Abstract
Between 15–20% of human cancers are associated with infection by oncogenic viruses. Oncogenic viruses, including HPV, HBV, HCV and HTLV-1, target mitochondria to influence cell proliferation and survival. Oncogenic viral gene products also trigger the production of reactive oxygen species which can elicit oxidative DNA damage and potentiate oncogenic host signaling pathways. Viral oncogenes may also subvert mitochondria quality control mechanisms such as mitophagy and metabolic adaptation pathways to promote virus replication. Here, we will review recent progress on viral regulation of mitophagy and metabolic adaptation and their roles in viral oncogenesis.
Keywords: mitochondria, mitophagy, virus, ROS, oncogenes
Introduction
Reactive oxygen species (ROS) are small molecules generated as by-products during normal cellular metabolism of oxygen. Cellular levels of ROS are crucial for cell fate (Pan, 2009; Martin and Barrett, 2002). In normal cells, ROS is now established to play key signaling roles in cell differentiation, autophagy and immune responses (Sena and Chandel, 2012; Ray et al., 2012). However, accumulation of intracellular ROS is potentially harmful to normal cells, causing oxidation of nucleic acids, proteins, and lipids, and can lead to oxidative stress (Cooke et al., 2003; Babusikova et al., 2013; Bernard et al., 2012; Sawada and Carlson, 1987). The deleterious effects of ROS have been associated with numerous human pathologies such as aging, cardiovascular diseases, neurodegenerative diseases and cancer (Brieger et al., 2012).
Intracellular ROS are produced in aerobic organisms as a result of enzyme reactions from various intracellular sources including peroxisomes, endoplasmic reticulum (ER) and mitochondria (Waris and Ahsan, 2006). Peroxisomes oxidize intracellular nicotinamide adenine dinucleotide phosphate (NADPH) to reduce oxygen (O2) to superoxide (O2−) by membrane-associated NADPH oxidase (NOX) and toxic molecules (e.g. ethanol) to hydrogen peroxide (H2O2) by catalase. The ER oxidizes unsaturated fatty acids and xenobiotics via cytochrome P450 and b5 enzymes to produce O2− and H2O2. (Bonekamp et al.; Schrader and Fahimi, 2006). Mitochondria produce ROS through incomplete reduction of oxygen (O2) to water (H2O) from the electron transfer chain (ETC) and by mitochondrial enzymes such as dehydrogenases. Mitochondria are considered the chief source of intracellular ROS production because mitochondrial ROS (mROS) have been directly linked to multiple physiologies including immunity, differentiation, autophagy and metabolic adaptation (Sena and Chandel, 2012), and diverse pathological conditions such as cancer, autoimmunity and cardiovascular diseases all share common characteristics of elevated mROS (Sena and Chandel, 2012; Li et al., 2013; Turrens, 2003).
The generation of mROS is tightly regulated in primary cells. Various cell stimuli, including immunoreceptor ligation and cytokine stimulation, pathogen infection and hypoxia increase mROS levels. In addition, elevated cytosolic calcium levels and activation of phosphatidylinositol 3-kinase lead to elevated mROS. However, due to the high reactivity and toxicity of mROS, mammalian cells have evolved a number of antioxidant enzymes including superoxide dismutases, peroxiredoxins, glutathione peroxidase, and catalase to neutralize mROS (Li et al., 2013; Sena and Chandel, 2012). Furthermore, mitophagy can also downregulate mROS by removing damaged mitochondria that secrete high levels of ROS (Tal et al., 2009; Wang et al., 2012; Lee et al., 2012a).
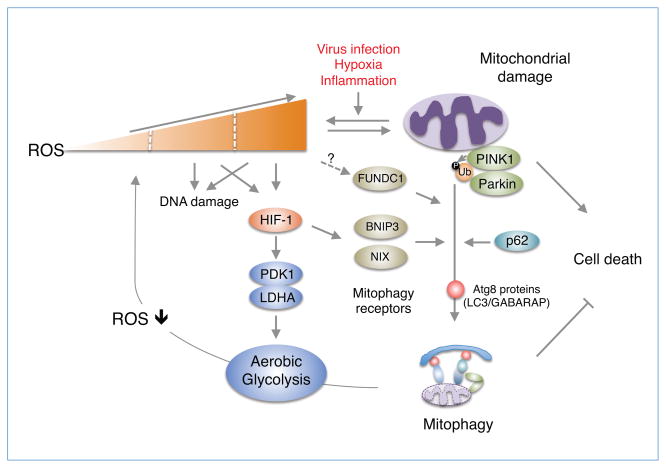
Mitophagy is the process by which dysfunctional mitochondria are selectively eliminated by the highly conserved autophagic machinery (Jin and Youle, 2012; Novak, 2012; Youle and Narendra, 2011). The best studied mitophagy pathway is the PTEN-induced putative kinase 1 (PINK1)/Parkin-mediated pathway (Fig. 1) (Jin and Youle, 2012). Mutations in PINK1 and Parkin cause early-onset neurodegenerative disease such as juvenile Parkinson’s disease (PD) (Kitada et al., 1998; Valente et al., 2004). PINK1 is a mitochondrial kinase that is imported into the matrix and the intermembrane space. In healthy polarized mitochondria, imported PINK1 is constitutively and rapidly degraded by sequential actions of mitochondrial proteases and the proteasome (Greene et al., 2012; Yamano and Youle, 2013; Jin et al., 2010). However, upon mitochondrial depolarization or damage induced by protonophores such as carbonyl cyanide 3-chlorophenylhydrazone (CCCP) or a short burst of mROS via photosensitizers (e.g. mitochondria-targeted KillerRed protein), the constitutive degradation of PINK1 is inhibited resulting in PINK1 accumulation on the outer mitochondrial membrane (Wang et al., 2012; Narendra et al., 2008), where it recruits Parkin and activates its ubiquitin ligase activity (Shiba-Fukushima et al., 2012). In recent studies, two groups independently demonstrated that PINK1 activates the enzyme activity of Parkin through PINK1-mediated ubiquitin phosphorylation at serine 65 (Kane et al., 2014; Koyano et al., 2014). Subsequently, activated Parkin ubiquitinates mitochondrial proteins mitofusin 1 and 2 (Mfn1/2), Drp1, Bcl-2 and voltage-dependent anion channel 1 (VDAC1) (Chen et al., 2010; Tanaka et al., 2010; Wang et al., 2011; Geisler et al., 2010). The adaptor protein p62/SQSTM1 is a ubiquitin-binding autophagy receptor that links ubiquitinated mitochondria to autophagosomes (Pankiv et al., 2007). However, it is unclear whether p62/SQSTM1 serves as a mitophagy receptor (Okatsu et al., 2010; Huang et al., 2011; Narendra et al., 2010), although it is plausible that p62/SQSTM1 may be required for Parkin-induced perinuclear clustering of damaged mitochondria (Okatsu et al., 2010; Narendra et al., 2010).
Figure 1. Regulation of ROS homeostasis through mitophagy and metabolic adaptation.
Mitochondrial ROS induced by virus infection can lead to mitochondrial dysfunction or damage. Accumulation of damaged mitochondria generates excessive mROS, which promotes oxidative stress, DNA damage and cell death. To mitigate the harmful effects of mROS and reduce mROS levels, cells induce mitophagy, by which damaged mitochondria are cleared, via the PINK1/Parkin complex (Youle and Narendra, 2011; Novak, 2012) and presumably HIF-1 activation of mitophagy receptors NIX and BINP3 or FUNDC1 (Ding and Yin, 2012). PINK1 phosphorylates ubiquitin (Ub) to activate Parkin (Kane et al., 2014; Shiba-Fukushima et al., 2012), which in turn induces the ubiquitination of mitochondrial proteins. The p62/SQSTM1 adaptor links the ubiquitinated mitochondria to the Atg8 family proteins (LC3/GABARAP) essential for the autophagosome maturation. Also, HIF-1 promotes metabolic adaptation (aerobic glycolysis) via the activation of PDK1 and LDHA (Kim et al., 2006; Cuninghame et al., 2014) to bypass mitochondrial oxidative phosphorylation generating mROS or presumably generate ATP in cells actively undergoing mitophagy or containing a few mitochondria.
