Abstract
B chromosomes are small, heterochromatic chromosomes that are transmitted in a non-Mendelian manner. We have identified a stock of Drosophila melanogaster that recently (within the last decade) acquired an average of 10 B chromosomes per fly. These B chromosomes are transmitted by both males and females and can be maintained for multiple generations in a wild-type genetic background despite the fact that they cause high levels of 4th chromosome meiotic nondisjunction in females. Most curiously, these B chromosomes are mitotically unstable, suggesting either the absence of critical chromosomal sites or the inability of the meiotic or mitotic systems to cope with many additional chromosomes. These B chromosomes also contain centromeres and are primarily composed of the heterochromatic AATAT satellite sequence. Although the AATAT sequence comprises the majority of the 4th chromosome heterochromatin, the B chromosomes lack most, if not all, 4th chromosome euchromatin. Presumably as a consequence of their heterochromatic content, these B chromosomes significantly modify position-effect variegation in two separate reporter systems, acting as enhancers of variegation in one case and suppressors in the other. The identification of B chromosomes in a genetically tractable organism like D. melanogaster will facilitate studies of chromosome evolution and the analysis of the mechanisms by which meiotic and mitotic processes cope with additional chromosomes.
Keywords: meiosis, supernumerary chromosomes, minichromosomes, mitotic instability
MANY organisms contain supernumerary chromosomes known as B chromosomes. Unlike the standard A chromosomes, B chromosomes are not required for the viability of an organism. B chromosomes are prevalent throughout eukaryotes and have been identified in >1300 plant and 500 animal species (Yoshida et al. 2011). They are typically smaller than A chromosomes, are inherited in a non-Mendelian fashion, and often undergo nondisjunction at anaphase in mitosis (Beukeboom 1994; Birchler and Han 2013). B chromosomes are also predominantly heterochromatic, such as those observed in the mealybug (Epstein et al. 1992). In cases where genes are present on these chromosomes, they are usually silenced (Beukeboom 1994; Martis et al. 2012; Banaei-Moghaddam et al. 2013), with the notable exception of ribosomal RNA (rRNA) genes. For example, Leach et al. (2005) demonstrated that rRNA genes were both present and transcribed in the plant Crepis capillaris.
B chromosomes are thought to arise through multiple mechanisms, including de novo formation, which is presumed to occur as a response to an interspecific hybridization event that alters the genome (Jones 2003). However, since B chromosomes are frequently found to contain A chromosome sequences, a more common hypothesis of B chromosome formation is that they arise from the A chromosomes. It has been hypothesized that the formation of B chromosomes in most species results either from some form of chromosome misdivision, as in maize (Kaszas and Birchler 1996), or from complex translocation events, as seen in the grasshopper Eyprepocnemis plorans (Henriques-Gil et al. 1983; Kaszas and Birchler 1996; Camacho et al. 2000)
In the genus Drosophila, B chromosomes were first identified in 1980 in a subpopulation of D. nasuta albomicana and later in subpopulations of D. subsilvestris, D. kikkawai, and D. malerkotliana (Ramachandra and Ranganath 1985; Sundaran and Gupta 1994; Gutknecht et al. 1995). We recently identified the first example of B chromosomes in D. melanogaster. These chromosomes were discovered by live imaging of meiotic prometaphase I in oocytes from one of our D. melanogaster stocks created ∼10 years ago. Because these B chromosomes are not present in the original stock used to create the line of interest, they appear to have arisen de novo in the last 10 years. The B chromosomes described here are primarily composed of heterochromatic sequences, specifically the AATAT satellite repeat. Although these chromosomes appear to possess centromeres, like many B chromosomes they are mitotically unstable (Voltolin et al. 2012).
Many lower vertebrate and invertebrate studies report minimal effects on the fitness of the organism, especially when B chromosomes are present in lower numbers (Beukeboom 1994). However, in some organisms, B chromosomes have been reported to cause sterility, and even lethality, to their host, although this is more common when B chromosomes are present in higher numbers (Beukeboom 1994; Camacho et al. 2000). For example, in D. nasuta albomicana, the upper limit of B chromosome accumulation was eight, indicating that they were detrimental to viability at higher numbers (Ramachandra and Ranganath 1987). Additionally, Masonbrink and Birchler (2012) demonstrated that increasing the number of B chromosomes and B chromosome derivatives eventually led to sterility in maize. Similar supernumerary chromosomes in humans have been linked to several developmental abnormalities, including delayed mental growth, sterility, and various nonspecific neurological disorders (Wisniewski et al. 1979; Wisniewski and Doherty 1985; Fuster et al. 2004). We find that the D. melanogaster B chromosomes induce very high levels of 4th chromosome nondisjunction in female meiosis and alter the effects of heterochromatin on gene expression. It is thus surprising that they appear to persist despite these presumably negative effects on fitness.
Because of their continued presence in the genomes of many species, B chromosomes may facilitate the introduction of diversity within genomes through the creation of new, functional chromosomes. For example, Hackstein et al. (1996) and Bernardo Carvalho et al. (2009) have postulated that the Drosophila Y chromosome evolved from a B chromosome. B chromosomes in D. melanogaster thus provide us with a new tool for understanding chromosome dynamics and problems associated with chromosomal abnormalities.
Materials and Methods
Stocks
Fly stocks were maintained at 25° on standard cornmeal medium. Unless otherwise noted, y w; spapol was used as a wild-type (WT) control and all B chromosomes were derived from y w; mtrm126/TM3; spapol. WT+B stocks were generated by crossing y w; mtrm126/TM3; spapol to y w; spapol flies and then selecting for TM3 in the first generation. FM7w; spapol was used to create the flies with nonexchange X chromosomes for nondisjunction assays. In larval neuroblasts, generation one was never scored, due to the inability to distinguish mtrm/+ larvae from TM3/+ larvae.
Chromosome cytology and immunostaining
Mitotic chromosome spreads of larval brains were prepared by dissecting third instar larvae in phosphate buffered saline (PBS) and incubating in 2 mL of PBS containing 100 μl of 10−3 M colchicine for 1 hr. Larval neuroblasts were then transferred to a hypotonic solution (0.5% sodium citrate) for 6 min, followed by 5 min in a fixation solution containing 45% acetic acid and 3% paraformaldehyde in PBS. Neuroblasts were squashed in 45% acetic acid between siliconized cover slips and glass slides and frozen in liquid nitrogen. The cover slips were then removed, and the slides were washed three times in room temperature PBS. The slides were mounted in Vectashield containing DAPI or treated with DAPI, washed three times in PBS, and mounted in ProLong Gold (Invitrogen). The number of B chromosomes was scored by carefully scanning through the z-stacks acquired during imaging. For the WT+B larval neuroblast experiments examining the number of B chromosomes, crosses were initiated at least two times for each and the results pooled. B chromosome size was approximated by measuring and comparing two B chromosomes to both 4th chromosomes in a minimum of 5 cells from 11 different brains for a total of 100 cells.
Indirect immunofluorescence assays of oocytes were done as described in Hughes et al. (2011). The primary antibodies were used at the following concentrations: rat anti-CID (1:2000) (Martins et al. 2009) and rabbit histone H3 trimethylated on K9 (H3K9m3) (1:250) (Abcam).
