Abstract
Fungal infections are often difficult to treat due to the inherent similarities between fungal and animal cells and the resulting host toxicity from many antifungal compounds. Cryptococcus neoformans is an opportunistic fungal pathogen of humans that causes life-threatening disease, primarily in immunocompromised patients. Since antifungal therapy for this microorganism is limited, many investigators have explored novel drug targets aim at virulence factors, such as the ability to grow at mammalian physiological temperature (37°C). To address this issue, we used the Agrobacterium tumefaciens gene delivery system to create a random insertion mutagenesis library that was screened for altered growth at elevated temperatures. Among several mutants unable to grow at 37°C, we explored one bearing an interruption in the URA4 gene. This gene encodes dihydroorotase (DHOase) that is involved in the de novo synthesis of pyrimidine ribonucleotides. Loss of the C. neoformans Ura4 protein, by targeted gene interruption, resulted in an expected uracil/uridine auxotrophy and an unexpected high temperature growth defect. In addition, the ura4 mutant displayed phenotypic defects in other prominent virulence factors (melanin, capsule and phospholipase) and reduced stress response compared to wild type and reconstituted strains. Accordingly, this mutant had a decreased survival rate in macrophages and attenuated virulence in a murine model of cryptococcal infection. Quantitative PCR analysis suggests that this biosynthetic pathway is induced during the transition from 30°C to 37°C, and that transcriptional regulation of de novo and salvage pyrimidine pathway are under the control of the Ura4 protein.
Keywords: pyrimidine biosynthesis, thermal tolerance, basidiomycete yeast
1. Introduction
Cryptococcus neoformans is a pathogenic fungus with a worldwide distribution. This yeast is found in the environment on rotting wood, and it is often associated with avian excreta. It can cause life-threatening respiratory and neurological infections, especially in immunocompromised patient populations. In fact, recent surveys estimate greater than 500,000 deaths due to C. neoformans each year, mainly in patients with AIDS and other immune compromising conditions (Park et al, 2009). The immunocompromised population has increased world-wide due to many factors including the AIDS pandemic and growing numbers of transplant patients. Together, these factors have turned this yeast into a major pathogen (Mitchell and Perfect, 1995; Singh and Husain, 2000).
Options for antifungal therapies for cryptococcosis are limited. Among the common agents used for this infection are polyenes (amphotericin B-based drugs), antimetabolites (flucytosine), and azoles (Perfect et al., 2010). However, drug toxicity and antifungal resistance remain important issues in the treatment of all fungal infections (Rex et al., 2001; Paiva and Pereira, 2013). To address potential novel strategies for antifungal therapy, we and others have studied virulence-associated phenotypes that allow C. neoformans to survive within the infected host and to cause disease. These factors include polysaccharide capsule, melanin, phospholipase, and growth at 37°C (Heitman et al., 2006, Brown et al., 2007; Li and Mody, 2010; Lam et al., 2013). The ability of this fungus to grow at human physiological temperature is controlled by several cellular factors, including the effectors of the calcineurin and Ras signal transduction pathways (Odom et al., 1997; Alspaugh, et al., 2000; Kozubowski et al., 2009). To identify additional elements that are required for high-temperature growth, we used a random insertion mutagenesis strategy mediated by Agrobacterium tumefaciens to generate mutants of C. neoformans unable to grow at 37°C (Idnurm et al., 2004; McClelland et al., 2005). Using this method, we identified temperature-sensitive strains containing mutations; one of these was defective in pyrimidine biosynthesis.
The de novo synthesis of uracil monophosphate (UMP) is essential for the synthesis of nucleic acids, and it is a common biosynthetic pathway for all organisms. Some cells can also make UMP using pyrimidine (Norager, et al. 2002). In addition to basic cell growth, many organisms, including pathogenic parasites, require efficient pyrimidine biosynthesis for rapid cell proliferation and adaptation to cell stress (Fairbanks et al. 1995; Fox and Bzik, 2010; Ali et al. 2013; Hegewald et al., 2013; Ong et al., 2013). In S. cerevisiae, transcription of genes linked to the uracil biosynthesis pathway can increase three to eight times the yeast cells are subjected to pyrimidine limitation (Roy et al. 1990). The enzyme dihydroorotate dihydrogenase (DHOase-URA1) converts dihydroooratate into orotate, and DHOase inhibitors have been successfully tested as antiproliferative agents in neoplastic growth, as suppressors of immunological responses (Chen et al. 1992; Greene et al. 1995; Liu et al., 2000, Khutornenko et al., 2010), and also as inhibitors of the growth of Toxoplasma gondii (Hegewald et al. 2013). In the basidiomycete Ustilago maydis, the pyr4 gene encodes dihydroorotate dehydrogenase, and its absence results in loss of pathogenicity and uracil auxotrophy (Banuett, 1995; Bölker, 2001; Zameitat, et al., 2007). Previous work by Morrow et al. (2012) showed that de novo synthesis of purine derivatives such as GTP is also important for virulence in C. neoformans.
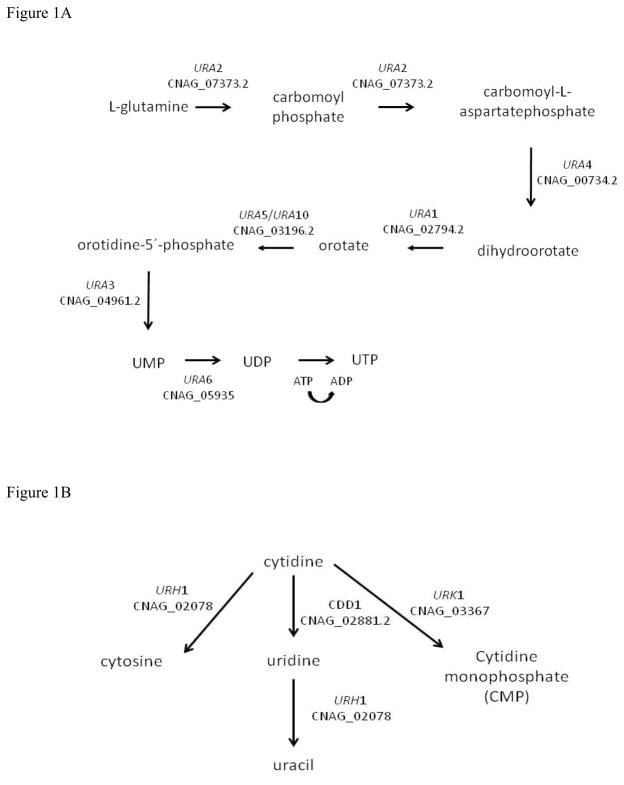
C. neoformans encodes genes for pyrimidine de novo (URA1 through URA6) and salvage (URH1, URK1 and CDD1) pathways (Figure 1A and B, respectively), suggesting this yeast can use both strategies to obtain this essential nutrient. One pyrimidine biosynthetic gene that has been previously characterized in C. neoformans is URA5, and this has been primarily used as selectable marker for transformation (Edman and Kwon-Chung, 1990; Kwon-Chung et al., 1992; Varma et al., 1992). However the virulence phenotypes associated with the ura5 mutation have not been extensively characterized. In this paper we studied the role of the URA4 gene in C. neoformans, the impact of its interruption on virulence factors and survival in vitro and in vivo. Also, the relationship between the pyrimidine de novo and salvage pathways and high temperature growth was established by transcription analysis. Our results showed that these pathways are not only important for the high temperature growth trait but also for the expression of several virulence factors, as well as efficient responses to cell stresses.
Figure 1.
Representation of the pymidine ribonucleotide de novo biosynthetic (A) and salvage (B) pathways. (Adapted from http://www.yeastgenome.org/).
2. Material and Methods
2.1. Strains, Media and Reagents
All wild-type, deletion and reconstituted strains were generated in the C. neoformans var. grubii KN99α background (MATalpha) donated by Dr Joseph Heitman. The ura5 mutans were obtained after selection of C. neoformans var. grubii strains baring spontaneous mutation of the URA5 gene after incubation on YPD medium containing 5-FOA (5-Fluoroorotic acid, 10μg/mL). Oligonucleotide primers are presented in Table 1 and 2. The media used were standard YPD medium (1% yeast extract, 2% peptone and 2% dextrose, 2% agar), Synthetic Medium (6.7 g/liter Sigma Yeast Nitrogen Base with amino acids and ammonium sulfate, 2% dextrose, 2% agar) with or without addition of 20μM uracil (Sigma). Geneticin (Invitrogen) and Hygromycin (Invitrogen) were added to the selection medium for transformation at 200 μg/mL final concentration, as indicated.
