Abstract
Purpose of review
Myocardial injury and disease often results in heart failure, the leading cause of death worldwide. To achieve myocardial regeneration and foster development of efficient therapeutics for cardiac injury, it is essential to uncover molecular mechanisms that will promote myocardial regeneration. In this review, we examine the latest progress made in elucidation of the roles of small non coding RNAs called microRNAs (miRs) in myocardial regeneration.
Recent findings
Promising progress has been made in studying cardiac regeneration. Several miRs, which includes miR-590, miR-199a, miR-17-92 cluster, miR-199a-214 cluster, miR-34a, and miR-15 family, have been recently shown to play an essential role in myocardial regeneration by regulating different processes during cardiac repair, including cell death, proliferation and metabolism. For example, miR-590 promotes cardiac regeneration through activating cardiomyocytes proliferation, while miR-34a inhibits cardiac repair through inducing apoptosis.
Summary
These recent findings shed new light on our understanding of myocardial regeneration and suggest potential novel therapeutic targets to treat cardiac disease.
Keywords: MicroRNA, cardiomyocyte, cardiac injury, regeneration
1. Introduction
Compromised myocardial function with heart failure is the worldwide leading cause of morbidity and mortality [1,2]. Heart transplantation remains the most effective treatment strategy for end-stage heart failure but can barely meet the increasing global demand because of the scarcity of donor hearts[3]. Unlike amphibians and fish [4-8], mammalian cardiomyocytes have limited renewal capacity compromising the ability of mammalian hearts to efficiently repair after injury [9-13]. Because of these limitations, researchers are constantly pursuing novel approaches towards therapies that could ameliorate heart failure. Currently, there is no available approach to reverse the loss of functional myocardium [14]. Efficacious regenerative therapeutics to reverse the progress of heart failure has become an urgent and critical goal of modern cardiovascular research.
Considerable effort has been extended to develop therapies based on transplantation of stem cells or different types of progenitor cells to help a failing heart repopulate with newly made cardiomyocytes [15-24]. An alternative and promising cell-free tactic is to use small molecules or paracrine factors to stimulate cardiomyocyte proliferation or differentiation of resident cardiac progenitor cells [25-31]. Recent progress made in reprogramming and trans-differentiation of non-cardiomyocytes also shows great promise to advance cardiac regeneration [32-45]. Moreover, additional investigations aim to dissect signaling pathways regulating endogenous cardiomyocytes regenerative capacity [46-51].
MicroRNAs (miRs) are small endogenous non-coding single-stranded RNAs that function in biologic processes primarily via post-transcriptional gene silencing in diverse organisms [52,53]. MiRs generally repress target gene expression by promoting mRNA degradation and/or inhibiting translation. Genes targeted by a miR have conserved Watson–Crick base pairing to the miR “seed” site, which is centered at the 5′end of the miR[54,55]. The essential roles of miRs have been shown in regeneration of different tissues and organs. For example, miR-133 promotes appendage regeneration in zebra fish [56], while miR-206 promotes the regeneration of neuromuscular synapses[57] and skeletal muscle[58] in mice. Though the mechanisms remain largely unknown, miRs have been shown to have critical functions in cardiac regeneration. For example, previous studies showed a combination of miRs (miR-1, miR-133, miR-208 and miR-499) has the capability of reprogramming cardiac fibroblasts into cardiomyocytes [40]. Previous reviews have summarized the previous progress made in studying reprograming and regenerating cardiac tissue, including critical miRs involved in cardiac development and homeostasis [59-69]. Here, we summarize the most recent investigations into the function of miRs in myocardial regeneration.