Fluorescent in situ hybridization (FISH) was performed as described in Xiang and Hawley (2006) with the following modifications: All FISH probes used were synthesized oligos conjugated to fluorophores. Part of the 359-bp repeat on the X chromosome was conjugated to Alexa Fluor 488 (Ferree and Barbash 2009); the AATAT repeat found primarily on the 4th chromosome was conjugated to Cy3 (Dernburg et al. 1996); the 2R (AACAC)6 sequence was conjugated to Alexa Fluor 488; and the 3R (Dodeca) sequence, (CCCGTACTGGT)4, was conjugated to Alexa Fluor 555 (Dernburg et al. 1996; Dernburg 2000).The hybridization temperature was 31° and denaturation temperature was 91° for the X and 4th chromosome probes and 37° and 91° for the 2R and Dodeca probes.
For all imaging, the DeltaVision microscopy system was used (Applied Precision, Issaquah, WA). The system is equipped with an Olympus 1670 inverted microscope and high-resolution charge-coupled device camera. The images were deconvolved using SoftWoRx v.25 software (Applied Precision).
Live imaging
Live imaging was carried out as described in Hughes et al. (2009). Briefly, stage 13 oocytes dissected from 2- to 3-day-old yeasted females were aligned in halocarbon oil 700 (Sigma), injected with Quant-iT OliGreen single-stranded DNA (ssDNA) Reagent (Invitrogen) diluted 0.7-fold with water and porcine rhodamine-conjugated tubulin minus glycerol (Cytoskeleton). Oocytes were covered with a YSI membrane and imaged using an LSM-510 META confocal microscope (Zeiss) with an alpha Plan-Fluar 100× (1.4 NA) with a 1.5 zoom. Images were acquired by taking a 10-series z-stack at 1-μm intervals with 20 sec between acquisitions using the AIM software v4.2. Images were transformed into 2D projections and concatenated into videos using AIM software v4.2.
Nondisjunction assays
Female chromosome nondisjunction (NDJ) at meiosis I was assayed by scoring the progeny from single virgins mated to attached XY, y+ v f B; C(4), ci eyR males, as previously described in Harris et al. (2003). For male NDJ, single males were crossed to y w; C(4), ci eyR females. For NDJ experiments of females with chiasmate Xs, wild-type flies were y w; spapol and WT+B flies were y w; spapol +B. For male NDJ, wild-type flies were y w/ y+Y; spapol and WT+B were y w/ y+Y; spapol +B. For experiments looking at NDJ of achiasmate Xs, wild type was FM7w/ y w; spapol and WT+B was FM7w/ y w; spapol +B. Statistical analysis of these data was then carried out as described in Zeng et al. (2010).
Computer modeling
Models were written in Perl version 5.16.2 and run on Mac OSX version 10.9. All code can be found in Supporting Information, File S3 and File S4.
Quantitative real-time PCR assays
Fluorescence-based quantitative real-time PCR (qPCR) was used to measure relative amounts of DNA between wild-type and WT+B stocks. DNA was prepared from five 1-day posteclosion virgin females using the DNeasy Blood and Tissue kit (Qiagen; 69504). Two-microliter aliquots of each DNA sample were added to 5 µl of 2× Sybr Select Master Mix (Applied Biosystems; 4472908), 0.5 µl each of 10 nm primer, and 2 µl of water. The reactions were performed in triplicate and spun down in a 96-well plate, sealed, and cycled on an ABI 7900HT according to ABI’s standard protocol.
Analysis of the fluorescence curves was done using ABI’s SDS2.4 software. All curves that showed errors as determined by the SDS2.4 software or that were >35 cycle threshold (Ct) were discarded. The remaining Ct values were exported and analyzed using Biogazelle qBase Plus version 2.4 software to generate normalized relative quantities using assays for yellow and ord as endogenous controls.
Position-effect variegation assays
For position-effect variegation (PEV) assays, virgins from the following stocks were crossed to wild-type and WT+B males: In(1)wm4, 39C-3 (centric region, chromosome 2L), 39C-4 (centric region, chromosome 2L), 118E-10 (centric region, chromosome 4), 118E-12 (centric region, chromosome 3R), 118E-32 (centric region, X chromosome), HS-2 (centric region, chromosome 3), and HS-5 (centric region, chromosome 2L) (Wallrath and Elgin 1995; Cryderman et al. 1998). In(1)wm4 females were also crossed to y w/y+ Y; mtrm126/TM3; spapol males. Both males and females were imaged at 3–5 days posteclosion from every stock with a minimum combined n = 10 per genotype.
Results
The discovery of B chromosomes in D. melanogaster
The B chromosomes described in this study were discovered in a stock containing the mtrm126 allele. matrimony (mtrm) is a haplo-insufficient gene encoding an inhibitor of Polo kinase specific to Drosophila female meiosis (Harris et al. 2003; Xiang et al. 2007; Bonner et al. 2013; Whitfield et al. 2013). When heterozygous, mtrm mutations cause high levels of achiasmate chromosome nondisjunction (Harris et al. 2003; Xiang et al. 2007). Females homozygous for the mtrm126 mutation are sterile, and cytological analysis of oocytes lacking functional Mtrm reveals that their chromosomes are highly fragmented (Xiang et al. 2007; Bonner et al. 2013). mtrm126 is a well-characterized allele of mtrm created by an imprecise P-element excision event resulting in a 203-bp deletion (Xiang et al. 2007). B chromosomes were not observed in the stock used to create the mtrm126 mutant line (mtrmKG08051), nor are B chromosomes present in stocks bearing the other known alleles of mtrm (see Discussion); thus it seems likely that these chromosomes arose de novo within the mtrm126 stock since its creation ∼10 years ago. The role that the mtrm126 allele played, if any, in the origin of these B chromosomes will be considered in the Discussion.
The B chromosomes were first discovered during live imaging of prometaphase I oocytes obtained from mtrm126/TM3 females, which revealed numerous extra chromosomes that were smaller than the 4th chromosomes and moved along the meiotic spindle in a dynamic fashion similar to the movements exhibited by the 4th chromosomes (File S1 and File S2). These extra chromosomes were observed clearly separated from the chromosomal mass in 19/28 such movies of oocytes from this stock.
