Summary
Allostery is an essential means for regulating biomolecular functions and provides unique opportunities for drug design, yet our ability to elucidate allosteric mechanisms remains limited. Here, based on extensive molecular dynamics simulations, we present an atomistic picture of the pathways mediating the allosteric regulation of the PPIase domain of Pin1 by its WW domain. Two pathways jointly propagate the action of substrate-WW binding to produce closure and rigidification of three PPIase catalytic-site loops. One pathway preexists in the apo protein but remains dormant until substrate-WW binding completes the second. The reduction in conformational entropy and preorganization of the catalytic-site loops observed here may explain why substrate-WW binding enhances ligand affinity and catalytic activity of the PPIase domain, and suggest a combination drug therapy for Pin1-related diseases. Whereas the traditional view of allostery has emphasized conformational transition, our study uniquely identifies a distinct role of conformational dynamics in eliciting allostery.
Introduction
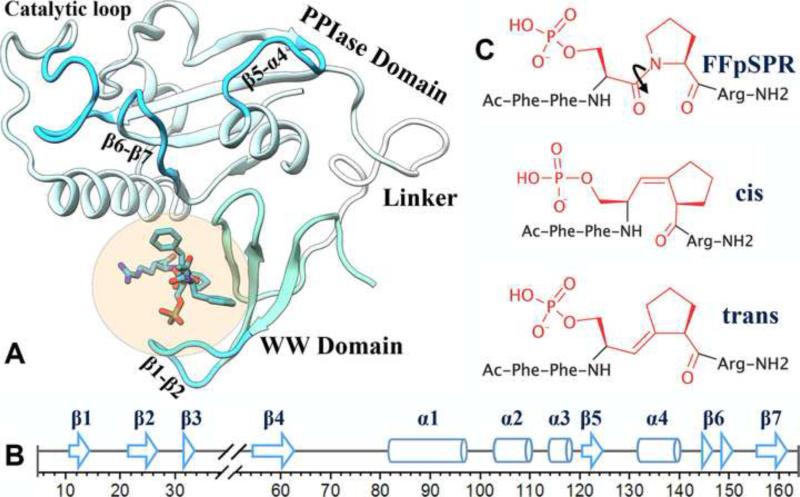
Pin1, a peptidyl-prolyl cis/trans isomerase (PPIase), acts on phosphoSer/Thr-Pro (pSer/Thr-Pro) motifs present in mitotic phosphoproteins (Lu et al., 1996), thereby controlling their fates (Liou et al., 2011). Pin1 dysregulation is implicated in various diseases, including cancer and Alzheimer's disease (Lu, 2004; Lu and Zhou, 2007; Lu et al., 1999b; Wulf et al., 2001). Therefore, Pin1 is an attractive therapeutic target, and a number of inhibitors have been designed (Moore and Potter, 2013; Wang and Etzkorn, 2006; Wang et al., 2004). The full-length Pin1 can be divided into an N-terminal WW domain (residues 1-39) and the C-terminal PPIase domain (residues 50-163) (Figure 1A,B). Both domains can selectively bind pSer/Thr-Pro containing substrates motifs, but only the PPIase domain can isomerize the peptidyl-prolyl bonds (Lu et al., 1999a; Zhou et al., 2000). The roles of the WW domain and, more specifically, substrate binding to it, have long been studied (Lu and Zhou, 2007; Lu et al., 1999a; Lu et al., 2002; Ranganathan et al., 1997; Verdecia et al., 2000). These roles may provide both better understanding of the functional mechanism of Pin1 and unique opportunities for designing Pin1-targeting drugs. Here we report a computational study on the conformational and dynamical effects of substrate-WW binding.
Figure 1. The protein and ligands in this study.
(A) Structure of Pin1 with FFpSPR bound to the WW domain; protein from 3TDB and substrate modeled after 1F8A. The β1-β2 loop and three loops around the catalytic site are highlighted in darker cyan.
(B) Secondary structures of Pin1.
(C) The three Pin1 ligands, from top to bottom: FFpSPR, cis ligand, and trans ligand.
Earlier studies have emphasized the potential of the WW domain as a non-catalytic binder in increasing local substrate concentration and in subcellular localization (Lu et al., 1999a; Lu et al., 2002). However, the substrate affinity and catalytic activity of the isolated PPIase domain are different from those of the full-length protein (Lu et al., 1999a; Namanja et al., 2011; Zhou et al., 2000), therefore suggesting that the WW domain can modulate substrate binding and catalysis. Indeed, numerous crystal structures of Pin1 have shown that the two domains are tightly packed against each other, although the linker between them is disordered (Ranganathan et al., 1997; Verdecia et al., 2000; Zhang et al., 2012). NMR studies have shown that binding of both substrates and a nonpeptidic ligand, polyethylene glycol (PEG), to the WW domain results in tighter coupling between the two domains (Jacobs et al., 2003; Vanwart et al., 2012). Side-chain methyl dynamics studies (Namanja et al., 2007; Namanja et al., 2011) have further shown that substrate binding to the WW domain leads to a loss of side-chain flexibility along a “conduit” of conserved hydrophobic residues linking the inter-domain interface and the catalytic site. Moreover, an I28A mutation in the inter-domain interface has been found to weaken inter-domain communication (Wilson et al., 2013).
Together the foregoing studies suggest that the WW domain may modulate the activity of the PPIase domain through allosteric regulation. However, the underlying mechanism remains poorly defined. Potentially, the large number of crystal structures of Pin1 in the Protein Data Bank (PDB) could provide clues to the various conformations accessible to the proteins. Unfortunately, in all these structures, both the WW site and the PPIase catalytic site are occupied, often by PEG, an additive for protein crystallization. The side-chain methyl dynamics studies (Namanja et al., 2007; Namanja et al., 2011) have given rise to the most detailed picture of the pathway for inter-domain communication, yet these studies are limited to methyl-containing side-chains and the possibility of other participating residues cannot be excluded. It is also unclear how ligand binding to the WW domain induces effects on substrate binding to and catalytic activity of the PPIase domain.
In recent years, computational studies have been found to be very useful in complementing experiments in elucidating allosteric mechanisms (Elber, 2011; Feher et al., 2014; Rousseau and Schymkowitz, 2005), including algorithms for identifying allosteric networks (Gerek and Ozkan, 2011; Ghosh and Vishveshwara, 2007; Kannan and Vishveshwara, 1999; Sethi et al., 2009; VanWart et al., 2014). In particular, molecular dynamics simulations have revealed two pathways and implicated a strong dynamic component in the allosteric regulation of thrombin by thrombomodulin (Gasper et al., 2012). This supports the growing emphasis on the possibility that allostery can be elicited through changes in protein dynamics without apparent conformational transitions (Cooper and Dryden, 1984; Petit et al., 2009; Tsai et al., 2008), departing from the classical view (Fischer et al., 2011; Monod et al., 1965).
In the present study we carried out extensive molecular dynamics simulations to elucidate the mechanism of allosteric regulation in Pin1. Our simulations show that substrate binding to the WW domain alone results in closure and rigidification of the three loops (referred to as β4-α1 (or catalytic), β5-α4, and β6-β7) around the catalytic site (Figure 1A,B), thus giving direct evidence for allosteric communication between the two distant binding sites. Two pathways are found to mediate the inter-domain allosteric regulation. Path1 emanates from the WW backside and propagates through the inter-domain interface and the PPIase domain core to the β5-α4 and β6-β7 loops; Path2 emanates from the WW front pocket and propagates through the bound substrate, the PPIase peripheral α1, and the α1-core interface to the catalytic loop. Path1 preexists in apo Pin1, but remains dormant until Path2 is completed by substrate-WW binding. Through restrained simulations and simulations of the I28A mutant, we further demonstrate that the two pathways must act in concert in order to elicit the allosteric effects. Moreover, our study suggests that substrate-WW binding, via rigidifying the catalytic-site loops, may enhance ligand affinity of the PPIase domain by reducing the conformational entropy cost for binding. Substrate-WW binding induced preorganization of the catalytic-site loops may also enhance the catalytic activity of the full-length Pin1. Finally the scenario of cooperative binding to the WW and PPIase sites presented here points to the possibility of a combination drug therapy for Pin1-related diseases.
