4 °C overnight. The mixture then was centrifuged at 4000g for 30 min at 4 °C.
Isolation, Reduction, and Purification of HMOs from Fecal Samples
Crude HMOs were isolated and purified from extracted feces through a series of liquid- and solid-phase extractions, as described previously.19,20 Briefly, four volumes of chloroform/methanol (2:1 v/v) were added to the decanted liquid, and the mixture was centrifuged at 4000g for 30 min at 4 °C. The upper layer was carefully transferred. Two volumes of ethanol were added, and the mixture was left at 4 °C overnight and then centrifuged for 30 min at 4 °C. The supernatant solution was evaporated to dryness using a centrifugal evaporator (Savant AES 2010).
HMOs were reduced to alditol form by adding sodium borohydride and incubating at 65 °C for 1 h, purified by solid-phase extraction using C8 and graphitized carbon columns,20 and evaporated to dryness. The sample was reconstituted in nanopure water prior to mass spectrometry.
Mass Spectrometric Analysis and Identification of Fecal HMOs
Oligosaccharides were profiled using matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry (MALDI FT-ICR MS, Agilent, formerly Varian, Palo Alto, CA, equipped with an external ProMALDI source) and nano-high-performance liquid chip/time-of-flight (nano-HPLC-Chip/TOF, Agilent 6200) MS, as described previously.19,20 For MALDI FT-ICR MS experiments, 2,5-dihydroxy-benzoic acid (DHB) was used as matrix (5 mg/100 μL in 50:50 ACN/H2O) and sodium chloride (0.01 M in 50:50 ACN/H2O) was used as cation dopant. The precursor ions were detected primarily as [M + Na]+ ions in the positive ionization mode. Identification of HMO compositions was done based on accurate masses using Glycan Finder software (written in-house in Igor Pro 5.04B, Wavemetrics), which filters experimental accurate masses to a specified mass tolerance (set at 20 ppm) based on accurate masses of theoretical oligosaccharides.
The nano-HLPC-Chip/TOF MS experiments, as described previously,19,20 were performed using a porous graphitized carbon HPLC microchip with a 40 nL enrichment column and 43 × 0.075 mm i.d./5 μm pore size analytical column. Separation was achieved using a binary gradient solvent system with A (3% ACN in 0.1% formic acid solution) and B (90% ACN in 0.1% formic acid solution). The chip column was equilibrated and eluted at a flow rate of 0.4 μL for nanopump and 4 μL for capillary pump. The gradient ran for 65 min and was programmed as follows: 2.5–20 min, 0–16% B; 20–30 min, 16–44% B; 30–35 min, B increased to 100%; 35–45 min, continue at 100% B; and 45–65 min, 0% B to allow equilibration of the column prior to the next sample injection. Data were acquired in the mass range of m/z 200–3000 using electrospray ionization (ESI) in the positive ionization mode. Deconvoluted peaks were extracted using the molecular feature of the Agilent Mass Hunter software at 20 ppm error. The resulting peak list was further analyzed using the LC–MS Searcher software21 (in-house, written in Java), which assigns structures to a peak based on retention time and accurate mass. LC–MS Searcher uses a library of 45 neutral HMOs and 30 acidic HMOs characterized using exoglycosidases and MS/MS by Wu et al.22,23 Around 2000 spectra are inputted in the program per run, and with the HMO library, the LC–MS Searcher software identifies HMOs from all of the scans based on retention times and m/z values. It then outputs intensities, structures, and deuterium/hydrogen (D/H) ratios of said HMOs. This library approach allows for separation of isomeric species and identification of individual structures based primarily on accurate masses and reproducible retention times, since extensive confirmation of HMOs by MS/MS and exoglycosidases were done previously.22,23
Quantification of Milk Oligosaccharides Using Deuterium-Labeled Internal Standard
Deuterium-labeled internal standards were used for the relative quantification of HMOs using MALDI FT-ICR MS, as described previously.20 This method gives a relative standard deviation of less than 15% and allows for a more linear quantitation over a larger dynamic range.20 Briefly, pooled HMOs isolated from milk of several donor mothers were reduced using sodium borodeuteride, purified by graphitized carbon solid-phase extraction, and used as reference solution. Both the deuterated reference solution and sample solution were spotted as one sample onto the MALDI plate and analyzed by MALDI FT-ICR MS in the positive mode (n = 3). The ratio of sample reduced by NaBH4 and standard reduced by NaBD4 (H/D ratio) was calculated using Microsoft Excel using the formula described below.
H/D ratios were used for quantitation as described by Ninonuevo et al.24
where m and n are the experimental intensities of A (monoisotopic peak) and A + 1, respectively, and p and q are calculated intensities of A and A + 1, respectively.
To compute the temporal changes in the H to D ratios across a set of samples, a reference sample is set and is used to compare with all the other samples
To quantitate oligosaccharide to the isomer level, both the deuterated reference solution and sample solution were also injected into the nano-HPLC-Chip/TOF MS. H/D ratios were calculated using LC–MS Searcher21 (the program outputs D/H ratios; we took the reciprocal) and verified manually using Microsoft Excel.
Bacterial DNA Extraction
Bacterial genomic DNA was extracted from fecal samples in order to analyze the infant gut microbial populations as previously described with a few modifications.25 Briefly, 200 mg of stool was resuspended 1:10 in ice-cold phosphate buffer solution (PBS) and centrifuged (8000g for 5 min at room temperature); then, the supernatant was decanted. This rinse was repeated twice, and the fecal pellet was resuspended in 200 μL of lysis buffer (2 mM EDTA, 1.2% TritonX-100, 20 mM Tris-HCl, pH 8.0) with freshly added 40 mg/mL lysozyme. The solution was incubated at 37 °C for 30 min. Buffer ASL from QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) was added to equal 2.0 mL, and the sample was then vortexed until it was thoroughly mixed. Samples were then homogenized by bead-beating in a FastPrep-24 instrument (MP Biomedicals, Solon, OH) for 2 min at 6.5 m/s. Homogenate was incubated for 5 min at 95 °C, vortexed, and centrifuged at 13 000g for 1 min to pellet stool particles. 1.2 mL of the supernatant was used to purify DNA with the Qiagen Stool Mini Kit according to the manufacturer’s instructions.
Pyrosequencing
Preparation of fecal DNA samples for pyrosequencing was carried out as previously described25 by the Core for Applied Genomics and Ecology (CAGE, University of Nebraska). The V1–V3 region of the 16S rRNA gene was PCR amplified using bar-coded universal primers with Roche-454 A or B Titanium adapter sequences, shown in italics, B-8F (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGAGTTTGATCMTGGCTCAG-3′) and A-518R (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNATTACCGCGGCTGCTGG-3′), where the N represents the 8 base barcode sequence unique to each sample. To ensure the representation of Bifidobacteria, Bifidobacteria-specific primer B-8Fbif (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGGGTTCGATTCTGGCTCAG-3′) was mixed with the 8F universal primer, which has a 3 base pair mismatch, at a concentration of 4:1. The PCR mixture and amplification conditions were as previously described.25 Sequencing was performed using the 454 Roche sequencing primer kit from the A end and carried out using the standard protocol on a Roche Genome Sequencer GS-FLX.
Illumina Sequencing
Samples were prepared for sequencing as previously described26 with some variations. The V4 region of the 16S rRNA gene was PCR amplified using universal barcoded primers with Illumina sequencing adapters, shown in italics, the N represents the 8 bp barcode sequence unique to each sample, and the linker is in bold, V4F (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNGTGTGCCAGCMGCCGCGGTAA-3′) and V4Rev (5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTCCGGACTACHVGGGTWTCTAAT-3′). PCR reactions contained 12.5 μL of 2× GoTaq Green Master Mix (Promega, Madison, WI), 1.0 μL of 25 mM MgCl2, 8.5 μL of water, 0.5 μL of forward and reverse primers (10 μM final concentration), and 2.0 μL of genomic DNA. PCR amplification was carried out in triplicate with conditions as previously described.26 Amplicons were combined and cleaned using the QIAquick 96 PCR Purification Kit (Qiagen, Valencia, CA). Amplicon DNA concentrations were quantified using the Quant-iT PicoGreen dsDNA Kit in 96-well microplates, and fluorescence detection, composite sample mixture, and gel purification were carried out as previously described.26 The sample was sent to the University of California DNA Technologies Core Facility for sequencing on an Illumina Genome Analyzer II sequencing platform.
