Abstract
Genetic manipulations of neuronal activity are a cornerstone of studies aimed to identify the functional impact of defined neurons for animal behavior. With its small nervous system, rapid life cycle, and genetic amenability, the fruit fly Drosophila melanogaster provides an attractive model system to study neuronal circuit function. In the past two decades, a large repertoire of elegant genetic tools has been developed to manipulate and study neural circuits in the fruit fly. Current techniques allow genetic ablation, constitutive silencing, or hyperactivation of neuronal activity and also include conditional thermogenetic or optogenetic activation or inhibition. As for all genetic techniques, the choice of the proper transgenic tool is essential for behavioral studies. Potency and impact of effectors may vary in distinct neuron types or distinct types of behavior. We here systematically test genetic effectors for their potency to alter the behavior of Drosophila larvae, using two distinct behavioral paradigms: general locomotor activity and directed, visually guided navigation. Our results show largely similar but not equal effects with different effector lines in both assays. Interestingly, differences in the magnitude of induced behavioral alterations between different effector lines remain largely consistent between the two behavioral assays. The observed potencies of the effector lines in aminergic and cholinergic neurons assessed here may help researchers to choose the best-suited genetic tools to dissect neuronal networks underlying the behavior of larval fruit flies.
Keywords: effector genes, locomotion, light avoidance, optogenetics, thermogenetics
THE binary GAL4/UAS system for targeted gene expression (Brand and Perrimon 1993) is widely used in Drosophila to manipulate or visualize neuronal networks and is an important tool that has largely contributed to the success of the fruit fly as a major model system in neuroscience. The availability of this expression system represents the starting point for the development of effector transgenes that allow researchers to dissect the function of genetically identifiable neurons with high spatial and temporal precision. This has turned the fly GAL4/UAS system into one of the most powerful neurogenetic tools available. Notably, the impact of this tool in various experiments is highly dependent on the selection of an appropriate effector line. For example, UAS-tetanus toxin (TNT) was successfully used in various studies to inhibit neurotransmitter release (Thum et al. 2006; Tripodi et al. 2008; Kong et al. 2010). TNT specifically cleaves neuronal Synaptobrevin (n-Syb), which is essential for synaptic vesicle release (Sweeney et al. 1995). In fly photoreceptors, however, TNT-resistant excitatory synapses exist along with TNT-sensitive ones (Rister and Heisenberg 2006). In addition, the potencies of the effector genes UAS-Kir2.1, UAS-TNT, UAS-Diphteria toxin A (UAS-DTA), and UAS-reaper (UAS-rpr) expressed in motor neurons, in mushroom body neurons, or pan-neuronally differed in the adult fly, depending on the properties of the defined target cells. For instance, adult-induced paralysis was more efficiently induced by effector genes silencing neuronal transmission than by effector genes causing cell ablation (Thum et al. 2006). Moreover, impairment of short-term memories was achieved by specific expression of UAS-shibirets (UAS-shits), while fly learning performance was not affected by the expression of UAS-TNT (Thum et al. 2006). Thus, it is crucial to choose effector genes that work robustly and reliably in the neuron type and behavior of interest.
In this study we used the Drosophila larva to systematically assess and compare the potency of 15 different effector lines in two distinct behaviors: 4 different effector genes causing cell ablation, 4 different effector genes that silence neuronal activity, and 7 different effector genes that increase neuronal excitability or intracellular signaling. In recent years, the Drosophila larva has emerged as a favorable model to investigate different neurobiological aspects based on its genetic accessibility, its reduced neuronal complexity in terms of cell numbers compared to adult flies, and its behavioral repertoire. Great advances were made in the understanding of neuronal networks required for larval learning and memory (Gerber et al. 2009; Selcho et al. 2009; Pauls et al. 2010; Von Essen et al. 2011; Selcho et al. 2014), olfaction (Vosshall and Stocker 2007; Stocker 2008; Gerber et al. 2009), vision (Keene et al. 2011; Kane et al. 2013), feeding (Cobb et al. 2009; Wang et al. 2013), and locomotor behavior (Saraswati et al. 2004; Selcho et al. 2012; Heckscher et al. 2012; Vogelstein et al. 2014), using the larva as a model system.
