Abstract
Virus budding is a complex, multistep process in which viral proteins make specific alterations in membrane curvature. Many different viral proteins can deform the membrane and form a budding virion, but very few can mediate membrane scission to complete the budding process. As a result, enveloped viruses have developed numerous ways of facilitating membrane scission, including hijacking host cellular scission machinery and expressing their own scission proteins. These proteins mediate scission in very different ways, though the biophysical mechanics underlying their actions may be similar. In this review, we explore the mechanisms of membrane scission and the ways in which enveloped viruses use these systems to mediate the release of budding virions.
Keywords: virus budding, ESCRT, membrane curvature, lipid phase segregation, line tension
OVERVIEW OF MEMBRANE SCISSION
Membrane scission, or fission, is the process in which the membrane neck of a budding vesicle or virus is severed and resealed into two separate membranes. The process of scission requires significant alterations of membrane curvature, typically caused by protein-lipid interactions and driven by the biophysical properties of lipid bilayers. The general process of membrane scission has three steps: the formation of a neck on a budding vesicle, the constriction of the membrane neck below a critical diameter, and the spontaneous fission and fusion of the membrane at the neck. This critical diameter varies based on the lipid-protein system; however, it is estimated to be in the range of 1–5 nm (Campelo & Malhotra 2012). Thus, the actual fission process is facilitated, but not mediated, by protein interactions and occurs spontaneously when the force put on a membrane is great enough to cause constriction beyond a critical threshold. The entire process, from alteration of membrane curvature to the final fission event, is a consequence of manipulation of lipid-lipid interactions within biological membranes.
Membrane Curvature and Scission
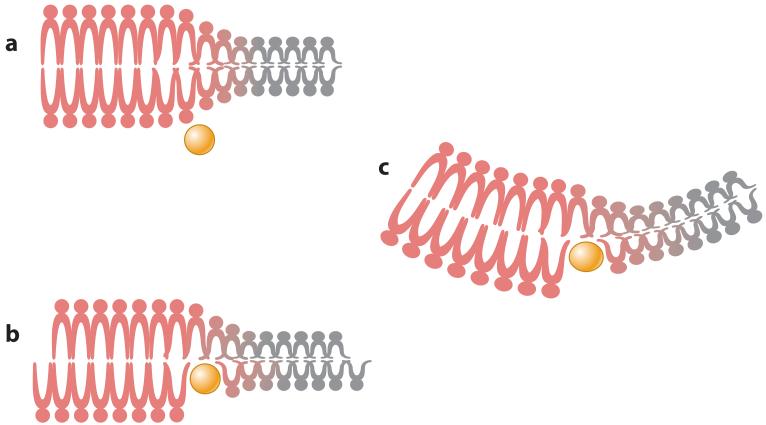
A lipid bilayer is organized such that it is resistant to the generation of curvature. The consequence of this resistance is that, if curvature is forced into a region of the bilayer, there will be a change in the elastic energy of the membrane (Helfrich 1973). Thus, by manipulating membrane curvature, it is possible to modify this elastic energy and use it to drive the process of membrane scission. Conversely, the induction of membrane curvature will require an input of energy greater than the cost of the elastic deformation. This reaction can be accomplished in different ways, but all involve manipulation of the arrangement of phospholipids in a bilayer. For example, the insertion of a protein into a lipid bilayer can cause a separation of the polar head groups, which changes the angle of the hydrophobic tails and causes a mismatch in surface area between the two leaflets of the bilayer (stacking defect) that can be energetically minimized by the induction of membrane curvature (Figure 1). Likewise, protein head group binding can cause a scaffolding effect, wrapping the membrane around a curved surface as a consequence of the attractive force between the protein and lipid. Curvature can also be induced as a direct consequence of lipid-domain formation within biological membranes.
Figure 1.
Induction of membrane curvature by protein insertion. (a) Amphipathic proteins (yellow) can bind to and insert into lipid bilayers, occasionally at the junction between two lipid phases (liquid-ordered- or raft-phase lipids, shown in red, and liquid-disordered- or bulk-phase plasma membrane lipids, shown in gray). (b) Protein insertion expands one leaflet of the lipid bilayer, which places the membrane under strain. (c) Membrane strain can be resolved by inducing curvature. Peptide insertion at the lipid phase boundary can also alter membrane curvature by modifying the line tension force between the two lipid phases.
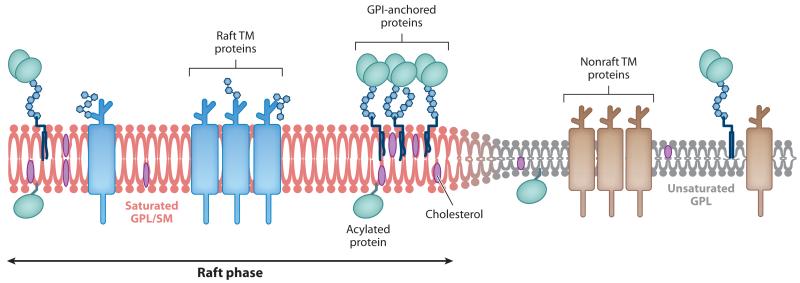
Biological membranes consist of a mixture of liquid-ordered (Lo) and liquid-disordered (Ld) phases (Figure 2). Lo lipid phases contain high levels of cholesterol, sphingolipids, and saturated phospholipids, whereas Ld lipid phases contain mainly unsaturated phospholipids (Lingwood & Simons 2010). In living cells, the Lo phase tends to form small, dynamic microdomains within the membrane, which have been termed lipid rafts (Lingwood & Simons 2010, Pike 2006). Because the saturated lipids of the Lo phase contain highly ordered acyl chains, the lipids tend to be more elongated and form a lipid domain that is thicker than an Ld domain. The resulting height difference between the two lipid phases causes the polar head groups of Ld-domain lipids to line up with the hydrophobic core of the Lo-domain lipids, an energetically unfavorable interaction that exerts a force, called line tension, on the membrane (Figure 2).
Figure 2.
Organization of lipid phases in the plasma membrane. Depiction of the lipid bilayer showing the differences between the raft and nonraft phases. Unsaturated glycophospholipids (GPL) represent the bulk (liquid-disordered) phase of the plasma membrane and contain mainly nonraft transmembrane (TM) proteins. Saturated GPL and sphingomyelin (SM) represent the raft (liquid-ordered) phase of the plasma membrane and are associated with higher levels of cholesterol and GPI-anchored, acetylated, and raft-localized TM proteins. The ordered packaging of the saturated lipids causes the raft phase to be thicker than the bulk plasma membrane phase, which leads to line tension between the lipid phases.