In addition to the PINK1/Parkin-mediated mitophagy pathway activated by mitochondrial damage, there are other mitophagy pathways that are developmentally programmed through specific receptors (Ding and Yin, 2012). For example, red blood cells (RBCs) lose their mitochondria in order to transport oxygen instead of consuming it by mitophagy (Ney, 2011; Fader and Colombo, 2006). The BH3-only protein (BOP) NIX plays an important role as a mitophagy receptor in maturing murine reticulocytes by recruiting Atg8 family members LC3 and GABARAP to damaged mitochondria (Novak et al., 2010). Also, in response to high oxidative phosphorylation activity, NIX regulates mitophagy elicited by the small GTPase Rheb (Melser et al., 2013). Another BOP member, BNIP3, also functions as a mitophagy receptor in mitophagy elicited by metabolic stress (Feng et al., 2013; Zhang and Ney, 2009). Furthermore, the mitochondrial protein FUNDC1 was reported to play a role as a mitophagy receptor in hypoxia-induced mitophagy (Fig. 1) (Liu et al., 2012). The expression of mitophagy-specific receptors are regulated during hypoxia through induction of hypoxia-inducible factor 1 (HIF-1) or suppression of microRNA-137 (Bruick, 2000; Li et al., 2014).
Hypoxia induces a change in mitochondrial redox and alters the production of mROS (Chandel et al., 1998; Guzy et al., 2005; Hamanaka and Chandel, 2009). Under normoxic conditions, HIF-1α subunits are hydoxylated on prolines by prolyl hydroxylases (PHDs) (Hägg and Wennström, 2005; Hirsilä et al., 2003). Hydroxylated HIF-1α is recognized by the von Hippel-Lindau (pVHL) tumor suppressor leading to the ubiquitination and degradation of HIF-1α. During hypoxia, mROS inhibits the activity of PHDs allowing for the stabilization of HIF-1α subunits and HIF-mediated transcription (Hamanaka and Chandel, 2009). Thus, hypoxia-induced mROS can be viewed as a double-edged sword by inducing mitochondrial damage and activating mitophagy pathways to remove damaged mitochondria. Under persistent hypoxic conditions, mROS levels could remain elevated and the overall content of mitochondria would be decreased by activated mitophagy resulting in insufficient ATP production for cell proliferation. Low mitochondrial content has been observed in solid tumors, which contain large regions associated with a low oxygen concentration, including hepatocellular carcinoma, renal cell carcinoma, and ovarian cancer (Cuezva et al., 2002; Simonnet et al., 2002; Wang et al., 2006).
In fact, most cancer cells do not rely primarily on mitochondrial oxidative phosphorylation to generate the energy required for cellular processes (Fantin et al., 2006; Moreno-Sánchez et al., 2007), but on aerobic glycolysis, which is known as “the Warburg effect” (Hsu and Sabatini, 2008; Vander Heiden et al., 2009). Aerobic glycolysis is a glycolytic process that occurs even in the presence of oxygen. HIF-1 mediates the metabolic adaptation by coordinately regulating genes encoding glycolytic enzymes (Semenza, 2011; Semenza et al., 1996; Seagroves et al., 2001), and activating expression of both pyruvate dehydrogenase kinase 1 (PDK1) and lactate dehydrogenase A (LDHA) (Kim et al., 2006; Papandreou et al., 2006; Stubbs and Griffiths, 2010; Semenza, 2007).
It is estimated that 15–20% of human cancers are associated with infection by oncogenic viruses (D’Agostino and Bernardi, 2005; McLaughlin-Drubin and Munger, 2008; Dayaram and Marriott, 2008). A key step in viral tumorigenesis is the specific interaction of a virus oncogenic gene product(s) with mitochondria (Chatterjee et al., 2011; Kroemer, 2006). An emerging body of evidence has demonstrated that diverse viruses target mitochondria as a mechanism to alter cell physiology, including cell survival (Ohta and Nishiyama, 2011; Galluzzi et al., 2008). Also, some oncogenic viruses can generate ROS via their oncogenic products: for example, hepatitis B virus (HBV) X protein (HBx); hepatitis C virus (HCV) core, E1 and NS3; human papillomavirus 18 (HPV-18) E2; human T-cell leukemia virus 1 (HTLV-1) p13 and Tax (Fig. 1). It is generally believed that virus infection-induced ROS triggers oxidative DNA damage, which can lead to cancer (Demple and Harrison, 1994; Lu et al., 2001; Dizdaroglu, 1992). However, other aspects of ROS-related physiology and pathology have not been extensively studied in viral oncogenesis. Furthermore, ROS-producing virus oncogene products play an essential role in virus replication, which may require high metabolic activity of host cells in addition to robust cell proliferation. Thus, it is likely that oncogenic viruses hijack mitochondria quality control and metabolic adaptation pathways to promote virus replication. This review will highlight recent progress on oncogenic viruses and their functional interactions with mitochondria that promote viral oncogenesis.
Hepatitis B virus (HBV)
More than 350 million people worldwide are chronically infected with HBV. The chronic infection is associated with the development of severe liver diseases including hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). HBV is a DNA virus belonging to the Hepadnaviridae family and has a highly compact genome of about 3200 bases in length. This genome contains four overlapping open reading frames that encode the viral core protein, surface proteins, reverse transcriptase and nonstructural X protein (HBx) (Koike, 2009). HBx is a multifunctional protein that regulates cellular signaling, transcription, proliferation, DNA repair, apoptosis and protein-degradation pathways. These activities may contribute not only to HBV replication but also to the development of HCC. One controversial aspect regarding HBx is whether this protein is anti-apoptotic or pro-apoptotic (Rawat et al., 2012).
HBx is predominantly located in the nucleus of HBV-infected hepatocytes at low expression levels, whereas at high expression levels it is mostly cytoplasmic. A portion of cytoplasmic HBx localizes to the mitochondrial outer membrane (Fig. 2) (Henkler et al., 2001; Huh and Siddiqui, 2002; Li et al., 2008). Interestingly, HBx is not observed in mitochondria at lower expression levels (Henkler et al., 2001). Mitochondrial localization of HBx triggers excessive mROS production, possibly by altering the expression of proteins involved in the oxidative phosphorylation pathway (Lee et al., 2004; Jung and Kim, 2013; Koike, 2009). HBx-induced mROS have been shown to be both pro-apoptotic and anti-apoptotic (Rawat et al., 2012). Excess mROS can induce apoptosis by mitochondrial membrane depolarization through modulation of the mitochondrial permeability transition pore (MTPC) (Shirakata and Koike, 2003). However, in primary hepatocytes, HBx regulation of the MPTC varies depending on the status of NF-κB activation. HBx activation of NF-κB suppressed mitochondrial membrane depolarization; however, when NF-κB signaling was blocked, HBx induced the MPTC (Clippinger and Bouchard, 2008). It is noteworthy that ROS can activate NF-κB (Li et al., 1998). Furthermore, recent studies demonstrate that HBx-induced ROS promotes HCC via dysregulation of the PTEN/Akt pathway (Ha, 2010), and HBx phosphorylation at serine 31 by Akt is essential for the anti-apoptotic activity of HBx (Lee et al., 2012b), indicating that cellular signaling events initiated by HBx-induced ROS accumulation could enhance cell survival and induce the development of HCC. Taken together, these results suggest that ROS levels or the availability of ROS-activated NF-κB or Akt, depending on the cellular context, may determine whether HBx is pro-apoptotic or anti-apoptotic.