Results
The design of the present work was largely inspired by the NMR studies of Peng and co-workers (Namanja et al., 2011). They measured the WW and PPIase binding affinities of a Pin1 substrate (FFpSPR) and two peptidomimetics in which the substrate pSP core is replaced by alkene isosteres to lock the imide as either cis or trans (Wang et al., 2004) (Figure 1C), as well as the effects of binding these ligands on Pin1 methyl-containing side-chain dynamics. FFpSPR mainly bound to the WW site, the cis ligand exclusively to the PPIase site, whereas the trans ligand bound to both sites. We modeled these three different binding modes onto Pin1 and then carried out molecular dynamics simulations in explicit solvent. The simulations present direct evidence for allosteric communication between the two distant binding sites and suggest two mediating pathways. The available experimental data (Jacobs et al., 2003; Namanja et al., 2011; Vanwart et al., 2012) provide validation, but our simulations lead to a more complete, atomistic picture for the allosteric regulation in Pin1.
To dissect the effects of substrate-WW binding and further validate the two putative pathways, we carried out simulations of apo Pin1 in which different regions of the protein were restrained. In addition, we carried out simulations of the I28A mutant, which perturbs the inter-domain interface (Wilson et al., 2013). These “control” simulations show that the allosteric effects are not fully elicited when the inter-domain links are perturbed. Therefore allosteric regulation is achieved through the concerted action of the two pathways.
Ligand Binding Results in Localized Conformational Changes
Based on the crystal structures of Pin1 bound with the cis or trans ligand at the catalytic site (PDB 3TCZ and 3TDB (Zhang et al., 2012)) and bound with a pSer-Pro containing peptide (PDB 1F8A (Verdecia et al., 2000)) at the WW site, we built initial models for Pin1 with FFpSPR bound at the WW site (Figure 1A), with the cis ligand bound at the catalytic site, and with two copies of the trans ligand bound, one at the WW site and one at the catalytic site. At the WW site, the ligands sit over a pocket lined by the concave front face of the WW domain β-sheet and fill a groove between the WW domain and the α1 helix of the PPIase domain. We simulated each of these systems as well as apo Pin1 in explicit solvent for 100 ns and used the last 40 ns for analysis.
In these simulations, the core structure of Pin1 is relatively well preserved, as indicated by a comparison of the conformations closest to the averages in the last 40 ns. Relative to apo Pin1, the Cα root-mean-square-deviations (RMSDs) on the secondary-structure core range from 1.1 to 1.5 Å for the three ligand-bound forms. There are subtle differences of the latter forms from the apo form, including movement of α1 toward both the PPIase central β-sheet and the WW domain (Figures 2A and S1A), in agreement with NMR data indicating tighter coupling between the domains upon ligand binding (Jacobs et al., 2003; Vanwart et al., 2012). On the other hand, RMSDs calculated on the rest of the protein (even after excluding the flexible inter-domain linker) are twice as large. For the three catalytic-site loops (residues 63-72 for the β5-α4 or catalytic loop, residues 126-132 for the β5-α4 loop, and residues 151-155 for the β6-β7 loop), the RMSDs of Pin1 bound with FFpSPR and cis and trans ligands from apo Pin1 are 2.6, 3.4, and 3.0 Å, respectively.
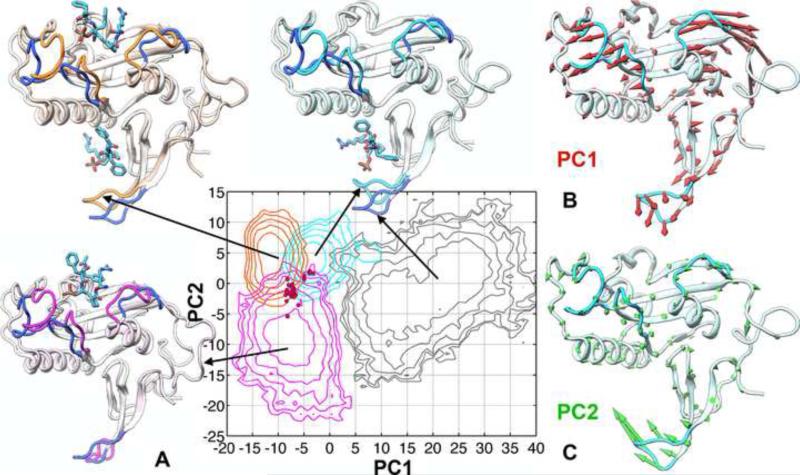
Figure 2. Conformational ensembles of apo Pin1 and the three ligand-bound forms.
(A) Free-energy surfaces over the first two PCs, contoured at 0.5 kcal/mol intervals; PC coordinates of 32 crystal structures are shown as red dots. The conformations closest to the simulation averages of the FFpSPR- (cyan), trans ligand- (orange), and cis ligand-bound (magenta) forms are shown superimposed to the corresponding conformation of the apo form (gray with four blue loops).
(B,C) Conformational differences represented by PC1 and PC2 are displayed as red and green arrows, respectively, on a Pin1 conformation with both PC1 and PC2 near 0.
See also Figures S1, S2, and S3.
It appears that the conformational changes induced by the ligand binding are largely found in the loops, especially those around the catalytic site. This finding is in line with the chemical shift perturbation (CSP) data of Peng and co-workers (Namanja et al., 2011). According to these data, binding of FFpSPR results in significant changes of backbone chemical shifts in the WW domain along with discernible changes in some of the residues in the 115-140 range; binding of the cis ligand results in significant changes in most of the residues in the 115-160 range; and binding of the trans ligand results in both the latter changes and additional changes in the WW domain. Similar CSP patterns are produced by chemical shifts predicted by SPARTA+(Shen and Bax, 2010) on the last 40 ns of the simulations of the four systems (Figure S2).
To further characterize the conformational differences among the four differently liganded forms of Pin1, we carried out principal component analysis on conformations pooled from the four independent simulations; in each simulation 20,000 conformations were evenly sampled over the last 40 ns. The results are presented as free-energy surfaces over the first two principal components (referred to as PC1 and PC2 hereafter; Figure 2A). The basins for the three ligand-bound forms are relatively closely positioned: the FFpSPR-bound form is separated from the trans ligand-bound form by a small difference along PC1, and the cis ligand-bound form is separated from those two mainly by a moderate difference along PC2. The FFpSPR-bound form also has a minor population, which is further separated along PC1. Free-energy contours of the three ligand-bound forms at 1.5 kcal/mol above the respective minima intersect, suggesting that thermal fluctuations allow Pin1 in any two of the ligand-bound forms to sample some overlapping conformations. On the other hand, the apo form is located in a separate basin, with a significant difference from the basins of the ligand-bound forms. The apo form likely has only rare conformational exchanges with the ligand-bound forms, except for the minor population of the FFpSPR-bound form. Whereas the basin for each ligand-bound form is well localized, the basin for the apo form is spread out, indicating that, at thermal equilibrium, the conformational ensemble sampled by the latter is not only distinct but also more expansive.
When 32 crystal structures are projected onto the PC1-PC2 plane, they are also located around the intersection region of the three ligand-bound forms and far removed from the basin of the apo form (Figure 2A). This is consistent with the fact that all these structures have both their WW site and catalytic site bound, either with actual ligands or with additives like PEG from the mother liquor for crystal growth.