2.2. Genetic manipulation
C. neoformans genomic DNA extraction was performed as published (Pitkin et al., 1996). General molecular biology manipulations were performed according to Sambrook et al. (1989). Southern blots were performed using Roche PCR DIG Probe Synthesis kit, DIG Wash and blot kit, CDP-star Ready according to the manufacturer’s instructions.
2.3. Agrobacterium tumefaciens random insertion mutagenesis
The C. neoformans mutant collection was created according to the method described by Idnurm et al. (2004). Briefly, Agrobacterium mediated-transformation was achieved by co-culture of the C. neoformans strain KN99α with A. tumefaciens LBA4404 strain harboring the plasmid pPZP-Hyg2 or pPZP-Neo1 (Walton et al, 2006, donated by Dr. J Heitman) and induced with 100 μM of acetosyringone. Co-culture was conducted for 48 hours and aliquots were plated on YPD medium supplemented with 200 μg/mL Geneticin or 200 μg/mL Hygromycin, and cefotaxime (100 μg/mL) to kill Agrobacterium. After 48 to 72 hours, antibiotic resistant colonies were transferred to YPD medium supplemented with the appropriate antibiotic and incubated at 30°C and 37°C. Strains that grew at 30°C but that failed to grow at 37°C were chosen for further analysis. The selectable marker insertion site was identified by inverse PCR methods as previously described (Idnurm et al., 2004). Briefly, we extracted genomic DNA from these strains, and digested with either HindIII or EcoRI, (Fermentas). The digested DNA was purified through a Qiagen column (Qiaquick PCR Purification Kit), and ligated to itself with T4 DNA ligase. PCR was performed using primers directed against selectable marker sequences (primers MAV30 and MAV32, MAV31 and MAV33 (Table 1). Gel purified PCR products obtained by inverse PCR were sequenced and compared to the Broad Institute C. neoformans strain H99 genome database to determine the position of the T-DNA insertion into the genome.
2.4. Gene deletion and reconstitution, and C. neoformans transformation
Independent ura4 mutants were generated by biolistic transformation and homologous recombination. The URA4 coding region was partially replaced with the Geneticin resistance cassette containing the NeoR gene under control of the actin ACT1 promoter and TRP1 terminator (Walton, et al., 2005). In vitro construction of the deletion cassette was performed using PCR overlap extension (Davidson et al., 2002) using the following primers (left arm: primers MAV067 and MAV097, right arm: MAV098 and MAV 072, and Geneticin resistance gene: MAV099 and MAV070) described in Table 1. The cassette was introduced into C. neoformans KN99α by the biolistic method as described (Toffaletti et al., 1993), and transformants were screened for the ura4 mutation by PCR and Southern blots. The ura4+URA4 reconstituted strain was generated by introducing the wild-type URA4 allele into the ura4 mutant strain by the biolistic method. Briefly, the reconstituted strain was selected on synthetic media (YNB) without uracil, and this strain also became sensitive to Geneticin. As a control, the same culture of mutant cells was subject to bombardment with gold particles without DNA and the selection on YNB without uracil did not yield any colony. The primers used to generate the deletion and reconstitution strains are described in Table 1.
2.5. High temperature growth ability, melanin and capsule production assay
Thermotolerance was evaluated on both rich (YPD) and synthetic media (YNB) at 30°C and 37°C. To assess melanin production, overnight cultures were incubated in YPD medium, spotted on Niger seed agar plates (75g/L Niger bird seed, 0.1% Dextrose, 2% agar, with 20μM uracil), and incubated at 30°C for up to 96 hours. Capsule was induced by inoculating fresh cells of each strain in CO2-independent medium (Gibco, Cat 18C45-088) with and without 20μM uracil at 30°C at 150 rpm rotation (O’Meara, et al., 2013). After 24 hours incubation, cells were stained with BactiDrop India Ink (Remel) and directly observed by light microscopy. Alternatively, induced cells were washed in 1x PBS, resuspended in PBS and 1% Bovine Serum Albumin, incubated with 1 μL of mAb 18B7 (Casadevall et al., 1998) under gentle agitation for 1 hour at room temperature. Cells were washed in PBS + 1% BSA and incubated with 1 μL of fluorescent Alexa Fluor® 594 Goat Anti-Mouse IgG (Life Technologies) for 1 hour at room temperature with gentle rotation. After 2 washes in PBS, cells were observed by fluorescent microscopy with red filter (Texas Red). Differential interference microscopy (DIC), bright field, and fluorescent images were obtained with a Zeiss Axio Imager.A1 fluorescent microscope and AxioCam MR digital camera. Also, we have cultured the strains on CO2-independent medium supplemented with uracil and uridine (20 μg/mL) up to 72 hours at 30°C and 37°C.
2.6. Multi stress sensitivity assay
Osmotic, ionic and cell wall stress were assessed by spotting cells onto YPD plates supplemented with 0.75 M KCl, 0.75 M NaCl, 0.5% Congo Red, or pH 8.0. All cultures were incubated at 30°C.
2.7. Minimal inhibitory concentration assay
Sensitivity to antifungal drugs was determined for mutant, reconstituted and wild type strains by standard minimal inhibitory concentration (MIC) assays according to Clinical and Laboratory Standards Institute (CLSI M27-A2) protocols with modifications. Briefly, fresh cultures were diluted and 100 cells were inoculated in YNB plus 20μM of uracil. The concentration of fluconazole ranged from 16 to 0.0625 μg/mL, and that of amphotericin B from 1 to 0.03 μg/mL. A control with cells and without drugs was included, and a blank containing only medium was also applied in the assay. MIC80 was defined as the drug concentration in which growth was at least 80% inhibited compared to the drug-free control following incubation for 72 h at 30°C. Growth was determined by measuring the turbidity of the cell suspension in a Multiskan Ascent (Lab System) at 620 and 540 nm. After absorbance reading, the cells were stained with 0.001% vital dye Rezasurine (Sigma) for 1 hour at 30°C and 150 rpms. All experiments were performed in triplicate for fluconazole and amphotericin B, and two biological replicates were applied.
2.8. Transcriptional analysis by real time PCR
Total RNA for transcriptional pattern analysis was obtained from cells incubated for 2 hours in YPD and YNB at 30°C and 37°C. Cells were harvested, washed three times in PBS, flash frozen on dry ice, and lyophilized. RNA extraction was performed according to Yang et al. (2002). cDNA synthesis was performed with RevertAid H minus First Strand cDNA synthesis kit (Thermo Scientific) with Oligo dT and random hexamer primers from 5 μg of total RNA. Real time PCR amplifications were made from diluted templates (1:10) with 800 nM target primers, 300 nM GPD1 (Glyceraldehyde-3-phosphate dehydrogenase, Varma and Kwon-Chung. 1999) as internal control primers, and 1X Power SYBR Green master mix (Life Technologies). Quantification of the transcript levels was performed using the ΔΔCT method (Livak and Schmittgen, 2001), normalizing against GPD1 as previously described by Vandesompele et al (2002).
2.9. Macrophage infection assay
Macrophage-like J744.A1 cells were used to assay survival of yeasts within mammalian cells according to the methods of Cox et al. (2003). In brief, mAb-opsonized (mAb 18B7 antibody (McFadden and Casadevall, 2004) C. neoformans cells were co-incubated in 96-well tissue culture plates with J774A.1 cells in 200 μL macrophage medium at a multiplicity of infection (MOI) of 10:1 (1×106 C. neoformans cells/200 μL, and 1×105 J774A.1 cells/200 μL). The J774A.1 cells were activated by adding LPS and INF-γ one day prior to introducing the fungal cells. After an initial co-culture for 1 hour, the non-internalized fungal cells were removed by washing 3 times with PBS, and fresh medium was added. After 24 hours of incubation, the mammalian cells were disrupted with 0.5% SDS for 5 minutes, and the number of viable C. neoformans cells was determined by quantitative culture. Statistical validation was calculated for each macrophage co-cultivation experiment, and CFU counts were compared among strains by one-way ANOVA followed by Tukey’s post-test for comparison of pairs. Differences were considered valid at a 95% confidence interval (p<0.05).