B chromosomes in D. melanogaster contain centromeres and 4th chromosome heterochromatic sequences
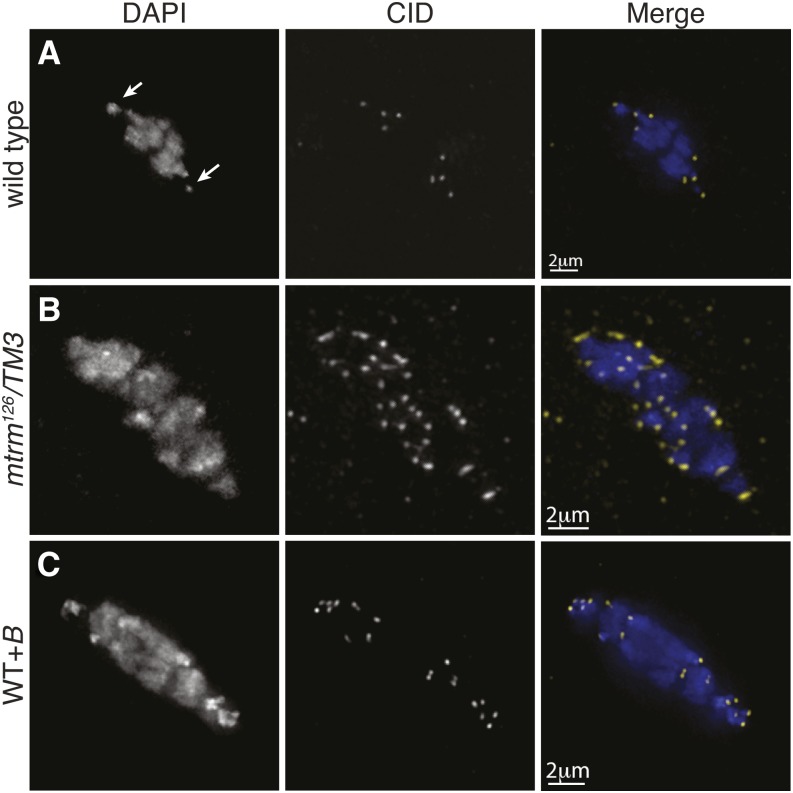
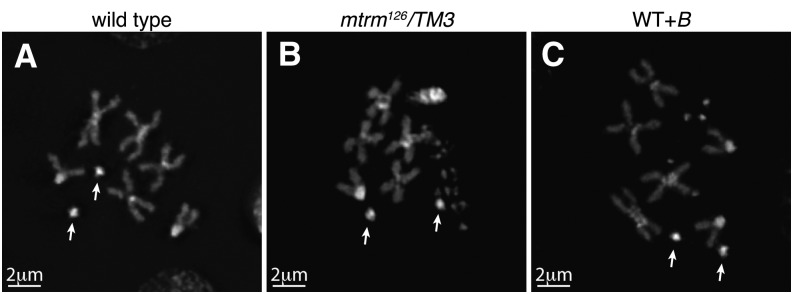
The presence of centromeres on these B chromosomes was investigated using an antibody recognizing centromere identifier (CID), the Drosophila CENP-A homolog. In wild-type prometaphase I oocytes, eight CID foci are present (Figure 1A). In prometaphase I oocytes from mtrm126/TM3 mothers, an average of 29.3 CID foci (SD 7.5, range of 10–43, n = 49) per nucleus were observed. Eight of these CID foci correspond to the centromeres of the normal chromosome complement, suggesting that the remaining DAPI-associated CID foci (∼21) correspond to the supernumerary B chromosomes (Figure 1B).
Figure 1.
B chromosomes contain centromeres. Fixed prometaphase I oocytes were prepared with antibodies against CID (yellow) and the DNA dye DAPI (blue). (A) Wild-type oocytes display the expected 8 CID foci. Arrows indicate the location of the 4th chromosomes. (B) A mtrm126/TM3 oocyte displaying 37 CID foci (mean of 29.3, SD 7.5, range 10–43, n = 49). (C) A WT+B oocyte with 19 CID foci.
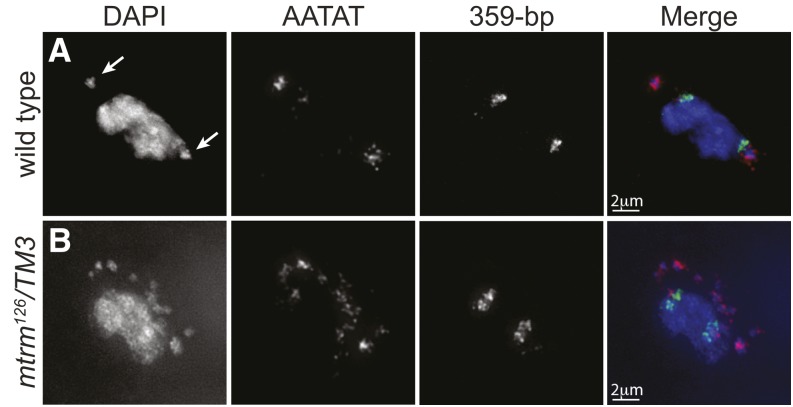
Next, the chromatin composition of the B chromosomes was examined. Indirect immunofluorescence using an antibody recognizing histone H3 trimethylated on K9 (H3K9m3), a marker of heterochromatin, showed it to be heavily associated with the B chromosomes, suggesting that they are highly heterochromatic (Figure S1). We then conducted FISH using four probes, each recognizing a different highly repeated block of heterochromatin on the Drosophila chromosomes (Figure 2 and Figure S2). Probes recognizing heterochromatic repeats of chromosomes 2R (AACAC), 3R (Dodeca), and the X (359 bp) failed to localize to the B chromosomes (Figure 2 and Figure S2). A FISH probe labeling the AATAT satellite repeat, which is found on a large region of the 4th chromosome, localized to both the 4th chromosomes and the B chromosomes (Figure 2 and Figure S2B).
Figure 2.
B chromosomes are composed of 4th chromosome heterochromatic sequences. Localization of FISH probes in fixed prometaphase I oocytes with DAPI (blue) and probes to the 359-bp heterochromatic repeat on the X (green) and to the AATAT heterochromatic repeats on a large region of the 4th chromosomes and a small region of the X chromosomes (red). (A) Wild-type oocyte showing the expected localization of both probes with arrows indicating the location of the 4th chromosomes. (B) A mtrm126/TM3 oocyte showing the AATAT probe displaying additional localization to B chromosomes.
These findings suggest that the B chromosomes are structurally similar to the 4th chromosomes, yet we cannot rule out the possibility that the B chromosomes are derived from the base of the X chromosome or from the long arm of the Y chromosome, both of which also carry small blocks of the AATAT satellite. However, we consider these alternatives unlikely because the B chromosomes do not also carry the 359-bp satellite repeat present on the X chromosome and because the block of AATAT staining seen for individual B chromosomes appears substantially more intense than does the AATAT staining at the base of the X or on the Y. Furthermore, the Y chromosome is an unlikely source because the AATAT block on the Y does not encompass the centromere (Bonaccorsi and Lohe 1991).
B chromosomes do not appear to contain 4th chromosome euchromatic genes
We next examined whether the B chromosomes also contained 4th chromosome euchromatic sequences. For this analysis, B chromosomes from the mtrm126 stock were introgressed into a wild-type genetic background, which we refer to as WT+B (described in detail below), to create a B chromosome-bearing line lacking the mtrm126 mutation (Figure 1C). This was done to eliminate any confounding effects of the mtrm126 mutation. Genetic assays revealed that the B chromosomes from the WT+B line were unable to rescue the recessive mutations cubitus interruptus (ci) and eyeless (ey), as well as the Rps3A Minute locus of the 4th chromosome.
While this result indicates that euchromatic genes are likely not present, it does not eliminate the possibility that 4th chromosome genes are present but somehow silenced. To investigate this option, qPCR was used to examine the levels of five euchromatic 4th chromosome genes in a line of WT+B flies carrying approximately six B chromosomes. We selected one gene in the most distal euchromatic region (Caps), two in the most proximal region (ci and Crk), and two mideuchromatic genes (Asator and mav) (Figure 3A) and did not find a higher copy number of any of these genes when compared to wild-type flies carrying only two structurally normal 4th chromosomes (Figure 3B). The wide distribution of these genes makes it unlikely that the B chromosomes contain functional or nonfunctional 4th chromosome euchromatic sequences.