The line tension force at a given lipid-phase boundary is determined by both the degree of saturation and the acyl-chain length of the Lo-phase lipids as well as the polarity of the Ld-phase lipid head groups. Thus, the presence of lipids with strongly charged head groups in the Ld phase, such as phosphatidylethanolamine and phosphatidylglycerol, can significantly affect the line tension energy, creating a highly energetically unfavorable interaction that can be resolved through membrane deformation (Bischof & Wilke 2012). Membrane deformation is also an energetically unfavorable condition, and the degree of deformation is directly related to the force of the line tension. If the line tension force surrounding a lipid phase is greater than the energy of deformation, then the membrane will curve and a bud will form (García-Sáez et al. 2007, Lipowsky 1993). In the presence of great enough levels of line tension, it is even possible for budding to progress through membrane scission without requiring any protein mediators (Döbereiner et al. 1993, Laradji & Sunil Kumar 2005, Lipowsky 1993, Minami & Yamada 2007, Sackmann & Feder 1995). However, in most biological systems, proteins are used to interact with and manipulate the membrane environment, facilitating and regulating the processes of both membrane budding and scission.
Scission Proteins
Protein-mediated alterations of membrane curvature have several advantages over lipid-only systems. First, proteins can catalyze modifications of membrane curvature, enhancing the efficiency of budding and scission. Second, by manipulating the expression, localization, or modification of a protein, organisms can regulate the specific location and timing of membrane deformation. Third, just as different lipid domains have different corresponding line tension energies, proteins also vary in their capacity to alter membrane curvature. This allows for an added level of regulation, wherein the full process of budding and scission requires the concerted action of multiple copies of a single protein or multiple proteins working together.
The ability of a protein to alter membrane curvature stems from the types of lipids the protein interacts with and the nature of those interactions. Many cellular proteins specifically interact with one defined lipid phase, altering its size, distribution, or behavior, and thus cause changes in line tension capable of driving alterations in membrane curvature. Other proteins elicit more direct alterations of membrane curvature, through lipid insertion or scaffolding, but in each example, it is the underlying lipid biophysical properties that drive membrane budding and scission.
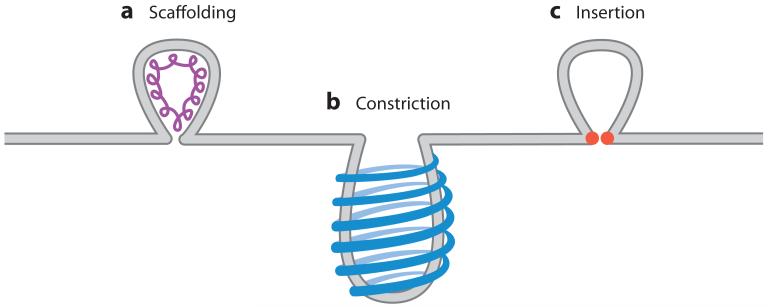
Vast arrays of cellular processes require membrane scission, and as such, several different protein-mediated membrane-scission systems have evolved. These scission processes can be largely grouped into two categories depending on which side of the membrane neck the scission proteins have access to (Figure 3). Many cellular processes involve vesicle budding into the cytoplasmic compartment, either from the plasma membrane or from internal membrane-bound compartments. Vesicle budding into the cytoplasm allows for protein access to the outer leaflet of the membrane neck. At this location, proteins have two main methods of inducing scission: constriction-mediated scission that occurs during endocytosis and insertion-mediated scission that occurs during vesicle budding from the endoplasmic reticulum (ER).
Figure 3.
Examples of the different types of membrane scission. (a) Assembly of scaffolding proteins on the inside of the membrane can deform the membrane sufficiently to cause membrane scission. (b) Constriction is accomplished by the formation of protein rings around the outside of a budding vesicle. Contraction of the rings then constricts the budding neck, which causes scission. (c) Lipid insertion can cause membrane scission after a bud has formed by protein insertion into the inner membrane leaflet (in the case of the influenza virus budding) or the outer membrane leaflet (in the case of cellular budding events) at the neck of the membrane bud. Stacking defects caused by the insertion event then alter membrane curvature, which leads to scission.
In contrast to endocytosis and ER trafficking, the orientation of budding vesicles from the endosome into the multivesicular body (MVB) prevents access of cytoplasmic proteins, such as dynamin, to the outside membrane neck of the budding vesicle (Figure 3). A MVB-localized dynamin homolog conceivably exists; however, none has been found to date. Instead, MVB scission occurs through a scaffolding process whereby a spiral assembly of peripheral membrane proteins pulls the neck of the budding vesicle, decreasing the pore diameter until scission occurs. Though each of these scission reactions progresses through a unique mechanism, they all involve interactions with specific lipid domains, they all exert specific effects on the interacting lipids, and they are all driven by the basic biophysical properties of biological membranes.
MECHANISMS OF MEMBRANE SCISSION
Constriction (Dynamin and Clathrin-Mediated Endocytosis)
Dynamin is a guanosine triphosphate (GTP)-ase that plays an essential role in membrane scission during clathrin-mediated endocytosis. Dynamin assembles into ringlike structures at the necks of budding vesicles (Figure 3). Assembly is thought to trigger GTPase activity, leading to a conformational change in the dynamin ring that constricts the vesicle neck below the critical diameter, which results in membrane scission (Faelber et al. 2012, Hinshaw & Schmid 1995, Warnock & Schmid 1996). Scission occurs at the boundary between the dynamin-coated membrane and the remaining membrane neck, because at this point the strain on the membrane is the greatest and the energy needed to cause scission is minimized (Morlot et al. 2012). The formation of the dynamin ring may also induce lipid-phase segregation at the membrane neck, a phenomenon that would further facilitate the process of membrane scission (Liu et al. 2006). Thus, assembly of the dynamin ring structure directly alters membrane curvature around the membrane neck and facilitates the creation of a high-tension lipid environment capable of causing spontaneous membrane scission.
Lipid Insertion (Arf1/Sar1 and Intracellular Vesicle Transport)
Membrane scission of coat protein complex-I and -II coated vesicles, which mediate vesicular traffic from the Golgi complex and the ER, respectively, is accomplished by the action of the ADP-ribosylation factor (Arf) 1 or the Sar1 protein (Barlowe et al. 1993, Beck et al. 2008, Bielli et al. 2005, Krauss et al. 2008, Lee et al. 2005, Spang 2008). Like dynamin, Arf1 and Sar1 are GTPases that undergo conformational changes upon GTP binding and hydrolysis. However, in contrast to dynamin, GTP binding by Arf1/Sar1 exposes an N-terminal amphipathic helix that binds membranes with high affinity (Beck et al. 2008, Bielli et al. 2005, Krauss et al. 2008, Lee et al. 2005, Seidel et al. 2004). It is thought that Arf1/Sar1 mediates membrane scission through amphipathic helix insertion into the outer leaflet of the lipid bilayer (Figure 3). This causes a stacking defect that results in expansion of the outer leaflet of the bilayer, the induction of membrane curvature, and scission (Sheetz & Singer 1974) (Figure 1).