Here, we first manipulated larval locomotion by effector gene expression in octopaminergic/tyraminergic (OA/TA) neurons, using a Tdc2-Gal4 driver (Cole et al. 2005). Several studies have previously shown that OA and TA act antagonistically on muscle contraction, resulting in reduced locomotion in larvae lacking OA, whereas hypomorphic TA receptor mutants show longer track distances (Kutsukake et al. 2000; Nagaya et al. 2002; Saraswati et al. 2004; Selcho et al. 2012). A small set of ∼40 OA/TA neurons within the ventral nerve cord is necessary to control normal locomotor activity in the larva (Selcho et al. 2012). Tdc2-positive cells within the central brain are dispensable for larval locomotion, but necessary for mediating nonnutritional sugar information during larval associative conditioning (Selcho et al. 2014).
In parallel, we compared the efficiency of the selected effector genes in light avoidance behavior by ectopic expression in photoreceptor neurons via the lGMR-Gal4 driver line (Moses and Rubin 1991; Keene and Sprecher 2012). In larvae, the visual system consists of two simple eyes [called Bolwig’s organ (BO)] that are much simpler than the adult compound eyes. Each eye includes 12 photoreceptors, which are subdivided into two types: 8 photoreceptors express green-sensitive rhodopsin6 (rh6) while 4 photoreceptors express blue-sensitive rhodopsin5 (rh5) (Helfrich-Förster et al. 2002; Sprecher et al. 2007; Sprecher and Desplan 2008). Neuronal projections of photoreceptor cells innervate the larval optic neuropile (LON), where they connect to their target cells (Sprecher et al. 2011; Keene and Sprecher 2012). Feeding Drosophila larvae perform a stereotypic photophobic behavior when they are confronted to choose between light and darkness. Interestingly, for this avoidance behavior only rh5 but not rh6 is required. In addition, also neuronal silencing of the second-order interneurons fifth lateral neuron (LN) and dorsal neurons (DN2s) strongly impairs rapid light avoidance behavior (Keene et al. 2011).
By investigating larval locomotion and rapid light avoidance, we obtained similar, but not the same results for the different effector genes when genetically manipulating aminergic Tdc2-Gal4-positive and cholinergic lGMR-Gal4-positive neurons. The observed potency of the effector lines in aminergic and cholinergic neurons assessed here may help researchers to choose the best-suited genetic tools to dissect neuronal networks underlying the larval behavior of Drosophila.
Materials and Methods
Fly strains
Flies were cultured according to standard methods. For the behavioral experiments, all UAS lines were crossed to either Tdc2-Gal4 or lGMR-Gal4 driver lines. Heterozygous controls were obtained by crossing Gal4-driver and UAS-effector to w1118. UAS lines included in this study were UAS-rpr, UAS-grim, UAS-hid,rpr, UAS-Kir2.1, UAS-ΔOrk, UAS-shits UAS-TNTE, UAS-DTI, UAS-TRPA1, UAS-3×TRPM8, UAS-NaChBac, UAS-2xChR2 (UAS-ChR2-wt), UAS-ChR2-XXL, UAS-Pacα, UAS-bPac, and 10xUAS-myr::GFP (Table 1).
Table 1. The genotypes, brief description of the mode of action, and the observed efficiencies of the effector genes used in this study.
| Name | Genotype | Insertion chromosome(s) | Reference | Mechanism | Efficiency in OA/TA neurons | Efficiency in photoreceptor neurons |
|---|---|---|---|---|---|---|
| Cell ablation | ||||||
| UAS-reaper (rpr) | w1118 P(UAS-rpr.C)27 | 1 | Zhou et al. (1997) | Binding to IAPs and thus inducing cell death | ++ | ++ |
| UAS-grim | w*; P{UAS-grim.N}2 | 2 | Wing et al. (1998) | Binding to IAPs and thus inducing cell death | ++ | ++ |