Figure 3.
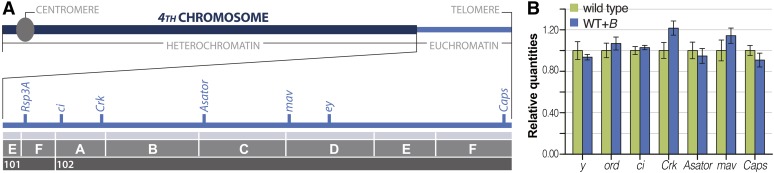
B chromosomes do not contain 4th chromosome genes. (A) Schematic of the 4th chromosome showing the distribution of the seven genes that were assayed by genetic approaches (Rsp3A, ci, and ey) or qPCR (ci, Crk, Asator, mav, and Caps). (B) qPCR analysis showing approximately equal relative quantities of the gene products tested in the wild-type and WT+B stocks. The 2nd chromosome gene ord and the X chromosome gene y were used as controls.
Determining the number of B chromosomes in the mtrm126 line in larval neuroblasts
Cytological examination of the B chromosomes in neuroblast metaphases (Figure 4) revealed them to be ∼64% the size of the 4th chromosome. However, given the difference in intensity of staining between the B and 4th chromosomes, it is possible that a simple measurement overestimates the size of the B chromosomes. We assessed the number of B chromosomes by two methods: counting CID foci in prometaphase I oocytes (Figure 1) and direct counting of B chromosomes in larval neuroblasts by careful analysis of z-stacks from acquired images (Figure 4). These two methods were employed because counting CID foci is difficult in the highly compacted configuration of a prometaphase I spindle, especially when we observe single DAPI masses that appear to be capped by CID foci on both ends (Figure S3), a complication not encountered in larval neuroblast imaging.
Figure 4.
B chromosomes in Drosophila melanogaster are smaller in size than the 4th chromosomes. Larval neuroblasts from 3rd instar larvae of (A) a normal wild-type female karyotype, (B) a male from the mtrm126 stock displaying 12 B chromosomes, and (C) a WT+B female displaying 5 B chromosomes. As measured during cytological examination, the B chromosomes are ∼0.36 µm in size, while the 4th chromosomes averaged 0.56 µm. Arrows indicate the location of 4th chromosomes.
Neuroblast preparations from the mtrm126 stock displayed an average of 9.9 (SD 2.3, range 6–15, n = 36) B chromosomes per cell. Figure 4B shows a neuroblast metaphase containing 12 B chromosomes. Similar numbers of B chromosomes were observed in neuroblasts from both males and females, demonstrating that B chromosomes are present in both sexes.
Assessment of the average number of B chromosomes in the mtrm126 stock yielded inconsistent results when counting CID foci in ovaries (21 B chromosomes) vs. direct counting in neuroblast preparations (10 B chromosomes). A possible explanation for this inconsistency is that the severe effects of the mtrm126 mutation on meiotic chromosome behavior may result in chromosome morphology or segregation defects, such as precocious sister centromere separation, that produce misleading estimates of B chromosome number in the oocytes of mtrm126 females (Harris et al. 2003; Xiang et al. 2007; Bonner et al. 2013). This explanation is supported by the observation that when B chromosomes are transferred into a mtrm+ line, the number of B chromosomes is similar in both neuroblasts and oocytes (see below).
B chromosomes can be maintained in the absence of the mtrm126 mutation
To examine the maintenance of the B chromosomes outside of the mtrm126 stock in which they arose and to assess the role, if any, of the mtrm126 mutation on B chromosome number, a series of crosses was performed to introduce the B chromosomes from the mtrm126 stock into an otherwise wild-type (i.e., mtrm+) background (Figure S4A). Briefly, mtrm126/TM3 males were crossed to females from a wild-type stock, and non-mtrm126 siblings were selected and crossed to each other to establish the WT+B stock.
To characterize this WT+B stock, indirect immunofluorescence was used to estimate the number of B chromosomes in prometaphase I oocytes from females from each generation indicated in Table 1 (for example, see Figure 1C). Using the CID antibody to mark centromeres, we found that the WT+B stock exhibited an average of ∼14 CID foci at each generation (Table 1), indicating the presence of about six B chromosomes (8 CID foci belong to the normal chromosome complement). While this number of CID foci is substantially less than that observed in the mtrm126 stock (perhaps due to both the initial outcrossing and the possible effects of the mtrm126 mutation noted above), the average number of CID foci did remain relatively constant throughout 18 generations in the WT+B stock, suggesting that the B chromosomes are well maintained in a wild-type background.
Table 1. Average number of CID foci in prometaphase I oocytes across generations of WT+B.
| Generation | 2 | 4 | 6 | 8 | 10 | 18 |
|---|---|---|---|---|---|---|
| CID focia | 15.4 | 13.0 | 14.5 | 14.6 | 14.0 | 14.9 |
| ±SD | 3.5 | 2.3 | 3.0 | 2.3 | 3.7 | 2.8 |
| n value | 23 | 24 | 28 | 30 | 20 | 16 |
Eight of the CID foci correspond to the autosomes and sex chromosomes.