Scaffolding (ESCRTs and Multivesicular Body Budding)
Budding of endosomes into the MVB, which occurs during endosomal sorting, represents an unusual topological challenge for membrane scission, because cellular scission machinery (such as dynamin) cannot access the site of scission on the outside of the budding vesicle neck. Instead, for MVB budding, membrane scission must occur from the inside of the vesicle neck. Membrane deformation and scission in this case require a cohort of cellular proteins referred to as the endosomal sorting complex required for transport (ESCRT) proteins. Many of these proteins were initially identified in vacuole trafficking in yeast and so were termed vacuolar protein-sorting proteins (VPSs), or charged multivesicular body proteins (CHMPs) in humans (Katzmann et al. 2001). The ESCRT proteins work together in four complexes: ESCRT-0, -I, -II, and -III. The ESCRT-0, -I, and -II complexes are involved in target binding, assembly, alterations of membrane curvature, and the recruitment of the ESCRT-III complex, which is required for membrane scission (for review see Hurley & Hanson 2010, Hurley et al. 2010).
Recent work has demonstrated that the ESCRT-III complex is sufficient to induce membrane scission (Adell & Teis 2011, Guizetti & Gerlich 2012). In these experiments, CHMP4 (Snf7) binds to the inner surface of the membrane neck, polymerizing in a spiral of gradually decreasing diameter (Fabrikant et al. 2009, Wollert et al. 2009). This constricting spiral bends the membrane and constricts the membrane neck below 50 nm. Further constriction could then lead to spontaneous membrane scission (Figure 3). Additionally, binding of CHMP2 (Vps2) and CHMP3 (Vps24) to the tip of the CHMP4 spiral could form a protein dome that may enhance the scaffolding effect, which would further narrow the membrane neck below the critical diameter (<2 nm) and enable membrane scission (Bodon et al. 2011, Fabrikant et al. 2009, Morita et al. 2011). Alternatively, palmitoylation of ESCRT-III components may enable lipid raft recruitment and the bridging of Lo and Ld domains, which would modify line tension and facilitate scission (Guizetti & Gerlich 2012, Hurley et al. 2010).
ESCRT-mediated scission may also require the activity of the Vps4 AAA+ ATPase (Babst et al. 1997, 1998). Vps4 is a dodecamer that assembles into a ring-shaped cylinder at the site of budding, hydrolyzing ATP to facilitate disassembly of the ESCRT complexes (Gonciarz et al. 2008, Lata et al. 2008, Saksena et al. 2009, Scott et al. 2005). Two models have been proposed for the mechanism by which Vps4 catalyzes membrane scission through ESCRT-III disassembly. First, the ESCRT-III subunits could be pulled through the pore of the Vps4 dodecamer, tightening the ring of CHMP4 subunits and constricting the membrane neck until scission occurs (Saksena et al. 2009, Scott et al. 2005). Second, rapid disassembly of the membrane-constraining CHMP3 ring could relax the membrane and trigger scission (Adell & Teis 2011). Alternatively, Vps4 may function only after the completion of membrane scission, disassembling and recycling ESCRT-III complexes so that multiple rounds of budding and scission can occur (Wollert & Hurley 2010).
The budding of endosomes into the multivesicular body is topologically comparable to the budding of enveloped viruses from the plasma membrane into the extracellular space. As such, the ESCRT machinery is co-opted for use by a variety of enveloped viruses. By recruiting the ESCRT machinery to sites of virus budding, it is possible to facilitate viral membrane scission via the ESCRT-III complex. This is one of the most common mechanisms of enveloped virus membrane scission.
OVERVIEW OF VIRUS ASSEMBLY, BUDDING, AND SCISSION
Enveloped virus assembly and budding is a complex, multistep process that begins with the recruitment of viral proteins to the plasma membrane. For several viruses, the binding of matrix proteins to the membrane is sufficient to induce vesiculation; however, other viruses require membrane-binding proteins to either initiate budding or recruit matrix proteins to the membrane, where they then alter membrane curvature. Much of the work investigating the roles of single viral proteins in budding is performed using a virus-like particle (VLP) system, in which cells express the protein(s) of interest and the capacity for VLP budding is assessed. In the VLP system, several viral proteins have been shown to cause membrane budding and scission; however, when investigated in vivo, most of the proteins induce bud formation but do not cause membrane scission (reviewed in Hurley et al. 2010). Thus, in the absence of additional proteins, the budding process stalls, and the budding virion remains attached to the membrane by a small membranous neck. It is at this point that membrane scission is necessary to release the budded virion.
Just as there are a multitude of different protein-based cellular scission mechanisms, viruses employ a variety of mechanisms to accomplish budding. Most viruses do not encode their own scission machinery; thus, recruitment of cellular proteins is an essential step in the completion of viral budding. However, as in MVB budding, the topology of enveloped virus budding prevents the access of many cytoplasmically located cellular scission proteins to the outside neck of the membrane bud. Because of this, the most common cellular scission protein, dynamin, does not appear to be involved in viral budding. Rather, the ESCRT proteins, which assemble on the inner surface of the membrane neck, may represent the standard scission pathway used in enveloped virus budding.
The prototypical example of viral membrane scission is the use of the ESCRT proteins by human immunodeficiency virus (HIV). Numerous other viruses, including many members of the Retroviridae family, exhibit a similar dependency on the host ESCRT proteins for the completion of membrane scission (reviewed in Martin-Serrano & Neil 2011). In addition, recent work has identified ESCRT-independent viruses that use alternate cellular scission machinery for budding, such as Rab11 family-interacting protein 2 (FIP2) (Utley et al. 2008). There is also one report of a virally encoded scission machine, the influenza virus M2 protein (Rossman et al. 2010b). Many of these proteins act at lipid-phase boundaries, and the energy stored as line tension between the two lipid phases may be sufficient to drive virus budding and membrane scission without requiring a dedicated scission factor.
VIRAL MECHANISMS OF MEMBRANE SCISSION
Scaffolding (ESCRT-Mediated Membrane Scission)
The ESCRT system
Several different enveloped viruses can recruit ESCRT components through late-domain motifs typically found in viral matrix proteins and use them to mediate membrane scission. Not surprisingly, there are several different variants of late-domain sequences, each of which mediates recruitment into the ESCRT pathway at a different stage. The P(T/S)AP motif found in HIV mediates binding to TSG101, facilitating recruitment of the ESCRT-I complex (Garrus et al. 2001, Gottlinger et al. 1991). The YPxL motif found in equine infectious anemia virus is responsible for binding to Alix, which bridges the ESCRT-I and ESCRT-III complexes (Fisher et al. 2007, Lee et al. 2007). Viruses containing the PPxY motif, such as Rous sarcoma virus, can bind to Nedd4-like proteins and are recruited into the ESCRT pathway at an as-yet-undefined point upstream of ESCRT-I assembly (Martin-Serrano et al. 2005).