| UAS-hid,rpr | w, P[UAS-hid], P[UAS-rpr] | 1 | Zhou et al. (1997) | Bindingto IAPs and thus inducing cell death | +++ | +++ |
| UAS-DTI | P{UAS-Cbβ\DT-A.I}14/TM3 | 3 | Bellen et al. (1992) | Inhibits protein synthesis | ++ | +++ |
| Neuronal silencing | ||||||
| UAS-shibirets (shits) | w; P{UAS-shits1.K}3 | 3 | Kitamoto (2001) | Temperature-sensitive dynamin impairs vesicle recycling | +++ | +++ |
| UAS-TNTE | w; P{UAS-TeTxLC.tnt}E2 | 2 | Sweeney et al. (1995) | Cleaves n-Syb and impairs vesicle docking | − | − |
| UAS-Kir2.1 | w*; P{UAS-Hsap\KCNJ2.EGFP}1 | 2 | Baines et al. (2001) | Inward rectifying K+ channel prevents membrane depolarization | +++ | +++ |
| UAS-ΔOrk | y1 w*; P{UAS-Ork1.Δ-C}2 | 2 | Nitabach et al. (2002) | Outward rectifying K+ channel prevents membrane depolarization | ++ | + |
| Neuronal activation | ||||||
| UAS-TRPM8 | yw; UAS-TRPM8 C4-D;UAS-TRPM8 C4-A, UAS-TRPM8 C1-A2) | 2; 3 | Peabody et al. (2009) | Temperature-dependent cation channel opens in response to ∼12°–16° → cell depolarization | − | − |
| UAS-TRPA1 | w; P{UAS-TRPA1(B).K}attP16 | 2 | Rosenzweig et al. (2005) | Temperature-dependent cation channel opens in response to ∼29° → cell depolarization | +++ | +++ |
| UAS-Pacα | P{UAS-Zzzz\PACα.SL} | 3 | Schröder-Lang et al. (2007) | Eukaryotic photoactivatable adenylyl cyclase increasing intracellular cAMP | − | ND |
| UAS-bPac | w*; P{UAS-bPAC.S} | 2 | Stierl et al. (2011) | Bacterial photoactivatable adenylyl cyclase increasing intracellular cAMP | − | ND |
| UAS-ChR2 | (a) P{UAS-ChR2.S}2; P{UAS-ChR2.S}3 (ChR2-wt) | (a) 2; 3 | (a) Nagel et al. (2003) | Photoactivatable cation channel (470 nm) → cell depolarization | (a) + | ND |
| (b) UAS-ChR2-XXL/cyo:GFP | (b) 2 | (b) Dawydow et al. (2014) | (b) +++ | |||
| UAS-NaChBac | y1 w*; P{UAS-NaChBac-EGFP}1/TM3, Sb1 | 3 | Nitabach et al. (2006) | Bacterial sodium channel increases sodium conductance → cell depolarization | ++ | ++ |
| Gal4 lines | ||||||
| Tdc2-Gal4 | w*; P{Tdc2-GAL4.C}2 | 2 | Cole et al. (2005) | Drives expression in octopaminergic/tyraminergic neurons | ||
| lGMR-Gal4 | y1 w*; wgSp-1/CyO; P{longGMR-GAL4}3/TM2 | 3 | Moses and Rubin (1991) | Drives expression in photoreceptor cells | ||
Behavioral assays
To analyze larval locomotor behavior we recorded single larvae for 1 min on 1.5% agarose in an 85-mm diameter petri dish under red light conditions. Recordings were made by a DMK22BUC03 video camera with a Pentax C2514-M objective in combination with IC capture software (www.theimagingsource.com). Offline tracking was done by the custom-made software package FlyTrace (J. P. Lindemann and E. Braun, http://web.biologie.uni-bielefeld.de/neurobiology/index.php/home) and a homemade MATLAB script to obtain crawling distances per minute for each larva. Experiments were performed at room temperature (∼24°) except for UAS-shits, UAS-TRPA1, and UAS-TRPM8, indicated respectively. To induce temperature-dependent cell manipulation, larvae kept at 25° were measured at restrictive temperature (33° for UAS-shits and UAS-TRPA1 and 16° for UAS-TRPM8) after 5 min incubation time. For optogenetic manipulation (UAS-ChR2-wt, UAS-ChR2-XXL, UAS-bPac, and UAS-Pacα) we used a 480-nm light-emitting diode (LED) with a light intensity of ∼0.14 mW/cm2. Although we used a cooling element, there was a slight increase in temperature (<1°) at the level of the arena due to the LED illumination. As published previously, all-trans-retinal (∼200 µM) was added to the standard medium to counter limited cellular availability of all-trans-retinal and thus increase efficiency of ChR2 expression for the UAS-ChR2-wt lines, but not for UAS-ChR2-XXL (Schroll et al. 2006; Ullrich et al. 2013; Dawydow et al. 2014).