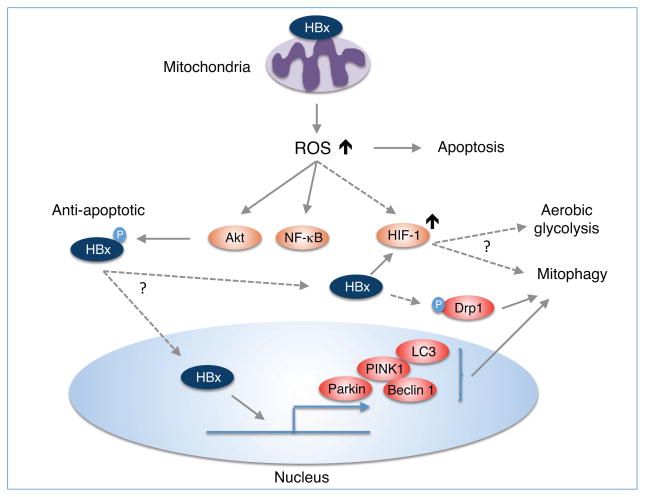
Figure 2. Possible role of HBV HBx-induced mROS in mitophagy and metabolic adaptation.
HBV infection induces the production of mROS via its gene product HBx and regulates mitophagy. Anti-apoptotic role of HBx may be dependent on the ability of HBx to eliminate damaged mitochondria via mitophagy. HBx, presumably the anti-apoptotic form phosphorylated by Akt (Lee et al., 2012b), can upregulate the expression of mitophagic proteins including PINK1, Parkin, Beclin, LC3 and also induce the phosphorylation of Drp1 to promote mitophagy (Kim et al., 2013b). However, the functional significance of HBx activation of HIF-1 is largely unknown.
However, there is no direct evidence that HBx-induced ROS is involved in HIF-1 activation or stabilization. Previous studies showed that HIF-1α was stabilized by direct HBx binding to the bHLH/PAS domain of HIF-1α, thereby preventing degradation of HIF-1α (Yoo and Lee, 2004; Yoo et al., 2003). HIF-1α was also up-regulated by HBx-stimulated transactivation through metastasis-associated protein 1 (MTA1), histone deacetylase 1 (HDAC1) and the mitogen-activated protein kinase pathway (Yoo et al., 2008). Furthermore, a recent study demonstrated that C-terminal mutations of HBx could regulate the ability of HBx to induce HIF-1α (Liu et al., 2014). Considering that the C-terminal region of HBx is involved in mitochondrial localization and oxidative stress (Jung and Kim, 2013; Huh and Siddiqui, 2002; Li et al., 2008), it is likely that mROS may mediate HBx activation of HIF-1α, possibly resulting in metabolic adaptation and activation of mitophagy.
HBx has recently been shown to utilize autophagy pathways for cell survival. Yi Mao et al reported that starvation-induced cell death was greatly increased in HBX-expressing hepatic and hepatoma cell lines treated with the autophagy inhibitor 3-methyladenine or transfected with an siRNA specific for the autophagy regulatory gene, beclin 1, indicating that HBx-induced autophagy was essential for cell survival (Mao et al., 2011). HBx can promote autophagy through the PI3K/Akt pathway (Wang et al., 2013a) and activates the autophagic lysosomal pathway in HepG2 cells via up-regulation of LC3-II, LC3-I, beclin 1 and lamp2a proteins (Wang et al., 2013b). Consistent with HBx activation of autophagy, HBx may also trigger mitophagy since it induced mitochondrial clustering in the perinuclear region (Kim et al., 2007). Indeed, HBV infection or HBx expression promoted the mitochondrial translocation of the dynamin-related protein (Drp1) by stimulating its phosphorylation at Ser616, thus leading to mitochondrial fission (Fig. 2) (Kim et al., 2013a). These events were also associated with increased gene expression of Parkin, PINK1 and LC3B and Parkin recruitment to the mitochondria, culminating in mitophagy (Kim et al., 2013a). These results suggest that HBV activates the mitophagy pathway via HBx protein to suppress virus-induced apoptosis. However, it remains unclear how HBx-induced ROS and HIF-1 activation are related to mitochondrial quality control.
It is well established that HBx plays a crucial role in HBV replication through several mechanisms (refer to recent review papers (Rawat et al., 2012; Feitelson et al., 2014)). However, it has not been elucidated how HBx-induced ROS and associated mitochondrial activities including metabolic adaptation, apoptosis and mitophagy contribute to HBV replication. A tight regulation of HBx-induced ROS is likely essential for efficient HBV replication, and chronic liver diseases and HCC.
Hepatitis C virus (HCV)
There are approximately 200 million people worldwide infected with HCV (Mohd Hanafiah et al., 2013; Gravitz, 2011), a chronic infection that frequently progresses to liver fibrosis and cirrhosis, various metabolic alterations (Adinolfi et al., 2011; Arrese et al., 2010) and the development of HCC or non-Hodgkin lymphoma (Jin, 2007; Hartridge-Lambert et al., 2012). HCV is an RNA virus belonging to the Flaviviridae family and has a genome of about 9600 bases long, consisting of a long open reading frame encoding a polyprotein precursor of 3010 amino acids (Kato, 2000). The HCV polyprotein is cleaved by cellular and viral proteases into ten different products, consisting of structural (core, E1 and E2) and nonstructural proteins (NS1, NS2, NS3, NS4A, NS4B, NS5A and NS5B) (Ashfaq et al., 2011).
A remarkable feature of HCV-associated pathogenesis is the ROS production in HCV-infected cells (refer to recent review papers for detail (Ivanov et al., 2013; Paracha et al., 2013)). In contrast to HBV, HCV encodes several ROS-producing proteins: core, E1, E2, NS3/4A, NS4B and NS5A. Among them, core is a potent ROS inducer and induces oxidative stress on par with that observed during chronic HCV infection (Korenaga et al., 2005; Okuda et al., 2002; Schwer et al., 2004; Paracha et al., 2013). HCV core enhances ROS production, mainly in mitochondria, by inhibiting oxidative phosphorylation complex I (Korenaga et al., 2005) and by stimulation of calcium uniporter activity (Fig. 3) (Li et al., 2007). Also, HCV core induces mitochondrial dysfunction by enhancing the expression of prohibitin, a chaperone of mitochondrial respiratory enzymes (Tsutsumi et al., 2009). HCV core is localized in mitochondria, and in mitochondria-associated membranes (MAM) where it elevates mitochondrial calcium concentration, which in turn is involved in MPTP opening, mitochondrial depolarization and subsequent mitochondrial damage (Benali-Furet et al., 2005). This oxidative stress triggers DNA damage and lipid peroxidation in HCV-infected cells (Machida et al., 2006).
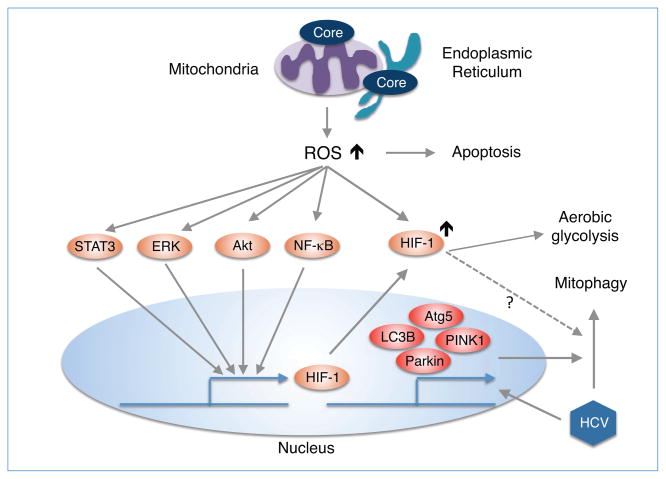
Figure 3. Possible role of HCV core-induced mROS in mitophagy and metabolic adaptation.
HCV infection induces the production of mROS mainly via its gene product core, and regulates mitophagy through the upregulation of PINK1 and Parkin (Kim et al., 2013c). However, there is no direct evidence that HCV core regulates mitophagy although it upregulates LC3B and Atg5 through ER-stress-induced signaling (Wang et al., 2014). HCV core activates and stabilizes HIF-1 by ROS or several cellular signaling pathways such as NF-κB, STAT3, Akt and Erk and then promotes metabolic adaptation (Ivanov et al., 2013).