Sequence Analysis
The QIIME software package (version 1.4.0) was used to analyze the results of both the pyrosequencing and Illumina sequencing runs.27 V1–V3 16S rRNA gene sequences from pyrosequencing reads were removed from analysis if they were <200 bp in length, contained >3 ambiguous bases, had a mean quality score <25, contained a homopolymer run greater than 6 nt, or did not contain a primer or a barcode sequence. Similar sequences were clustered into operational taxonomic units (OTUs) with UCLUST software28 and minimum identity of 97%. The most abundant sequence was chosen to represent each OTU. Taxonomy was assigned to each OTU with the Ribosomal Database Project (RDP) classifier,29 with a minimum support threshold of 80% and the RDP taxonomic nomenclature. OTU representatives were aligned against the Greengenes core set30 with PyNAST software31 with a minimum alignment length of 150 bp and a minimum identity of 75%. A phylogenetic tree was inferred using FastTree algorithm.32
Illumina V4 16S rRNA gene sequences were demultiplexed and quality-filtered using the QIIME software package as well.27 Reads were truncated after a maximum number of 3 consecutive low-quality scores. After quality trimming, reads were removed from analysis if they were <60 bp and the number of ambiguous bases was >3. OTU clustering, taxonomy assignment, phylogenetic tree-building, and alignment were carried out as stated above, with the exception that the minimum alignment length was 75 bp. OTUs with sequences that numbered less than 0.01% of the total reads were filtered out to reduce noise in the assignments.
Community Comparisons and Statistics
Alpha rarefaction was performed using the observed species metrics. Ten sampling repetitions were performed at each sampling depth, without replacement. Beta diversity was estimated by computing weighted and unweighted UniFrac distances between samples.
Pearson product–moment correlation coefficients were calculated in QIIME27 between the nano-HPLC-Chip/TOF MS D/H ratios of individual HMO isomers normalized to baseline intensity and the relative abundances of order-level bacterial taxa. Only bacterial orders detected in at least three samples per infant were included.
Real-Time Quantitative Polymerase Chain Reaction (qPCR)
SYBR green and TaqMan qPCR assays were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) with primers specific for universal Bacteria, Bacteroidales, and Bifidobacterium, and species-specific Bifidobacterium primers for B. longum group, B. adolescentis group, B. catenulatum group, B. breve, and B. bifidum. SYBR green assays contained 10 μL of 2× Takara Perfect Real Time master mix, 6 μL of water, 1 μL each of forward and reverse primers, and 2 μL of genomic DNA. Genomic DNA was diluted 1:100 for Bacteria, Bacteroidales, and Bifidobacterium assays. Cycling conditions were as previously described (Supporting Information Table 1). Bifidobacterium TaqMan qPCR assays contained 12.5 μL of 2× TaqMan Universal PCR master mix (Applied Biosystems), 2.5 μL each of forward and reverse primers, TaqMan probe, 3.75 μL of water, and 2.5 μL of diluted genomic DNA. B. longum group TaqMan assays contained 10 μL 2× TaqMan Universal PCR master mix (Applied Biosystems), 1 μL each forward and reverse primers and TaqMan probe, 5 μL water, and 2 μL genomic DNA. Reaction conditions and primer concentrations were as described in Supporting Information Table 1. All reactions were carried out in triplicate with a nontemplate control.
Bifidobacterium Species-Specific Terminal Restriction Fragment Length Polymorphism (Bif-TRFLP)
Bifidobacterium genus-specific primers (NBIF389, 5′-GCCTTCGGGTTGTAAAC; and NBIF1018R, 5′-GACCATGCACCCCTGTG-3′) were designed targeting a consensus alignment of 16S rRNA genes from all Bifidobacterium sequences deposited in the ribosomal database project (RDP) database33,34 aligned using ClustalX.35 Primer specificity and taxonomic coverage were determined using PrimerProspector,26 checking against a representative subset of the greengenes 16S rRNA database filtered at 97% identity.30 Samples were amplified by PCR in 50 μL reactions containing 5–100 ng of DNA template, 25 μL of 2× Promega GoTaq Green Master Mix (Promega, Madison, WI), 1 mM of MgCl2, and 2 pmol of each primer. Each PCR was performed in triplicate, and the products combined prior to purification. The PCR conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 51 °C for 1 min, and extension at 72 °C for 2 min, with a final extension at 72 °C for 5 min. Amplicons were digested using AluI and HaeIII following the manufacturers’ instructions for each enzyme. The digested DNA was submitted to the UC Davis College of Biological Sciences Sequencing Facility for fragment analysis. Traces were visualized using the program Peak Scanner v1.0 (Applied Biosystems) using a baseline detection value of 10 fluorescence units. Peak filtration and clustering were performed with R software using the IBEST script suite.36 OTU picking was based on an in silico digest database generated by the virtual digest tool from MiCA37 of good-quality 16S rRNA gene sequences compiled by the Ribosomal Database Project Release 10,33,34 allowing up to 3 nucleotide mismatches within 15 bp of the 5′ terminus of the forward primer.
Results
Fecal Profiling of Infant A
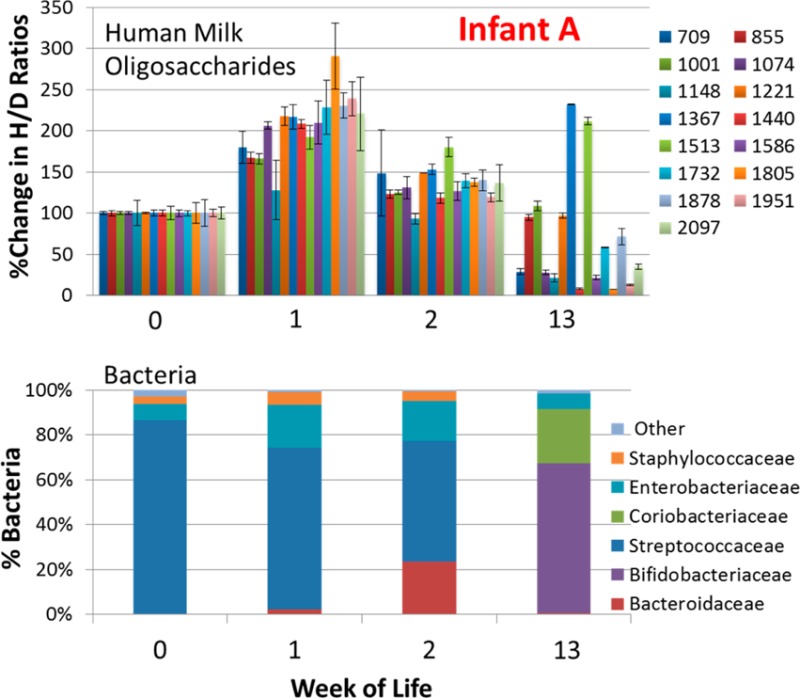
Fecal glycoprofiles and microbial profiles were obtained for weeks 0, 1, 2, and 13. Previous results have shown that the glycan composition is relatively constant for mature milk over a 6 month period.38 The amount of HMO ingested increases as milk consumption increases over the first several weeks of life. Figure 1 shows the fecal oligosaccharide compositions (analyzed by MALDI FT-ICR MS) and bacterial populations (analyzed by 16S rDNA pyrosequencing) of Infant A at the four time points. Oligosaccharides are listed in the legend as nominal masses of the reduced glycan. Corresponding monoisotopic masses and compositions of the nominal masses in Figure 1 are shown in Table 1. The table also shows monoisotopic masses of [M + Na]+ ion, the commonly observed ion in MALDI FT-ICR MS. Exact masses and elemental compositions were calculated using the NIST Glyco Mass Calculator (De Leoz and Stein, http://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:glycocalc).
Figure 1.