Each of these different late-domain motifs mediates a different ESCRT assembly reaction; however, the end stage of late-domain ESCRT recruitment is uniformly the activation of CHMP4 and Vps4 and the completion of membrane scission, as described above for MVB budding. A wide range of enveloped viruses, including RNA and DNA viruses, use this strategy for membrane scission and viral particle release; however, the best-documented use of ESCRT-mediated scission is that of the Retroviridae family of viruses.
HIV, retrovirus budding, and ESCRT-mediated scission
HIV budding is initiated through membrane binding of the Gag protein. Gag is a polyprotein that encodes the matrix, capsid, nucleocapsid, and p6 proteins, which are not separated until protease cleavage occurs in a post-budding maturation process. During viral assembly and budding, the matrix domain of the Gag polypeptide binds to the membrane (Hill et al. 1996), bringing the capsid domain to the site of virus budding. This enables the capsid to self-assemble into hexamers and induce the formation of a membrane bud that is connected to the plasma membrane by a narrow membrane neck in the range of 100 nm in diameter (Briggs et al. 2009, Morita et al. 2011, von Schwedler et al. 2003, Wright et al. 2007). The matrix and capsid domains cannot sufficiently constrict the membrane neck to cause scission without the additional action of the Gag protein p6 (Kozlovsky & Kozlov 2003). Gag-p6 contains a PTAP motif that serves as a binding and recruitment site for TSG101 and the ESCRT-I complex (Carlson & Hurley 2012, Garrus et al. 2001, Huang et al. 1995). Progression through the ESCRT pathway results in ESCRT-III-mediated membrane scission and the release of the budding virion.
Mutation of the p6 late-domain sequence, or inhibition of Vps4 activity, causes virion budding to stall at a terminal phase in which the viral protein assembly has occurred and a membrane bud has formed. The virion, however, fails to undergo membrane scission and remains attached to the host-cell plasma membrane by a small membrane neck. Through an as-yet-unidentified mechanism, budding is typically reinitiated at the site of failed scission, which results in the formation of a new virion attached to the host-cell plasma membrane, as before, but also attached to the previous virion by another small membrane neck. The repeated reinitiation of budding that occurs in the presence of late-domain or ESCRT defects results in the formation of a canonical scission-defective virion structure referred to as beads on a string (Yuan et al. 2000).
All three late-domain motifs [P(T/S)AP, YPxL, and PPxY] are found throughout the Retroviridae family. Equine infectious anemia virus contains a YPDL motif that can bind Alix and facilitate the assembly of the ESCRT-I and ESCRT-III complexes at the site of virus budding (Puffer et al. 1997, Strack et al. 2003). Another retrovirus, Rous sarcoma virus, contains a PPPPYV variant of the PPxY late-domain motif that facilitates binding to Nedd4-like proteins and recruitment into the early ESCRT complexes (Kikonyogo et al. 2001, Wills et al. 1994). In all cases, the late-domain sequences facilitate the recruitment of the host ESCRT proteins to the site of virus budding, a process that is essential for membrane scission and the release of the budding virus.
Given the essential role of the ESCRT proteins and late-domain interactions in retrovirus budding, it is not surprising that many viruses, including HIV, have evolved multiple late domains to facilitate redundant entry into the ESCRT pathway. The HIV Gag-p6 protein contains a second late domain (YPDL) capable of binding Alix (Strack et al. 2003). Interestingly, the Gag-Alix interaction also requires a second interaction, between Gag-nucleocapsid and the Alix Bro domain; this interaction uses RNA as a bridging factor and is required to enable the direct recruitment of CHMP4 and the mediation of membrane scission after the completion of Gag assembly (Carlson & Hurley 2012, Jouvenet et al. 2011, Sette et al. 2012).
For HIV, as well as for several other enveloped viruses, the combination of PTAP and YPDL late domains provides a redundant mechanism that ensures the ability of HIV to recruit the ESCRT machinery and undergo membrane scission, even if one of the domains undergoes mutation. Alternatively, the two late domains may also function in tandem, resulting in more effective ESCRT recruitment and activation than would be possible with only one late domain (Carlson & Hurley 2012).
Diversity of ESCRT-dependent viruses
ESCRT-dependent budding is not limited to the Retroviridae family; in fact, the three main late-domain sequences described above are found in many different virus families, including DNA viruses of the Poxviridae and Herpesviridae families and RNA viruses of the Rhabdoviridae, Filoviridae, and Paramyxoviridae families, among many others (Crump et al. 2007; Harty et al. 1999, 2000; Honeychurch et al. 2007; Schmitt et al. 2005). ESCRT-complex recruitment and membrane scission in these viruses appear to occur in a manner similar to retrovirus budding, with late-domain sequences mediating binding to designated ESCRT counterparts.
Herpesviridae family members also use the ESCRT system for membrane budding; however, in contrast to HIV budding, the herpes viruses are enveloped in an internal budding process. Human herpes virus 6 is thought to bud from the cytoplasm into the MVB, using host ESCRT proteins at their native location (Calistri et al. 2007, Das & Pellett 2011, Mori et al. 2008). Interestingly, Epstein-Barr virus (EBV) also uses the ESCRT machinery for its internal budding process; however, EBV buds through the nuclear membrane into the cytoplasm. This variant ESCRT-dependent budding requires the relocalization of Alix, CHMP4, and Vps4 to the inner nuclear membrane, a process that is dependent on the viral BFRF1 protein (Lee et al. 2012). Similarly, hepatitis B virus (HBV) uses Nedd4-like proteins to facilitate budding into the ER (Lambert et al. 2007). These budding systems raise the intriguing possibility that the ESCRT proteins can be relocalized to different cellular membranes, allowing for their uses in budding processes that are topologically distinct from MVB budding.
Virus budding and ESCRT-assisted membrane scission
Although ESCRT-mediated viral membrane scission has been demonstrated for many different viruses, there is a growing collection of viruses for which ESCRT involvement is less clear. In these cases, late-domain sequences have been found within viral proteins; however, disruption of these domains, or in some cases inhibition of Vps4 activity, reduces but does not eliminate viral budding. There are several possible explanations for this discrepancy. First, the viruses may contain other variant late-domain sequences, such as the FPIV sequence recently reported for PIV5 (Schmitt et al. 2005), that may compensate if the primary sequence is disrupted. Second, there may be other unknown ESCRT components, pathways, or viral protein substitutes that can function redundantly in certain circumstances and, thus, may compensate when the canonical late-domain sequences are disrupted or the activity of Vps4 is inhibited. This may explain the requirement for late-domain sequences in the budding of a collection of viruses that are insensitive to Vps4 inhibition, including Ebola virus, metapneumovirus (MPV), Newcastle disease virus, Sendai virus, and vesicular stomatitis virus (VSV) (Gosselin-Grenet et al. 2007, Irie et al. 2004, Neumann et al. 2005, Sabo et al. 2011, Schmitt et al. 2005, Shnyrova et al. 2007). Finally, these viruses may use another method of membrane scission in addition to the host ESCRT system, providing a measure of redundancy in the event that one of the scission pathways malfunctions (Figure 4). It is intriguing to speculate that this unidentified, redundant mechanism may be one of the primary scission mechanisms used by ESCRT-independent viruses, such as influenza virus (Rossman et al. 2010b), though further research is necessary to understand why these viruses would exhibit an intermediate level of dependency on the ESCRT system.