The phagocytic index was determined as follows: Separate plates containing macrophages where incubated with the three fungal strains under the same conditions, and after two hours of phagocytosis, the wells were washed twice with PBS to remove free yeast cells and stained with a commercial equivalent of Giemsa for fast processing of microscopic samples (Instant-Prov Quick Haematological Stain, New Prov Laboratory Products, Brazil) and observed in an inverted microscope for measurement of the phagocytic index. This was calculated as the percentage of cells containing at least one yeast to the total number of cells. For each strain, 900–1200 cells were counted in nine separate fields.
2.10. In vivo virulence assay
Virulence of the C. neoformans strains was assessed in the murine inhalation model of cryptococcal infection (Cox et al., 2000). Briefly, groups of 4- to 6-week-old C57/BL6 mice were anaesthetized with 7.5μL/g i.p. of a mixture of ketamine (10.25 mg/mL) and xylazine (0.86 mg/mL). The mice were then infected via nasal inhalation with the wild-type strain KN99α, mutant (ura4) or reconstituted (ura4Δ+URA4) strains. The inoculum (105 cells in a 25μL per strain) was prepared from cultures which were incubated overnight in YPD at 30°C. Ten mice were infected for each strain. Mice were examined twice daily and sacrificed according to clinical measures predicting mortality (weight loss >15%, inability to access food and water). Survival data were analyzed by the Kruskal–Wallis test. The ura4-infected mice were sacrificed at 40 days post-infection; lungs from two of the surviving mice were harvested, homogenized, serially diluted, and plated on YPD to access the number of viable cells in the lungs. These experiments were performed at Duke University in accordance with institutional guidelines for animal experimentation.
G. mellonella killing assay. The infection of Galleria mellonella by C. neoformans was performed according to Mylonakis et al (2005). This assessment of virulence was performed at a permissive temperature (30°C) for the ura4 mutant strain to assess temperature-independent aspects of survival in an infected host. Briefly, larvae with average body weight of 174 mg were separated in groups with 14 individuals. Each group was inoculated with 1×105 cells of wild type (KN99α), ura4 (FGC003) or the reconstituted strain ura4Δ+URA4 (FGC003R1) with a 10-μl Hamilton syringe. PBS (Phosphate buffered saline) was used as control. Ampicilin (20 mg/kg of body weight) was administrated with all inocula to avoid bacterial contamination. After infection, the G. mellonella larvae were incubated in a glass Petri dish (15mm diameter) at 30°C, and survival (as assessed by spontaneous or provoked motion) was accessed daily. Observation was completed when the larvae died or formed cocoons. Statistical analysis between treatment groups was preformed employing Prism (GraphPad Software, La Jolla, USA). A p value of <0.05 was considered significant by Tukey’s multiple comparison test.
3. Results
3.1. C. neoformans dihydroorotase activity is required for high temperature growth
To identify C. neoformans elements involved in high temperature growth, we used insertional mutagenesis to create a panel of C. neoformans mutant strains, screening for temperature sensitivity. Using the wild type strain KN99α as a recipient for mutagenesis, we identified 64 mutants unable to grow at 37°C. To define the sites of selectable marker insertion, we isolated genomic DNA from each strain and used inverse PCR to characterize the junction sequences between the inserted transfer DNA (t-DNA) and the interrupted C. neoformans DNA sequence. Typically, this method of transconjugation results in single, stable insertion mutations in the fungal genome (Idnurm et al, 2004). One of these strains, mutant G368, had an insertion present on chromosome I, which truncated a gene annotated as CNAG_00734 at amino acid residue 159 (Figure S2). BLAST search analysis at NCBI and SGD (Saccharomyces Genome Database) revealed that CNAG_00734 has similarity to the S. cerevisiae dihydroorotase (DHO) (EC 3.5.2.3) encoding gene URA4 (YLR420W). DHO catalyzes a step in pyrimidine ribonucleotide biosynthetic pathway (Figure 1A), converting N-carbamoyl-L-aspartate into L-dihydroorotate by cyclical dehydration (Guyonvarch et al., 1988). No human homolog of this gene was identified.
The predicted protein sequence encoded by the C. neoformans URA4 gene was aligned with orthologous sequences from Ustilago maydis, S. cerevisiae (S288c), Candida albicans, and Schizosaccharomyces pombe. The highest similarity is shared between the two basidiomycetes C. neoformans and U. maydis (61.2%), and the remaining similarities range between 47–59%. Moreover, the putative C. neoformans Ura4 protein has the two conserved signatures attributed to DHO enzymes (http://prosite.expasy.org/). The first domain spans residues 12-20, including two invariant histidines at positions 14 and 16. The second conserved domain spans residues 242-255 and has five highly conserved amino acids (D244, A246, P247, H248 and K253). Mehbood et al. (2010) described the crystal structure of Bacillus anthracis DHO. In this bacterium there is an active site, which includes a binuclear Zn center, where the H14 and H16 residues and the D224 and D244 residues all bind an α-Zinc atom, and two distant histidines and asparagines bind to a β-Zinc atom.
3.2 C. neoformans URA4 gene deletion and reconstitution
To ensure that the temperature-sensitive phenotype of this strain was due to the ura4 mutation, we created an independent ura4 mutant strain in which we replaced 181 base pairs from the wild type locus, encoding residues 126 to 151, with the neomycin resistance gene. The ura4 mutation and subsequent reconstitution were confirmed by Southern blot analysis (Supplemental Figure 1). These independently generated strains were used in all subsequent analyses.
3.3. Uracil auxotrophic mutants are sensitive to high temperature growth
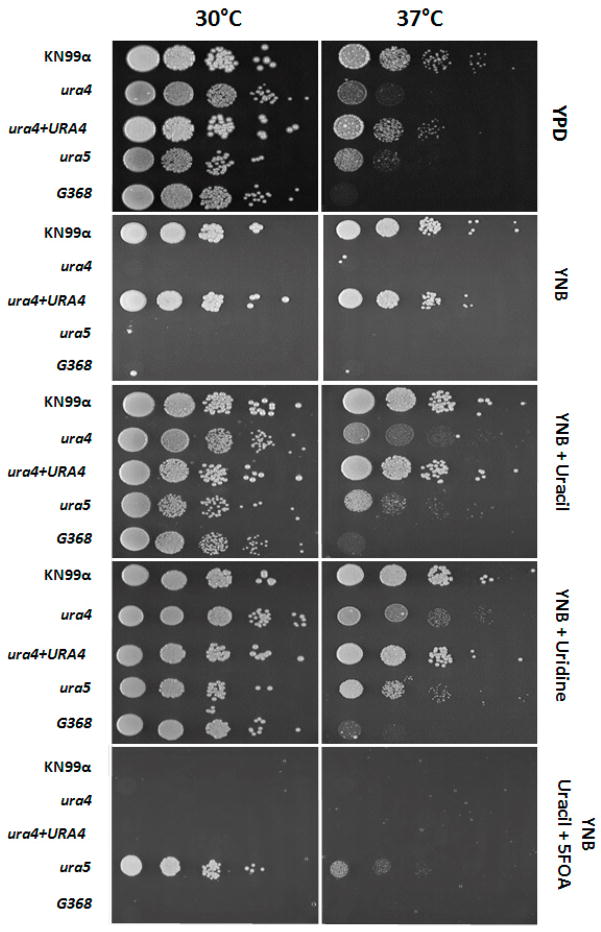
In order to confirm the high temperature growth sensitivity is due to absence of a functional dihydroorotase in C. neoformans, serial dilution of cells from the mutant FGC003 (ura4), wild type (KN99α) and reconstituted strain FGC003R1 (ura4Δ::URA4) were inoculated on YPD plates and incubated at 30°C and 37°C for 48h. Figure 2A shows that wild type and reconstituted strains grow at the same rate in both temperatures, whereas the ura4 mutant strain FGC003 is no longer able to grow at 37°C. We explored whether the URA4 gene itself or the pyrimidine biosynthetic pathway in general is required for high-temperature growth by testing two ura5 mutants for this phenotype. These strains have mutations in a distinct component of pyrimidine biosynthesis, and they were made in related but different strain backgrounds (KN99α and H99, both serotype A). Similar to the ura4 mutants, both ura5 strains were unable to grow at 37°C.
Figure 2.
High temperature growth and auxotrophic analysis. Wild type (KN99α), ura4::NeoR (FGC003), reconstituted strain ura4Δ+URA4 (FGC003R1), ura5 mutant and the original insertional mutant G368 were incubated on the following media at 30°C and 37°C for 48 hours: YPD; YNB supplemented with 2% glucose, 5 g/L ammonium sulfate, and amino acids (YNB), with or without addition of uracil (20μM) or uridine (20μM); and 5-FOA (5-fluoroorotic acid) medium (10 mg/mL). From left to right the amount of cells inoculated in each spot is: 1 × 104, 1 × 103, 1 × 102, 1 × 101 and 1 cell.