HCV has evolved several strategies including metabolic adaptation and mitophagy in response to oxidative stress. Although there is little evidence that HCV core activates HIF-1α (Liu et al., 2011), HCV infection leads to stabilization of HIF-1α through oxidative stress (Nasimuzzaman et al., 2007; Ripoli et al., 2010). HIF-1α stabilization requires the activation of NF-κB, STAT3, PI3K/AkT and p42/44 mitogen-activated protein kinase (Fig. 3), suggesting that HCV infection or HCV core-induced ROS may be involved in the activation of these signaling pathways akin to HBx activation of NF-κB and PI3K/Akt (Machida et al., 2006; Paracha et al., 2013). The HIF-1α protein promotes glycolytic adaptation in Huh-7.5 cells harboring infectious HCV (Ripoli et al., 2010) as well as HCV replication and metastasis properties of HCC by up-regulation of vascular endothelial growth factor (VEGF) and transforming growth factor-beta (TGF-β) (Wilson et al., 2012).
Moreover, it is likely that HCV-induced mitochondrial dysfunction leads to mitophagy. Indeed, HCV infection promotes the formation of PINK1/Parkin-mediated mitophagy by up-regulating the expression of PINK1 and Parkin (Kim et al., 2013c). Also, this same study demonstrated that HCV replication was diminished when PINK1 and Parkin were depleted, indicating that mitophagy was essential for HCV replication. However, there is currently no direct evidence that HCV core-induced mitochondrial dysfunction activates mitophagy. Because HCV core was recently reported to activate autophagy by up-regulating LC3B and Atg5 through ER stress-induced signaling (Wang et al., 2014), it is likely that HCV core is involved in the mitophagy pathway to remove damaged mitochondria.
Human papillomavirus (HPV)
HPVs have a DNA genome and are classified into two groups based on pathogenicity: low-risk and high-risk. At least 15 high-risk HPVs (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73 and 82) are linked to the development of several types of cancers, including cervical and oropharyngeal carcinomas (Gabriela et al., 2013; Ramqvist and Dalianis, 2010; Madkan et al., 2007). The genome of HPV consists of 8,000 bases and can be subdivided into three major regions: early, late and a long control region. The early region encodes six nonstructural regulatory proteins (E1, E2, E4, E5, E6 and E7) (Danos et al., 1982). E1 and E2 are involved in viral DNA replication and the regulation of early transcription. E4 is expressed during productive infection and associates with cytokeratin collapse. E5, E6 and E7 are viral oncogenes that are capable of cell immortalization and transformation. E6 and E7 inactivate two key tumor suppressor proteins p53 and pRb, respectively (Mu and Howley, 2002). Thus far, however, it is unclear whether E6 and E7 regulate redox regulation or mitochondrial metabolism in the development of cancer. Recently, HPV-16 E6 and E7 were shown to inhibit apoptosis by down-regulating the expression of the C1q receptor (gC1qR/p32/C1QBP/HABP1) (Gao et al., 2011), a mitochondrial surface protein overexpressed in certain cancers. Because down-regulation of gC1qR in human cancer cells strongly shifts metabolism from oxidative phosphorylation to glycolysis (Fogal et al., 2010), it is likely that E6 and E7 regulate metabolic balance between oxidative phosphorylation and aerobic glycolysis, probably in the absence of severe mitochondrial dysfunction (Fig. 4).
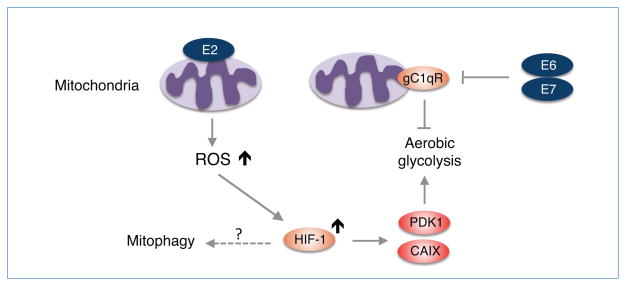
Figure 4. HPV-18 E2 regulates ROS production.
High-risk HPV-18 E2 localizes to mitochondria and induces ROS that stabilizes HIF-1, which in turn activates PDK1 and CAIX enzymes to promote aerobic glycolysis (Lai et al., 2013). HPV can also promote aerobic glycolysis by inhibition of gC1qR through E6 and E7 (Gao et al., 2011). There is no evidence that HPV infection and its gene products including E2, E6, and E7 induce mitophagy.
Historically, E2 was classified as a viral “anti-oncogene” because E2 can repress the transcription of E6 and E7 in the context of HPV (Soeda et al., 2006; Bernard et al., 1989; Francis et al., 2000). However, recent data challenge this notion and unambiguously indicate that E2 proteins from high-risk HPVs (HPV-16 and HPV-18) actually exhibit oncogenic characteristics in cervical cancer progression (Bellanger et al., 2011). For example, these E2 proteins induce abnormal mitosis and chromosomal instability, together with DNA breaks during anaphase. Furthermore, E2 down-regulates anti-sense mitochondrial non-coding RNAs (ASncmtRNAs) (Villota et al., 2012), which are down-regulated in cancer cells (Burzio et al., 2009). Most E2 proteins are predominantly retained in the nuclei of infected cells, but HPV-18 and 16 E2s can shuttle between the nucleus and cytoplasm. A mass spectrometry study revealed that HPV-18 E2 associates with inner mitochondrial membrane proteins including components of the respiratory chain (e.g. complex III proteins) (Lai et al., 2013). Furthermore, electron microscopy analysis revealed that HPV-18 E2 localizes to mitochondria and modifies the cristae morphology of mitochondria (Lai et al., 2013). This study also demonstrated that E2-induced mitochondrial ROS correlates with stabilization of HIF-1α and increased glycolysis by up-regulation of the HIF target glycolytic genes PDK1 and carbonic anhydrase IX (CAIX) (Fig. 4). These mitochondrial functions were not observed with the non-oncogenic (low-risk) HPV-E2 protein, suggesting that modification of cellular metabolism by high-risk HPV E2 proteins could play a role in the development of cancer by inducing metabolic adaptation.
On the other hand, little is known about whether high-risk E2 protein-induced ROS is involved in the initiation of mitophagy. Since HPV-18 E2-induced ROS did not induce apoptosis (Lai et al., 2013), the level of high-risk E2 protein-induced ROS may not be sufficient to induce mitochondrial damage.
Human T-cell leukemia virus type 1 (HTLV-1)
HTLV-1 is a complex delta-retrovirus that infects approximately 20 million people worldwide and is associated with an inflammatory neurological disease (HTLV-1-associated myelopathy/tropical spastic paraparesis [HAM/TSP]) and an aggressive T-cell leukemia (adult T-cell leukemia [ATL]). The HTLV-1 genome is flanked by long terminal repeats (LTRs) and contains Gag, Pol and Env genes that encode essential structural and enzymatic proteins typical of all retroviruses. In addition, the pX region encodes four open reading frames (ORF I, II, III and IV) that give rise to several regulatory and accessory proteins that influence viral replication, cell proliferation, survival and transformation. ORF I encodes the p12 accessory protein which contributes to cell proliferation and immune evasion by activation of JAK/STAT and NFAT pathways and downregulation of MHC class I proteins respectively (Bai and Nicot, 2012). ORF II yields p13 which can localize to either the nucleus or mitochondria and regulates cell proliferation and survival (Silic-Benussi et al., 2010b). ORF II also encodes p30, a nuclear protein that negatively regulates viral gene expression post-transcriptionally by retaining tax/rex mRNA in the nucleus (Nicot et al., 2004). p30 also regulates the cell cycle and the DNA damage response and is important for viral persistence in vivo (Anupam et al., 2013). ORF III encodes Rex which binds to the Rex response element on unspliced and singly spliced viral RNAs and shuttles these RNAs from the nucleus to the cytoplasm for translation by the host machinery (Bai et al., 2012). Tax is encoded by ORF IV and is essential for transactivation of viral gene expression by recruiting host transcription factors CREB and AP-1 and coactivators CBP/p300 to the LTR (Harrod et al., 1998). Tax is also a potent oncogene that drives cell transformation by inactivating tumor suppressors, modulating the cell cycle and constitutively activating pro-proliferative and anti-apoptotic signaling pathways such as NF-κB (Matsuoka and Jeang, 2007). Finally, the antisense strand of HTLV-1 encodes spliced and unspliced forms of HTLV-1 basic leucine zipper factor (HBZ). HBZ promotes T-cell proliferation and antagonizes Tax activation of viral transcription and NF-κB to establish viral latency (Zhao and Matsuoka, 2012).