The conformational differences represented by PC1 and PC2 are shown in Figure 2B,C. PC1 captures the main difference of the apo form from the FFpSPR- and trans ligand-bound forms (Figure 2A), and comprises an opening of the three catalytic-site loops in the PPIase domain and the β1-β2 loop in the WW domain. Evidently, the apo form favors more open conformations for these loops (opening of the β1-β2 loop was reported in a previous simulation of the apo protein (Vanwart et al., 2012)). For the FFpSPR- and trans ligand-bound forms, closure of the β1-β2 loop can be attributed to interaction with the WW-bound ligands. While closure of the catalytic-site loops in the trans ligand-bound form can be likewise explained, closure of these loops in the FFpSPR-bound form is an indirect effect, since in this case the catalytic site is empty. The apparent causal link between substrate binding at the WW site and loop closure at the catalytic site provides the first sign of allosteric communication between the two domains.
PC2 captures the main difference of the FFpSPR- and trans ligand-bound forms from the cis ligand-bound form, which is a closure of the β1-β2 loop. In other words, while the catalytic-site loops in all the three ligand-bound forms are closed, the β1-β2 loop in the cis ligand-bound form favors more open conformations, similar to the β1-β2 loop in the apo form.
The principal conformational differences involving the opening/closure of the three catalytic-site loops and the flapping of the β1-β2 loop are robustly obtained when calculated over different segments of the simulations. Moreover, these conformational differences can be straightforwardly demonstrated by using two explicit collective coordinates. To depict the opening/closure of the three catalytic-site loops, we use their radii of gyration (Rg); to depict the flapping of the β1-β2 loop, we use the distance between the centers of Cα atoms in the β1-β2 loop (residues 15-21) and the α1 helix (residues 82-97). For each system, we calculated the free energy surface over these two coordinates. When these free energy surfaces are overlaid (Figure S3), they paint essentially the same picture for the conformational differences among the four systems as indicated by the principal component analysis.
Substrate-WW Binding Leads to Significant Loop Rigidification Around the Catalytic Site
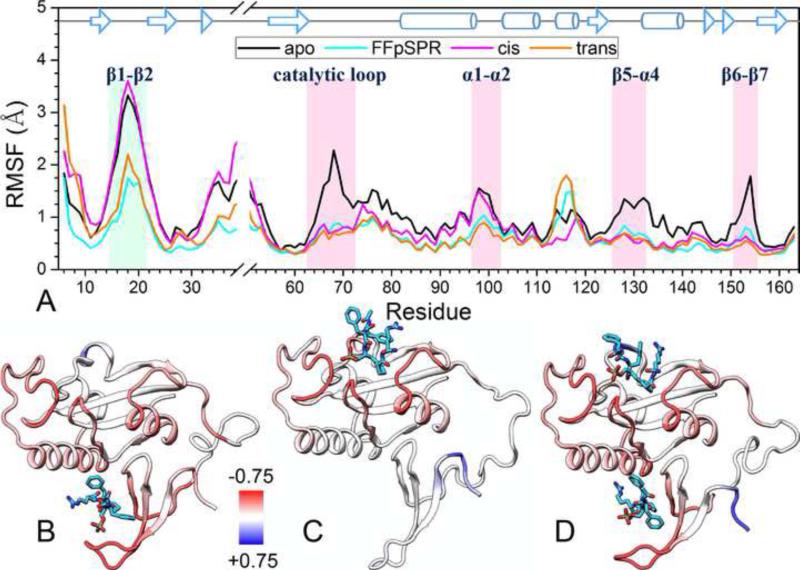
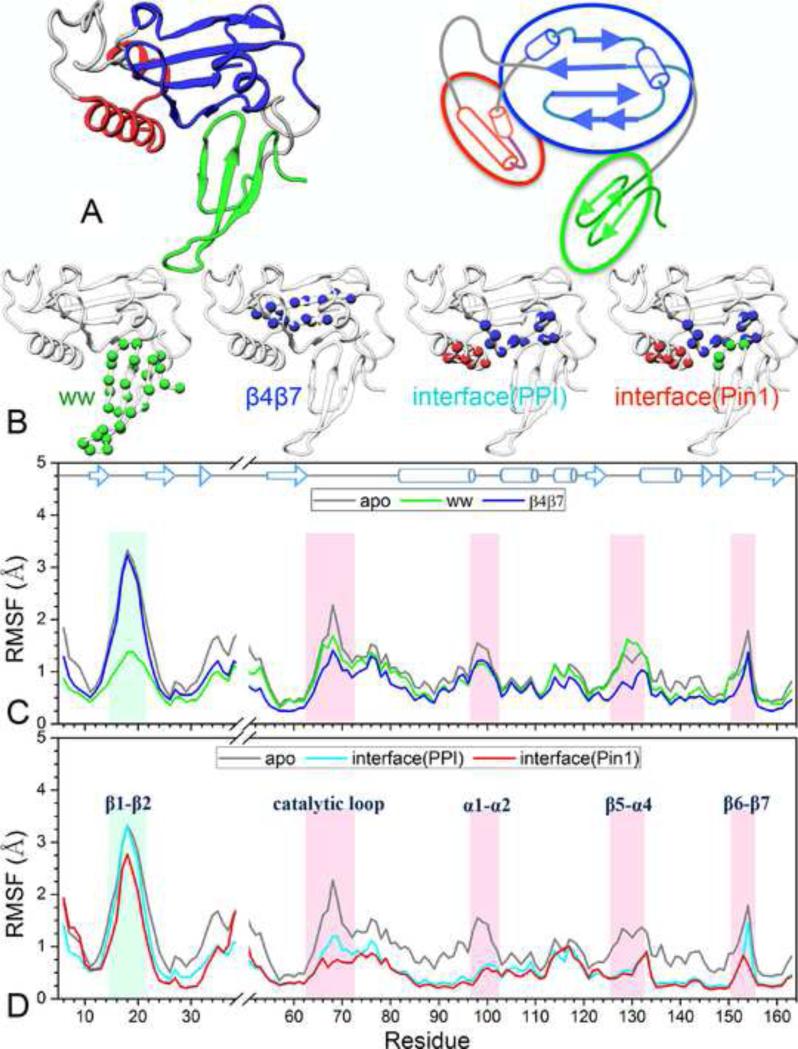
As alluded to above, the more localized free-energy basins of the ligand-bound forms (Figure 2A) indicate that, as a result of ligand binding, Pin1 samples a more restricted ensemble of conformations along PC1 and PC2. In other words, the protein molecule becomes less flexible. We calculated the Cα root-mean-square-fluctuation (RMSFs) of the four systems in the simulations to directly show the change in backbone flexibility (Figure 3). The apo form exhibits high flexibility in the three catalytic-site loops, the β1-β2 loop, and the α1-α2 loop. The catalytic-site loop flexibility is quenched with ligand binding, either at the WW site, at the PPIase site, or both. On the other hand, the flexibility of the β1-β2 and α1-α2 loops is quenched only when a ligand binds at the WW site. Together, the results in Figures 2 and 3 show that substrate-WW binding leads to both the closure and the more restricted conformational sampling of the catalytic-site loops. The allosteric communication is evidently one-way only, since ligand binding at the PPIase site does not lead to similar effects on the β1-β2 loop.
Figure 3. Backbone fluctuations in the absence and presence of ligands.
(A) The Cα RMSFs of individual residues in the last 40 ns. Loops with high flexibility in the apo form are highlighted by shading. The α3 helix, which is a less stable 310 helix, also shows high flexibility.
(B–D) Differences in RMSFs of the ligand-bound forms from those of the apo form are colored on the bound conformations of Pin1. Red and blue colors represent lower and higher flexibilities, respectively, in the bound forms.
See also Figures S4, S5, S6, and S8.