2. miRs play an essential role in myocardial regeneration
has-miR-590 and has-miR-199a
To identify miRs that function in cardiomyocytes proliferation, a recent study cultured neonatal rat ventricular cardiomyocytes and transfected them with a library of 875 human miR mimics in a high-throughput screening approach [70]. Based on that screening, 204 miRs significantly increased neonatal rat cardiomyocyte proliferation and 40 miRs from the original 204 miRs also increased cytokinesis and karyokinesis in neonatal mouse cardiomyocytes[70]. Among the 40 miRs, hsa-miR-590-3p, hsa-miR-199a-3p, hsa-miR-33b and hsa-miR-1825 can significantly increase the proliferation of 7-day postnatal cardiomyocyte, and even more remarkably, hsa-miR-590-3p and hsa-miR-199a-3p can significantly increase 2 month adult cardiomyocyte proliferation [70]. Hsa-miR-33b has been previously shown to have a role in regulating cell proliferation and fatty acid metabolism [71-73] while hsa-miR-1825 function was previously unclear. This study chose to focus on hsa-miR-590-3p and hsa-miR-199a-3p for further miR targets studies, given that these two miRs were the most effective at promoting proliferation in rat and mouse cardiomyocytes.
The miR target genes controlling cardiomyocytes proliferation were globally identified using a combined RNA deep-sequencing and siRNA screening approach [70]. The authors were able to identify three targets, Homer protein homolog 1 (Homer1), Homeodomain-only protein x (Hopx) and Chloride intracellular channel protein 5 (Clic5), that are miR regulated and also modulate cardiomyocyte cell proliferation. Further luciferase reporter assays indicated Homer1 and Hopx are targeted by both hsa-miR-590-3p and hsa-miR-199a-3p, while Clic5 is only targeted by hsa-miR-590-3p. Homer1 previously has been shown to interact with ryanodin receptor (RyR) to control intracellular calcium signaling and with PI3 kinase to prevent cell apoptosis [74-77]. Hopx, an atypical homeodomain-protein, regulates proliferation and differentiation of different cell types, including cadiomyocyte proliferation by modulating Gata4 acetylation and SRF-dependent gene expression [78-82]. Hopx is expressed in both embryonic and postnatal cardiomyocytes and was found to be significantly reduced in both human and mouse hearts with heart failure [78,79,83].
Consistent with the in vitro data, in vivo analysis using synthetic miRs indicated that overexpression of hsa-miR-590-3p and hsa-miR-199a-3p increased cardiomyocyte proliferation in neonatal mice [70]. After myocardial infarction (MI), mouse hearts transduced with AAV9-miR-590-3p and AAV9-miR-199a-3p had improved cardiac function and reduced fibrotic scar size compared to controls [70]. Together, this study suggested that hsa-miR-590-3p and hsa-miR-199a-3p can stimulate cardiac regeneration by promoting mature cardiomyocytes to re-enter cell cycle and progress through mitosis [70].
miR-199a-214 cluster
In addition to the work investigating miR-199a discussed above, other studies indicated that miR-199a repressed hypoxia-inducible factor-1alpha and Sirtuin 1 [84], as well as, the ubiquitin-proteasome system [85] in mouse hearts. Meanwhile, miR-199a was modulated by high glucose and hypoxia in heart failure patients [86] and miR-199a-214 cluster was down-regulated in explanted cardiac tissue from patients with dilated cardiomyopathy [87]. A recent study indicated that miR-199a-214 is cluster involved in heart failure by facilitating a cardiac metabolic shift from predominantly fatty acid utilization in healthy myocardium toward increased glucose metabolism in failing hearts [88]. Using a cardiac disease mouse model with transverse aortic constriction (TAC) pressure overload, the authors found that mice treated with antagomirs of miR-199a and miR-214 had improved cardiac function as well as normal arrangement of cardiomyocytes, significantly reduced cardiac fibrosis and hypertrophy, while vehicle-treated control hearts had impaired cardiac function and displayed cardiomyocyte disarray, interstitial fibrosis and hypertrophied myofibers [88]. The mechanistic studies indicated that both miR-199a and miR-214 directly repressed PPARδ, a critical regulator of mitochondrial fatty acid metabolism in heart, but didn't alter expression of genes involved in glucose metabolism [88].