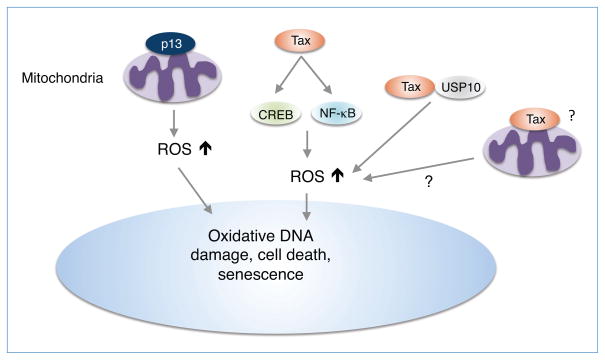
Of the HTLV-1 accessory and regulatory proteins, both p13 and Tax have been shown to induce ROS (Fig. 5). p13 contains an amphipathic alpha helix domain between amino acids 20–35 important for targeting p13 to the mitochondria where it inserts into the inner mitochondrial membrane and induces an inward K+ current that increases ROS production and promotes apoptosis (Silic-Benussi et al., 2010c, 2009; Biasiotto et al., 2010; Silic-Benussi et al., 2010a). p13 does not appear to be sufficient to induce cell death, but rather sensitizes cells to pro-apoptotic stimuli such as Fas ligand (Saggioro et al., 2009). Tax also induces ROS production, possibly through contributions from CREB and NF-κB, which correlates with DNA damage and cellular senescence (Kinjo et al., 2010). Tax may also induce ROS through interactions with ubiquitin-specific protease 10 (USP10), a component of stress granules (SGs) that plays critical roles in several SG-mediated activities, including inhibition of ROS production and ROS-dependent apoptosis (Takahashi et al., 2013). Tax inhibits arsenic-induced SG formation, stimulates ROS and enhances ROS-dependent apoptosis in T cells (Takahashi et al., 2013). Interestingly, cross-regulation between Tax and p13 has been reported that may impinge on production of ROS and cell survival (Silic-Benussi et al., 2010a). However, at this time the cellular source of Tax-induced ROS is unknown and requires further investigation.
Figure 5. HTLV-1 p13 and Tax proteins regulate ROS production.
HTLV-1 p13 localizes in the mitochondria and induces an inward K+ current that triggers mitochondrial depolarization, an increase in respiratory chain activation and ROS production (Silic-Benussi et al., 2009; Biasiotto et al., 2010). HTLV-1 Tax increases ROS production via CREB and NF-κB activation, interaction with USP10 (Takahashi et al., 2013) and/or localization in mitochondria. Increased ROS by p13 and Tax results in increased DNA damage, senescence and possibly cell death.
Recent mass spectrometry experiments from our lab showed that Tax may interact with several mitochondrial proteins including Hsp60, methylcrotonoyl-coenzyme A carboxylase subunit alpha, mitochondrial import inner membrane translocase subunit TIM50, mitochondrial phosphate carrier protein isoform B and propionyl-coenzyme A carboxylase alpha chain (Gao and Harhaj, 2013). Furthermore, using biochemical fractionation assays and confocal microscopy, we have found that a significant fraction of Tax localizes to mitochondria (submitted for publication). Whether Tax indeed interacts with these mitochondrial proteins to regulate ROS and/or cell death is an important topic for future investigation.
Herpesviruses
Human herpesviruses constitute a family of nine DNA viruses: Herpes simplex virus-1 (HSV-1/HHV-1), Herpes simplex virus-2 (HSV-2/HHV-2), Varicella zoster virus (VSV/HHV-3), Epstein-Barr virus (EBV/HHV-4), Cytomegalovirus (CMV/HHV-5), HHV-6A, HHV-6B, HHV-7 and Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8). Among them, only EBV and KSHV are oncogenic and frequently found to cause cancers in aged and immunosuppressed individuals. EBV infection is linked to the development of endemic Burkitt’s lymphoma, nasopharyngeal carcinoma, a subset of Hodgkin’s disease and other malignancies of lymphoid and epithelial cell origin (Young and Rickinson, 2004). The proteins encoded by EBV latency genes, including six EBV-encoded nuclear antigens (EBNA-1, -2, -3A, -3B, -3C, and -5) and three latent membrane proteins (LMP-1, -2A, and -2B), induce cell transformation by activating multiple signaling pathways that regulate proliferation and apoptosis. However, there is no direct evidence that EBV-encoded proteins induce mROS production. Rather, EBNA-1 promotes genomic instability via production of ROS by transcriptionally activating the leukocyte NADPH oxidase NOX2 (Gruhne et al., 2009). In addition, LMP-1 expression in the nasopharyngeal carcinoma cell line Ad-AH promotes HIF-1α accumulation by activation of the p42/p44 MAPK pathway through ROS production, particularly H2O2 (Cuninghame et al., 2014).
KSHV is the etiological agent of several neoplasms including Kaposi’s sarcoma, endothelial cell disease, and B cell malignancies primary effusion lymphoma and multicentric Castleman’s disease (Ganem, 2006; Carbone and Gloghini, 2008). Similar to EBV, it is unknown if KSHV gene products induce mROS. Instead, the KSHV early lytic gene viral G protein-coupled receptor (vGPCR) triggers ROS production in mECK36 cells via a Rac1-NADPH oxidase pathway (Ma et al., 2013). Thus, it remains to be determined whether EBV and KSHV infections induce mROS production and if so which viral gene products are responsible for mROS production.
CMV is a β-herpesvirus, with about 60–90% of adults having been infected by CMV at some time. CMV infections are usually asymptomatic but can be life-threatening for immunocompromised individuals such as HIV-1 infected patients and organ transplant recipients. Congenital CMV infection is also a major cause of morbidity in newly born infants and can lead to permanent disabilities. Thus far, CMV infection has not been clearly linked to the development of any human cancers, however CMV antigens have been detected in a large fraction of glioblastoma multiforme brain tumors. Whether CMV and its gene products regulate mROS production is currently unknown.
Conclusions
Several oncogenic viral gene products are known to induce oxidative stress in infected host cells or transfected cells. The best-known gene products are HBV HBx, HCV core, HPV E2 and HTLV-1 Tax (Table 1). These viral proteins generate ROS mainly in the mitochondria although it is unknown if HTLV-1 Tax can produce ROS in mitochondria. ROS have been recognized as a major player in the development of carcinoma, in particular, by inducing oxidative DNA damage that leads to oncogenic mutations. Generally, ROS induces cell death when produced in excessive amounts. Cells have established several strategies to survive in response to the deleterious effect of ROS: anti-apoptosis, autophagy and metabolic adaptation, resulting in malignancy along with genetic alterations. Similarly, ROS-producing oncogenic viral gene products hijack the survival mechanisms of host cells and together with ROS-induced genetic changes promote cellular transformation or viral replication. In this review, we focused on oncogenic viral regulation of mitophagy and metabolic adaptation by which mROS levels could be down-regulated. However, it remains largely unknown how oncogenic viruses precisely regulate mitophagy and metabolic adaptation pathways for oncogenesis or viral replication. In particular, little is known about the exact mechanisms underlying mitophagy pathways. On the other hand, EBV was shown to induce structural alterations of mitochondria after infection or expression of its gene products (LaJeunesse et al., 2005; Pal et al., 2014), but it is unknown whether EBV or EBV gene products induce mROS and mitophagy. Recently, Merkel cell polyomavirus (MCV) was identified as the causal agent for Merkel cell carcinoma, a rare skin cancer (Feng et al., 2008), but there is no evidence that MCV and its oncogenic gene product T antigen are involved in mROS production. Therefore, it would be of great interest to examine whether the oncogenic viruses EBV, KSHV and MCV regulate mROS-related mitochondrial physiology and metabolic adaptation during lytic productive replication and oncogenesis.