Changes in oligosaccharide intensities and bacterial population in the feces of Infant A. (A) Percent change in H/D ratios of HMOs in the feces of Infant A at weeks 0, 1, 2, and 13. Intensities were obtained using MALDI FT-ICR MS with n = 3. H/D ratios were normalized to week 0, set at 100%. Each bar represents an oligosaccharide nominal mass. (B) Corresponding fecal bacterial population of Infant A using 16S rDNA pyrosequencing.
Table 1. Nominal Masses, Monoisotopic Masses, and Compositions of the Reduced HMOs Observed in Infants A and Ba.
| reduced glycan, M |
monosaccharide
composition |
elemental composition |
[M + Na]+ ion | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| no. | nominal mass | monoisotopic mass | Hex | HexNAc | Fuc | NeuAc | C | H | N | O | monoisotopic mass |
| 1 | 709 | 709.2641 | 3 | 1 | 0 | 0 | 26 | 47 | 1 | 21 | 732.2533 |
| 2 | 855 | 855.3220 | 3 | 1 | 1 | 0 | 32 | 57 | 1 | 25 | 878.3112 |
| 3 | 1001 | 1001.3799 | 3 | 1 | 2 | 0 | 38 | 67 | 1 | 29 | 1024.3691 |
| 4 | 1074 | 1074.3963 | 4 | 2 | 0 | 0 | 40 | 70 | 2 | 31 | 1097.3855 |
| 5 | 1148 | 1147.4378 | 3 | 1 | 3 | 0 | 44 | 77 | 1 | 33 | 1170.4270 |
| 6 | 1221 | 1220.4542 | 4 | 2 | 1 | 0 | 46 | 80 | 2 | 35 | 1243.4434 |
| 7 | 1367 | 1366.5121 | 4 | 2 | 2 | 0 | 52 | 90 | 2 | 39 | 1389.5013 |
| 8 | 1440 | 1439.5285 | 5 | 3 | 0 | 0 | 54 | 93 | 3 | 41 | 1462.5177 |
| 9 | 1513 | 1512.5700 | 4 | 2 | 3 | 0 | 58 | 100 | 2 | 43 | 1535.5592 |
| 10 | 1586 | 1585.5864 | 5 | 3 | 1 | 0 | 60 | 103 | 3 | 45 | 1608.5756 |
| 11 | 1732 | 1731.6443 | 5 | 3 | 2 | 0 | 66 | 113 | 3 | 49 | 1754.63349 |
| 12 | 1805 | 1804.6607 | 6 | 4 | 0 | 0 | 68 | 116 | 4 | 51 | 1827.6499 |
| 13 | 1878 | 1877.7022 | 5 | 3 | 3 | 0 | 72 | 123 | 3 | 53 | 1900.6914 |
| 14 | 1951 | 1950.7186 | 6 | 4 | 1 | 0 | 74 | 126 | 4 | 55 | 1973.7078 |
| 15 | 2097 | 2096.7765 | 6 | 4 | 2 | 0 | 80 | 136 | 4 | 59 | 2119.7657 |
The monoisotopic masses of the [M + Na]+ precursor ion, the commonly observed ion in MALDI FT-ICR MS, are also shown. Hex, hexose; HexNAc, N-acetylhexosamine; Fuc, fucose; NeuAc, N-acetylneuraminic acid.
All HMOs were normalized to week 0. The graph in week 0 (Figure 1A) should not be interpreted as each composition having the same abundances. Each bar in the glycoprofile corresponds to a specific oligosaccharide composition. Each composition is composed of several isomers, as discussed in greater detail below. Infant A shows a general increase of all fecal HMOs from week 0 to week 1. At week 2, the amount of fecal HMOs decreases to nearly the original level of week 0. At week 13, the fecal HMOs decrease to a fraction of the peak amount at week 1. The decrease is not uniform; there are a number of compositions that increase or are unchanged at week 13.
Order-level fecal populations are shown in Figure 2A, and genus level, in Figure 2B. Multiplexed pyrosequencing and Illumina sequencing of Infant A’s samples yielded 12 779 and 288 390 partial 16S rRNA gene sequence reads (mean length 492 and 143 bp), respectively. The mean number of reads per sample was 5458 for pyrosequencing and 72 098 for Illumina. The longer pyrosequencing reads allowed taxonomic identification down to the genus level. The observed early predominance of Streptococcus in the gut (Figure 1B) is consistent with previous observations in breast fed infants.39 Marcobal et al.40 previously demonstrated that tested species of streptococci consumed HMO poorly or not at all. During the first week the fecal oligosaccharides increase; we hypothesize that they are being provided by the mother but are not being consumed by the gut microbes. The amount of oligosaccharides in feces peaks during week 1; however, by week 2, there is a noticeable decrease in intensities of HMOs corresponding with a small increase in fecal Bacteroides sp. population (Figure 1A,B). Members of Bacteroides and Bifidobacterium sp. are known to be strong consumers of HMOs.7,8 Illumina sequencing shows a constant presence of low levels of Bifidobacterium sp. in the first 3 weeks that is not detected in the pyrosequencing results. Interestingly, by week 13, the Bacteroides are replaced by Bifidobacterium sp., which dominate the fecal bacterial population, and the levels of most fecal HMO compositions have dropped dramatically.
Figure 2.
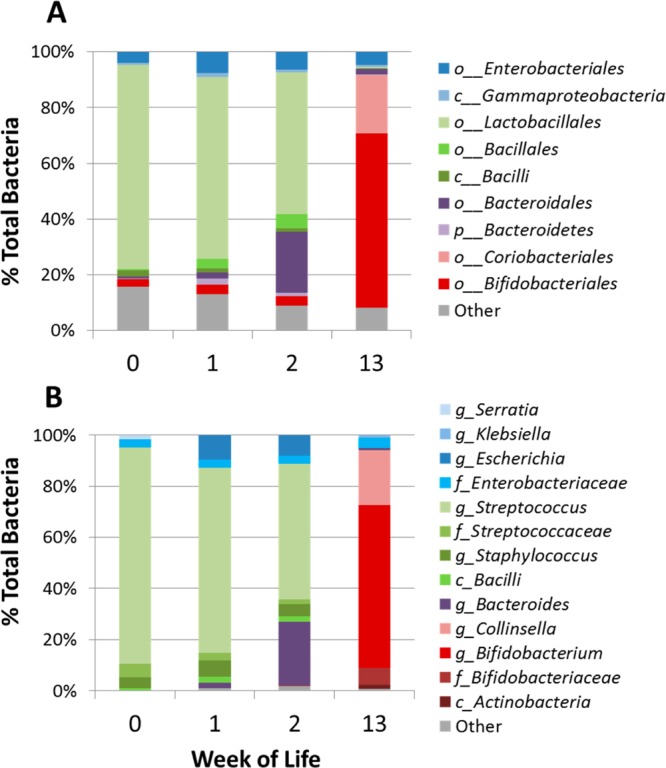
Infant A microbial population changes over the first 13 weeks based on next-generation sequencing. (A) Illumina and (B) pyrosequencing profile of the V4 and the V1–V3 regions of the 16S rRNA gene, respectively. The letters preceding the taxon are taxonomy identifiers: p (phylum), c (class), o (order), f (family), and g (genus).
Bifidobacterium-specific TRFLP and qPCR revealed a succession of bifidobacterial species over time (Figure 3). B. longum (includes two subspecies: B. longum subsp. longum and B. longumssp. infantis) dominated these samples from weeks 0–13, followed by an increase in B. adolescentis, B. bifidum, and B. breve at weeks 13–26 (data not shown for weeks > 13).
Figure 3.