It is worth noting that the ligand-induced change in backbone flexibility of the loops occurs on the 10s of ns timescale of the simulations. The opening/closure of the three catalytic-site loops can be monitored by their Rg during the course of the 100-ns simulations (Figure S4). The four systems all start with a low Rg value (about 10 Å). When a ligand molecule is bound at the catalytic site (as in the trans and cis ligand-bound forms), the loops are locked at this low Rg value. The apo form deviates from the low Rg value after about 50 ns and remains at 20-30% enlarged Rg values for the rest of the simulation. The FFpSPR-bound form, also without a ligand at the catalytic site, is able to break away from the low Rg value during the first 40 ns but then returns to its apparently preferred low Rg value for nearly all of the remaining time.
The order parameters of sidechain methyls, which are dominated by ps-ns dynamics (Chatfield et al., 1998), were measured by Peng and co-workers (Namanja et al., 2011) for the four systems. Relative to the apo form, the ligand-bound forms consistently had higher values, signifying reduced flexibility, for about 10 methyl-containing sidechains in the PPIase domain (Figure S5A), distributed around the catalytic site (Leu61, Leu122, Ala124, and Met130), the α1-PPIase core interface (Leu60, Ile89, Ile93, and Ile156), and the PPIase-WW interface (Leu141 and Val150) (Figure S5B). The order parameters calculated from our simulations largely reproduce this pattern (Figure S5C,D), although some of the values are not as low as the measured ones, for insufficient conformational sampling in the simulations (Gasper et al., 2012). Notably, the catalytic loop (residues 63-72) does not have any methyl-containing sidechains, and therefore order parameters could not be measured to provide information for the effects of ligand binding on the dynamics of this important loop. It is also interesting that, for some residues (e.g., Leu60 and Leu61), the reduction in flexibility upon ligand binding is limited to the sidechains only (as reported by ), as their Cα RMSFs are unaffected.
In addition to simulations of the four systems described above, we also carried simulations of two “control” systems (Figure S6). The RMSF results for Pin1 with the trans ligand bound only to the catalytic site are similar to those for the cis ligand-bound form, thus confirming that the inter-domain communication is one-way only. In addition, a simulation of the isolated PPIase domain in apo form yields lower RMSFs for the catalytic and β6-β7 loops than the counterparts in the full-length apo Pin1, suggesting that, in apo form, the WW domain amplifies the loop flexibility of the PPIase domain.
Pathways of Inter-Domain Communication
How is the action of substrate-WW binding propagated to affect the catalytic-site loop conformations and dynamics? Several algorithms (Gerek and Ozkan, 2011; Ghosh and Vishveshwara, 2007; Kannan and Vishveshwara, 1999; Sethi et al., 2009; VanWart et al., 2014) have been developed for identifying allosteric networks, i.e., clusters of linked residues that putatively propagate allosteric signals. Here we used the original algorithm of Kannan and Vishveshwara (1999), in which residues are linked into clusters when tertiary contacts between sidechains persist during a simulation.
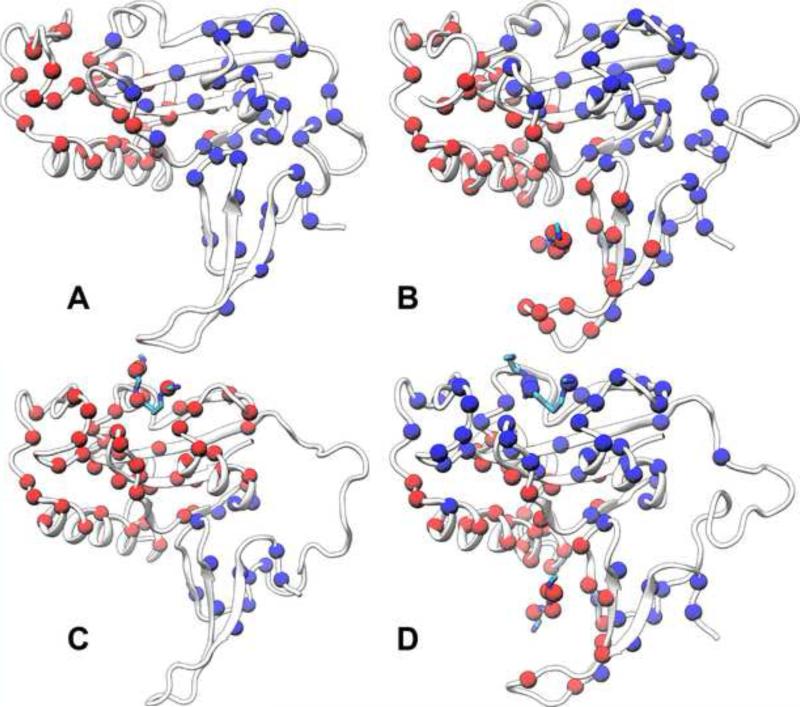
For apo Pin1, the simulation produces two major clusters of linked residues (Figure 4A). The first cluster consists of the WW backside, the WW-PPIase domain interface, the PPIase domain core (i.e., α4, β5, β6, β7, and α4), and the β5-α4 and β6-β7 loops. The second cluster consists of the peripheral β1, the β1-PPIase core interface, the α1-α2 loop, α2, and the catalytic loop. Cluster 1 is largely maintained in the FFpSPR-Pin1 complex, but cluster 2 now extends to the WW front pocket via the bound substrate (Figure 4B). We propose that these two clusters form two pathways, referred to as Path1 and Path2, respectively, that propagate the allosteric signal from the WW domain to the PPIase catalytic site. Specifically, Path1 emanates from the WW backside and propagates through the inter-domain interface and the PPIase domain core to the β5-α4 and β6-β7 loops; Path2 emanates from the WW front pocket and propagates through the bound substrate, α1, and the α1-core interface to the catalytic loop. Path1 preexists in apo Pin1, but apparently remains dormant until Path2 is completed by substrate-WW binding.
Figure 4. Allosteric networks of four systems.
(A) Apo Pin1.
(B) FFpSPR-Pin1 complex.
(C) cis ligand-Pin1 complex.
(D) trans ligand-Pin1 complex.
Clusters of residues that persistently form tertiary contacts between sidechains in the simulations are shown as spheres, either in red or in blue, at Cα positions. The main chains of the protein and the ligands are displayed in cartoon and stick, respectively. Clusters with no more than 10 residues are not displayed.
See also Figures S7 and S9.
In the cis ligand-bound complex, Path1 is broken at the WW-PPIase domain interface because of weak links to the PPIase domain core; all the remaining PPIase residues coalesce into a single cluster (Figure 4C). As a result, there is no path that connects the cis ligand to the WW domain, providing an explanation for the one-way allosteric communication noted above. The allosteric networks of the trans ligand-bound complex are very similar to those of the FFpSPR-Pin1 complex, except that the catalytic loop is now part of the first cluster, due to links provided by the ligand molecule at the catalytic site (Figure 4D). Again, two pathways tightly couple the two domains.
To demonstrate that the allosteric pathways identified above are robust, we carried out a community network analysis as introduced by Sethi et al. (2009). This method accounts for motional correlation between residues. When applied to the apo Pin1 simulation, six communities were obtained (Figure S7A). The WW domain is represented by two communities, numbered 0 and 1, respectively, for the β1-β2 hairpin and the rest of WW; the PPIase core is represented by community 2; and the α1-α2 appendage is represented by communities 3-5, respectively, for α1 N-terminus/catalytic loop, α1 C-terminus, and α2/α3. For the FFpSPR-Pin1 complex (Figure S7B), communities 0 and 1 coalesce into a single one representing the full WW domain. Moreover, this community is joined by the N-terminus of the bound substrate, whose C-terminus joins community 4. Therefore the substrate reinforces the connection between community 1 (i.e., WW domain) and 4 (i.e., α1 C-terminus). Finally a new connection is generated between community 1 and community 3 (i.e., α1 N-terminus/catalytic loop). The emergence of this new connection corresponds nicely to the completion of Path2 by substrate-WW binding.