Table 1.
Oncogenic viral gene products triggering the production of mitochondrial reactive oxygen species.
| Virus | Viral protein | Mechanism to induce mROS production | Reference |
|---|---|---|---|
| HBV | HBX | May alter the expression of proteins involved in oxidative phosphorylation. | Jung & Kim, 2013, Koike, 2009, Lee et al., 2004 |
| HCV | Core | Inhibits oxidative phosphorylation complex I. | Korenaga et al., 2005 |
| Stimulates calcium uniporter activity. | Li et al, 2007 | ||
| HPV | E2 | Unknown | |
| HTLV-1 | p13 | Induces an inward potassium current at the mitochondrial inner membrane. |
Silic-Benussi et al., 2009 Biasiotto et al., 2010 |
| Tax | Unknown |
Acknowledgments
The laboratories of E.W.H and Y.B.C. are funded by National Institutes of Health grants RO1CA135362 and R21AI103379 respectively.
References
- Adinolfi LE, Restivo L, Zampino R, Lonardo A, Loria P. Metabolic alterations and chronic hepatitis C: treatment strategies. Expert Opin Pharmacother. 2011;12:2215–34. doi: 10.1517/14656566.2011.597742. [DOI] [PubMed] [Google Scholar]
- Anupam R, Doueiri R, Green PL. The need to accessorize: molecular roles of HTLV-1 p30 and HTLV-2 p28 accessory proteins in the viral life cycle. Front Microbiol. 2013;4:275. doi: 10.3389/fmicb.2013.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese M, Riquelme A, Soza A. Insulin resistance, hepatic steatosis and hepatitis C: a complex relationship with relevant clinical implications. Ann Hepatol. 2010;9:112–118. [PubMed] [Google Scholar]
- Ashfaq Ua, Javed T, Rehman S, Nawaz Z, Riazuddin S. An overview of HCV molecular biology, replication and immune responses. Virol J. 2011;8:161. doi: 10.1186/1743-422X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babusikova E, Evinova A, Hatok J. Oxidative Changes and Possible Effects of Polymorphism of Antioxidant Enzymes in Neurodegenerative Disease 2013 [Google Scholar]
- Bai XT, Nicot C. Overview on HTLV-1 p12, p8, p30, p13: accomplices in persistent infection and viral pathogenesis. Front Microbiol. 2012;3:400. doi: 10.3389/fmicb.2012.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XT, Sinha-Datta U, Ko NL, Bellon M, Nicot C. Nuclear export and expression of human T-cell leukemia virus type 1 tax/rex mRNA are RxRE/Rex dependent. J Virol. 2012;86:4559–65. doi: 10.1128/JVI.06361-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger S, Tan CL, Xue YZ, Teissier S, Thierry F. Tumor suppressor or oncogene? A critical role of the human papillomavirus (HPV) E2 protein in cervical cancer progression. Am J Cancer Res. 2011;1:373–389. [PMC free article] [PubMed] [Google Scholar]
- Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–33. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- Bernard Ba, Bailly C, Lenoir MC, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J Virol. 1989;63:4317–24. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, Martinez L, Greidinger EL, Yu BD, Gallo RL. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18:1286–90. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasiotto R, Aguiari P, Rizzuto R, Pinton P, D’Agostino DM, Ciminale V. The p13 protein of human T cell leukemia virus type 1 (HTLV-1) modulates mitochondrial membrane potential and calcium uptake. Biochim Biophys Acta. 2010;1797:945–51. doi: 10.1016/j.bbabio.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Bonekamp Na, Völkl A, Fahimi HD, Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. Biofactors. 35:346–55. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- Brieger K, Schiavone S, Miller FJ, Krause K-H. Reactive oxygen species: from health to disease. Swiss Med Wkly. 2012;142:w13659. doi: 10.4414/smw.2012.13659. [DOI] [PubMed] [Google Scholar]
- Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A. 2000;97:9082–7. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzio Va, Villota C, Villegas J, Landerer E, Boccardo E, Villa LL, Martínez R, Lopez C, Gaete F, Toro V, et al. Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc Natl Acad Sci U S A. 2009;106:9430–4. doi: 10.1073/pnas.0903086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A, Gloghini A. KSHV/HHV8-associated lymphomas. Br J Haematol. 2008;140:13–24. doi: 10.1111/j.1365-2141.2007.06879.x. [DOI] [PubMed] [Google Scholar]
- Chandel N, Maltepe E, Goldwasser E, Mathieu C, Simon M, Schumacker P. Mitochondrial reactive oxygen species trigger hypoxia- induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila) 2011;4:638–54. doi: 10.1158/1940-6207.CAPR-10-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, Wang G. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–23. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger AJ, Bouchard MJ. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J Virol. 2008;82:6798–811. doi: 10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Cuezva JM, Krajewska M, Heredia ML, De Cuezva M, Lo M, Heredia D, Krajewski S, Santamari G, Kim H, Zapata JM, et al. The Bioenergetic Signature of Cancer: A Marker of Tumor Progression The Bioenergetic Signature of Cancer: A Marker of Tumor Progression 1. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- Cuninghame S, Jackson R, Zehbe I. Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology. 2014;456–457:370–383. doi: 10.1016/j.virol.2014.02.027. [DOI] [PubMed] [Google Scholar]
- D’Agostino D, Bernardi P. Mitochondria as functional targets of proteins coded by human tumor viruses. Adv cancer. 2005 doi: 10.1016/S0065-230X(04)94003-1. [DOI] [PubMed] [Google Scholar]
- Danos O, Katinka M, Yaniv M. Human papillomavirus 1a complete DNA sequence: genome organization among Papovaviridae novel type of. EMBO J. 1982;1:231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram T, Marriott SJ. Effect of transforming viruses on molecular mechanisms associated with cancer. J Cell Physiol. 2008;216:309–14. doi: 10.1002/jcp.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–48. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Ding W-X, Yin X-M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–64. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res. 1992;275:331–42. doi: 10.1016/0921-8734(92)90036-o. Available at: http://www.ncbi.nlm.nih.gov/pubmed/1383774. [DOI] [PubMed] [Google Scholar]
- Fader CM, Colombo MI. Multivesicular Bodies and Autophagy in Erythrocyte Maturation Addenda ES CE ACKNOWLEDGEMENTS RIB. Autophagy. 2006:122–125. doi: 10.4161/auto.2.2.2350. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Feitelson Ma, Bonamassa B, Arzumanyan A. The roles of hepatitis B virus-encoded X protein in virus replication and the pathogenesis of chronic liver disease. Expert Opin Ther Targets. 2014;18:293–306. doi: 10.1517/14728222.2014.867947. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol. 2010;30:1303–18. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis Da, Schmid SI, Howley PM. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J Virol. 2000;74:2679–86. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriela A, Adriana P, Coralia B, Anca B, Mariana A, Lorelei IB, Mihai S. Human Papillomaviruses Oncoproteins. InTech; 2013. [Google Scholar]
- Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu Rev Pathol. 2006;1:273–96. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- Gao L, Harhaj EW. HSP90 protects the human T-cell leukemia virus type 1 (HTLV-1) tax oncoprotein from proteasomal degradation to support NF-κB activation and HTLV-1 replication. J Virol. 2013;87:13640–54. doi: 10.1128/JVI.02006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L-J, Gu P-Q, Fan W-M, Liu Z, Qiu F, Peng Y-Z, Guo X-R. The role of gC1qR in regulating survival of human papillomavirus 16 oncogene-transfected cervical cancer cells. Int J Oncol. 2011;39:1265–72. doi: 10.3892/ijo.2011.1108. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gravitz L. A smouldering public- health crisis. Nature. 2011;474:S2–S4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta Ma, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon Ea. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–85. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein–Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc Natl Acad Sci U S A. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–8. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Ha H-L. HBx-induced reactive oxygen species activates hepatocellular carcinogenesis via dysregulation of PTEN/Akt pathway. World J Gastroenterol. 2010;16:4932. doi: 10.3748/wjg.v16.i39.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägg M, Wennström S. Activation of hypoxia-induced transcription in normoxia. Exp Cell Res. 2005;306:180–91. doi: 10.1016/j.yexcr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin Cell Biol. 2009;21:894–9. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrod R, Tang Y, Nicot C, Lu HS, Vassilev A, Nakatani Y, Giam CZ. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol Cell Biol. 1998;18:5052–61. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartridge-Lambert SK, Stein EM, Markowitz AJ, Portlock CS. Hepatitis C and non-Hodgkin lymphoma: the clinical perspective. Hepatology. 2012;55:634–41. doi: 10.1002/hep.25499. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkler F, Hoare J, Waseem N, Goldin RD, McGarvey MJ, Koshy R, King Ia. Intracellular localization of the hepatitis B virus HBx protein. J Gen Virol. 2001;82:871–82. doi: 10.1099/0022-1317-82-4-871. [DOI] [PubMed] [Google Scholar]
- Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–80. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb Ra. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh KW, Siddiqui A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion. 2002;1:349–59. doi: 10.1016/s1567-7249(01)00040-x. [DOI] [PubMed] [Google Scholar]
- Ivanov AV, Bartosch B, Smirnova Oa, Isaguliants MG, Kochetkov SN. HCV and oxidative stress in the liver. Viruses. 2013;5:439–69. doi: 10.3390/v5020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DY. Molecular pathogenesis of hepatitis C virus-associated hepatocellular carcinoma. Front Biosci. 2007;12:222–233. doi: 10.2741/2060. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane La, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–42. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–9. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S-Y, Kim Y-J. C-terminal region of HBx is crucial for mitochondrial DNA damage. Cancer Lett. 2013;331:76–83. doi: 10.1016/j.canlet.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Kane La, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf Sa, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5:129–151. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- Kim J, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H-Y, Lee S, Kim SW, Sohn S, Kim K, Cho H. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol. 2007;81:1714–26. doi: 10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013a;9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013b;9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Syed GH, Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013c;9:e1003285. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo T, Ham-Terhune J, Peloponese J-M, Jeang K-T. Induction of reactive oxygen species by human T-cell leukemia virus type 1 tax correlates with DNA damage and expression of cellular senescence marker. J Virol. 2010;84:5431–7. doi: 10.1128/JVI.02460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009;286:60–8. doi: 10.1016/j.canlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Korenaga M, Wang T, Li Y, Showalter La, Chan T, Sun J, Weinman Sa. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481–8. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014 doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kroemer G. Mitochondria in cancer. Oncogene. 2006;25:4630–2. doi: 10.1038/sj.onc.1209589. [DOI] [PubMed] [Google Scholar]
- Lai D, Tan CL, Gunaratne J, Quek LS, Nei W, Thierry F, Bellanger S. Localization of HPV-18 E2 at mitochondrial membranes induces ROS release and modulates host cell metabolism. PLoS One. 2013;8:e75625. doi: 10.1371/journal.pone.0075625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse DR, Brooks K, Adamson AL. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 alter mitochondrial morphology during lytic replication. Biochem Biophys Res Commun. 2005;333:438–42. doi: 10.1016/j.bbrc.2005.05.120. [DOI] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012a;441:523–40. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-P, Lan K-H, Li C-P, Chao Y, Lin H-C, Lee S-D. Pro-apoptotic or anti-apoptotic property of X protein of hepatitis B virus is determined by phosphorylation at Ser31 by Akt. Arch Biochem Biophys. 2012b;528:156–62. doi: 10.1016/j.abb.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Lee YI, Hwang JM, Im JH, Lee YI, Kim NS, Kim DG, Yu DY, Moon HB, Park SK. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460–71. doi: 10.1074/jbc.M309280200. [DOI] [PubMed] [Google Scholar]
- Li SK, Ho SF, Tsui KW, Fung KP, Waye MYM. Identification of functionally important amino acid residues in the mitochondria targeting sequence of hepatitis B virus X protein. Virology. 2008;381:81–8. doi: 10.1016/j.virol.2008.07.037. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang X, Zhuang H, Chen H-G, Chen Y, Tian W, Wu W, Li Y, Wang S, Zhang L, et al. MicroRNA-137 Is a Novel Hypoxia-responsive MicroRNA That Inhibits Mitophagy via Regulation of Two Mitophagy Receptors FUNDC1 and NIX. J Biol Chem. 2014;289:10691–701. doi: 10.1074/jbc.M113.537050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Mai J, Choi ET, Wang H, Yang X. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Boehning DF, Qian T, Popov VL, Weinman Sa. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. FASEB J. 2007;21:2474–85. doi: 10.1096/fj.06-7345com. [DOI] [PubMed] [Google Scholar]
- Li YP, Schwartz RJ, Waddell ID, Holloway BR, Reid MB. Skeletal muscle myocytes undergo protein loss and reactive oxygen-mediated NF-kappaB activation in response to tumor necrosis factor alpha. FASEB J. 1998;12:871–80. doi: 10.1096/fasebj.12.10.971. [DOI] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Liu LP, Hu BG, Ye C, Ho RLK, Chen GG, Lai PBS. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br J Cancer. 2014;110:1066–73. doi: 10.1038/bjc.2013.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou X, Zhu C, Song H, Liu F. Effects of HCV core protein on the expression of hypoxia-inducible factor 1 alpha and vascular endothelial growth factor. Zhonghua Gan Zang Bing Za Zhi. 2011;19:751–754. doi: 10.3760/cma.j.issn.1007-3418.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Lu aL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys. 2001;35:141–70. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- Ma Q, Cavallin LE, Leung HJ, Chiozzini C, Goldschmidt-clermont PJ, Mesri EA. A Role for Virally Induced Reactive Oxygen Species in Kaposi’ s Sarcoma Herpesvirus Tumorigenesis. Antioxid Redox Signal. 2013;18:80–90. doi: 10.1089/ars.2012.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Cheng KT-H, Lai C-K, Jeng K-S, Sung VM-H, Lai MMC. Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J Virol. 2006;80:7199–207. doi: 10.1128/JVI.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madkan VK, Cook-Norris RH, Steadman MC, Arora a, Mendoza N, Tyring SK. The oncogenic potential of human papillomaviruses: a review on the role of host genetics and environmental cofactors. Br J Dermatol. 2007;157:228–41. doi: 10.1111/j.1365-2133.2007.07961.x. [DOI] [PubMed] [Google Scholar]
- Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415:68–74. doi: 10.1016/j.bbrc.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Martin K, Barrett J. Reactive oxygen species as double-edged swords in cellular processes: low-dose cell signaling versus high-dose toxicity. Hum Exp Toxicol. 2002;21:71–75. doi: 10.1191/0960327102ht213oa. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–80. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782:127–50. doi: 10.1016/j.bbadis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S, Chatelain EH, Lavie J, Mahfouf W, Jose C, Obre E, Goorden S, Priault M, Elgersma Y, Rezvani HR, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013;17:719–30. doi: 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–42. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- Mu K, Howley PM. Human papilloma virus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane La, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen D-F, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasimuzzaman M, Waris G, Mikolon D, Stupack DG, Siddiqui A. Hepatitis C virus stabilizes hypoxia-inducible factor 1alpha and stimulates the synthesis of vascular endothelial growth factor. J Virol. 2007;81:10249–57. doi: 10.1128/JVI.00763-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ney Pa. Normal and disordered reticulocyte maturation. Curr Opin Hematol. 2011;18:152–7. doi: 10.1097/MOH.0b013e328345213e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot C, Dundr M, Johnson JM, Fullen JR, Alonzo N, Fukumoto R, Princler GL, Derse D, Misteli T, Franchini G. HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication. Nat Med. 2004;10:197–201. doi: 10.1038/nm984. [DOI] [PubMed] [Google Scholar]
- Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxid Redox Signal. 2012;17:794–802. doi: 10.1089/ars.2011.4407. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta A, Nishiyama Y. Mitochondria and viruses. Mitochondrion. 2011;11:1–12. doi: 10.1016/j.mito.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M, Li K, Beard MR, Showalter La, Scholle F, Lemon SM, Weinman Sa. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- Pal AD, Basak NP, Banerjee AS, Banerjee S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis. 2014;00:1–10. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- Pan J-S. Reactive oxygen species: A double-edged sword in oncogenesis. World J Gastroenterol. 2009;15:1702. doi: 10.3748/wjg.15.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns Ra, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. doi: 10.1186/1743-422X-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramqvist T, Dalianis T. Oropharyngeal cancer epidemic and human papillomavirus. Emerg Infect Dis. 2010;16:1671–7. doi: 10.3201/eid1611.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat S, Clippinger AJ, Bouchard MJ. Modulation of apoptotic signaling by the hepatitis B virus X protein. Viruses. 2012;4:2945–72. doi: 10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–90. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoli M, D’Aprile A, Quarato G, Sarasin-Filipowicz M, Gouttenoire J, Scrima R, Cela O, Boffoli D, Heim MH, Moradpour D, et al. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation. J Virol. 2010;84:647–60. doi: 10.1128/JVI.00769-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggioro D, Silic-Benussi M, Biasiotto R, D’Agostino D, Ciminale V. Control of cell death pathways by HTLV-1 proteins. Front Biosci. 2009;14:3338–3351. doi: 10.2741/3456. [DOI] [PubMed] [Google Scholar]
- Sawada M, Carlson JC. Changes in superoxide readical and lipid peroxide formation in the brain, heart and liver during the lifetime of the rat. Mech Ageing Dev. 1987;41:125–137. doi: 10.1016/0047-6374(87)90057-1. [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–66. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Schwer B, Ren S, Pietschmann T, Kartenbeck J, Bartenschlager R, Yen T, Ott M. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagroves TN, Ryan HE, Lu H, Bradly G, Knapp M, Thibault P, Laderoute K, Johnson RS, Lu HAN, Wouters BG. Transcription Factor HIF-1 Is a Necessary Mediator of the Pasteur Effect in Mammalian Cells Transcription Factor HIF-1 Is a Necessary Mediator of the Pasteur Effect in Mammalian Cells. 2001 doi: 10.1128/MCB.21.10.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39:231–4. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–53. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Jiang B, Leung SW, Passantino R, Concordet J-P, Maire P, Giallongo A. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A Gene Promoters Contain Essential Binding Sites for Hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Sena La, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–67. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071–8. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- Silic-Benussi M, Biasiotto R, Andresen V, Franchini G, D’Agostino DM, Ciminale V. HTLV-1 p13, a small protein with a busy agenda. Mol Aspects Med. 2010a;31:350–8. doi: 10.1016/j.mam.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silic-Benussi M, Cannizzaro E, Venerando A, Cavallari I, Petronilli V, La Rocca N, Marin O, Chieco-Bianchi L, Di Lisa F, D’Agostino DM, et al. Modulation of mitochondrial K(+) permeability and reactive oxygen species production by the p13 protein of human T-cell leukemia virus type 1. Biochim Biophys Acta. 2009;1787:947–54. doi: 10.1016/j.bbabio.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Silic-Benussi M, Cavallari I, Vajente N, Vidali S, Chieco-Bianchi L, Di Lisa F, Saggioro D, D’Agostino DM, Ciminale V. Redox regulation of T-cell turnover by the p13 protein of human T-cell leukemia virus type 1: distinct effects in primary versus transformed cells. Blood. 2010b;116:54–62. doi: 10.1182/blood-2009-07-235861. [DOI] [PubMed] [Google Scholar]
- Silic-Benussi M, Marin O, Biasiotto R, D’Agostino DM, Ciminale V. Effects of human T-cell leukemia virus type 1 (HTLV-1) p13 on mitochondrial K+ permeability: A new member of the viroporin family? FEBS Lett. 2010c;584:2070–5. doi: 10.1016/j.febslet.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonnet H, Alazard N, Pfeiffer K, Gallou C, Béroud C, Demont J, Bouvier R, Schägger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–68. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- Soeda E, Ferran MC, Baker CC, McBride Aa. Repression of HPV16 early region transcription by the E2 protein. Virology. 2006;351:29–41. doi: 10.1016/j.virol.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Stubbs M, Griffiths JR. The altered metabolism of tumors: HIF-1 and its role in the Warburg effect. Adv Enzyme Regul. 2010;50:44–55. doi: 10.1016/j.advenzreg.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Higuchi M, Makokha GN, Matsuki H, Yoshita M, Tanaka Y, Fujii M. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood. 2013;122:715–725. doi: 10.1182/blood-2013-03-493718.The. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–5. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen D-F, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–80. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi T, Matsuda M, Aizaki H, Moriya K, Miyoshi H, Fujie H, Shintani Y, Yotsuyanagi H, Miyamura T, Suzuki T, et al. Proteomics analysis of mitochondrial proteins reveals overexpression of a mitochondrial protein chaperon, prohibitin, in cells expressing hepatitis C virus core protein. Hepatology. 2009;50:378–86. doi: 10.1002/hep.22998. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–44. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Villota C, Campos A, Vidaurre S, Oliveira-Cruz L, Boccardo E, Burzio Va, Varas M, Villegas J, Villa LL, Valenzuela PDT, et al. Expression of mitochondrial non-coding RNAs (ncRNAs) is modulated by high risk human papillomavirus (HPV) oncogenes. J Biol Chem. 2012;287:21303–15. doi: 10.1074/jbc.M111.326694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–58. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kang R, Huang H, Xi X, Wang B, Wang J, Zhao Z. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression. Autophagy. 2014;10:766–84. doi: 10.4161/auto.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Guo Q-S, Wang Z-W, Qian H-X. HBx induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway. Mol Cell Biochem. 2013a;372:161–8. doi: 10.1007/s11010-012-1457-x. [DOI] [PubMed] [Google Scholar]
- Wang P, Wang Z, Qian H, Guo Q. Role of autophagy in HepG-2 cells induced by hepatitis B virus x protein. Zhonghua Yi Xue Za Zhi. 2013b;93:3556–3558. [PubMed] [Google Scholar]
- Wang Y, Liu VWS, Xue WC, Cheung aNY, Ngan HYS. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95:1087–91. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8:1462–76. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GK, Brimacombe CL, Rowe Ia, Reynolds GM, Fletcher NF, Stamataki Z, Bhogal RH, Simões ML, Ashcroft M, Afford SC, et al. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration. J Hepatol. 2012;56:803–9. doi: 10.1016/j.jhep.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–69. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y-G, Lee M-O. Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J Biol Chem. 2004;279:36242–9. doi: 10.1074/jbc.M401290200. [DOI] [PubMed] [Google Scholar]
- Yoo YG, Na TY, Seo HW, Seong JK, Park CK, Shin YK, Lee MO. Hepatitis B virus X protein induces the expression of MTA1 and HDAC1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene. 2008;27:3405–13. doi: 10.1038/sj.onc.1211000. [DOI] [PubMed] [Google Scholar]
- Yoo Y-G, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu D-Y, Seong JK, Lee M-O. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:39076–84. doi: 10.1074/jbc.M305101200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Rickinson A. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ney Pa. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Matsuoka M. HBZ and its roles in HTLV-1 oncogenesis. Front Microbiol. 2012;3:247. doi: 10.3389/fmicb.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]