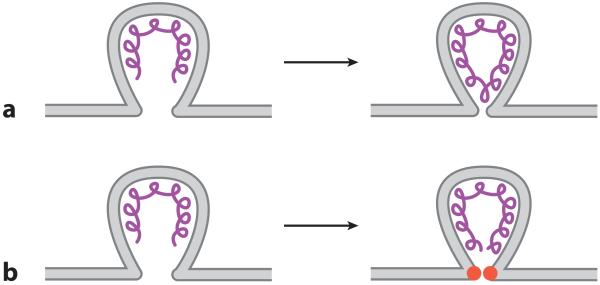
Figure 4.
Capacity for scaffold proteins to cause membrane scission during virus budding. (a) In vitro viral matrix protein assembly can cause both membrane budding and scission; however, (b) in vivo matrix assembly may be sufficient to deform the membrane but could require a dedicated scission protein (red) to mediate membrane scission.
Lipid Insertion (Virus-Mediated Membrane Scission)
Influenza virus M2-mediated membrane scission
Influenza virus is the only virus known to encode its own scission protein (Rossman et al. 2010b). The budding of influenza viruses is ESCRT independent (Bruce et al. 2009, Watanabe & Lamb 2010) and, in a VLP system, can be mediated by each of several different viral proteins, though in vivo budding appears to be more complex. Virus budding is thought to begin with concentration of the viral glycoproteins hemagglutinin and neuraminidase in lipid raft domains, a process that may be sufficient to alter membrane curvature (Leser & Lamb 2005, Rossman & Lamb 2011, Schmitt & Lamb 2005, Takeda et al. 2003). In fact, hemagglutinin has been shown to mediate VLP budding by itself, which demonstrates its intrinsic membrane curvature–altering capacity (Chen et al. 2007). Previous results have also suggested that the influenza virus M1 protein is sufficient for budding, despite the absence of any late-domain sequences (Gómez-Puertas et al. 2000). However, further studies have shown that the capacity of M1 to mediate budding requires membrane anchoring and the presence of the M2 protein (Chen et al. 2007, Wang et al. 2010). During virus budding, M1 may act as a scaffolding protein, providing a docking site for the influenza virus scission protein M2 (Chen et al. 2008; Rossman et al. 2010a,b; Rossman & Lamb 2011) (Figure 4).
M2 is a homotetrameric ion-channel protein, the activity of which is essential for viral uncoating during cellular entry (Lamb et al. 1994). Recent work has shown that the M2 cytoplasmic tail contains two essential domains: a membrane-proximal amphipathic helix and an M1-binding motif (Chen et al. 2008, McCown & Pekosz 2006, Rossman et al. 2010a). Binding to the M1 protein is thought to mediate M2 recruitment to the sites of virus budding, where the amphipathic helix is able to alter membrane curvature, which causes membrane scission and the release of budding virions (Chen et al. 2008; Rossman et al. 2010a,b; Rossman & Lamb 2011).
Interestingly, M2-mediated scission appears to be regulated by membrane cholesterol. M2 can bind cholesterol through its amphipathic helix, though when expressed in the absence of any other viral protein, M2 is located in a Ld lipid phase and binds only minimal amounts of cholesterol (Leser & Lamb 2005, Schroeder et al. 2005, Thaa et al. 2011). During viral infection, M2 is recruited to the Lo sites of virus budding through association with the M1 protein, which places M2 in a cholesterol-rich environment at the boundary between the Ld and Lo lipid phases, a property that may explain its poor incorporation into budding virions (Nayak et al. 2009, Rossman et al. 2010a, Schroeder 2010, Schroeder et al. 2005, Thaa et al. 2011). M2 may form a ring around the Lo site of virus budding that is progressively constricted as the virus assembles (Rossman et al. 2010b, Rossman & Lamb 2011, Schroeder 2010). As the Lo phase is incorporated into the budding virion, the M2 ring is proposed to constrict, forming a small focus of M2 at the neck of the budding virion, a site that would contain lower levels of cholesterol then the virion itself (Rossman et al. 2010b, Rossman & Lamb 2011). In this low-cholesterol Ld environment, the M2 amphipathic helix may insert deeply into the membrane, which would cause a pronounced lipid-stacking defect that induces positive membrane curvature at the neck of the budding virion, reduce the diameter of the membrane neck below the critical threshold, and allow scission to occur (Rossman et al. 2010b). Mutation of the M2 amphipathic helix results in virions stalling in the final stages of budding, which results in the characteristic beads-on-a-string morphology indicative of a failure to undergo membrane scission (Rossman et al. 2010b).
The action of M2 in mediating membrane scission may be more complex than originally thought. Mutation of the putative M2 cholesterol-binding site affected protein localization but did not affect the ability of the influenza virus to bud (Thaa et al. 2011, 2012), and individual mutations within the amphipathic helix had only moderate effects on virus replication (Rossman et al. 2010a, Stewart & Pekosz 2011). The reason for this effect is unclear; however, M2 may associate with Lo membranes through multiple protein domains, and its scission activity may be mediated through both direct alteration of membrane curvature (via amphipathic helix insertion) and modulation of lipid-phase segregation. This hypothesis is supported by a recent structural study that showed that the M2 amphipathic helix binds more strongly to regions of high membrane curvature, such as the neck of budding virions, and that its ability to alter membrane curvature depends on the presence of raft-type lipids but not necessarily cholesterol (Wang et al. 2012). These authors also speculated that the alteration of membrane curvature by the M2 protein will force further lipid-phase segregation, a process that may enhance curvature at the membrane neck and facilitate membrane scission.
Rab11-FIP-mediated membrane scission
Rab11 is a small GTP-binding protein that plays numerous important roles in cellular membrane trafficking, including plasma membrane recycling and cytokinesis (reviewed in Prekeris 2003). The many activities of Rab11 are mediated through specific interactions with the Rab11 family–interacting proteins (FIPs). The FIPs interact with Rab11 through an amphipathic helix domain in their C terminus, termed the Rab11-binding domain (Hales et al. 2001, Prekeris et al. 2001). Recent results suggest that Rab11-FIP2 may play an essential role in mediating membrane scission during the budding of respiratory syncytial virus(RSV) (Utley et al. 2008), which is the first indication that Rab11-FIP2 may be involved in scission reactions.