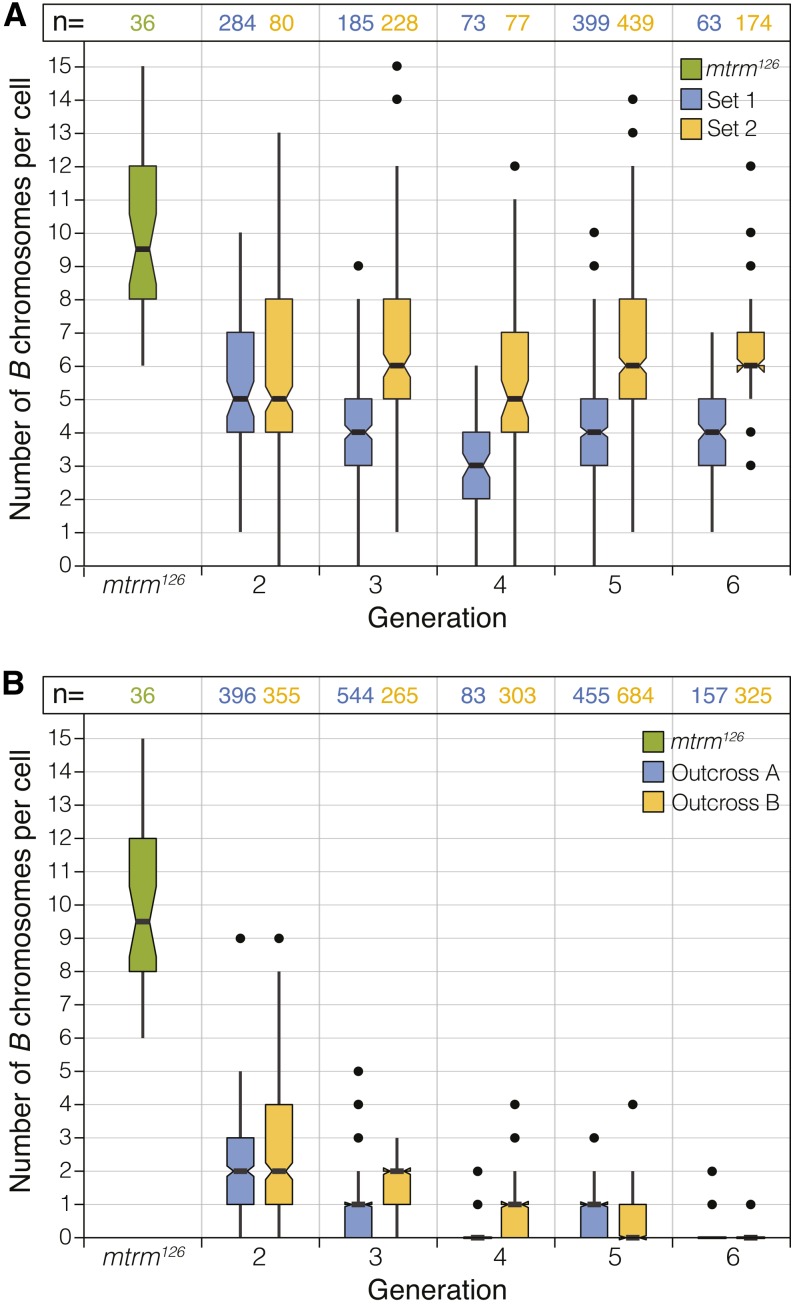
The number of B chromosomes was also assayed by direct analysis of larval neuroblast metaphases (Figure 4C). Two versions of this experiment were performed, referred to hereafter as set 1 and set 2. Set 1 was set up exactly like the WT+B stock used in the experiment estimating B chromosome number by CID foci described above, which started with mtrm126/TM3 males. Set 2 followed the same procedure but began with mtrm126/TM3 females crossed to wild-type males (Figure S4B). Both stocks were maintained by crossing siblings for multiple generations, and the number of B chromosomes was analyzed in neuroblast preparations.
For both set 1 and set 2, we examined larval neuroblast metaphases from generations two through six and observed similar numbers of B chromosomes in WT+B flies from both sets (Figure 5A). The mean number of B chromosomes for each generation ranged from 3 to 5 in set 1 and from 5 to 6 in set 2 (Figure 5A), which is similar to the number estimated by CID foci as described above. Although variation was observed at each generation, the average number of B chromosomes remained fairly constant in both set 1 and set 2. These studies allowed us to show that the B chromosomes can be propagated through both the male and female germlines and that B chromosomes do not require the mtrm126 allele for their transmittance or maintenance. Additionally, the variation in B chromosome number observed at each generation indicates that their segregation does not follow the classical Mendelian paradigm.
Figure 5.
B chromosomes are maintained in a wild-type background. (A) Plot of the number of B chromosomes in individual metaphase preparations of WT+B larval neuroblasts from generations 2–6 in set 1 and set 2. Set 1 was initiated from crosses using mtrm126/TM3 males, and set 2 was initiated with mtrm126/TM3 females. For each set, the experiment was performed twice and the data were pooled. Larval neuroblast data from the mtrm126 stock are shown for comparison (green). (B) Plot of the number of B chromosomes in individual metaphase preparations of larval neuroblasts from mtrm126/TM3 flies outcrossed to wild type six consecutive times. Of the 52 larvae scored (482 cells total) in generation 6, only 12 still contained at least one cell with at least one B chromosome. Of these, only two larvae contained at least one cell with more than one B chromosome. Outcross A was started with mtrm126/TM3 males, and outcross B was started with mtrm126/TM3 females. The mtrm126 stock data shown in A are again included for comparison. For each outcross, the experiment was started four times and the data were pooled. (A and B) In each experiment, generation 1 was not scored. For each plot, the centerline indicates the mean, with the shaded area containing the middle 50% of the observations. Notches represent the 95% confidence interval for each observation, and points represent outliers.
D. melanogaster B chromosomes are mitotically unstable
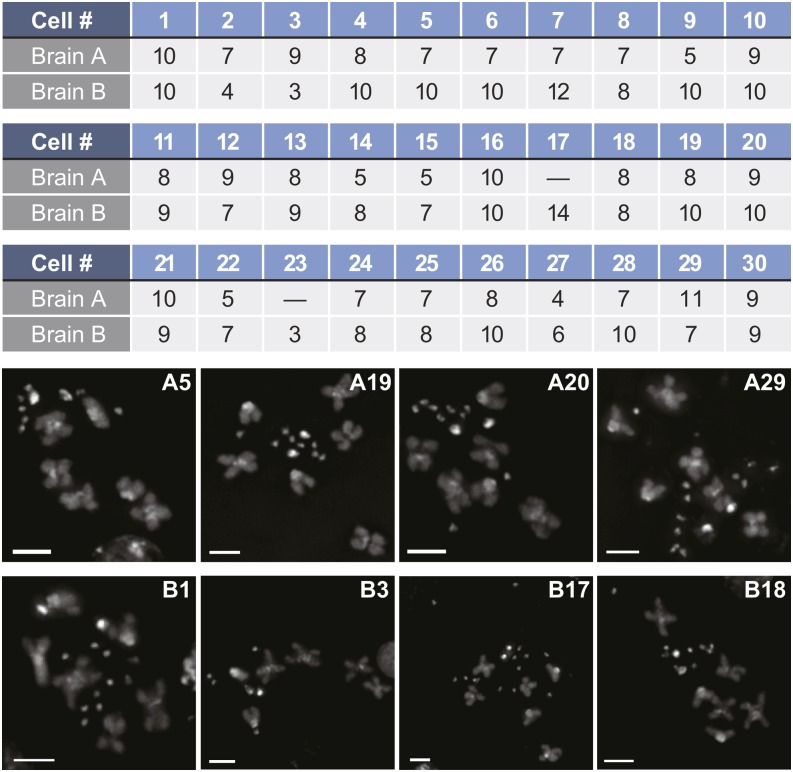
The neuroblast studies revealed that the average number of B chromosomes within a given brain was variable from cell to cell. Figure 6 shows the variation observed within different metaphase cells for two larval brains (labeled brain A and brain B). Note that in brain A, the number of B chromosomes in a given cell varied from 5 to 11, while in brain B the number of B chromosomes varied from 3 to 14. Variation within a given brain was observed in 102 of 105 brains from which we were able to quantify four or more cells. This variability among cells of the same brain demonstrates that the B chromosomes are mitotically unstable.
Figure 6.
B chromosomes are mitotically unstable. Number of CID foci for 28 (brain A) and 30 (brain B) cells from two neuroblast preparations illustrating the mitotic instability of the B chromosomes. Dashes represent cells that we imaged but were unable to score. Bars, 2 µm.