WW-α1 Links Are Essential for Propagation of Allosteric Signal
The preceding pathway analyses suggest that Pin1 is a tripartite molecule, consisting of the WW domain, the PPIase core, and the α1-α2 appendage (Figure 5A), and that ligand binding serves to create links between these modules. This reasoning led us to further hypothesize that allosteric effects may be elicited by adding inter-module links missing in the apo protein, but not by reinforcing intra-module links. We sought to test this hypothesis and identify such essential inter-module links, by creating intra- and inter-module links in the form of RMSD restraints to a representative conformation from the apo Pin1 simulation. In separate simulations, we introduced restraints on the WW domain; on β4 and β7 of the PPIase core; on a set of residues, hereafter referred to as interface(PPI), that includes the WW-facing side of β1 and the WW-facing and β1-facing sides of the α4-β6 region; and on another set, referred to as interface(Pin1), that further includes the β2-β3 loop (Figure 5B).
Figure 5. Design and results of the restrained simulations.
(A) Pin1 as a tripartite molecule, with its three modules, the WW domain, the PPIase core, and the α1-α2 appendage, shown in green, blue, and red, respectively. Left: 3-dimensioanl structure; right: schematic representation.
(B) Four sets of intra-module (shown as Cα spheres in single color) and inter-module (shown as Cα spheres in mixed color) restraints. The restrained residues are: WW, residues 6-34; β4β7, residues 55-62 and 156-161; interface(PPI), residues 86, 87, 90, 91, 93, 94, 97, 136-138, 140-142, and 145-151; interface(Pin1), all interface(PPI) residues and residues 28-31.
(C,D) The Cα RMSFs of the four restrained simulations, compared to those of the unrestrained apo Pin1 simulation.
The effects of these intra- and inter-module restraints on backbone flexibility are illustrated by the RMSF results displayed in Figure 5C,D; the RMSFs of apo Pin1 without restraints are also displayed as a reference. Restraining the WW domain has no effect on the catalytic-site loop flexibility. The same is largely true of restraining β4 and β7, though the RMSF amplitudes of the catalytic-site loops are now somewhat suppressed, perhaps due to their proximity to the restrained region. These results confirm that reinforcing intra-module links is ineffective in eliciting allosteric effects.
On the other hand, adding inter-module links proves more effective. With the interface(PPI) restraints, which introduce links between the PPIase core and the α1-α2 module, the RMSF amplitudes of the catalytic-site loops are further suppressed (while the WW β1-β2 loop remained flexible). Finally the interface(Pin1) restraints, which include just four more residues, Ile28-Ala31, from the WW domain, produce effects on the catalytic-site loop flexibility that are indistinguishable from those by substrate-WW binding. It appears that, in the triangular relation of the WW domain, the PPIase core, and the α1-α2 appendage, WW-α1 links are essential for the propagation of the allosteric signal.
Note that the links are created by restraining to a representative conformation from the apo Pin1 simulation. Consequently there is very little difference between the average conformations of the unrestrained and restrained simulations (Figure S1B). The simulation with the interface(Pin1) restraints uniquely demonstrates that allosteric effects can be produced through changes in protein dynamics without accompanying conformational changes.
I28A Mutation Weakens Inter-Domain Links
Peng and co-workers (Wilson et al., 2013) introduced the I28A mutation, located at the tip of the β2-β3 loop and within the interface with the PPIase domain core (Figure S8A). Their NMR data showed that this mutation weakened the WW-PPIase core contact, reduced substrate affinities for both the WW site and the catalytic site, and enhanced sub-ns mobility of the catalytic loop. To explain these observations and provide further validation of our allosteric pathways, we carried out simulations of the I28A mutant in both apo and FFpSPR-bound forms.
Our simulations reveal that the mutation results in a rotation of the WW domain, with the mutation site as the pivot point, in both the apo form (Figure S8A) and the FFpSPR-bound form (Figure S8B). As a result of this rotation, the WW domain is farther separated from α1 and the WW-α1 groove is widened, such that the WW-bound substrate cannot effectively interact with α1 and neighboring residues, potentially leading to a reduced substrate affinity for the WW site. The larger WW-α1 separation also hinders the ability of the substrate in bridging between the WW and α1-α2 modules. Consequently, Path2 for the mutant is incomplete not only in the apo form but also in the substrate-bound form (Figure S9). Moreover, even Path1 is broken in the substrate-bound form, due to weak links to the PPIase domain core (similar to the situation in the cis ligand-bound Pin1).
The effects of the I28A mutation on the backbone flexibilities of the apo and substrate-bound forms are illustrated by a comparison of mutant and wild-type RMSFs (Figure S8C,D). In the apo form, the mutation amplifies the flexibility of the β1-β2 loop and extends the high flexibility of the catalytic loop to the α1 N-terminus. High loop flexibilities persist even after substrate-WW binding. The high catalytic-site loop flexibilities likely reflect the breakage of the allosteric pathways and potentially explain the reduced substrate affinity for this site in the mutant.
Docking to Catalytic Site Indicates Loop Preorganization Upon Substrate-WW Binding
As will be elaborated in Discussion, catalytic-site loop rigidification upon substrate-WW binding may yield enhanced affinity for the distal site, by reducing the conformational entropy cost for binding. The argument for this mechanism would be strengthened if the loop conformations sampled by the substrate-WW bound form are more favorable for ligand binding than those sampled by the apo form. We sought to test whether this is indeed true, by docking the trans and cis ligands to the catalytic site using representative conformations from the simulations of the apo and FFpSPR-bound forms.
Running RosettaLigand (Davis and Baker, 2009) on 10 conformations from the simulation of each system, the trans ligand-Pin1 interaction energies are −6.8 ± 2.7 and −11.7 ± 1.4 kcal/mol for the apo and FFpSPR-bound forms, respectively. For comparison, the interaction energy calculated on the trans ligand-bound form after RosettaLigand refinement is −15.8 ± 2.2 kcal/mol, which represents the optimal value for binding the trans ligand at the catalytic site. That the interaction energy for the FFpSPR-bound form is much closer to the optimal value than for the apo form confirms that substrate-WW binding indeed preorganizes the catalytic-site loops for ligand binding. This preorganization is illustrated by the docked poses generated by RosettaLigand (Figure 6). Similar results are obtained for docking the cis ligand (Figure S10).
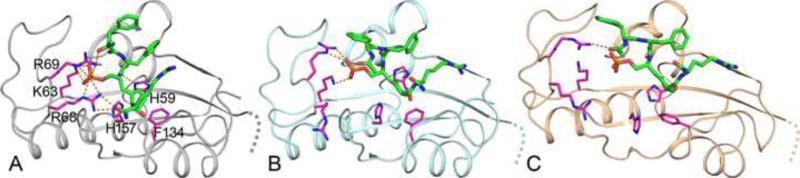
Figure 6. Docked poses for the trans ligand at the catalytic site, generated using RosettaLigand on conformations from our simulations.
(A) The optimal pose of the ligand, refined from the simulation of Pin1 with this ligand bound at the catalytic site (as well as the WW site). The ligand Pro residue is properly positioned relative to Pin1 His59, Phe134, and His157; the ligand pSer sidechain forms salt bridges with Lys63, Arg68, and Arg69 of the catalytic loop.
(B) The ligand docked to the empty catalytic site of a conformation from the simulation of Pin1 with FFpSPR bound at the WW site. The ligand Pro residue is also properly positioned, and the pSer sidechain still forms some of the salt bridges in (A).
(C) The ligand docked to the catalytic site of a conformation from the simulation of apo Pin1. The ligand Pro residue is shifted to the right, and the pSer sidechain loses all the salt bridges.
See also Figure S10.