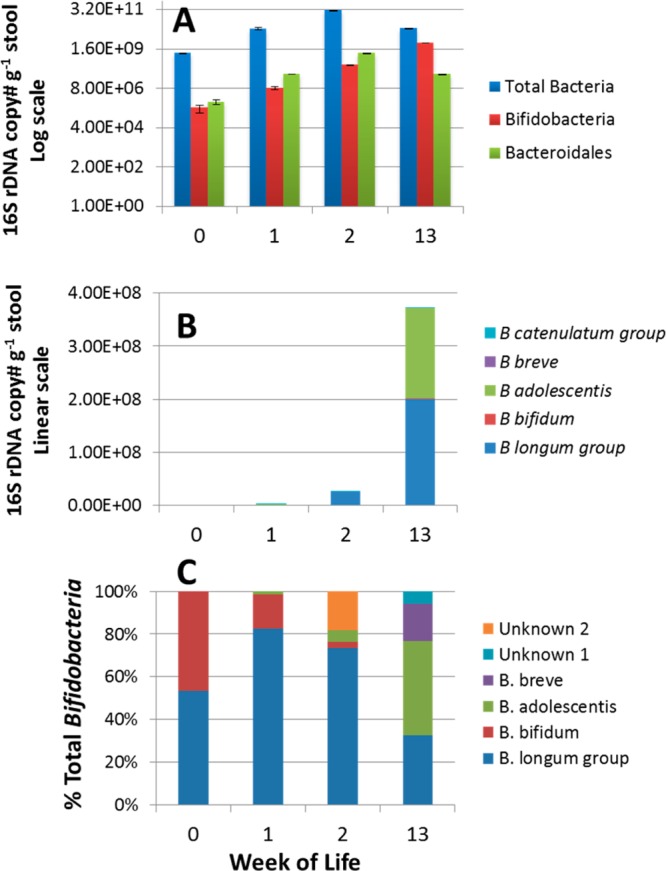
Infant A microbial population changes over the first 13 weeks based on qPCR and Bif-TRFLP analyses. qPCR of (A) Total bacteria, Bifidobacteria, and Bacteroidales and (B) species-specific Bifidobacteria populations; (C) Bif-TRFLP profile of the total bifidobacteria community.
The correlation of bifidobacterial growth with HMO consumption was confirmed by calculating Pearson product–moment correlation coefficients between normalized HMO D/H ratios and the relative abundances of all order-level bacterial taxa detected in at least three samples. Bifidobacteriales was the only clade yielding a significant, negative correlation coefficient with five HMOs, as shown in Table 2 (negative correlation implying that as fecal bifidobacteria numbers increase, fecal HMOs decrease, consistent with consumption of the HMOs). Other bacteria demonstrate a significant, positive correlation, indicating that as these microbes increased fecal HMOs increased as well (consistent with lack of consumption of HMOs by these bacteria, e.g., Enterobacteriales and Bacillales).
Table 2. Pearson Product–Moment Correlation between Order-Level Taxa and HMO Abundance in Infant A.
| substrate | prob | r | order |
|---|---|---|---|
| 5130a | 0.083 | 0.829 | Lactobacillales |
| 0.044 | 0.889 | Bacillales | |
| pLNH | 0.008 | 0.963 | Lactobacillales |
| IFLNH IIII | 0.042 | 0.891 | Clostridiales |
| MFLNH III | 0.023 | 0.929 | Enterobacteriales |
| MFLNH I | 0.019 | 0.937 | Lactobacillales |
| 0.029 | –0.916 | Bifidobacteriales | |
| IFLNH I | 0.020 | –0.933 | Bifidobacteriales |
| LNT | 0.041 | 0.894 | Bacillales |
| 0.046 | –0.885 | Bifidobacteriales | |
| LnNH | 0.014 | –0.948 | Bifidobacteriales |
| 0.010 | 0.958 | Lactobacillales | |
| TFLNH | 0.007 | 0.967 | Other |
| 5130a | 0.014 | 0.949 | Enterobacteriales |
| DFLNHc | 0.012 | 0.954 | Bacillales |
| LNH | 0.032 | 0.909 | Other |
| 5230b | 0.013 | –0.951 | Bifidobacteriales |
| 5230a | 0.044 | 0.888 | Enterobacteriales |
Fecal Profiling of Infant B
A second infant, Infant B, also shows development of a saccharolytic microbiota. Figure 4A,B shows the glycoprofiles (analyzed by MALDI FT-ICR MS) and bacterial populations (analyzed by 16S rDNA pyrosequencing) over the infant’s first 14 weeks of life, respectively. We observe the fecal glycans to increase dramatically from week 0 to week 1. Note that the y axis in this figure is logarithmic and that the increases vary from 14-fold to >1 million-fold. At week 2, there is little change in HMO abundance in the feces, whereas at week 14, all of the measured fecal HMOs have decreased (range 2-fold to 7000-fold higher than baseline).
Figure 4.
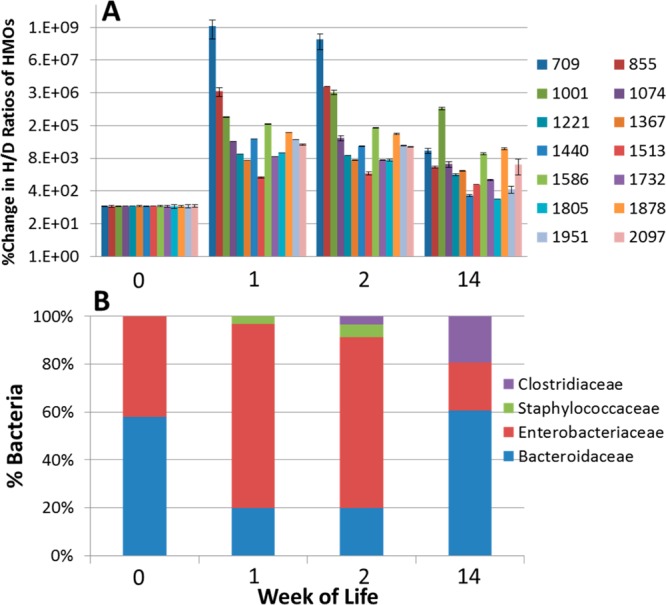
Changes in oligosaccharide intensities and bacterial population in the feces of Infant B. (A) Percent change in H/D ratios of HMOs in the feces of Infant B at weeks 0, 1, 2, and 14. Intensities were obtained using MALDI FT-ICR MS in (+) ion mode with n = 3. H/D ratios were normalized to week 0, set at 100%. Each bar represents an oligosaccharide nominal mass. (B) Corresponding fecal bacterial population of Infant B using 16S rDNA pyrosequencing.
Multiplexed pyrosequencing and Illumina sequencing of Infant B’s samples yielded 30 886 and 447 149 partial 16S rRNA gene sequence reads, respectively. Order level fecal populations are shown in Figure 5A, and genus level, in Figure 5B. The fecal bacterial population of this infant is initially populated by nearly equal amounts of Enterobacteriaceae (primarily Escherichia) and Bacteroides in week 0. By week 1, Escherichia increases to be the dominant species as the amount of Bacteroides decreases. At week 2, there is little change in either the fecal microbiota or the fecal HMO composition. By week 14, Bacteroidales dominate the population at 60%, and the corresponding HMO fecal profile shows a decrease in intensity of nearly all HMO compositions. These results correlate with our previous in vitro observations that members of Bacteroidaceae, e.g., Bacteroides fragilis, are consumers of HMOs.40
Figure 5.
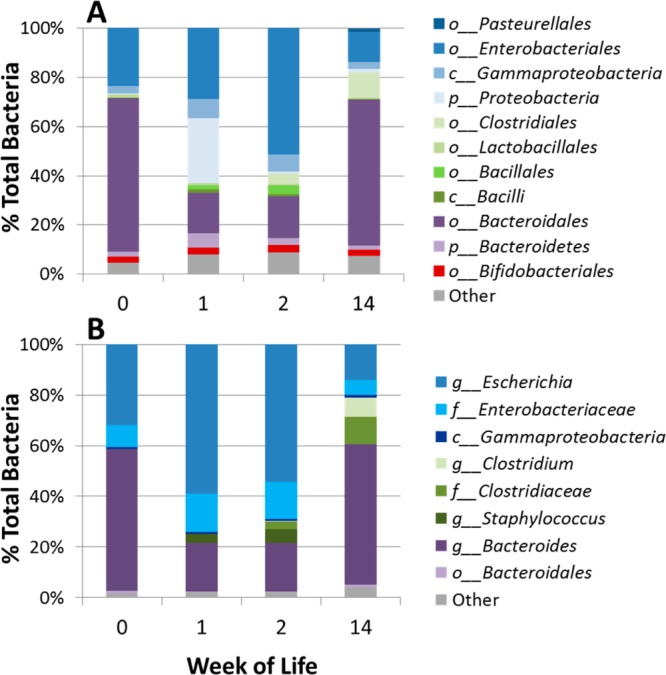
Infant B microbial population changes over the first 14 weeks of life based on next-generation sequencing. (A) Illumina and (B) pyrosequencing profile of the V4 region and V1–V3 regions of the16S rRNA gene, respectively.