Given the variation in B chromosome number observed in the sibling crosses, we were curious whether random segregation of the B chromosomes in both sexes could stably maintain B chromosomes over multiple generations at the numbers we observed. To test this, we created a computer simulation seeded with 1000 males and 1000 females, each carrying 6 B chromosomes (File S3). After the first generation, a sperm and an egg, both carrying a random number of B chromosomes ranging from 0 to 6, are randomly matched, and one offspring is created carrying the sum of the B chromosomes from those gametes. (For example, the fusion of an egg carrying 6 B chromosomes with a sperm carrying 2 B chromosomes would result in an offspring with 8 B chromosomes. This offspring would then go on to create gametes with any number of B chromosomes from 0 to 8 and mate at random with another offspring.) The model assumes that each random mating results in a random number of offspring up to 20. The 1000 males and 1000 females are then randomly chosen from all offspring to continue to the next generation. This was repeated for 1000 generations (Figure S5). Using this model, we find that the majority of offspring retain at least 2 and at most 11 B chromosomes per generation, a result consistent with our data.
B chromosomes can be lost by repeated outcrossing
Next, a series of successive outcrosses was initiated to determine how quickly the B chromosomes could be eliminated from a population. mtrm126/TM3 flies were crossed to wild-type flies (Figure S4C), and the non-mtrm126 progeny were again mated to wild-type flies for five more generations. Larval neuroblast preparations showed that B chromosomes were present in 65 of 66 larvae scored at the second generation of outcrossing, but by the sixth generation were detectable in only 12 of 52 larvae scored (Figure 5B). Of these 12 larvae, two contained at least one cell carrying more than one B chromosome. This indicates that B chromosomes can be diluted and subsequently eliminated from a stock with multiple outcrosses. The reduction at each generation also suggests that the B chromosomes do not yet possess a strong drive mechanism for their maintenance in a wild-type genetic background.
We then modeled the rate of B chromosome loss using a computer simulation of our outcross experiments (File S4), starting with six B chromosomes per mother and zero per father. In this model, each B chromosome segregates at random, regardless of the movement of any other B or 4th chromosome. One resulting chromosome complement is selected at random and allowed to continue to the next generation. WT+B females are again crossed to wild-type males. One thousand crosses were modeled for each generation. By generation seven, >90% of the progeny would have zero B chromosomes, and they would be essentially lost from the stock after 10 generations (Figure S6).
B chromosomes in D. melanogaster cause female-specific 4th chromosome nondisjunction
Proper segregation of the achiasmate 4th chromosomes during female meiosis is known to be dependent upon their heterochromatic sequences (Hawley et al. 1992; Karpen et al. 1996). Knowing that the B chromosomes are enriched for the AATAT satellite repeat that primarily comprises the 4th chromosome heterochromatin, we performed a NDJ assay in a wild-type X chromosome background to test the levels of 4th chromosome NDJ in WT+B females (Table 2). The results of these experiments showed that WT+B females displayed an average of 27.0% 4th chromosome NDJ, compared to only 0.3% NDJ in wild type. This observation is not surprising in light of the finding by Hawley et al. (1992) that any 4th chromosome derivative possessing the proximal 4th chromosome heterochromatin (which is rich in the AATAT satellite sequence) is capable of inducing high levels of 4th chromosome NDJ in females.
Table 2. 4th chromosome nondisjunction in females.
| Maternal genotype | Wild type | WT+B |
|---|---|---|
| Normal | ||
| 4 | 1385 | 1640 |
| 4th nondisjunction | ||
| 44 | 4 | 420 |
| 0 | 0 | 191 |
| Adjusted total | 1389 | 2251 |
| % diplo-4 | 0.3 | 18.7 |
| % nullo-4 | 0.0 | 8.5 |
| Total % 4th NDJ | 0.3 | 27.1 |
Female WT+B flies were crossed to males bearing an attached 4th chromosome. Their progeny displayed 27.1% 4th nondisjunction, compared to 0.3% in wild type.
Because the B chromosomes caused a high frequency of NDJ of the achiasmate 4th chromosomes, the effect of the B chromosomes on the segregation of achiasmate X chromosomes was also examined. To create WT+B flies containing achiasmate X chromosomes, crosses were initiated as described in Figure S4A, except the wild-type females were homozygous for the X chromosome balancer FM7w. At the second generation, females heterozygous for the FM7w balancer chromosome and lacking the TM3 balancer were used for the NDJ assay. The WT+B stock with achiasmate X chromosomes exhibited 4.6% X chromosome NDJ, which is a small but significant increase from the wild-type level of 0.8%. This effect was much smaller than the 29.6% NDJ observed for the 4th chromosome in this experiment (Table S1). The small effect on X chromosome NDJ may be due to the presence of the AATAT satellite at the base of the X chromosome (see Hawley et al. 1992).
Further analysis showed that the observed NDJ was specific to females (Table 3), a result that was not surprising given that males use different mechanisms to segregate achiasmate chromosomes (Hawley and Theurkauf 1993; McKee 2009). Because we have demonstrated that males are able to transmit B chromosomes to their progeny, the absence of NDJ in WT+B males is not likely to be due simply to a lack of B chromosome transmission.
Table 3. 4th chromosome nondisjunction in males.
| Paternal genotype | Wild type | WT+B |
|---|---|---|
| Normal | ||
| 4 | 737 | 964 |
| 4th nondisjunction | ||
| 44 | 0 | 1 |
| 0 | 0 | 3 |
| Adjusted total | 737 | 968 |
| % diplo-4 | 0.0 | 0.1 |
| % nullo-4 | 0.0 | 0.3 |
| Total % 4th NDJ | 0.0 | 0.4 |
Male WT+B flies were crossed to females bearing an attached 4th chromosome. Their progeny displayed 0.4% 4th nondisjunction, compared to 0.0% in wild type.