Discussion
Through extensive molecular dynamics simulations, we have presented direct evidence for allosteric communication between the WW and PPIase domains of Pin1, and identified two pathways for mediating the allosteric regulation. The simulation results are in broad agreement with available experimental data (Jacobs et al., 2003; Namanja et al., 2011; Vanwart et al., 2012; Wilson et al., 2013), and present a more complete, atomistic picture for the allosteric regulation. In addition, our illustrative study on Pin1 has implications for a wide-range of issues, including a distinct role of conformational dynamics in eliciting allostery, conformational entropy as a determinant of binding affinity, and cooperative ligand binding.
Pin1 as a Three-Module Allosteric Enzyme
Our network analyses reveal that the PPIase domain can be further divided into the domain core and the α1-α2 appendage. Together with the WW domain, Pin1 can be viewed as a tripartite enzyme (Figure 5A). Interfacial interactions provide links between the WW domain and the PPIase core and between the PPIase core and the α1-α2 module, but WW-α1 links are largely missing in the apo protein. The substrate-binding pocket on the WW domain borders the WW-α1 groove. When a substrate binds, this groove is filled and a bridge between the WW and α1-α2 modules is formed. The WW-bound substrate thus provides essential links between the WW and α1-α2 modules to complete the second allosteric pathway from the WW domain to a catalytic-site loop.
The first allosteric pathway, which connects the WW domain with two other catalytic-site loops via the WW-PPIase core interface, preexists in apo Pin1, but remains dormant until the second pathway is completed by substrate-WW binding. That is, allosteric regulation is achieved only through the concerted action of the two pathways. Indeed, the two pathways appear to reinforce each other, since in some cases (e.g., the cis ligand-bound Pin1 and the substrate-bound I28A mutant; Figures 4C and S9B), incompletion of the second pathway is accompanied by breakage of the first pathway. The essential role of the second pathway is demonstrated not only by the absence of allosteric communication in the apo protein, but also by the observation that the communication is one-way only, i.e., ligand binding at the PPIase site does not lead to allosteric effects on the β1-β2 loop of the WW domain.
Peng and co-workers (Namanja et al., 2007; Namanja et al., 2011) previously proposed a hydrophobic conduit, consisting of about 10 methyl-containing residues which lost sidechain flexibility upon substrate binding (Figure S5B), for inter-domain communication. Our study suggests that these methyl-containing sidechains only provide a partial picture for the allosteric mechanism. The complete mechanism involves two mutually reinforcing pathways, one leading to two of the catalytic-site loops and the other leading to the third. The latter, i.e., the catalytic loop (residues 63-72), does not have any methyl-containing sidechains and serves as a reminder of the limitation of order parameter measurements. Molecular dynamics simulations also have limitations, such as the range of accessible timescales. Fortunately for Pin1, much of the conformational dynamics important for allosteric communication appears to occur at timescales up to 10s of ns and have been captured by our simulations.
Restrained Molecular Dynamics as a Tool for Dissecting Allosteric Effects
Here we introduced restrained molecular dynamics simulations to help dissect the allosteric effects of substrate binding and identify essential links along allosteric pathways. Our restrained simulations provide further support to the division of Pin1 into three modules and to the role of substrate-WW binding in furnishing essential WW-α1 links. This approach can be applied to allosteric proteins in general for revealing their modular designs and ascertaining allosteric pathways.
The restrained simulations also provide a unique opportunity to isolate the role of conformational dynamics in eliciting allostery. The classical view of allostery (Fischer et al., 2011; Monod et al., 1965) has emphasized the role of conformational transition. However, there is growing appreciation of the importance of the change in dynamics, as opposed to the change in conformation, even to the extent that perhaps allostery can be elicited by the former alone (Cooper and Dryden, 1984; Gasper et al., 2012; Petit et al., 2009; Tsai et al., 2008; Vashisth et al., 2013). However, in practice, a change in dynamics is always accompanied by some change in conformation and vice versa. It has thus been difficult to cleanly separate dynamical effects from conformational effects. Our application of restrained simulations demonstrates their utility in this regard. In these simulations, the mobility of a selected region of the protein is limited while imposing the apo conformation, thus generating a change in dynamics without an accompanying change in conformation. We produced allosteric effects similar to those by substrate-WW binding when the dynamically affected region included the essential WW-α1 links, thus providing a clear example of allostery by dynamics alone.
Conformational Entropy Reduction and Preorganization
By CSP mapping, Peng and co-workers (Namanja et al., 2011) measured the binding affinities of FFpSPR and the trans and cis ligands for the WW and catalytic sites of Pin1 and the isolated PPIase domain. The results for the catalytic site were puzzling: while the cis ligand, which does not bind to the WW site, had a 4-fold lower affinity for the full-length protein than for the PPIase, both FFpSPR and the trans ligand had 2 to 3-fold higher affinities for the full-length protein. We can now provide an explanation, based on the notion that catalytic-site loop rigidification can enhance affinity by reducing the entropy cost of binding. Conformational entropy has been recognized as a determinant of binding affinity in general (Frederick et al., 2007; Zhou and Gilson, 2009) and invoked to support the possibility of allostery by dynamics in particular (Petit et al., 2009).
Figure S6 shows that, compared to the isolated PPIase domain, the WW domain in full-length Pin1 amplifies the conformational flexibility of the three catalytic-site loops. We suggest that there is a corresponding increase in entropy cost for binding, thus explaining the 4-fold lower affinity of the cis ligand for the full-length protein than for the PPIase.
We expect the same 4-fold reduction in affinity for the trans ligand and FFpSPR, were they only able to bind the catalytic site. However, these two ligands also bind to the WW site, and our simulations show that FFpSPR-WW binding results in significant reduction in the catalytic-site loop flexibility (Figure 3). Therefore, after the WW site is occupied, the conformational entropy cost for binding to the catalytic site of Pin1 is even lower than for binding to the catalytic site of the isolated PPIase domain. This explains the 2 to 3-fold higher affinities of FFpSPR and the trans ligand.
The argument based on reduction in entropy cost is strengthened if the rigidified loop conformations of the substrate-WW bound form are more favorable for ligand binding than the flexible ensemble of the apo form. Our calculations using RosettaLigand show that this is indeed true (Figures 6B and S10B). Substrate-WW binding thus preorganizes the catalytic-site loops for ligand binding, presenting a unique form of cooperativity.
If substrate-WW binding preorganizes the catalytic-site loops for binding, the preorganization could lead to enhanced catalytic activity. This might explain the slightly higher kcat/KM value of the full-length Pin1 relative to the isolated PPIase domain (Lu et al., 1999a; Zhou et al., 2000).
The above argument can also explain the effects of the I28A mutation on binding affinity and catalytic activity. Peng and co-workers (Wilson et al., 2013) found lowered substrate binding affinities for both the WW site and the catalytic site as well as lowered kcat/KM. We attribute the reduced affinity for the WW site to a widened WW-α1 groove, due to a rotation of the WW domain away from α1. Furthermore, we suggest that the reduced affinity for the catalytic site is due to a disruption of the allosteric pathways in the WW-substrate bound form of the mutant (Figure S9B). As a result, the catalytic-site loop flexibility is not quenched as much as in the wild-type protein (Figure S8D), leading to reduced affinity at the catalytic site and lowered kcat/KM.
Reliability of the Allosteric Picture and Further Tests
We have used multiple analysis methods to characterize the differences in conformations, dynamics, and allosteric networks among the various liganded forms of Pin1. The principal component analysis (Figure 2) and Rg (Figures S3 and S4) and RMSF (Figure 3) results show that substrate binding to the WW domain leads to the closure of and mobility loss in the three catalytic-site loops of the PPIase domain. These remote effects demonstrate allosteric communication between the domains. Then, the two network analysis methods both reveal that the WW-bound substrate acts as a bridge between the WW domain and the α1 helix to complete the second allosteric pathway toward the catalytic loop. Lastly our restrained simulations confirm that the WW-α1 links are essential for the allosteric regulation. These various analyses of the many simulations are consistent and complementary in generating a robust, detailed picture of the allosteric behavior of Pin1.