Similar to PIV5, the budding of RSV appears to require multiple viral proteins, though no late-domain motifs have yet been uncovered within RSV protein sequences and virus budding is ESCRT independent (Utley et al. 2008). Interestingly, RSV budding required the activity of Rab11-FIP2 for the efficient release of viral particles, an activity that depended on its N-terminal C2 lipid-binding domain. Mutation of this domain resulted in an inability to trigger membrane scission and release budding virions (Lindsay & McCaffrey 2004, Utley et al. 2008). However, a capacity to alter membrane curvature has not yet been demonstrated for Rab11-FIP2, and thus the mechanism of action remains unknown. Interestingly, budding of the closely related avian and human MPV is also ESCRT independent (Sabo et al. 2011, Weng et al. 2011), though no dependence on Rab11-FIP2 has yet been found for MPV.
Both Rab11 and Rab11-FIP3 have also been implicated in the assembly and budding of influenza viruses (Bruce et al. 2010). Disruption of Rab11-FIP3 caused defects in virus assembly, whereas disruption of Rab11 itself significantly reduced viral particle release and appeared to inhibit membrane scission (Bruce et al. 2010). Inhibition of the receptor for activated C kinase (RACK)-1 protein also appeared to inhibit influenza virus membrane scission (Demirov et al. 2012). Rab11, Rab11-FIP3, and RACK-1 are all involved in other cellular scission reactions, specifically abscission during cellular cytokinesis. During abscission, however, the proteins mediate vesicle transport from the apical recycling endosome to the cleavage furrow and do not directly mediate alternations of membrane curvature (reviewed in Ai & Skop 2009, Simon & Prekeris 2008). Recent results have shown that Rab11 facilitates the transport of viral proteins and nucleocapsids to the plasma membrane during the assembly and budding of influenza virus, hantaviruses, and paramyxoviruses (Amorim et al. 2011, Chambers & Takimoto 2010, Eisfeld et al. 2011, Momose et al. 2011, Rowe et al. 2008). This raises the intriguing possibility that Rab11, Rab11-FIP3, and RACK-1 do not directly affect membrane scission but rather play an essential role in delivering necessary components to the sites of virus budding, whether proteins for viral assembly, as in the case of hantavirus, or components for membrane scission, as in the case of RSV and influenza virus. Further studies are necessary to determine the direct effects of these proteins on viral assembly and scission.
UNANSWERED QUESTIONS IN VIRAL MEMBRANE SCISSION
Despite the extensive amount of research investigating virus budding and membrane scission, there are many enveloped viruses whose mechanism of scission remains unknown. Several of these viruses, such as MPV, VSV, Sendai virus, and Newcastle disease virus, are known to bud through an ESCRT-independent mechanism (Gosselin-Grenet et al. 2007, Irie et al. 2004, Shnyrova et al. 2007, Weng et al. 2011; reviewed in Chen & Lamb 2008). It is intriguing to speculate about the mechanism by which these viruses affect membrane scission. Are there late-domain-type sequences that can be indentified for binding to the Rab11-FIP2 machinery? Do any of these viruses encode proteins that are known to modify the lipid environment or bridge lipid domains, and so facilitate lipid-mediated scission? Are there other viral proteins that directly interact with membranes and mediate membrane scission, like the influenza virus M2 protein? One of the most intriguing possibilities is that there may be other scission machinery at work in the cell. Given the myriad of essential scission processes that occur within the cell, there are likely additional scission factors that have not yet been discovered. However, given the topological problem of mediating scission from the inside of the budding vesicle, the number of cellular scission factors capable of mediating viral budding may be limited.
Two unusual examples of viral use of cellular scission proteins have recently been reported. One study examining the budding of ESCRT-dependent HBV showed the existence of an unusual ESCRT-type budding pathway (Bardens et al. 2011). Although enveloped HBV uses Nedd4-like proteins to facilitate ESCRT-dependent scission (Lambert et al. 2007), HBV can also bud naked capsids by using only part of the ESCRT machinery (Bardens et al. 2011). Capsid budding requires the ESCRT protein Alix but is not sensitive to Vps4 inhibition. This result is intriguing, because many studies establish the ESCRT independence of virus budding by inhibiting Vps4. There may be other ESCRT-mediated pathways leading to viral membrane scission that do not require the activity of Vps4, and thus have been overlooked, though HBV capsid budding is the only documented example of this putative alternate ESCRT pathway.
Another study of VSV budding has determined that dynamin is involved in the scission process (Raux et al. 2010). The authors found that dynamin interacts with the VSV matrix (M) protein and that VSV membrane scission is sensitive to dynamin inhibition. It is not clear if dynamin is acting directly to mediate membrane scission and, if so, how it functions on the opposite side of the membrane from its typical site of action. However, given that dynamin is one of the primary players in cellular membrane scission, it is not surprising that some viruses may have found a way to use this protein to affect their own budding.
The budding and membrane scission of VSV virions may also involve a lipid-mediated mechanism. The VSV M protein appears capable of driving membrane budding, though efficient virus budding only occurs when the VSV glycoprotein (G) is also expressed (Mebatsion et al. 1996, Schnell et al. 1998). The VSV G and M proteins localize to different lipid domains on the surface of infected cells (Swinteck & Lyles 2008), which raises the possibility that the interaction between G and M proteins alters phase segregation and line tension at the sites of virus budding, facilitating membrane scission. This activity could provide a redundant mechanism for membrane scission as well as an additional explanation for why VSV contains late-domain sequences but does not require ESCRT activity for membrane scission. The modification of lipid domains and use of line tension energy to drive membrane scission may also function in the budding of viruses with known mechanisms of scission.
Influenza virus assembly occurs at Lo-raft-type plasma membrane domains, and the expression of hemagglutinin and neuraminidase causes a coalescence of these domains into larger lipid domains referred to as a barge of rafts (Leser & Lamb 2005, Schmitt & Lamb 2005). The recruitment of Ld-localized M2 tetramers to the Lo site of virus budding may serve to locally enhance the line tension between the Lo and Ld phases of the plasma membrane. Thus, M2 alterations of membrane curvature may exploit an existing propensity for curvature, which could explain the energetics of the membrane-scission reaction elicited by the M2 protein or could serve as a redundant minimal mechanism for scission (Rossman et al. 2010b). HIV also buds from lipid raft domains, and the expression of Env causes a coalescence of lipid rafts similar to that observed during influenza virus budding (Hogue et al. 2011, Waheed & Freed 2009). This coalescence of lipid domains may serve to increase the local line tension between the two lipid phases at the site of virus budding, facilitating ESCRT-dependent membrane scission through modification of line tension, as has been proposed for ESCRT-dependent MVB budding (Hurley et al. 2010).