Consistent with its predicted function, mutation of the C. neoformans URA4 gene caused the mutant strain to be auxotrophic for uracil, similar to the ura5 mutant strain (Figure 2B). Similar to the wild type, the ura4 mutant strain FGC003 was inhibited by the addition of 5-Fluoroorotic acid (5-FOA) to the growth medium. In contrast, the ura5 mutant was able to grow well on 5-FOA-containing media, as expected considering that the two genes are predicated to act in different steps of the pathway, with the C. neoformans Ura5 converting 5-FOA to fluorodeoxyuridine, which is toxic to cells (Edman and Kwon-Chung, 1990; Kwon-Chung et al. 1992).
Taken together these observations suggest that: (i) the URA4 gene (CNAG_00734) is part of the pyrimidine biosynthetic pathway encoding DHO in C. neoformans; and (ii) blocking pyrimidine ribonucleotide biosynthesis affects the ability of C. neoformans serotype A to grow at mammalian physiological temperature.
3.4. The ura4 mutation affects multiple virulence factors in C. neoformans
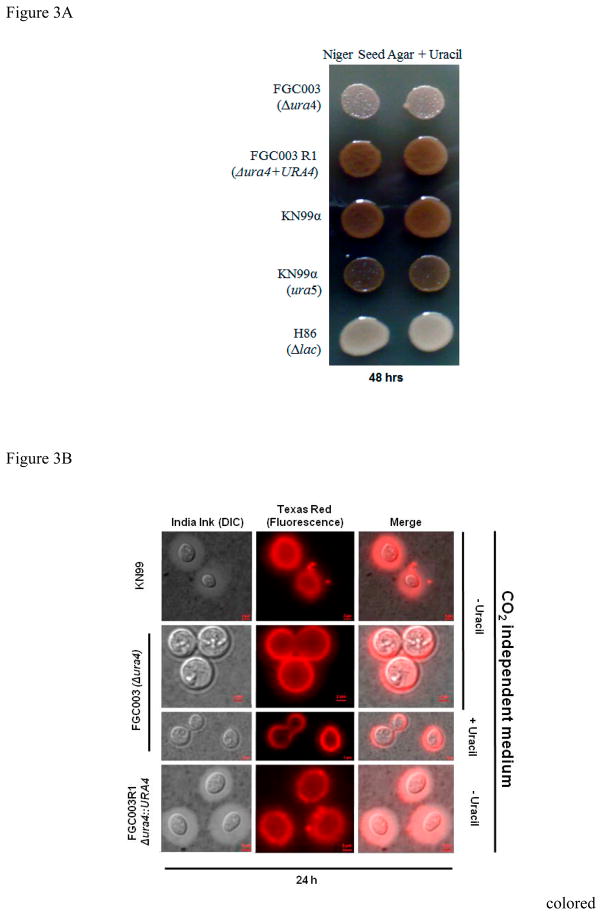
Besides high temperature growth, “classical” virulence factors for C. neoformans include the production of melanin, capsule and phospholipase B activity. Melanized cells are more resistant to reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI) than non-melanized cells (Wang and Casadevall, 1994). Strains that do not produce this pigment are reported to have attenuated virulence in animal models (Kwon-Chung et al. 1982; 1986). The ura4 mutant strain produced melanin, however in a delayed fashion. While colonies of the wild type (KN99α), reconstituted strain (ura4Δ+URA4) and ura5 mutant produce melanin within 48 hours of incubation (Figure 3A), the ura4 strain colonies are pigmented only after 72 hours (data not shown). This was observed regardless of whether the medium was supplemented with uracil.
Figure 3.
Melanin and capsule production in ura4 mutant. (A) 1 × 105 cells were spotted on Niger Seed Agar medium with or without uracil (20μM) and incubated at 30°C for up to 96 hours. Melanin production is represented by a dark brown pigment. Strains used are: Wild type (KN99α), ura4::NeoR (FGC003), reconstituted strain ura4+URA4 (FGC003R1), ura5 mutant and H86 (laccase-deficient mutant) as negative control. (B) Capsule production detected by India ink staining and DIC microscopy in bright field, immune fluorescence microscopy and both images merged. Medium used was CO2-independent medium with and without uracil. Image captures were performed after 24 hours post-capsule induction. Strains used: Wild type (KN99α), ura4::NeoR (FGC003), reconstituted strain ura4Δ+URA4 (FGC003R1). Refer to Material and Methods for immunofluorescence details.
Another well-studied virulence factor is the production of a polysaccharide capsule which allows C. neoformans to evade phagocytosis by macrophages (Kozel and Gotschlich, 1982; Chang and Kwon-Chung, 1994, 1998; Chang et al. 1996). We evaluated this phenotypic trait by direct observation of India ink staining and light microscopy, as well as with immunofluorescence. Both methods reveal a dramatic decrease in capsule production by the ura4 mutant in comparison to the wild type (KN99α) and reconstituted (FGC003R1) strains (Figure 3B), regardless of uracil supplementation. Interestingly, two independent ura5 mutants presented a similar capsule-poor phenotype as observed for the ura4 mutant strain, irrespective of uracil addition to the medium (data not shown), however, addition of uridine (20μM) reverted this condition for the ura4 mutant (Figure S3) [ANOVA, t-Student’s and Scott Knout, p < 0.05]. At 30°C there was no significant variation in the capsular volume between KN99α, ura4Δ, or ura4Δ+URA4. Together, these data suggest that an intact pyrimidine biosynthesis pathway is required for the efficient induction of surface capsule expression
Phospholipase B is involved in tissue invasion by breaking down the host cell membrane leading to cell lysis (Ghannoum, 2000). Previous studies revealed that the C. neoformans phospholipase B mutant (plb1) has a slower intracellular growth inside macrophages (Cox et al. 2001). Phospholipase B activity can be assessed by measuring the zone of precipitation (Pz) on egg yolk-containing media (Price et al., 1982). Nine independent colonies were spotted for each strain to calculate an average Pz index. Statistical analysis showed that the Pz index, while not significantly different between the wild type (1.76±0.26) and the ura4+URA4 reconstituted strain (1.56±0.32), was significantly smaller for the ura4 mutant (1.17±0.73, p<0.05), consistent with Ura4-dependent phospholipase B production. Figure S4 illustrates the capsule production of these strains. Addition of uracil (20μM) or uridine (20μM) in the medium did not rescue the phospholipase activity for the mutant.
3.5. URA4 gene is involved in multi-stress resistance
The C. neoformans cell wall is an important organelle which provides protection from the environment and determines the multiple cell morphologies that this yeast assumes during its development (reviewed in Casadevall and Perfect, 1998). To assess the role of Ura4 in cell integrity and stress response, we incubated the wild type, ura4 mutant and ura4+URA4 strains on media containing various compounds known to affect cell wall phenotypes. The ura4 mutant displayed a marked sensitivity to Congo Red and high salt concentrations (Figure 4A). Cell wall assembly is impaired by Congo Red (CR), which interferes with cell morphology and mother cell-bud separation in S. cerevisiae (for review see Kopecká and Gabriel, 1992; Ram and Klis, 2006). Additionally, resistance to salt stress has been correlated to C. neoformans survival in vivo, and genes involved and/or regulated by the HOG pathway must be expressed during osmotic stress (Idnurm et al., 2009; Ko et al., 2009; Jung et al., 2012).
Figure 4.