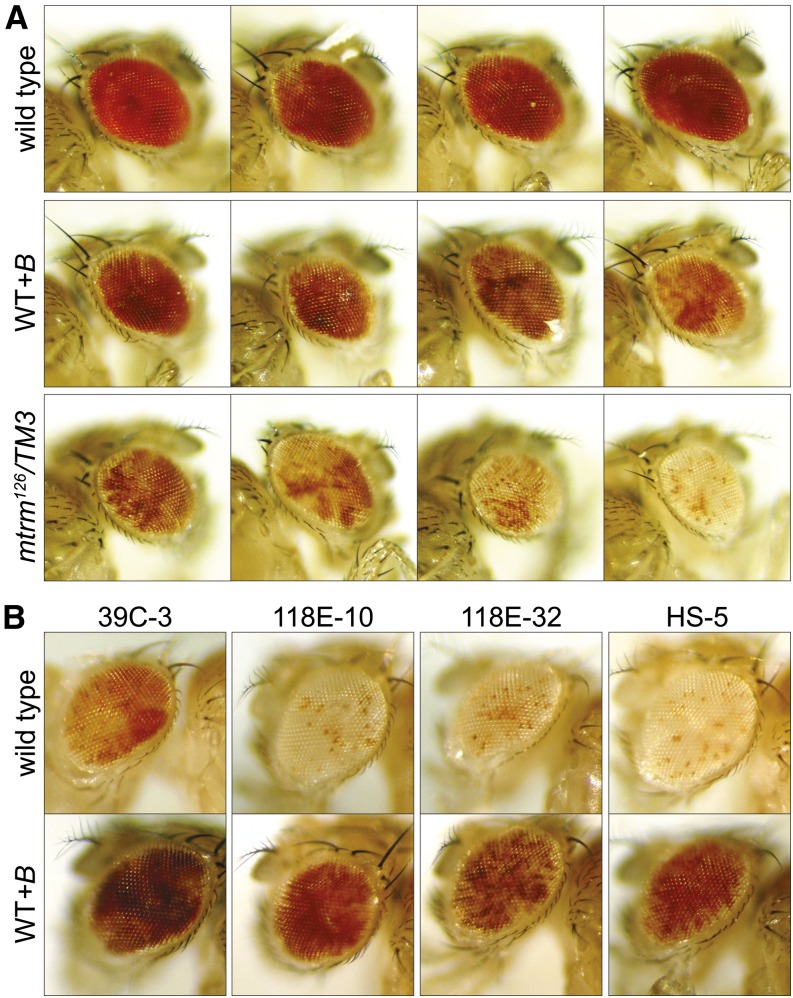
B chromosomes in D. melanogaster significantly affect position-effect variegation
PEV is a phenomenon that occurs when a change in the heterochromatin state causes a change in gene expression (Sun et al. 2004). While the exact mechanism of PEV function is not fully understood, it is well established that introducing extra heterochromatin into a genome has a distinct impact on PEV (Sun et al. 2004). This is commonly assayed by use of the white (w) gene, which, when in close proximity to heterochromatin, is differentially expressed to varying degrees within each cell due to the silencing effect of heterochromatin. Variegation is assessed by the degree of mosaicism in the eye (Schotta et al. 2003), and it has been noted that the 4th chromosomes have the capacity to induce PEV (Haynes et al. 2007). Because the B chromosomes appear to be composed primarily of 4th chromosome heterochromatic sequences, we determined the effect of B chromosomes on PEV as assayed by In(1)wm4, an inversion of the X chromosome that places the w gene in proximity to pericentric heterochromatin, and by several pericentric insertion reporter lines of the w gene for the X, 2nd, 3rd, and 4th chromosomes (Wallrath and Elgin 1995; Cryderman et al. 1998).
We compared flies derived by crossing In(1)wm4 females or the variegating transgenic females (Wallrath and Elgin 1995; Cryderman et al. 1998) to males from a wild-type stock and a WT+B stock. In(1)wm4, females were also crossed to mtrm126 males. Given that the addition of heterochromatin is generally expected to suppress variegation in a fashion that correlates with the amount of heterochromatin added (Dimitri and Pisano 1989), it was surprising to observe that while a few of the tested lines showed no variegation (Figure S7), several lines showed strong, yet contrasting PEV phenotypes (Figure 7). For the In(1)wm4 translocation, we observed a notable enhancement of variegation in the presence of the WT+B genetic background that was further increased in the mtrm126 genetic background, which contains additional B chromosomes (Figure 7A). We also observed a strong suppression of variegation in four of the variegating transgenic lines (Figure 7B). This is not an unprecedented result, as there have been reports of other genes differentially affecting various PEV reporters, such as in the case of the kinase JIL-1, where some mutant forms of the protein can act as suppressors of PEV while other forms of the protein act as enhancers of PEV depending on the level of kinase activity (Wang et al. 2012). It is clear from these experiments that the heterochromatic B chromosomes have the ability to alter transcription at genetic loci sensitive to changes in heterochromatic boundaries.
Figure 7.
B chromosomes significantly modify position-effect variegation. For each line, females carrying the reporter gene were crossed to males of the genotypes listed. (A) In(1)wm4 flies were examined for their degree of PEV from crosses with a wild-type control, WT+B, or mtrm126/TM3. (B) Four insertion reporter lines with the w+ gene inserted in the pericentric region of chromosomes 2 (39C-3 and Hs-5), 4 (118E-10), and the X (118E-32), were examined when crossed to a wild-type control or WT+B.
Discussion
The data presented above report the presence of B chromosomes in D. melanogaster. These supernumerary chromosomes display a number of defining characteristics of B chromosomes. Specifically, they are morphologically distinguishable from the A chromosomes, contain their own centromeres, are highly heterochromatic, are mitotically unstable, and appear to lack euchromatin. It is not known whether all of the B chromosomes observed in the mtrm126 stock arose from a single occurrence or arose multiple times during the 10-year life of this stock.
Because these B chromosomes arose in a stock bearing the mtrm126 mutation, we must consider the possibility that they arose as a consequence of the mtrm126 mutation. Oocytes from mtrm null mothers are known to display fragmented chromosomes during meiosis (Bonner et al. 2013), although such homozygotes are invariably sterile and thus unlikely to transmit those fragments. Still, it is possible that some low degree of chromosome breakage or fragmentation does occur in mtrm/+ heterozygotes, and that one such event produced the B chromosomes studied here. However, it should be noted that we did not find B chromosomes in two other independently derived alleles of mtrm (Df(3L)66CT2-T10 and mtrmz-35931) or in the progenitor of the mtrm126 allele (mtrmKG08051) (Figure S8) and do not believe that B chromosome production is a general property of stocks bearing mtrm mutations. Perhaps the simplest explanation for the origin of the B chromosomes would be a centromere misdivision event of the 4th chromosomes yielding an isochromosome with two copies of the short arm of chromosome 4 whose transmission was enhanced by the mtrm126/+ background.
It is also possible that the event that created the original B chromosome is a common event, but that in wild-type stocks a univalent is quickly diluted out. This would indicate that these B chromosomes within the mtrm126 stock were maintained and amplified, possibly because of the defects in the distributive system that cause the missegregation of achiasmate chromosomes in mtrm126 heterozygous females. It is known that mtrm126/+ females display high levels of 4th chromosome nondisjunction without observable 4th chromosome loss. Indeed, in many crosses involving mtrm/+ females, the number of diplo-4 exceptional progeny actually exceeds the number of nullo-4 reciprocal exceptions (Harris et al. 2003). A related phenomenon could account for the accumulation of B chromosomes.
We are equally as ignorant with regard to the mechanisms of meiotic transmission in a wild-type genetic background. Our data fit a model in which B chromosomes are transmitted at random. However, because the high meiotic nondisjunction rate induced by the B chromosomes is specific to achiasmate 4th chromosomes in females, we suspect that at least some mechanism for homology-dependent pairing might exist. Additionally, the mitotic instability indicates improper segregation of B chromosomes during mitosis. The mitotic instability could be the consequence of either some defect in the B chromosomes themselves or strain placed on mitotic spindles by a near doubling of kinetochore number. We feel that this issue is best approached using a marked B chromosome to study mitotic loss in individuals heterozygous for known mitotic mutants.
The discovery of B chromosomes in D. melanogaster will be of real utility for a wide range of studies. Significant insight can be gained from understanding how a mitotic spindle that typically handles eight centromeres is able to somewhat faithfully segregate up to twice that number, as well as from understanding the etiology of mitotic instability and meiotic transmission (Masonbrink and Birchler 2012; Masonbrink et al. 2012).
Supplementary Material
Acknowledgments
We thank Claudio Sunkel, Sarah Elgin, and Lori Wallrath for providing reagents; Jay Unruh for helpful discussion; and two anonymous reviewers for helpful comments. We also acknowledge all members of the Hawley lab for valuable input and suggestions, especially Cathleen Lake for critical reading of the manuscript; Angela Miller for assistance with writing and figure preparation; Clarissa Smith for DNA preparation; and Amanda Bonner for assistance with data analysis. R.S.H. is an American Cancer Society Research Professor.