Interestingly, the drop in abundances of oligosaccharide for Infant B is not as large as that for Infant A, where the majority of bacteria at week 13 was Bifidobacterium sp. In vitro studies using strains of both Bifidobacterium longum ssp. infantis (B. infantis) and Bacteriodes thetaiotaomicron show that the former is a stronger consumer of HMO than the latter.7,40Bifidobacterium sp. populations were low in Infant B, detectable by Illumina sequencing (Figure 5A) but not by pyrosequencing (Figure 5B) or qPCR (Figure 6). B. longum group was the only clade detected by qPCR, although higher populations of B. bifidum were detected by TRFLP as well as small populations of B. breve and B. adolescentis (data not shown).
Figure 6.
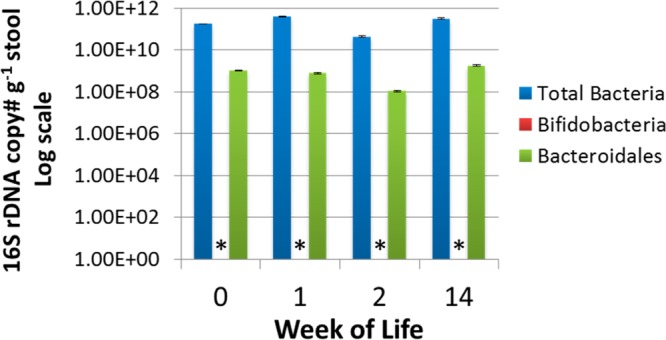
InfantB microbial population changes over the first 14 weeks of life based on qPCR analysis. qPCR of total bacteria, Bifidobacteria, and Bacteroidales. * Bifidobacteria are below the limit of detection (1 × 104 16S rRNA genes per gram of stool).
Isomer-Specific Temporal Changes in Fecal HMOs
A more extensive analysis supports the hypothesis of structure/isomer-specific consumption of HMOs in the infants’ gut. The fecal profile of Infant A is dominated by Bifidobacterium spp. by week 13 (Figure 1), and the fecal profile of Infant B is dominated by Bacteroides spp. by week 14 (Figure 4). Although the patterns are not equivalent, suggesting differential consumption, there are some similarities in the consumption or nonconsumption of certain HMOs.
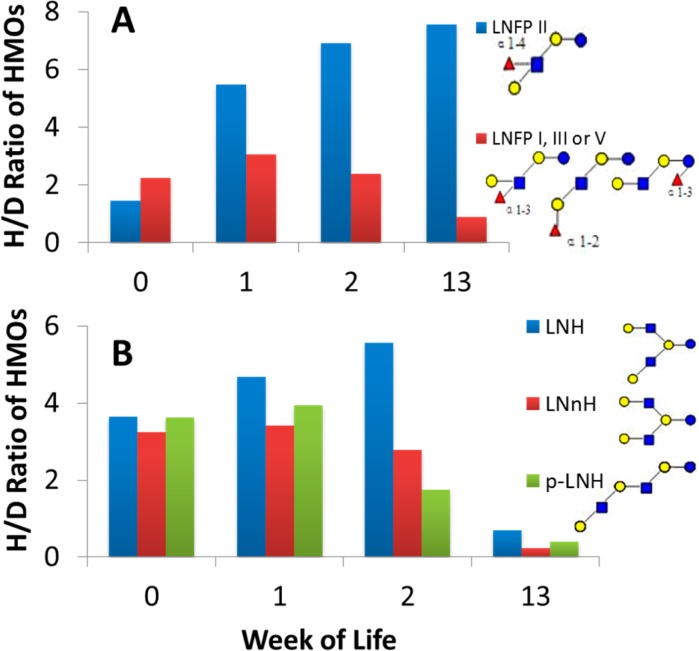
In the fecal profile of Infant A, for example, a pentasaccharide with monoisotopic mass of 855.3220 having the composition 3 hexose (galactose or glucose), 1 N-acetylglucosamine, and 1 fucose was found to have four isomers corresponding to the common name lacto-N-fucopentaose: LNFP I, LNFP II, LNFP III, and LNFP V. The fecal profiles are shown in Figure 7A as quantified by the H/D ratio using nano-HLPC chip/TOF MS data. One of these four isomers behaved unlike the other three structurally very similar isomers. LNFP II (structure inset Figure 7A, note the terminal α-1,4-fucosylation) continually increased from week 0 to week 13 (suggesting a lack of consumption by intestinal bacteria). The other three isomers, LNFP I, III, and V, all increased in fecal intensity during week 1 but decreased dramatically during week 2 and stayed very low through week 13 (suggesting consumption). These isomers bear the α-1,2-fucosyl and α-1,3-fucosyl residues.
Figure 7.
H/D ratios of two isomeric groups of oligosaccharides in the fecal HMO profile of Infant A. H/D ratios were calculated using nano-HLPC chip/TOF MS data. (A) Four isomers of m/z 856 ([M + H]+, M = 855.3220, second bar from the left in each week in Figure 1A). (B) Three isomers of m/z 538 ([M + 2H]2+, z = 2, M = 1074.3963, fourth bar from the left in Figure 1A). M = monoisotopic (neutral) mass.
Figure 7B demonstrates another group of isomers from Infant A with an overall hexasaccharide structure with a common monoisotopic mass of 1074.3963 (m/z 538, z = 2) and composed of 4 hexose and 2 N-acetylglucosamine with at least three linear or branched structures containing terminal β(1,3)- and β(1,4)-galactose residues. All three isomers follow a similar profile increasing in week 1 and decreasing thereafter (consistent with consumption). The extracted ion chromatograms of these isomers are shown in Figure 8. All isomers significantly decreased by week 13 (Figure 8D). The results suggest little variation between β-galactosidase linkage isomers in this infant.
Figure 8.
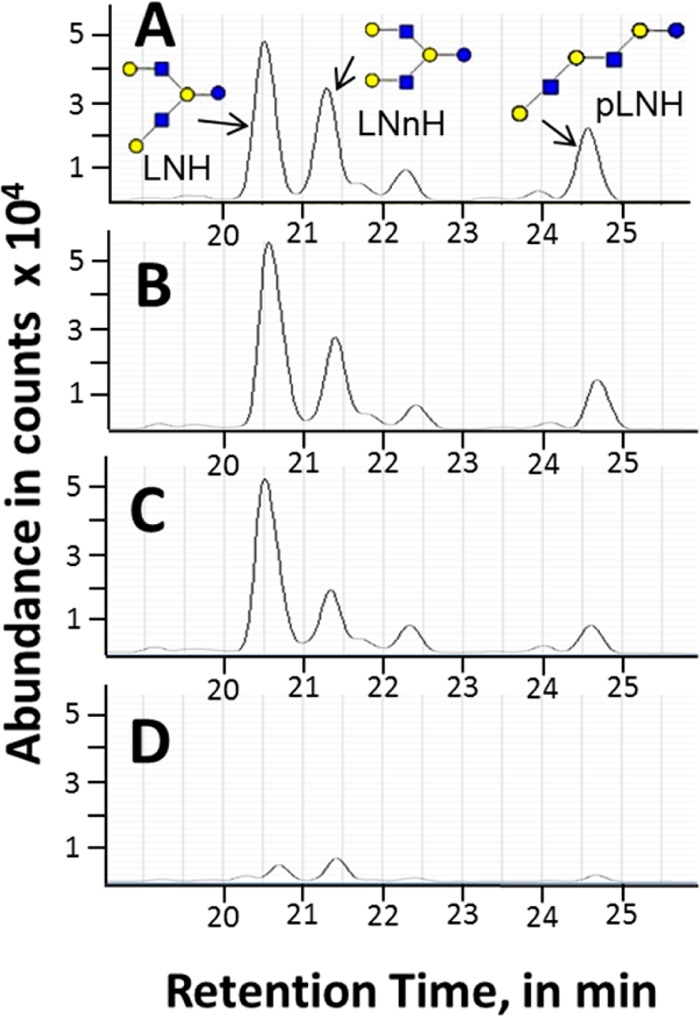
LC–MS extracted ion chromatograms of m/z 538 ([M + 2H]2+, z = 2, M = 1074.3963) in the fecal HMO profile of Infant A using nano-HLPC chip/TOF MS. Chromatograms at (A) week 0, (B) week 1, (C) week 2, and (D) week 13. M = monoisotopic (neutral) mass.