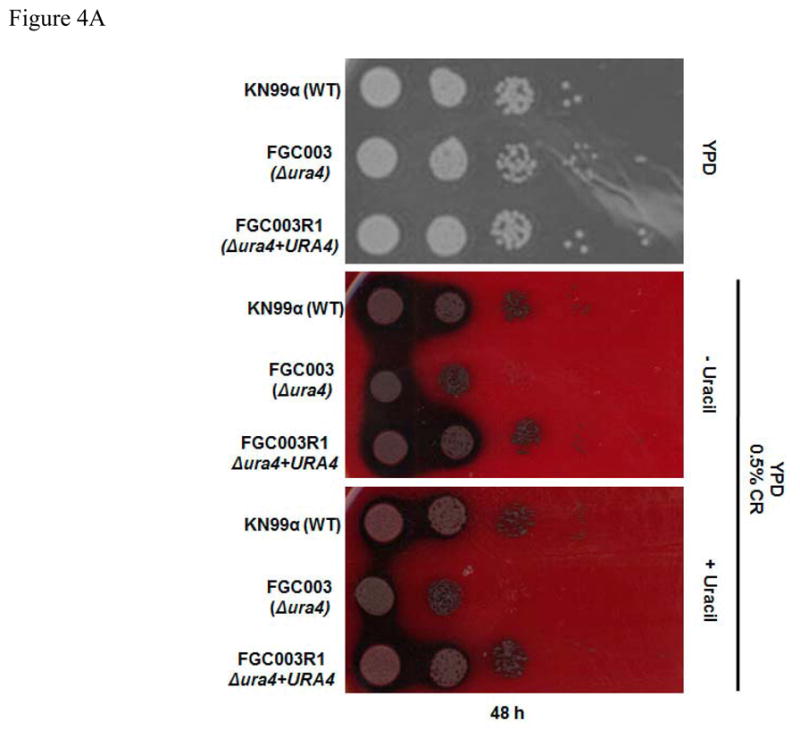
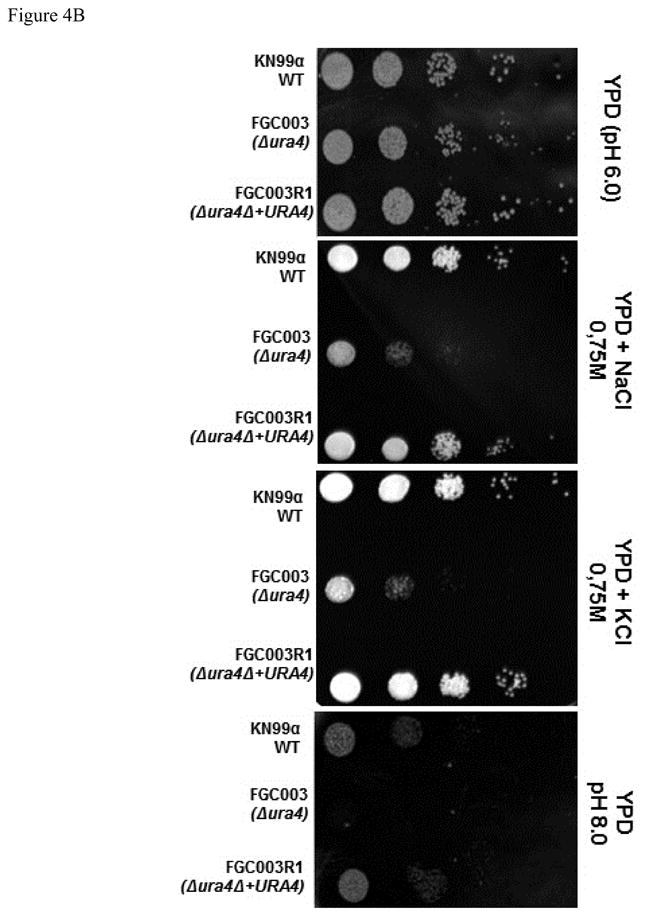
The ura4 mutation renders cells susceptible to multi stress. Dilutions (104 to 1 cell) of wild type (KN99α), ura4::NeoR (FGC003) and reconstituted strain ura4Δ+URA4 (FGC003R1) were spotted on (A) YPD, YPD + 0.5% Congo Red and YPD + 0.5% Congo Red + Uracil and (B) YPD, YPD + 0.75 M NaCl, YPD + 0.75 M KCl and YPD (pH 8.0). Plates were incubated at 30°C from 48 to 72 hours.
Another stress that C. neoformans is subject to during the infection of the host is altered pH. The fungal cells have a preference for acidic growth conditions, and during infection the mammalian tissues offer a slightly alkaline environment. Therefore, an array of genes must be transcribed to respond to this new environment (Casadevall, 2005). We observed that the ura4 mutant has a dramatic growth defect at elevated pH compared to wild type and reconstituted strains (Figure 4B).
3.6. Uracil deficiency increases drug susceptibility
Antifungal agents with activity against cryptococcosis act mainly upon the cell membrane (van der Horst et al., 1997). The azoles (e.g., fluconazole) interferes with the ergosterol biosynthesis, whereas the polyenes (e.g., amphotericin B) binds the ergosterol in the fungal cell membrane leading to leakage of uni- and divalent cations resulting in cell death (for reviews see Bolard, 1986, Fromtling, 1988). To assess whether any of these drugs would act in association with the pyrimidine biosynthesis pathway defect, we tested them against the wild type, ura4 mutant and ura4+URA4 reconstituted strains. The three strains were equally sensitive to fluconazole in rich medium (YPD) as well as in synthetic medium (YNB supplemented with uracil). MIC testing with amphotericin B demonstrated that the ura4 mutant strain had an increased sensitivity to this drug leading to growth arrest at 0.06 μg/mL, compared to the MIC for the wild type KN99α of 0.5 μg/mL. The reconstituted strain (uraΔ+URA4) demonstrated a higher MIC than the mutant, though not fully to the level of wild type. We also tested whether other defects in pyrimidine biosynthesis would result in similar changes in amphotericin susceptibility, and we noted that the ura5 mutant strain behaved similar to the ura4 mutant being inhibited by amphotericin B at 0.06 μg/mL. Banerjee et al (2014) tested the susceptibility of C. neoformans var grubii (H99) ura5 mutant against fluconazole and amphotericin B. They also observed that mutants with a depleted pyrimidine pathway are more susceptible to amphotericin B compared to wild type and reconstituted, whereas fluconazole affected the wild type and mutant equally. This information suggests that inhibition of the pyrimidine biosynthetic pathway, in association with cell membrane-active agents, might have a positive impact on the treatment of cryptococcosis.
3.7. The URA4 gene is essential for fungal survival within macrophages
All the results described above indicate that the ura4 mutant strain would not likely succeed in avoiding macrophage phagocytosis and subsequent fungal killing. Therefore, we tested for intracellular survival in the macrophage lineage J744.A1. When co-cultured with these murine macrophages, the ura4 mutant strain did not survive as well as the wild type (KN99α) or reconstituted (ura4Δ+URA4) strains. After 24 hours of co-incubation with J774.A1 cells, the ura4 mutant strain demonstrated a marked decrease in survival (435.3±37.4 CFU/ml) compared to wild type (735.9±42.3 CFU/ml) or the reconstituted strain (814±82 CFU/ml) (p < 0.05 for each comparison). The ura5 mutant similarly does not survive well inside the macrophage (214.5±74.2 CFU/ml). The altered survival was not attributable to varying rates of engulfment since the phagocytosis index was identical among these strains (data not shown). Taken together, these results link defects in the pyrimidine biosynthetic pathway with alterations in C. neoformans intracellular survival.
3.8. URA4 gene is essential for in vivo survival
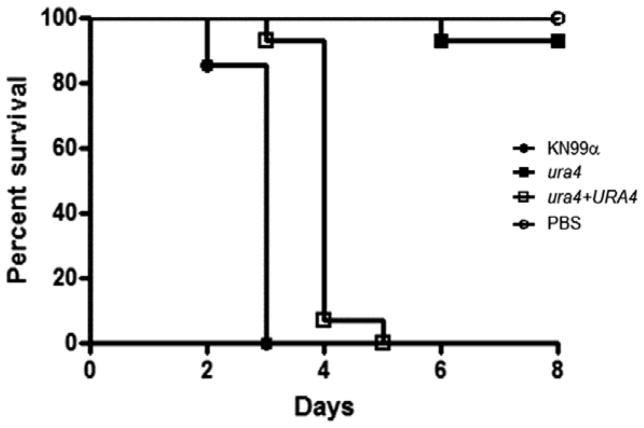
We compared the relative virulence of the wild type, ura4, and ura4 + URA4 reconstituted strains in a murine inhalational model of cryptococcosis (Cox et al., 2000). As presented in Figure 5, infections with the wild type and reconstituted strains resulted in an average survival of 17.5 (±1.87) and 17.5 (±1.58) days post-infection, respectively. On the contrary, the ura4 mutant strain-infected mice survived until the end of the experiment (40 days post-infection). Lungs from two of the surviving mice were harvested and homogenized, followed by serial dilution and quantitative cultures. Even though these animals appeared clinically healthy, there were still viable yeast cells in their lungs (2.04×105 ±5.6×103 CFU/mg).
Figure 5.