We have already validated our computational results against a range of experimental data, including CSPs (Figure S2), order parameters (Figure S5), and the effects of the I28A mutation. Additional tests can be designed to further interrogate the allosteric picture presented here. For example, it is known that a different substrate, a 10-residue phosphothreonine peptide from the mitotic phosphatase Cdc25C, has less effect than FFpSPR in inducing Pin1 inter-domain coupling (Jacobs et al., 2003; Namanja et al., 2007). The Cdc25C peptide lines the binding pocket on the WW domain in a mode distinct from that of FFpSPR (Wintjens et al., 2001). Though its affinity for the WW domain is higher, CSP mapping suggests that the Cdc25C peptide interacts less intimately with the PPIase domain than FFpSPR (Namanja et al., 2007). The present study leads us to predict that a reduced ability in forming Path2 explains why the Cdc25C peptide is less effective in inducing Pin1 inter-domain coupling. It will be interesting to test this prediction in molecular dynamics simulations.
Another way to directly test the putative essential role of WW-α1 links in eliciting the allosteric effects is to see whether a constitutively active variant can be produced by crosslinking the WW domain and the α1 helix. The Cα-Cα distance between residues 31 (WW domain) and 93 (α1 helix) is slightly under 10 Å, and therefore it may be possible for a disulfide bond or metal coordination to form when these residues are mutated to cysteines. Such a mutant would be a tantalizing system for future simulations and experiments.
Potential for Combination Drug Therapy
The cooperative effects delineated above suggest that a WW-directed ligand can be a positive allosteric modulator for a catalytic site-directed ligand. Combinations of allosteric and orthosteric drugs have been noted for their abilities to enhance pharmacological action, reduce side effects, and combat drug-resistant mutants (Epping-Jordan et al., 2007; Nussinov and Tsai, 2013; Zimmermann et al., 2007). Pin1 dysregulation is implicated in various diseases (Lu, 2004; Lu and Zhou, 2007; Lu et al., 1999b; Wulf et al., 2001), and a number of orthosteric inhibitors have been designed (Moore and Potter, 2013; Wang and Etzkorn, 2006). The study here suggests the potential for a combination drug therapy against Pin1-related diseases, whereby allosteric inhibitors at the WW site enhance the binding affinities of orthosteric inhibitors at the catalytic site.
Methods
System Preparations
Initial models of the systems studied were prepared in Discovery Studio (Accelrys Software Inc., San Diego), based on the crystal structures of Pin1 bound with the cis or trans ligand at the catalytic site (PDB 3TCZ and 3TDB (Zhang et al., 2012)) and bound with a pSer-Pro containing peptide (PDB 1F8A (Verdecia et al., 2000)) at the WW site. The systems included Pin1 with FFpSPR bound at the WW site; Pin1 with the cis ligand bound at the catalytic site; and Pin1 with two copies of the trans ligand bound, one at the WW site and one at the catalytic site. For the Pin1-FFpSPR complex, the protein was from 3TDB and the substrate was modeled from one in 1F8A (after superimposing the protein molecules in these two PDB entries). The Pin1-cis ligand complex was taken directly from 3TCZ. The Pin1-trans ligand complex both retained the catalytic site-bound trans ligand in 3TDB and had a second copy of the trans ligand modeled after the WW-bound substrate in the Pin1-FFpSPR complex. Missing residues of the ligands and the protein (residues 39-50 in the latter case, which form the inter-domain linker) were added.
The above systems were modified straightforwardly to yield others. For example, the substrate was removed from the Pin1-FFpSPR complex to produce apo Pin1; the WW domain and the inter-domain linker were further deleted to produce the isolated PPIase domain in apo form; and the WW-bound ligand was removed from the Pin1-trans ligand complex to yield a complex with only one copy of the trans ligand bound, at the catalytic site.
Molecular Dynamics Simulations
All simulations were carried out using the AMBER software with the AMBER99SB force field (Hornak et al., 2006). The pSer residue was modeled as singly protonated, with atomic charges taken from Homeyer et al. (2006). The cis-locked and trans-locked imide fragments were treated as non-standard residues, denoted as CIS and TRA, respectively. The atomic charges of ACE-CIS/TRA-NME were calculated by the R.E.D. server (http://q4md-forcefieldtools.org/RED/) using Gaussian 03. The net charges of ACE and NME were set to 0, and those of CIS/TRA to -1. Other missing force-field parameters of the non-standard residues were taken from the general AMBER force field (gaff) and parm99 data set with minor modifications (Homeyer et al., 2006).
Each complex (or protein) was solvated in a cubic box with TIP3P waters (Jorgensen et al., 1983), with at least 10 Å between the solute and nearest side of the box. Sodium and chloride ions were added to neutralize the system and yield a 30 mM salt concentration. The whole system was first energy-minimized, with a series of position restraints on the solute (all atoms, backbone atoms, Cα atoms, and finally no atoms). Subsequently, the system was heated from 0 K to 295 K with backbone atoms restrained for 50 ps, and then equilibrated with Cα atoms restrained for 50 ps. The restraints excluded the inter-domain linker. The simulation was continued at constant pressure (1 bar, maintained by isotropic position scaling with a 2-ps relaxation time) and constant temperature (using the Langevin thermostat with a 2-ps−1 collision frequency) for 100 ns. The SHAKE algorithm (Ryckaert et al., 1977) was used to constrain all bonds involving hydrogens, allowing for a 2-fs timestep. Electrostatic interactions were treated by the particle mesh Ewald sum method (Essmann et al., 1995), with a 10 Å cutoff for non-bonded interactions in direct space.
We also carried out restrained simulations, in which a subset of Pin1 residues was restrained to their conformation in the frame closest to the average in the last 40 ns of the apo Pin1 simulation. The restraint was imposed on the RMSD calculated on the heavy atoms of the restraint set, with a harmonic force constant of 10 kcal/mol/Å2. All restrained simulations started from the last frame of the apo Pin1 simulation and ran for 100 ns.
The I28A mutant in apo form and in FFpSPR-bound form were also simulated. After introducing the mutation in the last frame of the corresponding wild-type simulation, each mutant simulation ran for 100 ns.
Principal Component Analysis and RMSF Calculation
Except for the radii of gyration shown in Figure S4, all analyses were made on the last 40 ns of each simulation. Principal component analysis was carried out using the Ptraj module in AMBER, over a total of 80,000 Pin1 conformations (20,000 conformations evenly sampled from the simulation of each of the four systems shown in Figure 2). For each system, the conformations were binned into a histogram over the first two principal components, and the histogram was converted into a free energy surface according to the Boltzmann relation.
Cα RMSFs were calculated for each system after superimposing all of its conformations to the simulation average, over the secondary-structure core.
Chemical Shift Prediction
SPARTA+ (Shen and Bax, 2010) was used to predict chemical shifts of backbone N and H nuclei, using 2,000 conformations for each system. Then the NH CSP of each residue (in ppm) was calculated as (Vashisth et al., 2013), where Δδ denotes the difference in chemical shift between a ligand-bound form and the apo form.
Order Parameter Calculation
values were calculated for all sidechain methyls in 40 1-ns windows. For each window, was calculated as (Chatfield et al., 1998)
where x, y, and z are Cartesian coordinates of a unit vector along a carbon-methyl bond, and 〉...〉 denotes the average over the conformations in the 1-ns window, after aligning to the initial model using all Cα atoms to remove translation and rotation. The averages of values over the 40 windows are reported (Figure S5C).