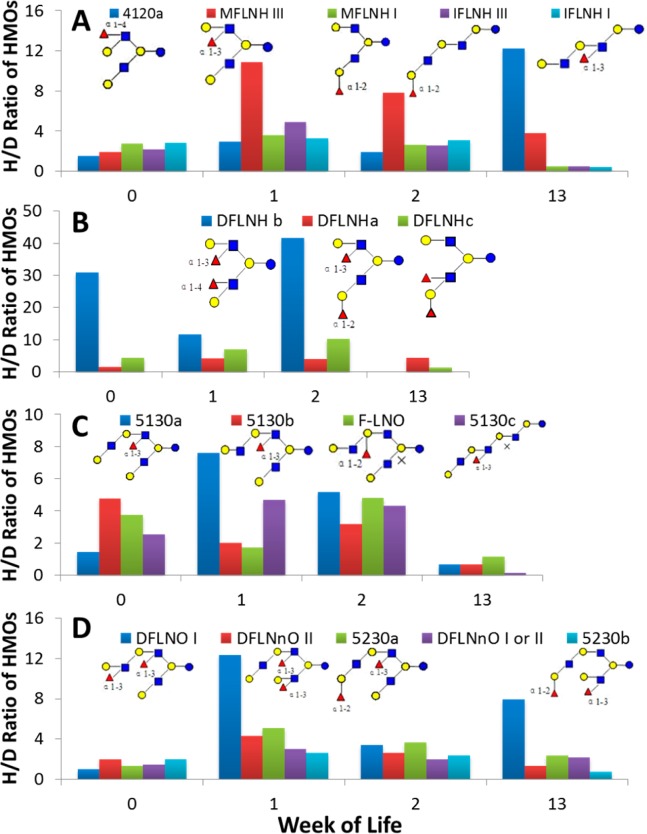
Figure 9 demonstrates marked differences in individual fecal HMO structures of Infant A over time, consistent with the hypothesized highly selective nature of consumption of HMOs by the intestinal bacteria. For example, four HMOs (MFLNH I, MFLNH III, IFLNH I, and IFLNH III (MW 1220.4542)) are present in the feces until week 2 but diminish rapidly by week 13 (concurrent with the bloom of bifidobacteria), suggesting preferential consumption; meanwhile, isomer 4120a increases at week 13, suggesting that the fecal microbes may be unable to digest this structure (Figure 9A). Isomers DFLNHb, and DFLNHc (MW 1366.512) both show a decrease concomitant with the bloom of bifidobacteria at week 13; however, this pattern is not seen with isomer DFLNHa (Figure 9B). Isomers 5130a, 5130b, F-LNO, and 5130c (MW 1585.5864) all diminished at week 13, suggesting consumption (Figure 9C). Isomers 5230a, 5230b, and DFLNnO II (MW 1731.6443) demonstrate the pattern consistent with consumption, but DFLNnO I is markedly increased, suggesting a lack of consumption by the fecal microbes at 13 weeks (Figure 9D).
Figure 9.
H/D ratios of four isomeric groups of oligosaccharides in the fecal HMO profile of Infant A. H/D ratios were calculated using nano-HLPC chip/TOF MS data. (A) Five isomers of m/z 611 ([M + 2H]2+, z = 2, M = 1220.4542, sixth bar from the left in each week in Figure 1A). (B) Three isomers of m/z 684 ([M + 2H]2+, z = 2, M = 1366.5121, seventh bar from the left in Figure 1A). (C) Four isomers of m/z 794 ([M + 2H]2+, z = 2, M = 1585.5864, tenth bar from the left in Figure 1A). (D) Five isomers of m/z 867 ([M + 2H]2+, z = 2, M = 1731.6443, 11th bar from the left in Figure 1A). M = monoisotopic (neutral) mass.
Isomer analyses of fecal HMOs for Infant B as quantified by the H/D ratios using nano-HPLC chip/TOF MS data are shown in Supporting Information Figures 1–3. Fecal profiles of Infant B for lacto-N-fucopentaose isomers (MW 855.3220) are shown in Supporting Information Figure 1A. The α-1,4-fucosylated LNFP II increased slightly from week 0 to week 1 and then plateaued in weeks 2–14 (5.7753 ± 0.3185), suggesting nonconsumption by intestinal bacteria. The other three isomers, LNFP I, III, and V, increased dramatically in fecal intensity during week 1 but decreased in intensity at week 14, suggesting consumption at the fecal profile shift to Bacteroides spp. at week 14.
Supporting Information Figure 1B shows fecal profiles of another group of isomers with MW 1074.3963. All three isomers, LNH, LNnH, and p-LNH, show a decrease concomitant with the bloom of bacteroides at week 14, suggesting consumption. The extracted ion chromatograms of said isomers are shown in Supporting Information Figure 2. All isomers increased from week 0 to week 2 and then decreased in week 14, consistent with consumption. Similar to Infant A, the results suggest little variation between β-galactosidase linkage isomers in Infant B.
Other individual fecal HMO structures and isomers are shown in Supporting Information Figure 3. For example, all five isomers of MW 1220.4542 (4120a, MFLNH I, MFLNH III, IFLNH I, and IFLNH III) decrease over time (Supporting Information Figure 3A). Isomers DFLNH a, b, and c all show a decrease at week 14 (Supporting Information Figure 3B). Isomers 5130a and 5130c (MW 1585.5864) both show a decrease at week 14, suggesting consumption, whereas 5130b and F-LNO increased at week 14 (Supporting Information Figure 3C). Isomers 5230a and DFLNnO I and II (MW 1731.6443) show a pattern consistent with consumption, but 5230b is markedly increased, suggesting a lack of consumption by the fecal microbes at 14 weeks (Supporting Information Figure 3D).
Discussion
Rationale for Quantitation Method
Quantitation of milk oligosaccharides is complicated by the different ionization properties of the different structures. Suppression effects are observed when the sample is analyzed as a mixture by mass spectrometry. For example, sialylated oligosaccharides are often suppressed by neutral oligosaccharides in the positive mode, whereas the reverse happens in the negative mode.41 Separation of the oligosaccharides eliminates suppression effects; however, there are still slight variations in the ionization of the individual structures. To minimize these effects, each compound was used as its own control. In addition, the internal standards from the pooled milk sample contain more structures than any individual specimen, allowing the analysis of large biological diversity.
Fecal HMO Profiles Correlate with Changes in Bacterial Population
The two infants studied were both healthy, breast-fed infants, but the colonization of their intestinal tract differed remarkably. The fecal microbiota of Infant A was initially dominated by Streptococcus spp., whereas the gut microbiota of Infant B was dominated by Escherichia spp. These bacteria are common commensals of the birth canal and are not HMO consumers, i.e., do not have the genetic capability to produce the glycosidases necessary to break down the linkages of HMO. The increase in fecal HMOs in weeks 0–2 in both infants is consistent with the hypothesized lack of HMO consumption during this period. However, by week 13, Bifidobacterium spp. dominated in Infant A, and levels of most fecal HMOs dropped dramatically. The decline in HMO intensity is significantly negatively correlated to Bifidobacteriales abundance and positively correlated with several other bacteria, notably, Enterobacteriales and Bacillales. For Infant B, Bacteroides spp. were dominant by week 14, at which time levels of many fecal HMOs were decreased compared to those at weeks 1 and 2.