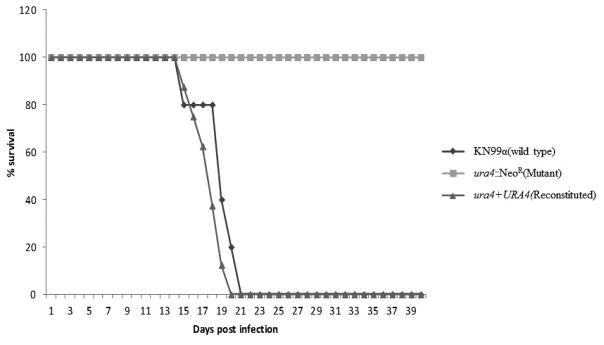
Virulence test. Groups of ten 4- to 6-week-old 57BL/6 mice were infected by nasal inhalation with wild type (KN99α), ura4::NeoR (FGC003) and reconstituted strain ura4Δ::URA4 (FGC003R1). Survival was followed during the course of the infection. P values were <0.001 for the comparisons between ura4::NeoR (FGC003), and wild type and ura4Δ+URA4 (FGC003R1). Five mice were inoculated with the wild type (KN99α).
Also, we checked whether the ura4 mutant would display attenuated virulence at the permissive growth temperature (30°C) in a Galleria mellonella killing assay. The wild-type strain and the reconstituted (ura4+URA4) strains resulted in killing of all Galleria larvae at 3 and 5 days, respectively. In contrast, infection with the ura4 mutant strain did not result in host killing in this assay, statistically similar to PBS treatment (p<0.05 between mutant and wild-type). (Figure 6). Therefore, we conclude that the ura4 mutant is attenuated for virulence at both permissive and restrictive temperatures for its growth.
Figure 6.
C. neoformans ura4 mutant is avirulent in an invertebrate infection model at 30°C. Galleria mellonella larvae were inoculated with either the wild type (KN99α), ura4::NeoR (FGC003) or reconstituted strain ura4Δ+URA4 (FGC003R1). The infected larvae were monitored for survival
3.9. Pyrimidine de novo and salvage pathway genes respond to high temperature growth
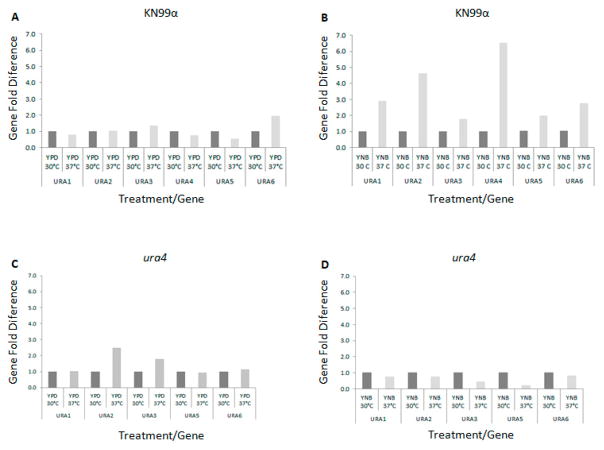
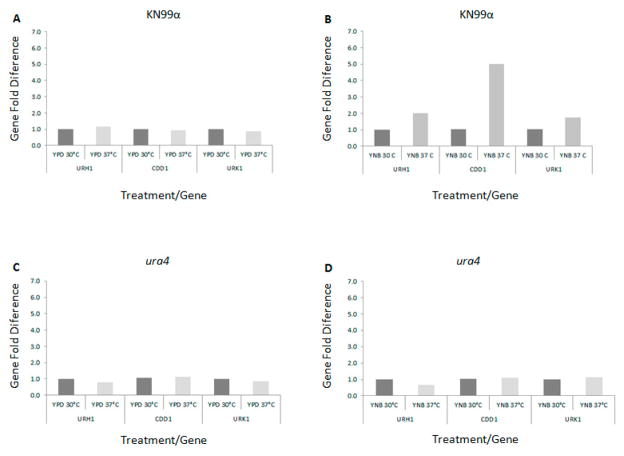
We have demonstrated that several of the major virulence phenotypes are affected by both ura4 and ura5 mutations. Therefore, we analyzed the transcriptional pattern of the genes involved in the pyrimidine biosynthetic and salvage pathways by real time PCR in complex medium (YPD) and synthetic medium (YNB) at 30°C and 37°C. Figures 7A and 8A show that the expression pattern for genes in both pathways does not change substantially in rich medium (YPD) regardless of the temperature. All the genes tested, in both de novo and salvage pathways, were induced when incubated in YNB medium not supplemented with uracil at 37°C compared to 30°C (Figure 7B and 8B, respectively). The highest levels of induction in these experiments are for URA4, URA2 and URA1, which have 4.6-, 6.5- and 2.9-fold increases in expression after 2 hours at 37°C in YNB. In the salvage pathway there is a 5-fold increase in CDD1 expression at 37°C in YNB and a 2-fold increase for URK1 and URH1. These results indicate that there is increased transcript abundance for multiple genes in the pyrimidine de novo biosynthesis and also the salvage pathway during growth at human physiological temperature in poor nutritional condition, which is a situation likely encountered during the host infection.
Figure 7.
Real time PCR. Expression pattern of the pyrimidine de novo pathway genes URA1, 2, 3, 4, 5 and 6 in the wild type strain KN99 cultivated in (A) rich medium YPD and (B) synthetic medium YNB (no uracil added) and the mutant strain ura4 cultivated in (C) rich medium YPD and (D) synthetic medium YNB (no uracil added) at 30°C and 37°C.
Figure 8.
Real time PCR. Expression pattern of the pyrimidine salvage pathway genes CDD1, URK1, URH1 in the wild type strain KN99 cultivated in (A) rich medium YPD and (B) synthetic medium YNB (no uracil added) and the mutant strain ura4 cultivated in (C) rich medium YPD and (D) synthetic medium YNB (no uracil added) at 30°C and 37°C.
We also tested the role of the ura4 mutation on the transcriptional induction of other genes in these pathways. URA2 and URA3 are 2.5- and 1.8-fold induced, respectively, in complex medium (YPD) at 37°C in the ura4 mutant compared to YPD medium at 30°C. Under the same conditions we observed no change in expression for URA1, URA5 and URA6 (Figure 7C).
A pronounced shift in expression pattern of the genes in the de novo biosynthetic pathway was observed when the ura4 mutant strain was cultivated in synthetic medium (YNB) at the restrictive temperature (37°C): all 5 genes tested are down-regulated at 37°C (Figure 7D). URA1, URA2 and URA6 had a 20% reduction of their expression on YNB at 37°C compared to 30°C. URA3 and URA5 genes also had their transcription pattern dramatically repressed (40% and 20%) compared to YNB at 30°C (Figure 8D).
Comparatively, all 5 genes (URA1, URA2, URA3, URA5 and URA6) were induced in the wild type strain incubated in YNB at 37°C (2.9-, 4.6-, 1.8-, 2.0- and 2.8-fold respectively, Figure 7B). These data suggest that the dihydroorotase gene may play a regulatory role in the pyrimidine biosynthetic pathway where the lack of its function impairs the transcriptional response necessary during heat stress. No significant alterations in transcript abundance were noted for the genes in the uracil salvage pathway in the ura4 mutant background, at either temperature, or in either rich or poor growth media (Figure 8C and D). Taken together these observations suggest that dihydroorotase may act as a regulatory point in both the de novo and the salvage pathways, being necessary to respond to the stresses encountered at high temperatures with nutrient depletion.
4. Discussion
Insertional mutagenesis mediated by Agrobacterium tumefaciens in C. neoformans has been a useful tool for gene function discovery and analysis (Idnurm et al., 2004; Idnurm and Heitman, 2005; Walton et al., 2005; 2006; Feretzaki and Heitman, 2013; Lee et al., 2013). In this paper, this technique was employed to identify genes involved in high temperature growth, an important virulence factor for this yeast. Among the transformants unable to grow at high temperature, we identified one that has a truncated copy of the URA4 gene. This enzyme, which is involved in the third step on the pyrimidine pathway, catalyzes the conversion of carbamoyl-L-aspartate to dihydroorotate. The role of the C. neoformans pyrimidine biosynthetic pathway in high temperature growth was confirmed with independently generated ura4 mutants, as well as by documenting temperature-restricted growth of the ura5 mutant strain, with a defect in a separate gene in this pathway. Transcriptional profiling data also support the correlation between pyrimidine availability and heat stress, since both biosynthetic and salvage pathways are highly induced at 37°C. Another relevant finding was the role of this pathway in general stress tolerance. Moreover, strains with loss of pyrimidine biosynthesis had attenuated virulence.