miR-17-92 cluster
The miR-17-92 cluster encodes six polycistronic miRs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a miR-92a), of which some miRs have the same seed site. MiR-17-92 germ-line loss-of-function resulted in abnormal myocardial differentiation from second heart field cardiac progenitors, by repressing the cardiac progenitor gene Isl1 during embryonic cardiac development [89]. MiR-17-92 also represses T-box genes during cardiac and craniofacial morphogenesis [89,90]. MiR-92a inhibits endothelial cell migration and angiogenesis in adult mice, while inhibition of miR-92a improves heart function and angiogenesis after MI or vascular injury [91,92]. A recent study reported that miR-17-92, particularly miR-19 as a key component, can induce proliferation of cardiomyocytes and help protect the heart from ischemic injury caused by MI [93]. Compared with controls, proliferation of cardiomyocytes was decreased in miR-17-92 loss-of-function hearts and increased in miR-17-92 gain-of-function hearts. Importantly, after MI, miR-17-92 gain-of-function hearts had improved cardiac function, reduced scar size and more proliferating cardiomyocytes at the border zone when compared with controls, suggesting an essential role in cardiac regeneration. In vitro analysis suggests that miR-17-92 induces cardiomyocyte proliferation through direct repression of PTEN by miR-19, a tumor suppressor previously shown to be a miR-17-92 target in tumorgenesis [94].
miR-34a
Aging is a critical risk factor for heart diseases and old patients with cardiac injury usually have worse outcome than young patients [95]. Compared to young mice, the aged mouse heart has increased cardiomyocyte apoptosis, fibrosis and hypertrophy, with decreased telomere length [96]. The expression levels of miR-34a, which previously had been shown as a regulator in apoptosis and senescence [97-101], were higher in older human and mouse hearts compared to young hearts [96]. In vitro data indicated this age induced miR promoted H2O2-induced apoptosis in rat neonatal cardiomyocytes [96]. In vivo assays using miR-34a antagomir treatment further indicated miR-34a induced cell death [96]. Moreover, miR-34a knock-out mice had less cell death and hypertrophy, as well as better cardiac contractile function compared to wild-type mice [96]. Notably, after acute MI, miR-34a expression was significantly increased at the border zone and treatment with miR-34a antagomirs or locked nucleic acid (LNA) based anti-miRs significantly improved cardiac function [96]. A key target of miR-34a identified in this study is Pnuts (also known as Ppp1r10), which is a gene previously reported in modulating apoptosis, telomere shortening and DNA repair [102].
miR-15 family
Mouse neonatal hearts can regenerate after injury, but this ability is gradually lost by postnatal day (P) 7 [49]. The expression levels of miR-15, miR-30 and let-7 families were increased in P10 compared to P1 mouse heart, suggesting a functional role in the transition to non-regenerative myocardium [103]. Transfection data in rat cardiomyocytes indicated that miR-518 and miR-302 family promoted cardiomyocyte proliferation while Let-7 and miR-15 family inhibited proliferation [70].
The miR-15 family, consisting of 6 closely related miRs (miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497), was up-regulated in different heart diseases [104]. Cardiac-specific overexpression of miR-195 (MYH7-miR-195 TG) resulted in premature cell cycle arrest at G2 phase leading to reduced heart size and congenital heart abnormalities such as ventricular hypoplasia and ventricular septal defects [103]. Mouse hearts treated with LNA anti-miR15 to inhibit miR-15 had reduced infarct size and enhanced cardiac function after ischemia reperfusion surgery [105]. Wild type (WT) P1 mouse hearts regenerate after MI and the newly formed cardiomyocytes are derived mainly from pre-existing cardiomyocytes [106]. However, after MI, MYH7-miR-195 TG hearts fail to regenerate and had significantly impaired cardiac function compared to WT hearts, perhaps due to induction of inflammatory genes and repression of mitochondrial and cell cycle genes [106]. Conversely, LNA anti-miR15 treatment increased proliferation of cardiomyocytes and improved left ventricular systolic function after adult MI [106].
It has been reported that miR-195 contributed to the repression of a number of cell cycle genes including checkpoint kinase 1 (Chek1), cyclin-dependent kinase 1 (Cdk1), baculoviral IAP repeat-containing 5 (Birc5), nucleolar and spindle associated protein 1 (Nusap1), and sperm associated antigen 5 (Spag5) [103]. Among these cell cycle genes, Chek1 has the miR-195 seed site conserved between mice and humans and was directly targeted by miR-195 based on the luciferase reporter analysis [103]. Chek1 is a cell cycle gene that coordinates mitotic progression with spindle checkpoints [103]. Recently a hepatocellular carcinoma study found miR-195 suppressed cancer cell proliferation and led to reduced tumor size through directly targeting NF-κB signaling related genes IKKα and TAB3 [107-109].