Calculation of Allosteric Networks
The algorithm of Kannan and Vishveshwara (1999), implemented in the Wordom analysis tool (Seeber et al., 2011), was used to identify allosteric networks. In each network, residues are linked by persistent tertiary contacts between sidechains during a simulation. Whether two residues, i and j, are linked is determined by their interaction percentage, defined as
where nij is the number of sidechain-sidechain heavy atom pairs within a 5 Å cutoff, and Ni and Nj are the normalization factors specified by residue types. Links are excluded between a residue and its four nearest neighbors in sequence. In each conformation sampled from a simulation, a provisional link is formed if Iij exceeds a threshold, Icritic. If a provisional link is formed in at least 48% of all the conformations from a simulation, then an actual link is formed. Icritic is chosen so that the largest cluster is approximately half of the total number of residues.
Docking to Catalytic Site by RosettaLigand
RosettaLigand (Davis and Baker, 2009) was used to dock the trans and cis ligands into the catalytic site of Pin1 in conformations sampled from the simulations of the apo form and the FFpSPR-bound form, and to refine the poses of the ligands obtained in the simulations of Pin1 with the trans and cis ligands bound at the catalytic site. Full flexibility was allowed for the ligands, but only sidechain flexibility was allowed for the protein thereby preserving the backbone conformations. 5,000 poses were generated in each docking run.
For each system, 10 conformations were selected from the simulation by clustering based on Cα RMSD. For the two systems with a ligand at the catalytic site, each of these conformations both provided the initial pose and served as the reference for selecting the refined pose. The top scoring (i.e., lowest protein-ligand interaction energy) pose with a ligand RMSD < 2 Å from the reference was selected as the refined pose. The centroid of the 10 refined poses is shown in Figure 6A for the trans ligand-bound system and in Figure S10A for the cis ligand-bound system.
To dock a ligand to a system that was free of the ligand in the simulation, one selected conformation of the corresponding ligand-bound system was used to generate the initial pose, based on aligning the protein Cα atoms within 10 Å of the ligand. After the docking run, a refined pose for the ligand-bound system was used as the reference for selecting the best pose for the ligand-free system. The upper bound on ligand RMSD was increased to 2.3 Å since a 2 Å bound did not produce any pose in three of the 20 docking runs for apo Pin1. The centroid of the 10 best poses is shown in Figure 6B,C for the trans ligand docked to FFpSPR-bound Pin1 or apo Pin1; corresponding results for the cis ligand are shown in Figure S10B,C.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey Peng for commenting on the manuscript. This work was supported by National Institutes of Health Grant GM58187.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions H.-X.Z. designed the research. J.G. and X.P. performed the research and analyzed the data. J.G. and H.-X.Z. wrote the manuscript.
References
- Chatfield DC, Szabo A, Brooks BR. Molecular dynamics of staphylococcal nuclease: comparison of simulation with 15N and 13C NMR relaxation data. J Am Chem Soc. 1998;120:5301–5311. [Google Scholar]
- Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Elber R. Simulations of allosteric transitions. Curr Opin Struct Biol. 2011;21:167–172. doi: 10.1016/j.sbi.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Epping-Jordan M, Le Poul E, Rocher J-P. Allosteric modulation: a novel approach to drug discovery. Innov Pharm Technol. 2007;24:24–26. [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- Feher VA, Durrant JD, Van Wart AT, Amaro RE. Computational approaches to mapping allosteric pathways. Curr Opin Struct Biol. 2014;25:98–103. doi: 10.1016/j.sbi.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Olsen KW, Nam K, Karplus M. Unsuspected pathway of the allosteric transition in hemoglobin. Proc Natl Acad Sci U S A. 2011;108:5608–5613. doi: 10.1073/pnas.1011995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper PM, Fuglestad B, Komives EA, Markwick PR, McCammon JA. Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities. Proc Natl Acad Sci U S A. 2012;109:21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning. PLoS Comput Biol. 2011;7:e1002154. doi: 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Vishveshwara S. A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc Natl Acad Sci U S A. 2007;104:15711–15716. doi: 10.1073/pnas.0704459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer N, Horn AH, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12:281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DM, Saxena K, Vogtherr M, Bernado P, Pons M, Fiebig KM. Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J Biol Chem. 2003;278:26174–26182. doi: 10.1074/jbc.M300796200. [DOI] [PubMed] [Google Scholar]
- Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Kannan N, Vishveshwara S. Identification of side-chain clusters in protein structures by a graph spectral method. J Mol Biol. 1999;292:441–464. doi: 10.1006/jmbi.1999.3058. [DOI] [PubMed] [Google Scholar]
- Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl–prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lu P-J, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine-or phosphothreonine-binding modules. Science. 1999a;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999b;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Moore JD, Potter A. Pin1 inhibitors: pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. 2013;23:4283–4291. doi: 10.1016/j.bmcl.2013.05.088. [DOI] [PubMed] [Google Scholar]
- Namanja AT, Peng T, Zintsmaster JS, Elson AC, Shakour MG, Peng JW. Substrate recognition reduces side-chain flexibility for conserved hydrophobic residues in human Pin1. Structure. 2007;15:313–327. doi: 10.1016/j.str.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson KA, Etzkorn FA, Peng JW. Stereospecific gating of functional motions in Pin1. Proc Natl Acad Sci U S A. 2011;108:12289–12294. doi: 10.1073/pnas.1019382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai C-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz J. A systems biology perspective on protein structural dynamics and signal transduction. Curr Opin Struct Biol. 2005;15:23–30. doi: 10.1016/j.sbi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- Seeber M, Felline A, Raimondi F, Muff S, Friedman R, Rao F, Caflisch A, Fanelli F. Wordom: a user-friendly program for the analysis of molecular structures, trajectories, and free energy surfaces. J Comput Chem. 2011;32:1183–1194. doi: 10.1002/jcc.21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci U S A. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWart AT, Durrant J, Votapka L, Amaro RE. Weighted implementation of suboptimal paths (WISP): an optimized algorithm and tool for dynamical network analysis. J Chem Theory Comput. 2014;10:511–517. doi: 10.1021/ct4008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwart AT, Eargle J, Luthey-Schulten Z, Amaro RE. Exploring residue component contributions to dynamical network models of allostery. J Chem Theory Comput. 2012;8:2949–2961. doi: 10.1021/ct300377a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisth H, Storaska AJ, Neubig RR, Brooks CL., 3rd Conformational dynamics of a regulator of G-protein signaling protein reveals a mechanism of allosteric inhibition by a small molecule. ACS Chem Biol. 2013;8:2778–2784. doi: 10.1021/cb400568g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Etzkorn FA. Peptidyl-prolyl isomerase inhibitors. Biopolymers. 2006;84:125–146. doi: 10.1002/bip.20240. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Xu B, Mullins AB, Neiler FK, Etzkorn FA. Conformationally locked isostere of phosphoSer-cis-Pro inhibits Pin1 23-fold better than phosphoSer-trans-Pro isostere. J Am Chem Soc. 2004;126:15533–15542. doi: 10.1021/ja046396m. [DOI] [PubMed] [Google Scholar]
- Wilson KA, Bouchard JJ, Peng JW. Interdomain interactions support interdomain communication in human Pin1. Biochemistry. 2013;52:6968–6981. doi: 10.1021/bi401057x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintjens R, Wieruszeski JM, Drobecq H, Rousselot-Pailley P, Buee L, Lippens G, Landrieu I. 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides. J Biol Chem. 2001;276:25150–25156. doi: 10.1074/jbc.M010327200. [DOI] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang XJ, Chen X, Bowman ME, Luo Y, Noel JP, Ellington AD, Etzkorn FA, Zhang Y. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem Biol. 2012;7:1462–1470. doi: 10.1021/cb3000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Gilson MK. Theory of free energy and entropy in noncovalent binding. Chem Rev. 2009;109:4092–4107. doi: 10.1021/cr800551w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu P-J, Shen M, Stoller G, Küllertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann GR, Lehar J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.