Previously, C. albicans URA3 has been used as a selectable marker (URA-blaster), generating over time a large mutant collections (Fonzi and Irwin, 1993). Brand et al. (2004) reported a genome position effect upon URA3 expression: lower URA3 transcriptional levels determined by location correlated with attenuated virulence. In C. neoformans the URA5 gene was used to assist molecular genetic manipulation of this fungus as a selectable marker for transformation (Edman and Kwon-Chung, 1990; Varma et al 1992). The in vitro phenotypes of this mutant strain were not explored in depth. The ura5 mutant was noted to be highly attenuated in a murine model of systemic cryptococcosis, although this strain eventually caused lethal disease in mice (Varma et al., 1992). This finding is similar to the studies presented here: although the ura4 mutant caused no mortality after 40 days of infection, the strain was still viable and recoverable from infected tissue at the end of the experiment.
In similar studies, an adenine auxotrophic mutant was recovered from a patient with systemic cryptococcosis (Schiappa et al., 2002). Prior studies had suggested that defects in adenine biosynthesis resulted in profound virulence defects in a rabbit model of cryptococcal meningitis (Perfect et al., 1993). It is possible that different microenvironments in the host may offer different nutrient availability for these strains. Therefore, an adenine auxotrophic strain may be able to grow in the bone marrow, a tissue in which adenine is not predicted to be a limiting nutrient given the high cell turnover at this site. However, the same auxotrophic strain may be unable to grow in cerebral spinal fluid, which is more nutrient-poor.
Other studies have suggested that the analysis of metabolic pathways has the potential to identify targets for novel therapeutic interventions for cryptococcal disease. C. neoformans mutants with defects in methionine, threonine, isoleucine, valine and lysine biosynthesis display reduced growth rates, high temperature growth defects, sensitivity to the inhibitory effects of antifungal drugs and reduced virulence (Yang et al., 2002; Pascon et al., 2004; Kingsbury et al., 2004; Kingsbury et al., 2004a; Kingsbury and McCusker, 2008; Kingsbury and McCusker, 2010; Kingsbury and McCusker, 2010a; Kingsbury and McCusker, 2010b). Therefore, defects in metabolic pathways may offer more attenuation in survival than merely inducing specific nutritional auxotrophies.
Recently, the purine biosynthetic pathway in C. neoformans was studied as a novel target for antifungal intervention (Morrow et al., 2012). Defects in de novo purine biosynthesis, and not purine salvage, resulted in strains with growth defects and reduced virulence. Moreover, fungal-specific features were identified in the predicted structure of the inosine monophosphate dehydrogenase enzyme involved in guanine nucleotide synthesis. Together, these studies suggest that nucleic acid synthesis pathways offer potential, unexplored targets for intervention in treating fungal diseases.
Reduced growth caused by dihydroorotase defects have been reported in other organisms. For example, in Salmonella typhimurium lack of pyrC (URA4 homolog) prevents the conversion of carbamoyl aspartate to dihydroorotate, leading to the accumulation of toxic intermediaries, which may cause growth reduction (Turnbough and Bochner, 1985). In S. cerevisiae the ura4 mutant has reduced vegetative growth at 30°C (Yoshikawa et al., 2011). In a clinical isolate of S. cerevisiae, lack of the Ura3 protein led to a reduced rate of growth for the mutant and also renders a strain with attenuated virulence in a murine model of infection (Goldstein and McCusker, 2001). In the parasite Trypanosoma brucei it was found that alterations in pyrimidine biosynthesis yielded strains with low infectivity in animal model (Ali et al. 2013). Also in Toxoplasma gondii, strains with a uracil auxotrophy elicit protective immunity to subsequent infection (Fox and Bzik, 2010). Moreover, Hegewald et al (2013) demonstrated that dihydroorotate dehydrogenase (URA1) is a relevant drug target for 1-hydroxyquuinolones in T. gondii. Taken together, this work implicates the importance of the pyrimidine biosynthetic pathways in the virulence of multiple microbial pathogens, suggesting that inhibitors of gene products within this pathway could help to treat diverse infections, perhaps in association with other therapeutic drugs. We showed that lack of functional Ura4 in C. neoformans affects other virulence factors and reduced stress response; however, at this point our results do not allow us to explain the mechanism of this interference, and new experiments such as global gene expression under pyrimidine depletion could be performed to shed some light on this issue.
Supplementary Material
Acknowledgments
The authors would like to thank Bianca Trama for technical support and Dr. Patricia Albuquerque for providing the Galleria mellonella larvae. This work was sponsored by a FAPESP grant (2007/50536-3) to MAV and NIH grants (AI050128 and AI074677) to JAA.
References
- Ali JA, et al. Pyrimidine biosynthesis is not an essential function for Trypanosoma brucei bloodstream forms. PLoS One. 2013;8(3):e58034. doi: 10.1371/journal.pone.0058034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36(2):352–65. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Burkard L, Panepinto JC. Inhibition of Nucleotide Biosynthesis Potentiates the Antifungal Activity of Amphotericin B. PLoS ONE. 2014;9(1):e87246. doi: 10.1371/journal.pone.0087246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta. 1986;864(3–4):257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- Bölker M. Ustilago maydis--a valuable model system for the study of fungal dimorphism and virulence. Microbiology. 2001;147(Pt 6):1395–401. doi: 10.1099/00221287-147-6-1395. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3(4):900–9. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Campbell LT, Lodge JK. Cryptococcus neoformans, a fungus under stress. Curr Opin Microbiol. 2007;10(4):320–5. doi: 10.1016/j.mib.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; Washington, DC: 1998. [Google Scholar]
- Casadevall A. Fungal virulence, vertebrate endothermy, and dinosaur extinction: is there a connection? Fungal Genet Biol. 2005;42(2):98–106. doi: 10.1016/j.fgb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14(7):4912–9. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun. 1998;66(5):2230–6. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Penoyer LA, Kwon-Chung KJ. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64(6):1977–83. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Perrella FW, Behrens DL, Papp LM. Inhibition of dihydroorotate dehydrogenase activity by brequinar sodium. Cancer Res. 1992;52(13):3521–7. [PubMed] [Google Scholar]
- Cox GM, et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71(1):173–80. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39(1):166–75. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68(2):443–8. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148(Pt 8):2607–15. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10(9):4538–44. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270(50):29682–9. [PubMed] [Google Scholar]
- Feretzaki M, Heitman J. Genetic Circuits that Govern Bisexual and Unisexual Reproduction in Cryptococcus neoformans. PLoS Genet. 2013;9(8):e1003688. doi: 10.1371/journal.pgen.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134(3):717–28. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Bzik DJ. Avirulent uracil auxotrophs based on disruption of orotidine-5′-monophosphate decarboxylase elicit protective immunity to Toxoplasma gondii. Infect Immun. 2010;78(9):3744–52. doi: 10.1128/IAI.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromtling RA. Overview of medically important antifungal azole derivatives. Clin Microbiol Rev. 1988;1(2):187–217. doi: 10.1128/cmr.1.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13(1):122–43. doi: 10.1128/cmr.13.1.122-143.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Development of Saccharomyces cerevisiae as a model pathogen: a system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics. 2001;159:499–513. doi: 10.1093/genetics/159.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene S, Watanabe K, Braatz-Trulson J, Lou L. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochem Pharmacol. 1995;50(6):861–7. doi: 10.1016/0006-2952(95)00255-x. [DOI] [PubMed] [Google Scholar]
- Guyonvarch A, Nguyen-Juilleret M, Hubert JC, Lacroute F. Structure of the Saccharomyces cerevisiae URA4 gene encoding dihydroorotase. Mol Gen Genet. 1988;212(1):134–41. doi: 10.1007/BF00322456. [DOI] [PubMed] [Google Scholar]