3. Conclusions and Perspectives
Important recent progress has been made in the field of cardiac regeneration research. The success of cell-based therapies for heart repair, although measurable, has been modest to date likely because the infused cells fail to efficiently integrate into the heart. Moreover, reprogramming and trans-differentiation of non-cardiomyocytes are limited by poor efficiency and other technical challenges [15-20,32-40,64]. Thus, new innovative strategies are needed to enhance cardiac regeneration and one of the alternative compelling strategies is to trigger the endogenous cardiomyocyte regenerative capacity. Exciting new findings, revealed in the last few years, indicate that resident cardiomyocytes can be induced to reenter the cell cycle and undergo cytokinesis. More work is needed to investigate the underlying molecular mechanisms for induced cardiomyocyte cell cycle reentry. The studies summarized in this review indicate that miR-based therapeutics, using miR antagonists or mimics, has strong potential to be used to promote cardiomyocyte cell cycle re-entry and improve cardiac function after cardiac injury.
A majority of myocardial regeneration related miRs, or regenerative miRs, play essential roles in cell proliferation (Figure 1), not only in cardiomyocyte but also many other cell types. Moreover, their functions in proliferation are conserved between species from mouse to human. For examples, miR-17-92 cluster was the first described oncogenic miR [110,111], while miR-15a and miR-16 are the first identified tumor suppressor miRs [112]. Notably, regenerative miRs could share the same target genes during myocardial regeneration as they do in other contexts. For example, individual miR-15 family members, that have the same seed site, have characterized targets in contexts other than heart regeneration [107-109]. These characteristics would enable cardiac researchers to investigate candidate target genes for the miR-15 family in the context of cardiac regeneration.
Figure 1.
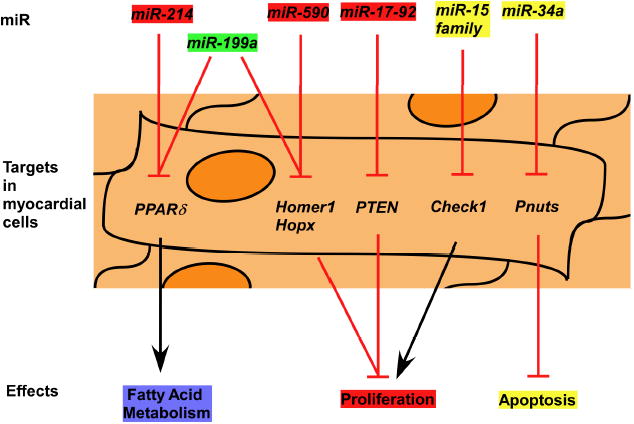
Summary of recently reported miRs that play essential roles in myocardial regeneration. MiRs could promote (in red) or inhibit (in yellow) myocardial regeneration, or play a dual-role (in green) in myocardial regeneration.
Other regenerative miRs have different mechanisms of modulating cardiac function after injury (Figure 1). As an example, miR-34a mainly regulated apoptosis and senescence during cardiac repair [96]. Notably, within the same cluster, miRs may modulate cardiac function via independent mechanisms, like miR-19 induced cardiomyocytes proliferation while miR-92a inhibited angiogenesis [91,92]. Moreover, the same miR could play different roles after different types of cardiac injury. Take miR-199a as an example, in mice with transvers aortic constriction (TAC) pressure overload, miR-199a inhibition with antagomir-199a improved cardiac contractility [88]. In contrast, overexpression of miR-199a using AAV9-199a induced cardiac regeneration in mice after MI [70]. A potential explanation, worthy of further investigation, for those observations is that miRs have multiple targets and repress different targets in the context of different injury types.
Although progress is promising, current miR studies in myocardial regeneration are limited to some extent due to the lack of cardiomyocyte specificity of in vivo anti-miR administration protocols, therefore new technologies or further studies are still needed to address these limitations. For example, LNA anti-miR15 treatment increased cardiomyocytes proliferation after MI, but also robustly induced proliferation of the non-cardiomyocytes compartment [106]. It is unknown if this non-cardiomyocyte effect is necessary for cardiac repair and so has therapeutic relevance. Moreover, treatment using anti-miR chemistries could efficiently improve cardiac function in mouse and some cases even pig, but whether these approaches could sufficiently repair injured human hearts remains to be demonstrated. Studies in non-human primates and eventually human patients are needed.