The enrichment of Bifidobacterium spp. and Bacteroides spp. in the gastrointestinal tract of infants has been previously demonstrated.42 Moreover, metagenomic analysis has revealed that this enrichment also corresponds with increased expression of many genes involved in consumption of complex oligosaccharides.43 We have previously shown in vitro that Bacteroides are good consumers of HMOs40 and that B. infantis can grow with HMOs as the sole carbon source.4 In spite of the complexity of this ecological niche, the in vivo data here presented match closely the in vitro results for the single species analysis. These results further support the concept that one function of HMOs is to selectively enrich a saccharolytic bacterial consortium despite the variety of bacteria introduced into the infant in the early days of life.
Enzymatic Activity: A Signature of the Gut Microbiota
Our method allows simultaneous analysis of up to 75 structurally elucidated oligosaccharides and the monitoring of hundreds of structures simultaneously.44 Profiles of human milk across 6 months of lactation show the constant delivery of HMOs with minor fluctuation particularly for the smaller, more abundant oligosaccharides (degree of polymerization < 7).38 This level of detail allows the elucidation of gross glycosidic enzyme actions by the microbiota, i.e., the consumption of known structures is an indicator of the presence of specific active bacterial enzymes in the gut.
The whole genome sequencing of B. infantis revealed a number of exoglycosidases including galactosidases, fucosidases, and sialidases.45,46 This high degree of enzymatic activity is not seen in several other bifidobacterial species and in part explains the dominance of the B. longum group in Infant A (B. infantis is a member of the B. longum group).
Of particular interest is the catabolism of oligosaccharides containing (α1,2)-fucose. The presence of 2′FL and other (α1,2)-fucose-containing HMOs such as lacto-N-fucopentaose (LNFP) indicates that the mother has at least one functional fucosyl transferase (FUT2) allele and is therefore a secretor (i.e., able to express specific fucosylated structures in secretions such as tears, milk, or saliva).19,47,48 In adults, secretor status is associated with increased fecal bifidobacteria49 and with alterations in susceptibility to viral and bacterial infections.23,50 The observation that these special fucosylated oligosaccharides are preferentially consumed in healthy infants suggests the hypothesis that certain bifidobacteria thrive in a milieu that contains either milk from a secretor mom or intestinal secretions from a secretor infant. This may have particular value to the fragile premature infant where human milk is partially protective against sepsis and necrotizing enterocolitis51 and nonsecretor status may predispose to these common complications.52
Limitations of Present Study and Avenues for Future Research
The analysis of fecal samples from two infants was designed as a proof-of-concept exercise to demonstrate the potential utility of analyzing the fecal microbiota and fecal glycomics in tandem. The small sample size was helpful in generating hypotheses, but future studies with larger sample sizes would be necessary to confirm and replicate the findings.
Analysis of milk specimens from the mothers of these infants would have been helpful to confirm consistent delivery of HMOs over time (demonstrated in other mothers, but not in these two cases). Inclusion of maternal dietary analysis and details regarding the mothers’ health status (e.g., obesity, diabetes, medications) would have been useful. The major strength of these observations is the detailed analyses performed including differentiation of isomers with differing patterns of possible consumption by the intestinal microbes. Diagnostic testing of the stool of infants, children, and adults for rapid determination of secretor status or to identify patterns of HMOs or bacterial enzymes suggestive of increased risk for necrotizing enterocolitis, Clostridium difficile colitis, or inflammatory diseases of the gut may be possible with approaches similar to those demonstrated herein. Such an approach may be of value in high-risk populations.
Conclusions
This proof-of-concept study combines glycomics and genomics to monitor the changes in HMOs and gut microbiota in breastfeeding infants. The observed changes over time in two infants suggest the need for more extensive studies of a larger cohort to test the hypothesis that HMOs enrich a cognate oligosaccharide-consuming microbial population in the infant gut by orchestrating a shift in the infant fecal microbiota from a nonsaccharolytic population dominated by commensals of the birth canal to a population dominated by saccharolytic microbes concurrent with a decrease in fecal HMO intensities. Further studies in nonhuman primates and other mammals would be valuable to determine whether feedback evolutionary mechanisms link the infant gut and maternal milk oligosaccharide production.
Acknowledgments
We are grateful for funds from the U.S. Department of Agriculture (NRI-CSREES 2008-35200-18776 to D.A.M.), the National Institutes of Health (GM049077, HD061923, P42 ES02710, R01AT007079, and HD059127), and California Dairy Research Foundation Grant 06 LEC-01-NH. The Agilent QTOF MS instrument was obtained through a grant (S10RR027639). D.A.M. acknowledges funding from the Shields Endowed Chair in Dairy Science. M.L.A.D. thanks Milady R. Ninonuevo and John S. Strum for helpful discussions on data analysis and Stephanie C. Gaerlan for help in data filtering.
Glossary
Abbreviations
- Bif-TRFLP
Bifidobacterium species-specific terminal restriction fragment length polymorphism
- CID
collision-induced dissociation
- H/D
hydrogen to deuterium ratio
- HMO
human milk oligosaccharide
- IRMPD
infrared multiphoton dissociation
- LC
liquid chromatography
- LNT
lacto-N-tetraose
- MALDI FT-ICR
matrix-assisted laser desorption/ionization Fourier transform-ion cyclotron resonance
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- qPCR
real-time quantitative polymerase chain reaction
- TOF
time-of-flight
Supporting Information Available
qPCR primers and probes; H/D ratios of two isomeric groups of oligosaccharides in the fecal HMO profile of Infant B; LC–MS extracted ion chromatograms of m/z 538 ([M + 2H]2+, z = 2, M = 1074.3963) in the fecal HMO profile of Infant B using nano-HLPC chip/TOF MS; and H/D ratios of four isomeric groups of oligosaccharides in the fecal HMO profile of Infant B. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
○ (Maria Lorna A. De Leoz) Mass Spectrometry Data Center, National Institute of Standards and Technology, 100 Bureau Drive, Gaithersburg, Maryland 20899-8362, United States.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- German J. B.; Dillard C. J.; Ward R. E. Bioactive components in milk. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 653–8. [DOI] [PubMed] [Google Scholar]
- Zivkovic A. M.; German J. B.; Lebrilla C. B.; Mills D. A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninonuevo M. R.; Park Y.; Yin H.; Zhang J.; Ward R. E.; Clowers B. H.; German J. B.; Freeman S. L.; Killeen K.; Grimm R.; Lebrilla C. B. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 2006, 54, 7471–80. [DOI] [PubMed] [Google Scholar]
- LoCascio R. G.; Ninonuevo M. R.; Freeman S. L.; Sela D. A.; Grimm R.; Lebrilla C. B.; Mills D. A.; German J. B. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007, 55, 8914–9. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T.; Rey F. E.; Manary M. J.; Trehan I.; Dominguez-Bello M. G.; Contreras M.; Magris M.; Hidalgo G.; Baldassano R. N.; Anokhin A. P.; Heath A. C.; Warner B.; Reeder J.; Kuczynski J.; Caporaso J. G.; Lozupone C. A.; Lauber C.; Clemente J. C.; Knights D.; Knight R.; Gordon J. I. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D.; Dallas D. C.; Mills D. A. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 2013, 159, 649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A.; Barboza M.; Sonnenburg E. D.; Pudlo N.; Martens E. C.; Desai P.; Lebrilla C. B.; Weimer B. C.; Mills D. A.; German J. B.; Sonnenburg J. L. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 2011, 10, 507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D.; Barile D.; Mills D. A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr 2012, 3, 415S–21S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P.; Warren C. D.; Altaye M.; Morrow A. L.; Ruiz-Palacios G.; Pickering L. K.; Newburg D. S. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 2001, 11, 365–72. [DOI] [PubMed] [Google Scholar]