- Hegewald J, Gross U, Bohne W. Identification of dihydroorotate dehydrogenase as a relevant drug target for 1-hydroxyquinolones in Toxoplasma gondii. Mol Biochem Parasitol. 2013;190(1):6–15. doi: 10.1016/j.molbiopara.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Heitman JF, SG, John E, Edwards JE, Jr, Aaron P, Mitchell AP. Molecular Principles of Fungal Pathogenesis. ASM Press; Washington, DC: 2006. p. 700. [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3(4):e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Reedy JL, Nussbaum JC, Heitman J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell. 2004;3(2):420–9. doi: 10.1128/EC.3.2.420-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Walton FJ, Floyd A, Reedy JL, Heitman J. Identification of ENA1 as a virulence gene of the human pathogenic fungus Cryptococcus neoformans through signature-tagged insertional mutagenesis. Eukaryot Cell. 2009;8(3):315–26. doi: 10.1128/EC.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KW, Strain AK, Nielsen K, Jung KH, Bahn YS. Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet Biol. 2012;49(4):332–45. doi: 10.1016/j.fgb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khutornenko AA, et al. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci U S A. 2010;107(29):12828–33. doi: 10.1073/pnas.0910885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Threonine biosynthetic genes are essential in Cryptococcus neoformans. Microbiology. 2008;154(Pt 9):2767–75. doi: 10.1099/mic.0.2008/019729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Fungal homoserine kinase (thr1Δ) mutants are attenuated in virulence and die rapidly upon threonine starvation and serum incubation. Eukaryot Cell. 2010;9(5):729–37. doi: 10.1128/EC.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1 Δ) mutants. Eukaryot Cell. 2010a;9(5):717–28. doi: 10.1128/EC.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2{Δ}) mutants is influenced by the carbon source and rapamycin. Microbiology. 2010b;156(Pt 3):929–39. doi: 10.1099/mic.0.034348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. Novel chimeric spermidine synthase-saccharopine dehydrogenase gene (SPE3-LYS9) in the human pathogen Cryptococcus neoformans. Eukaryot Cell. 2004;3(3):752–63. doi: 10.1128/EC.3.3.752-763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology. 2004a;150(Pt 5):1547–58. doi: 10.1099/mic.0.26928-0. [DOI] [PubMed] [Google Scholar]
- Ko YJ, et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell. 2009;8(8):1197–217. doi: 10.1128/EC.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecká M, Gabriel M. The influence of congo red on the cell wall and (1–3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol. 1992;158(2):115–26. doi: 10.1007/BF00245214. [DOI] [PubMed] [Google Scholar]
- Kozel TR, Gotschlich EC. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J Immunol. 1982;129(4):1675–80. [PubMed] [Google Scholar]
- Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11(3):370–80. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Polacheck I, Popkin TJ. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982;150(3):1414–21. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Rhodes JC. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51(1):218–23. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A, Edman JC, Bennett JE. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J Med Vet Mycol. 1992;30(1):61–9. [PubMed] [Google Scholar]
- Lam WC, Gerik KJ, Lodge JK. Role of Cryptococcus neoformans Rho1 GTPases in the PKC1 signaling pathway in response to thermal stress. Eukaryot Cell. 2013;12(1):118–31. doi: 10.1128/EC.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IR, et al. Characterization of the complete uric acid degradation pathway in the fungal pathogen Cryptococcus neoformans. PLoS One. 2013;8(5):e64292. doi: 10.1371/journal.pone.0064292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Mody CH. Cryptococcus. Proc Am Thorac Soc. 2010;7(3):186–96. doi: 10.1513/pats.200907-063AL. [DOI] [PubMed] [Google Scholar]
- Liu S, Neidhardt EA, Grossman TH, Ocain T, Clardy J. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure. 2000;8(1):25–33. doi: 10.1016/s0969-2126(00)00077-0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McClelland CM, Chang YC, Kwon-Chung KJ. High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens. Fungal Genet Biol. 2005;42(11):904–13. doi: 10.1016/j.fgb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- McFadden DC, Casadevall A. Unexpected diversity in the fine specificity of monoclonal antibodies that use the same V region gene to glucuronoxylomannan of Cryptococcus neoformans. J Immunol. 2004;172(6):3670–7. doi: 10.4049/jimmunol.172.6.3670. [DOI] [PubMed] [Google Scholar]
- Mehboob S, Mulhearn DC, Truong K, Johnson ME, Santarsiero BD. Structure of dihydroorotase from Bacillus anthracis at 2.6 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 11):1432–5. doi: 10.1107/S1744309110037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8(4):515–48. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow CA, et al. De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2012;8(10):e1002957. doi: 10.1371/journal.ppat.1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73(7):3842–50. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norager S, Jensen KF, Bjornberg O, Larsen S. E. coli dihydroorotate dehydrogenase reveals structural and functional distinctions between different classes of dihydroorotate dehydrogenases. Structure. 2002;10(9):1211–23. doi: 10.1016/s0969-2126(02)00831-6. [DOI] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16(10):2576–89. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Xu W, Selvig KM, O’Meara JM, Mitchell AP, Alspaugh JA. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 2013;34(4):673–84. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong HB, Sienkiewicz N, Wyllie S, Patterson S, Fairlamb AH. Trypanosoma brucei (UMP synthase null mutants) are avirulent in mice, but recover virulence upon prolonged culture in vitro while retaining pyrimidine auxotrophy. Mol Microbiol. 2013;90(2):443–55. doi: 10.1111/mmi.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva JA, Pereira JM. New antifungal antibiotics. Curr Opin Infect Dis. 2013;26(2):168–74. doi: 10.1097/QCO.0b013e32835ebcb7. [DOI] [PubMed] [Google Scholar]
- Park BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–30. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Pascon RC, Ganous TM, Kingsbury JM, Cox GM, McCusker JH. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology. 2004;150(Pt 9):3013–23. doi: 10.1099/mic.0.27235-0. [DOI] [PubMed] [Google Scholar]
- Perfect JR, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2010;50(3):291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Toffaletti DL, Rude TH. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61(10):4446–51. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkin JW, Panaccione DG, Walton JD. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology. 1996;142 (Pt 6):1557–65. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- Price MF, Wilkinson ID, Gentry LO. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20(1):7–14. doi: 10.1080/00362178285380031. [DOI] [PubMed] [Google Scholar]
- Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1(5):2253–6. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- Rex JH, et al. Need for alternative trial designs and evaluation strategies for therapeutic studies of invasive mycoses. Clin Infect Dis. 2001;33(1):95–106. doi: 10.1086/320876. [DOI] [PubMed] [Google Scholar]
- Roy A, Exinger F, Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990;10(10):5257–70. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JF, EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1989. [Google Scholar]
- Schiappa D, et al. An auxotrophic pigmented Cryptococcus neoformans strain causing infection of the bone marrow. Med Mycol. 2002;40(1):1–5. doi: 10.1080/mmy.40.1.1.5. [DOI] [PubMed] [Google Scholar]
- Singh N, Husain S. Infections of the central nervous system in transplant recipients. Transpl Infect Dis. 2000;2(3):101–11. doi: 10.1034/j.1399-3062.2000.020302.x. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175(5):1405–11. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL, Jr, Bochner BR. Toxicity of the pyrimidine biosynthetic pathway intermediate carbamyl aspartate in Salmonella typhimurium. J Bacteriol. 1985;163(2):500–5. doi: 10.1128/jb.163.2.500-505.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst CM, et al. Treatment of cryptococcal meningitis associated with the acquired immunodeficiency syndrome. National Institute of Allergy and Infectious Diseases Mycoses Study Group and AIDS Clinical Trials Group. N Engl J Med. 1997;337(1):15–21. doi: 10.1056/NEJM199707033370103. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Kwon-Chung KJ. Characterization of the glyceraldehyde-3-phosphate dehydrogenase gene [correction of glyceraldehyde-3-phosphate gene] and the use of its promoter for heterologous expression in Cryptococcus neoformans, a human pathogen. Gene. 1999;232(2):155–63. doi: 10.1016/s0378-1119(99)00132-8. [DOI] [PubMed] [Google Scholar]
- Varma A, Edman JC, Kwon-Chung KJ. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect Immun. 1992;60(3):1101–8. doi: 10.1128/iai.60.3.1101-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell. 2006;17(9):3768–80. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57(5):1381–96. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun. 1994;62(7):3004–7. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Pascon RC, Alspaugh A, Cox GM, McCusker JH. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology. 2002;148(Pt 8):2617–25. doi: 10.1099/00221287-148-8-2617. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, et al. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast. 2011;28(5):349–61. doi: 10.1002/yea.1843. [DOI] [PubMed] [Google Scholar]
- Zameitat E, Freymark G, Dietz CD, Loffler M, Bolker M. Functional expression of human dihydroorotate dehydrogenase (DHODH) in pyr4 mutants of Ustilago maydis allows target validation of DHODH inhibitors in vivo. Appl Environ Microbiol. 2007;73(10):3371–9. doi: 10.1128/AEM.02569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.