Table 1.
Recently published reports of miRs that function in cardiomyocyte regeneration
| Ref | miR | Function in cardiomyocyte (CM) regeneration | Target genes in heart |
|---|---|---|---|
| 34 | miR-590 | promote 7-day and 2 month postnatal CMs to re-enter cell cycle and progress through mitosis | Homer1,Hopx and Clic5 |
| 34 | miR-1825 | increase the proliferation of 7-day postnatal CMs | unclear |
| 34 | miR-33b | increase the proliferation of 7-day postnatal CMs | unclear |
| 34, 52 | miR-199a | promote 7-day and 2 month CMs to re-enter cell cycle and progress through mitosis; facilitate a cardiac metabolic shift from fatty acid toward glucose metabolism | Homer1, Hopx and PPARδ |
| 52 | miR-214 | facilitate a cardiac metabolic shift from fatty acid toward glucose metabolism | PPARδ |
| 53, 57 | miR-17-92 | induce proliferation of CMs | PTEN, Isl1 and Tbx1 |
| 60 | miR-34a | induce cell death and hypertrophy of CMs | Pnuts |
| 67, 69, 70, | miR-15 family | repress proliferation of CMs | Chek1 |
Key Points.
-
#
MiRs have the capabilities to regulate cardiac reprogramming and regeneration
-
#
MiRs function in cardiac repair by regulating proliferation, apoptosis, metabolism, angiogenesis and senescence.
-
#
MiRs have multiple targets, making the molecular mechanisms more complicated such that the same miR may function differently after different types of cardiac injuries.
-
#
Manipulation of miR levels can be achieved by antagomirs/LNA anti-miR and miR mimics/AAV9-miR, making miR-based therapeutics feasible.
Acknowledgments
The authors would like to acknowledge funding supports 2012-2013 Michael Bilitch Fellowship in Cardiac Pacing and Electrophysiology from HRS (to J. Wang); NIH grants 5R01HL093484-05 and R01 DE 023177-01 (to J.F.Martin).
Footnotes
Conflicts of interest: There are no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
**of outstanding interest
- 1.Bui AL, Horwich TB, Fonarow GC. Epidemiology and risk profile of heart failure. Nat Rev Cardiol. 2011;8:30–41. doi: 10.1038/nrcardio.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stehlik J, Edwards LB, Kucheryavaya AY, Benden C, Christie JD, Dobbels F, Kirk R, Rahmel AO, Hertz MI. The Registry of the International Society for Heart and Lung Transplantation: Twenty-eighth Adult Heart Transplant Report--2011. J Heart Lung Transplant. 2011;30:1078–1094. doi: 10.1016/j.healun.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4 (+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajstura J, Rota M, Cappetta D, Ogorek B, Arranto C, Bai Y, Ferreira-Martins J, Signore S, Sanada F, Matsuda A, et al. Cardiomyogenesis in the aging and failing human heart. Circulation. 2012;126:1869–1881. doi: 10.1161/CIRCULATIONAHA.112.118380. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107:305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. [DOI] [PubMed] [Google Scholar]
- 15.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 17.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 18.Ptaszek LM, Mansour M, Ruskin JN, Chien KR. Towards regenerative therapy for cardiac disease. Lancet. 2012;379:933–942. doi: 10.1016/S0140-6736(12)60075-0. [DOI] [PubMed] [Google Scholar]
- 19.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, et al. NKX2-5 (eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 20.Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Malliaras K, Zhang Y, Seinfeld J, Galang G, Tseliou E, Cheng K, Sun B, Aminzadeh M, Marban E. Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol Med. 2013;5:191–209. doi: 10.1002/emmm.201201737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–398. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beigi F, Schmeckpeper J, Pow-Anpongkul P, Payne JA, Zhang L, Zhang Z, Huang J, Mirotsou M, Dzau VJ. C3orf58, a novel paracrine protein, stimulates cardiomyocyte cell-cycle progression through the PI3K-AKT-CDK7 pathway. Circ Res. 2013;113:372–380. doi: 10.1161/CIRCRESAHA.113.301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 31.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 32.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava D, Ieda M. Critical factors for cardiac reprogramming. Circ Res. 2012;111:5–8. doi: 10.1161/CIRCRESAHA.112.271452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inagawa K, Ieda M. Direct reprogramming of mouse fibroblasts into cardiac myocytes. J Cardiovasc Transl Res. 2013;6:37–45. doi: 10.1007/s12265-012-9412-5. [DOI] [PubMed] [Google Scholar]