- Hong P.; Ninonuevo M. R.; Lee B.; Lebrilla C.; Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br. J. Nutr. 2009, 101, 482–6. [DOI] [PubMed] [Google Scholar]
- Huang P.; Xia M.; Tan M.; Zhong W.; Wei C.; Wang L.; Morrow A.; Jiang X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol 2012, 86, 4833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppa G. V.; Zampini L.; Galeazzi T.; Facinelli B.; Ferrante L.; Capretti R.; Orazio G. Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr. Res. 2006, 59, 377–82. [DOI] [PubMed] [Google Scholar]
- Morrow A. L.; Ruiz-Palacios G. M.; Jiang X.; Newburg D. S. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 2005, 135, 1304–7. [DOI] [PubMed] [Google Scholar]
- Andersson B.; Porras O.; Hanson L. A.; Lagergard T.; Svanborg-Eden C. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J. Infect. Dis. 1986, 153, 232–7. [DOI] [PubMed] [Google Scholar]
- Bode L.; Jantscher-Krenn E. Structure–function relationships of human milk oligosaccharides. Adv. Nutr 2012, 3, 383S–91S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M.; Cervantes L. E.; Ramos P.; Chavez-Munguia B.; Newburg D. S. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003, 278, 14112–20. [DOI] [PubMed] [Google Scholar]
- Martin-Sosa S.; Martin M. J.; Hueso P. The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J. Nutr. 2002, 132, 3067–72. [DOI] [PubMed] [Google Scholar]
- Kunz C.; Rudloff S.; Baier W.; Klein N.; Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr 2000, 20, 699–722. [DOI] [PubMed] [Google Scholar]
- De Leoz M. L.; Gaerlan S. C.; Strum J. S.; Dimapasoc L. M.; Mirmiran M.; Tancredi D. J.; Smilowitz J. T.; Kalanetra K. M.; Mills D. A.; German J. B.; Lebrilla C. B.; Underwood M. A. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J. Proteome Res. 2012, 11, 4662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz M. L.; Wu S.; Strum J. S.; Ninonuevo M. R.; Gaerlan S. C.; Mirmiran M.; German J. B.; Mills D. A.; Lebrilla C. B.; Underwood M. A. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. J. Anal. Bioanal. Chem. 2013, 405, 4089–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum J. S.; Kim J.; Wu S.; De Leoz M. L.; Peacock K.; Grimm R.; German J. B.; Mills D. A.; Lebrilla C. B. Identification and accurate quantitation of biological oligosaccharide mixtures. Anal. Chem. 2012, 84, 7793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.; Tao N.; German J. B.; Grimm R.; Lebrilla C. B. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 2010, 9, 4138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza M. W.; Blackwell C. C.; Molyneaux P.; James V. S.; Ogilvie M. M.; Inglis J. M.; Weir D. M. Association between secretor status and respiratory viral illness. BMJ 1991, 303, 815–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninonuevo M. R.; Ward R. E.; LoCascio R. G.; German J. B.; Freeman S. L.; Barboza M.; Mills D. A.; Lebrilla C. B. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal. Biochem. 2007, 361, 15–23. [DOI] [PubMed] [Google Scholar]
- Martinez I.; Kim J.; Duffy P. R.; Schlegel V. L.; Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 2010, 5, e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Lauber C. L.; Walters W. A.; Berg-Lyons D.; Lozupone C. A.; Turnbaugh P. J.; Fierer N.; Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Kuczynski J.; Stombaugh J.; Bittinger K.; Bushman F. D.; Costello E. K.; Fierer N.; Pena A. G.; Goodrich J. K.; Gordon J. I.; Huttley G. A.; Kelley S. T.; Knights D.; Koenig J. E.; Ley R. E.; Lozupone C. A.; McDonald D.; Muegge B. D.; Pirrung M.; Reeder J.; Sevinsky J. R.; Turnbaugh P. J.; Walters W. A.; Widmann J.; Yatsunenko T.; Zaneveld J.; Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–1. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Garrity G. M.; Tiedje J. M.; Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z.; Hugenholtz P.; Larsen N.; Rojas M.; Brodie E. L.; Keller K.; Huber T.; Dalevi D.; Hu P.; Andersen G. L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Bittinger K.; Bushman F. D.; DeSantis T. Z.; Andersen G. L.; Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N.; Dehal P. S.; Arkin A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010, 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R.; Chai B.; Farris R. J.; Wang Q.; Kulam-Syed-Mohideen A. S.; McGarrell D. M.; Bandela A. M.; Cardenas E.; Garrity G. M.; Tiedje J. M. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007, 35, D169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R.; Wang Q.; Cardenas E.; Fish J.; Chai B.; Farris R. J.; Kulam-Syed-Mohideen A. S.; McGarrell D. M.; Marsh T.; Garrity G. M.; Tiedje J. M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A.; Blackshields G.; Brown N. P.; Chenna R.; McGettigan P. A.; McWilliam H.; Valentin F.; Wallace I. M.; Wilm A.; Lopez R.; Thompson J. D.; Gibson T. J.; Higgins D. G. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–8. [DOI] [PubMed] [Google Scholar]
- Abdo Z.; Schuette U. M.; Bent S. J.; Williams C. J.; Forney L. J.; Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 2006, 8, 929–38. [DOI] [PubMed] [Google Scholar]
- Shyu C.; Soule T.; Bent S. J.; Foster J. A.; Forney L. J. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 2007, 53, 562–70. [DOI] [PubMed] [Google Scholar]
- Ninonuevo M. R.; Perkins P. D.; Francis J.; Lamotte L. M.; LoCascio R. G.; Freeman S. L.; Mills D. A.; German J. B.; Grimm R.; Lebrilla C. B. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J. Agric. Food Chem. 2008, 56, 618–26. [DOI] [PubMed] [Google Scholar]
- Adlerberth I.; Wold A. E. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009, 98, 229–38. [DOI] [PubMed] [Google Scholar]
- Marcobal A.; Barboza M.; Froehlich J. W.; Block D. E.; German J. B.; Lebrilla C. B.; Mills D. A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leoz M. L.; Young L. J.; An H. J.; Kronewitter S. R.; Kim J.; Miyamoto S.; Borowsky A. D.; Chew H. K.; Lebrilla C. B. High-mannose glycans are elevated during breast cancer progression. Mol. Cell. Proteomics 2011, 10, M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M.; Young D.; Scott J.; Norin E.; Amarri S.; Adam R.; Aguilera M.; Khanna S.; Gil A.; Edwards C. A.; Dore J.; Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 77–84. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T.; Rey F. E.; Manary M. J.; Trehan I.; Dominguez-Bello M. G.; Contreras M.; Magris M.; Hidalgo G.; Baldassano R. N.; Anokhin A. P.; Heath A. C.; Warner B.; Reeder J.; Kuczynski J.; Caporaso J. G.; Lozupone C. A.; Lauber C.; Clemente J. C.; Knights D.; Knight R.; Gordon J. I. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz M. L. A.; Wu S.; Strum J. S.; Niñonuevo M. R.; Gaerlan S. C.; Mirmiran M.; German J. B.; Mills D. A.; Lebrilla C. B.; Underwood M. A. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. J. Anal. Bioanal. Chem. 2013, 405, 4089–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A.; Chapman J.; Adeuya A.; Kim J. H.; Chen F.; Whitehead T. R.; Lapidus A.; Rokhsar D. S.; Lebrilla C. B.; German J. B.; Price N. P.; Richardson P. M.; Mills D. A. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 18964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A.; Garrido D.; Lerno L.; Wu S.; Tan K.; Eom H. J.; Joachimiak A.; Lebrilla C. B.; Mills D. A. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 2012, 78, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman E. F.; Ginsburg V. Correlation between secretor status and the occurrence of 2′-fucosyllactose in human milk. Biochem. Biophys. Res. Commun. 1967, 28, 50–3. [DOI] [PubMed] [Google Scholar]
- Shen L.; Grollman E. F.; Ginsburg V. An enzymatic basis for secretor status and blood group substance specificity in humans. Proc. Natl. Acad. Sci. U.S.A. 1968, 59, 224–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P.; Makivuokko H.; Alakulppi N.; Nikkila J.; Tenkanen H.; Rabina J.; Partanen J.; Aranko K.; Matto J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 2011, 6, e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell C. C. The role of ABO blood groups and secretor status in host defences. FEMS Microbiol. Immunol. 1989, 1, 341–9. [DOI] [PubMed] [Google Scholar]
- Underwood M. A. Human milk for the premature infant. Pediatr. Clin. North Am. 2013, 60, 189–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow A. L.; Meinzen-Derr J.; Huang P.; Schibler K. R.; Cahill T.; Keddache M.; Kallapur S. G.; Newburg D. S.; Tabangin M.; Warner B. B.; Jiang X. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J. Pediatr. 2011, 158, 745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.