The modification of line tension to drive membrane scission may be a common feature in the activity of any putative, virally encoded scission protein, similarly to the influenza virus M2 protein. The ESCRT-independent alphaviruses encode a 6K protein that contains a palmitoylated amphipathic helix, which increases the efficiency of budding but is not specifically required (Liljeström et al. 1991, Sanz et al. 2003, Taylor et al. 2007). Although the mechanism of alphavirus membrane scission is currently unknown, it is intriguing to speculate that scission may be facilitated by the association of 6K with lipid-raft-type membrane domains, as seen with the influenza virus M2 protein and the VSV G protein (Rossman et al. 2010b, Yao et al. 1996). This may cause similar lipid-stacking defects or alterations in lipid-phase segregation and may provide the energy, if not the mechanism, for membrane scission. Future work will undoubtedly elucidate the mechanism of scission for many of these ESCRT-independent viruses. It is hoped that further discoveries of novel scission mechanisms will lead to a better understanding of the process of scission itself and the ways in which viruses use these different mechanisms to achieve a similar goal.
CONCLUSIONS
Proteins of the ESCRT-III complex (Wollert et al. 2009), as well as a multitude of different viral proteins (Chen et al. 2007, Fang et al. 2007, Gómez-Puertas et al. 2000, Rossman et al. 2010b, Shnyrova et al. 2007), can cause membrane budding in vitro without requiring any other factors. This indicates that the proteins can alter membrane curvature sufficiently to cause the formation of a membrane bud and are further able to trigger scission. These results are surprising, because in vivo budding appears to require a cohort of proteins, each of which mediates a single step of the budding and scission process. Thus, these results call into question the validity of in vitro budding model systems and pose the question of how virus assembly affects the ability of different proteins to cause membrane scission.
It has been shown that the assembly of curvature-inducing proteins on a lipid bilayer generates an attractive force between the protein scaffolding components, which causes further scaffold assembly, membrane curvature, and scission (Reynwar et al. 2007). However, at biologically relevant attractive forces, budding may be initiated, but there would be insufficient force to complete the scission process (Ruiz-Herrero et al. 2012, Smith et al. 2007). In this case, the completion of scission may require an additive force, either a dedicated scission factor, a force-generating machine such as the cytoskeleton, or line tension developed through lipid-phase segregation (Figure 4). The requirement for this additive force may represent a regulatory step, ensuring that proper viral assembly has occurred before membrane scission is initiated.
These observations raise the question of what the exact contributions of viral proteins are to the membrane budding process and whether the role of scission proteins is to modify the lipid environment, using lipid biophysical properties to drive the membrane scission reaction. Thus, there is likely a broad range of scission proteins for both cellular processes and viral budding, proteins that act by constriction, by membrane insertion, by scaffolding, or by clustering different lipids or lipid domains. Further studies will likely reveal the effects of line tension on membrane scission, the prevalence of line tension alterations during membrane scission, and the mechanism by which viral proteins can alter and use line-tension energy.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adell MA, Teis D. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 2011;585:3191–96. doi: 10.1016/j.febslet.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai E, Skop AR. Endosomal recycling regulation during cytokinesis. Commun. Integr. Biol. 2009;2:444–47. doi: 10.4161/cib.2.5.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim MJ, Bruce EA, Read EK, Foeglein A, Mahen R, et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 2011;85:4143–56. doi: 10.1128/JVI.02606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–31. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–93. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardens A, Doring T, Stieler J, Prange R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell. Microbiol. 2011;13:602–19. doi: 10.1111/j.1462-5822.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, d’Enfert C, Schekman R. Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 1993;268:873–79. [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, Rutz C, Bassler J, et al. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. USA. 2008;105:11731–36. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J. Cell Biol. 2005;171:919–24. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof AA, Wilke N. Molecular determinants for the line tension of coexisting liquid phases in monolayers. Chem. Phys. Lipids. 2012;165:737–44. doi: 10.1016/j.chemphyslip.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J. Biol. Chem. 2011;286:40276–86. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. USA. 2009;106:11090–95. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010;84:5848–59. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, et al. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–78. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Calistri A, Sette P, Salata C, Cancellotti E, Forghieri C, et al. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 2007;81:11468–78. doi: 10.1128/JVI.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, Malhotra V. Membrane fission: the biogenesis of transport carriers. Annu. Rev. Biochem. 2012;81:407–27. doi: 10.1146/annurev-biochem-051710-094912. [DOI] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl. Acad. Sci. USA. 2012;109:16928–33. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PLoS ONE. 2010;5:e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–32. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008;82:10059–70. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007;81:7111–23. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Yates C, Minson T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 2007;81:7380–87. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Pellett PE. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 2011;85:5864–79. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov D, Gabriel G, Schneider C, Hohenberg H, Ludwig S. Interaction of influenza A virus matrix protein with RACK1 is required for virus release. Cell. Microbiol. 2012;14:774–89. doi: 10.1111/j.1462-5822.2012.01759.x. [DOI] [PubMed] [Google Scholar]
- Döbereiner HG, Käs J, Noppl D, Sprenger I, Sackmann E. Budding and fission of vesicles. Biophys. J. 1993;65:1396–403. doi: 10.1016/S0006-3495(93)81203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 2011;85:6117–26. doi: 10.1128/JVI.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Held M, Gao S, Posor Y, Haucke V, et al. Structural insights into dynamin-mediated membrane fission. Structure. 2012;20:1621–28. doi: 10.1016/j.str.2012.08.028. [DOI] [PubMed] [Google Scholar]
- Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung H-Y, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–52. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- García-Sáez AJ, Chiantia S, Schwille P. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 2007;282:33537–44. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gómez-Puertas P, Albo C, Pérez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 2000;74:11538–47. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, et al. Biochemical and structural studies of yeast Vps4 oligomerization. J. Mol. Biol. 2008;384:878–95. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin-Grenet A-S, Marq J-B, Abrami L, Garcin D, Roux L. Sendai virus budding in the course of an infection does not require Alix and VPS4A host factors. Virology. 2007;365:101–12. doi: 10.1016/j.virol.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA. 1991;88:3195–99. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–40. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, et al. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001;276:39067–75. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA. 2000;97:13871–76. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 1999;73:2921–29. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA. 1996;93:3099–104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–92. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 2011;85:9749–66. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeychurch KM, Yang G, Jordan R, Hruby DE. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J. Virol. 2007;81:7310–15. doi: 10.1128/JVI.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–18. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell. 2010;143:875–87. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–66. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 2004;78:2657–65. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–55. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, et al. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA. 2001;98:11199–204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky Y, Kozlov MM. Membrane fission: model for intermediate structures. Biophys. J. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Jia JY, Roux A, Beck R, Wieland FT, et al. Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J. Biol. Chem. 2008;283:27717–23. doi: 10.1074/jbc.M804528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Holsinger LJ, Pinto LH. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Receptor-Mediated Virus Entry into Cells. Cold Spring Harb. Lab.; Cold Spring Harbor, NY: 1994. pp. 303–21. [Google Scholar]