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Banaei-Moghaddam A. M., Meier K., Karimi-Ashtiyani R., Houben A., 2013. Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell 25: 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo Carvalho A., Koerich L. B., Clark A. G., 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L. W., 1994. Bewildering Bs: an impression of the 1st B-chromosome conference. Heredity 73: 328–336. [Google Scholar]
- Birchler J. A., Han F., 2013. Meiotic behavior of small chromosomes in maize. Front Plant Sci 4: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Lohe A., 1991. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics 129: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner A. M., Hughes S. E., Chisholm J. A., Smith S. K., Slaughter B. D., et al. , 2013. Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc. Natl. Acad. Sci. USA 110: E1222–E1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho J. P., Sharbel T. F., Beukeboom L. W., 2000. B-chromosome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D. E., Cuaycong M. H., Elgin S. C. R., Wallrath L. L., 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107: 277–285. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., 2000 In situ hybridization to somatic chromosomes, pp. 25–55 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner, and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [DOI] [PubMed] [Google Scholar]
- Dernburg A. F., Sedat J. W., Hawley R. S., 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Dimitri P., Pisano C., 1989. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics 122: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H., James T. C., Singh P. B., 1992. Cloning and expression of Drosophila HP1 homologs from a mealybug, Planococcus citri. J. Cell Sci. 101(Pt 2): 463–474. [DOI] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster C., Rigola M. A., Egozcue J., 2004. Human supernumeraries: Are they B chromosomes? Cytogenet. Genome Res. 106: 165–172. [DOI] [PubMed] [Google Scholar]
- Gutknecht J., Sperlich D., Bachmann L., 1995. A species specific satellite DNA family of Drosophila subsilvestris appearing predominantly in B chromosomes. Chromosoma 103: 539–544. [DOI] [PubMed] [Google Scholar]
- Hackstein J. H., Hochstenbach R., Hauschteck-Jungen E., Beukeboom L. W., 1996. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? Bioessays 18: 317–323. [DOI] [PubMed] [Google Scholar]
- Harris D., Orme C., Kramer J., Namba L., Champion M., et al. , 2003. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 165: 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., Irick H., Zitron A. E., Haddox D. A., Lohe A., et al. , 1992. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 13: 440–467. [DOI] [PubMed] [Google Scholar]
- Hawley R. S., Theurkauf W. E., 1993. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 9: 310–317. [DOI] [PubMed] [Google Scholar]
- Haynes K. A., Gracheva E., Elgin S. C., 2007. A Distinct type of heterochromatin within Drosophila melanogaster chromosome 4. Genetics 175: 1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques-Gil N., Arana P., Santos J. L., 1983. Spontaneous translocations between B chromosomes and the normal complement in the grasshopper Eyprepocnemis plorans. Chromosoma 88: 145–148. [Google Scholar]
- Hughes S. E., Beeler J. S., Seat A., Slaughter B. D., Unruh J. R., et al. , 2011. Gamma-tubulin is required for bipolar spindle assembly and for proper kinetochore microtubule attachments during prometaphase I in Drosophila oocytes. PLoS Genet. 7: e1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Gilliland W. D., Cotitta J. L., Takeo S., Collins K. A., et al. , 2009. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 5: e1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N., 2003. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 8: 417–423. [DOI] [PubMed] [Google Scholar]
- Karpen G. H., Le M. H., Le H., 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. [DOI] [PubMed] [Google Scholar]
- Kaszas E., Birchler J. A., 1996. Misdivision analysis of centromere structure in maize. EMBO J. 15: 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Leach C. R., Houben A., Field B., Pistrick K., Demidov D., et al. , 2005. Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics 171: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T., Maia A. F., Steffensen S., Sunkel C. E., 2009. Sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J. 28: 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martis M. M., Klemme S., Banaei-Moghaddam A. M., Blattner F. R., Macas J., et al. , 2012. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 109: 13343–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masonbrink R. E., Birchler J. A., 2012. Accumulation of multiple copies of maize minichromosomes. Cytogenet. Genome Res. 137: 50–59. [DOI] [PubMed] [Google Scholar]
- Masonbrink R. E., Gaeta R. T., Birchler J. A., 2012. Multiple maize minichromosomes in meiosis. Chromosome Res. 20: 395–402. [DOI] [PubMed] [Google Scholar]
- McKee B. D., 2009. Homolog pairing and segregation in Drosophila meiosis. Genome Dyn. 5: 56–68. [DOI] [PubMed] [Google Scholar]
- Ramachandra N. B., Ranganath H. A., 1985. Supernumerary chromosomes in Drosophila nasuta albomicana. Experientia 41: 680–681. [DOI] [PubMed] [Google Scholar]
- Ramachandra N. B., Ranganath H. A., 1987. Characterization of heterochromatin in the B chromosomes of Drosophila nasuta albomicana. Chromosoma 95: 223–226. [Google Scholar]
- Schotta G., Ebert A., Dorn R., Reuter G., 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14: 67–75. [DOI] [PubMed] [Google Scholar]
- Sun F. L., Haynes K., Simpson C. L., Lee S. D., Collins L., et al. , 2004. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24: 8210–8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaran A. K., Gupta J. P., 1994. Accumulation of B-chromosomes in Drosophila kikkawai Burla. Cytobios 80: 211–215. [PubMed] [Google Scholar]
- Voltolin D. T., Paes A. D., Foresti F., Bortolozzi J., Porto-Foresti F., 2012. Cytogenetic analysis of B chromosomes in one population of the fish Moenkhausia sanctaefilomenae (Steindachner, 1907) (Teleostei, Characiformes). Comp Cytogenet 6: 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L. L., Elgin S. C., 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277. [DOI] [PubMed] [Google Scholar]
- Wang C., Cai W., Li Y., Girton J., Johansen J., et al. , 2012. H3S10 phosphorylation by the JIL-1 kinase regulates H3K9 dimethylation and gene expression at the white locus in Drosophila. Fly (Austin) 6: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield Z. J., Chisholm J., Hawley R. S., Orr-Weaver T. L., 2013. A meiosis-specific form of the APC/C promotes the oocyte-to-embryo transition by decreasing levels of the Polo kinase inhibitor matrimony. PLoS Biol. 11: e1001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski L. P., Doherty R. A., 1985. Supernumerary microchromosomes identified as inverted duplications of chromosome 15: a report of three cases. Hum. Genet. 69: 161–163. [DOI] [PubMed] [Google Scholar]
- Wisniewski L., Hassold T., Heffelfinger J., Higgins J. V., 1979. Cytogenetic and clinical studies in five cases of inv dup(15). Hum. Genet. 50: 259–270. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Hawley R. S., 2006. The mechanism of secondary nondisjunction in Drosophila melanogaster females. Genetics 174: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Takeo S., Florens L., Hughes S. E., Huo L. J., et al. , 2007. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 5: e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Terai Y., Mizoiri S., Aibara M., Nishihara H., et al. , 2011. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 7: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Li H., Schweppe N. M., Hawley R. S., Gilliland W. D., 2010. Statistical analysis of nondisjunction assays in Drosophila. Genetics 186: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.