- 39.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. This paper showed a combination of miRs could directly converting fibroblasts to cardiomyocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian L, Berry EC, Fu JD, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- 46.Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 55.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 56.Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seeger FH, Zeiher AM, Dimmeler S. MicroRNAs in stem cell function and regenerative therapy of the heart. Arterioscler Thromb Vasc Biol. 2013;33:1739–1746. doi: 10.1161/ATVBAHA.113.300138. [DOI] [PubMed] [Google Scholar]
- 61.Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol. 2012;52:949–957. doi: 10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boettger T, Braun T. A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res. 2012;110:1000–1013. doi: 10.1161/CIRCRESAHA.111.247742. [DOI] [PubMed] [Google Scholar]
- 63.Ounzain S, Crippa S, Pedrazzini T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim Biophys Acta. 2013;1833:923–933. doi: 10.1016/j.bbamcr.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 64.Addis RC, Epstein JA. Induced regeneration--the progress and promise of direct reprogramming for heart repair. Nat Med. 2013;19:829–836. doi: 10.1038/nm.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christoffels VM, Pu WT. Developing insights into cardiac regeneration. Development. 2013;140:3933–3937. doi: 10.1242/dev.096867. [DOI] [PubMed] [Google Scholar]
- 66.Zhu H, Fan GC. Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc Res. 2012;94:284–292. doi: 10.1093/cvr/cvr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70**.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. This is a landmark paper for function of miRs in myocardial regeneration. [DOI] [PubMed] [Google Scholar]
- 71.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11:922–933. doi: 10.4161/cc.11.5.19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rong R, Ahn JY, Huang H, Nagata E, Kalman D, Kapp JA, Tu J, Worley PF, Snyder SH, Ye K. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat Neurosci. 2003;6:1153–1161. doi: 10.1038/nn1134. [DOI] [PubMed] [Google Scholar]
- 76.Hwang SY, Wei J, Westhoff JH, Duncan RS, Ozawa F, Volpe P, Inokuchi K, Koulen P. Differential functional interaction of two Vesl/Homer protein isoforms with ryanodine receptor type 1: a novel mechanism for control of intracellular calcium signaling. Cell Calcium. 2003;34:177–184. doi: 10.1016/s0143-4160(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 77.Feng W, Tu J, Yang T, Vernon PS, Allen PD, Worley PF, Pessah IN. Homer regulates gain of ryanodine receptor type 1 channel complex. J Biol Chem. 2002;277:44722–44730. doi: 10.1074/jbc.M207675200. [DOI] [PubMed] [Google Scholar]
- 78.Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 79.Kook H, Epstein JA. Hopping to the beat Hop regulation of cardiac gene expression. Trends Cardiovasc Med. 2003;13:261–264. doi: 10.1016/s1050-1738(03)00107-5. [DOI] [PubMed] [Google Scholar]
- 80.Obarzanek-Fojt M, Favre B, Kypriotou M, Ryser S, Huber M, Hohl D. Homeodomain-only protein HOP is a novel modulator of late differentiation in keratinocytes. Eur J Cell Biol. 2011;90:279–290. doi: 10.1016/j.ejcb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19:450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang JM, Sim SM, Kim HY, Park GT. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol. 2010;89:537–546. doi: 10.1016/j.ejcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 83.Trivedi CM, Cappola TP, Margulies KB, Epstein JA. Homeodomain only protein x is down-regulated in human heart failure. J Mol Cell Cardiol. 2011;50:1056–1058. doi: 10.1016/j.yjmcc.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haghikia A, Missol-Kolka E, Tsikas D, Venturini L, Brundiers S, Castoldi M, Muckenthaler MU, Eder M, Stapel B, Thum T, et al. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J. 2011;32:1287–1297. doi: 10.1093/eurheartj/ehq369. [DOI] [PubMed] [Google Scholar]