- Lambert C, Doring T, Prange R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and γ2-adaptin. J. Virol. 2007;81:9050–60. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laradji M, Sunil Kumar PB. Domain growth, budding, and fission in phase-separating self-assembled fluid bilayers. J. Chem. Phys. 2005;123:224902. doi: 10.1063/1.2102894. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–57. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Liu PT, Kung HN, Su MT, Chua HH, et al. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–17. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–99. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–27. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 1991;65:4107–13. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 2004;117:4365–75. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lipowsky R. Domain-induced budding of fluid membranes. Biophys. J. 1993;64:1133–38. doi: 10.1016/S0006-3495(93)81479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl. Acad. Sci. USA. 2006;103:10277–82. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011;9:519–31. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 2006;80:8178–89. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T, Konig M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–51. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- Minami A, Yamada K. Domain-induced budding in buckling membranes. Eur. Phys. J. E. 2007;23:367–74. doi: 10.1140/epje/i2006-10198-5. [DOI] [PubMed] [Google Scholar]
- Momose F, Sekimoto T, Ohkura T, Jo S, Kawaguchi A, et al. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS ONE. 2011;6:e21123. doi: 10.1371/journal.pone.0021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, et al. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic. 2008;9:1728–42. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–42. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot S, Galli V, Klein M, Chiaruttini N, Manzi J, et al. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–29. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–61. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Ebihara H, Takada A, Noda T, Kobasa D, et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J. Virol. 2005;79:10300–7. doi: 10.1128/JVI.79.16.10300-10307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006;47:1597–98. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- Prekeris R. Rabs, Rips, FIPs, and endocytic membrane traffic. Sci. World J. 2003;3:870–80. doi: 10.1100/tsw.2003.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R, Davies JM, Scheller RH. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–70. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 1997;71:6541–46. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux H, Obiang L, Richard N, Harper F, Blondel D, Gaudin Y. The matrix protein of vesicular stomatitis virus binds dynamin for efficient viral assembly. J. Virol. 2010;84:12609–18. doi: 10.1128/JVI.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–64. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010a;84:5078–88. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. The influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010b;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–36. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RK, Suszko JW, Pekosz A. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology. 2008;382:239–49. doi: 10.1016/j.virol.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrero T, Velasco E, Hagan MF. Mechanisms of budding of nanoscale particles through lipid bilayers. J. Phys. Chem. B. 2012;116:9595–603. doi: 10.1021/jp301601g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo Y, Ehrlich M, Bacharach E. The conserved YAGL motif in human metapneumovirus is required for higher-order cellular assemblies of the matrix protein and for virion production. J. Virol. 2011;85:6594–609. doi: 10.1128/JVI.02694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E, Feder T. Budding, fission and domain formation in mixed lipid vesicles induced by lateral phase separation and macromolecular condensation. Mol. Membr. Biol. 1995;12:21–28. doi: 10.3109/09687689509038491. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Madan V, Carrasco L, Nieva JL. Interfacial domains in Sindbis virus 6K protein. Detection and functional characterization. J. Biol. Chem. 2003;278:2051–57. doi: 10.1074/jbc.M206611200. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 2005;79:2988–97. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Buonocore L, Boritz E, Ghosh HP, Chernish R, Rose JK. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–96. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C. Cholesterol-binding viral proteins in virus entry and morphogenesis. Subcell. Biochem. 2010;51:77–108. doi: 10.1007/978-90-481-8622-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C, Heider H, Moncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–69. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel RD, III, Amor JC, Kahn RA, Prestegard JH. Conformational changes in human Arf1 on nucleotide exchange and deletion of membrane-binding elements. J. Biol. Chem. 2004;279:48307–18. doi: 10.1074/jbc.M402109200. [DOI] [PubMed] [Google Scholar]
- Sette P, Dussupt V, Bouamr F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J. Virol. 2012;86:11608–15. doi: 10.1128/JVI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc. Natl. Acad. Sci. USA. 1974;71:4457–61. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyrova AV, Ayllon J, Mikhalyov II, Villar E, Zimmerberg J, Frolov VA. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J. Cell Biol. 2007;179:627–33. doi: 10.1083/jcb.200705062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GC, Prekeris R. The role of FIP3-dependent endosome transport during cytokinesis. Commun. Integr. Biol. 2008;1:132–33. doi: 10.4161/cib.1.2.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA, Jasnow D, Balazs AC. Designing synthetic vesicles that engulf nanoscopic particles. J. Chem. Phys. 2007;127:084703. doi: 10.1063/1.2766953. [DOI] [PubMed] [Google Scholar]
- Spang A. The life cycle of a transport vesicle. Cell. Mol. Life Sci. 2008;65:2781–89. doi: 10.1007/s00018-008-8349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SM, Pekosz A. Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication. J. Virol. 2011;85:12179–87. doi: 10.1128/JVI.05970-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–99. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Swinteck BD, Lyles DS. Plasma membrane microdomains containing vesicular stomatitis virus M protein are separate from microdomains containing G protein and nucleocapsids. J. Virol. 2008;82:5536–47. doi: 10.1128/JVI.02407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA. 2003;100:14610–17. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 2007;81:13631–39. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaa B, Levental I, Herrmann A, Veit M. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem. J. 2011;437:389–97. doi: 10.1042/BJ20110706. [DOI] [PubMed] [Google Scholar]
- Thaa B, Tielesch C, Moller L, Schmitt AO, Wolff T, et al. Growth of influenza A virus is not impeded by simultaneous removal of the cholesterol-binding and acylation sites in the M2 protein. J. Gen. Virol. 2012;93:282–92. doi: 10.1099/vir.0.038554-0. [DOI] [PubMed] [Google Scholar]
- Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, et al. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc. Natl. Acad. Sci. USA. 2008;105:10209–14. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, et al. The protein network of HIV budding. Cell. 2003;114:701–13. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–76. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harmon A, Jin J, Francis DH, Christopher-Hennings J, et al. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J. Virol. 2010;84:4673–81. doi: 10.1128/JVI.02306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cady SD, Hong M. NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide. Biophys. J. 2012;102:787–94. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Schmid SL. Dynamin GTPase, a force-generating molecular switch. Bioessays. 1996;18:885–93. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Lamb RA. Influenza virus budding does not require a functional AAA+ ATPase, VPS4. Virus Res. 2010;151:58–63. doi: 10.1016/j.virusres.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Lu W, Harmon A, Xiang X, Deng Q, et al. The cellular endosomal sorting complex required for transport pathway is not involved in avian metapneumovirus budding in a virus-like-particle expression system. J. Gen. Virol. 2011;92:1205–13. doi: 10.1099/vir.0.029306-0. [DOI] [PubMed] [Google Scholar]
- Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 1994;68:6605–18. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–69. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–77. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–26. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JS, Strauss EG, Strauss JH. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol. 1996;70:7910–20. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 2000;74:7250–60. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]