- 86.Greco S, Fasanaro P, Castelvecchio S, D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC, Menicanti L, Martelli F. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61:1633–1641. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baumgarten A, Bang C, Tschirner A, Engelmann A, Adams V, von Haehling S, Doehner W, Pregla R, Anker MS, Blecharz K, et al. TWIST1 regulates the activity of ubiquitin proteasome system via the miR-199/214 cluster in human end-stage dilated cardiomyopathy. Int J Cardiol. 2013;168:1447–1452. doi: 10.1016/j.ijcard.2012.12.094. [DOI] [PubMed] [Google Scholar]
- 88*.el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MM, et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab. 2013;18:341–354. doi: 10.1016/j.cmet.2013.08.009. This paper showed miR-199a-214 cluster inhibits mitochondrial fatty acid metabolism during cardiac repair, mainly through repressing PPARdelta. [DOI] [PubMed] [Google Scholar]
- 89.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J, Bai Y, Li H, Greene SB, Klysik E, Yu W, Schwartz RJ, Williams TJ, Martin JF. MicroRNA- 17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003785. e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iaconetti C, Polimeni A, Sorrentino S, Sabatino J, Pironti G, Esposito G, Curcio A, Indolfi C. Inhibition of miR-92a increases endothelial proliferation and migration in vitro as well as reduces neointimal proliferation in vivo after vascular injury. Basic Res Cardiol. 2012;107:296. doi: 10.1007/s00395-012-0296-y. [DOI] [PubMed] [Google Scholar]
- 92.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 93*.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, et al. mir-17-92 Cluster Is Required for and Sufficient to Induce Cardiomyocyte Proliferation in Postnatal and Adult Hearts. Circ Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. This paper showed mir-17-92 cluster, especially miR-19, promotes cardiomyocyte proliferation and PTEN is a major cardiac target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lakatta EG. Age-associated cardiovascular changes in health: impact on cardiovascular disease in older persons. Heart Fail Rev. 2002;7:29–49. doi: 10.1023/a:1013797722156. [DOI] [PubMed] [Google Scholar]
- 96*.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. This study showed miR-34a could induce cell death of cardiomyocytes, and one of its main target gene is pnuts. [DOI] [PubMed] [Google Scholar]
- 97.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 98.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 100.Xu Q, Seeger FH, Castillo J, Iekushi K, Boon RA, Farcas R, Manavski Y, Li YG, Assmus B, Zeiher AM, et al. Micro-RNA-34a contributes to the impaired function of bone marrow-derived mononuclear cells from patients with cardiovascular disease. J Am Coll Cardiol. 2012;59:2107–2117. doi: 10.1016/j.jacc.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 101.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:E110–116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Leon G, Sherry TC, Krucher NA. Reduced expression of PNUTS leads to activation of Rb-phosphatase and caspase-mediated apoptosis. Cancer Biol Ther. 2008;7:833–841. doi: 10.4161/cbt.7.6.5839. [DOI] [PubMed] [Google Scholar]
- 103.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW, 2nd, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, Dalby CM, Robinson K, Stack C, Latimer PA, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106*.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110:187–192. doi: 10.1073/pnas.1208863110. This work showed miR-195, a member of miR-15 family, represses cardiomyocytes proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 108.Jin G, Klika A, Callahan M, Faga B, Danzig J, Jiang Z, Li X, Stark GR, Harrington J, Sherf B. Identification of a human NF-kappaB-activating protein, TAB3. Proc Natl Acad Sci U S A. 2004;101:2028–2033. doi: 10.1073/pnas.0307314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ding J, Huang S, Wang Y, Tian Q, Zha R, Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al. Genome-wide screening reveals that miR-195 targets the TNF-alpha/NF-kappaB pathway by down-regulating IkappaB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology. 2013;58:654–666. doi: 10.1002/hep.26378. [DOI] [PubMed] [Google Scholar]
- 110.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 112.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]