Light avoidance was performed under red light conditions. The behavioral arena is made of a petri dish with a cover shading two of four quarters of the arena, thus consisting of a dark side and light-exposed side. Illumination intensity from a white-light LED lamp was 780 lux. The dark side clouds everything. A group of 30 larvae was collected from a food vial that was kept in darkness for 30 min before the experiment. During the 5-min preference test larvae freely move on the plate. After 5 min, larvae were counted on lit and dark quarters to calculate a dark preference index:
Immunofluorescence
Central nervous systems of third instar larvae were dissected in phosphate-buffered saline (PBS, pH 7.4) (Selcho et al. 2009). Afterward, the specimens were fixated in 4% paraformaldehyde in PBS for 40 min, washed four times in PBS with 0.3% Triton-X 100 (PBT), and blocked with 5% normal goat serum in PBT. Specimens were incubated with either antityrosine decarboxylase 2 (anti-Tdc2) [pab0822-p, Covalab; dilution 1:200 (Pech et al. 2013)] or anti-GFP [A6455, Molecular Probes (Eugene, OR); dilution 1:1000] in blocking solution for 1 night at 4°. Preparations were washed six times with PBT and incubated for 1 night at 4° with the secondary antibody goat anti-rabbit IgG DyLight488 (111-485-144, Jackson ImmunoResearch; dilution 1:250). Finally, specimens were rinsed six times in PBT and mounted in 80% glycerol in PBS. Until scanning with a Leica TCS SPE confocal microscope, specimens were stored in darkness at 4°.
BOs of third instar larvae were dissected in PBS and immediately fixed in 4% paraformaldehyde in PBS for 20 min. Samples were washed 8–10 times with PBT and subsequently incubated with primary antibodies overnight at 4°. The following primary antibodies were used: rat anti-Elav 1:20, mouse anti-Chaoptin 1:20 (both from Developmental Studies Hybridoma Bank), and rabbit anti-GFP (Molecular Probes; dilution 1:1000). The next day samples were washed every 30 min in PBT and subsequently incubated with secondary antibodies overnight at 4°. The following secondary antibodies were used: anti-rat Alexa-647 (Jackson ImmunoResearch), anti-mouse Alexa-488, and anti-rabbit Alexa-488 (both from Molecular Probes; 1:200). Next, samples were washed two times for 15 min in PBT and two times for 15 min in PBS before mounting in 50% glycerol. Images were taken with a Leica SP5 confocal microscope.
Statistical methods
For statistical comparison between genotypes, a Wilcoxon rank sum test was used. To compare single genotypes against chance level, we used the Wilcoxon signed-rank test. All statistical analyses and visualizations were done with R version 3.0.2 (www.r-project.org). In Figure 1, Figure 2, Figure 3, and Figure 4, data are presented as box plots, with 50% of the values of a given genotype being located within the box, and whiskers represent the entire set of data. Outliers are indicated as open circles. The median performance index is indicated as a thick line within the box plot. Significance levels between genotypes shown in the figures (Figure 1, Figure 2, Figure 3, and Figure 4) refer to the raw P-values obtained in the statistical tests.
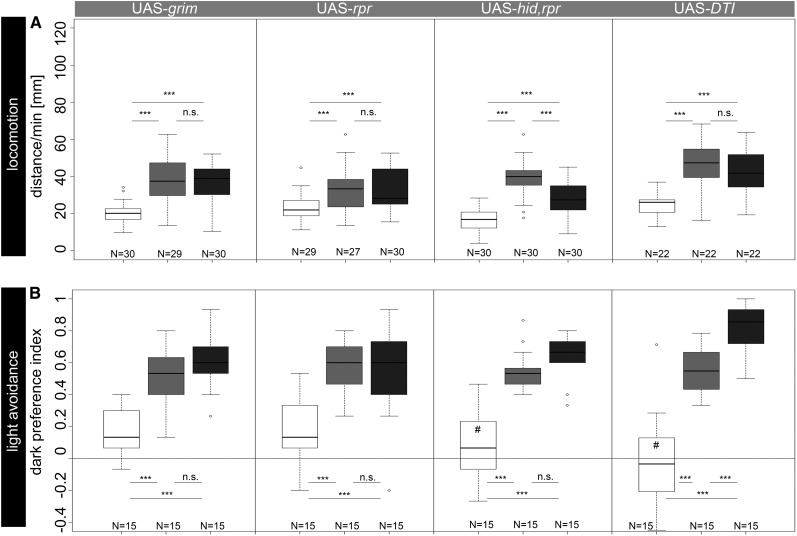
Figure 1.
(A) Locomotor behavior after Tdc2-Gal4-driven ablation of OA/TA neurons with UAS-grim, UAS-rpr, UAS-hid,rpr, or UAS-DTI. Experimental larvae showed significantly reduced performances compared to controls. (B) Rapid light avoidance behavior after specific ablation of photoreceptor neurons using lGMR-Gal4. Expression of Grim, Rpr, and Hid together with Rpr or DTI significantly reduced larval dark preferences compared to those of controls. Open box, experimental larvae; box with light shading, Gal4/+ larvae; box with dark shading, UAS/+ larvae. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05. #, not significantly different from chance level.
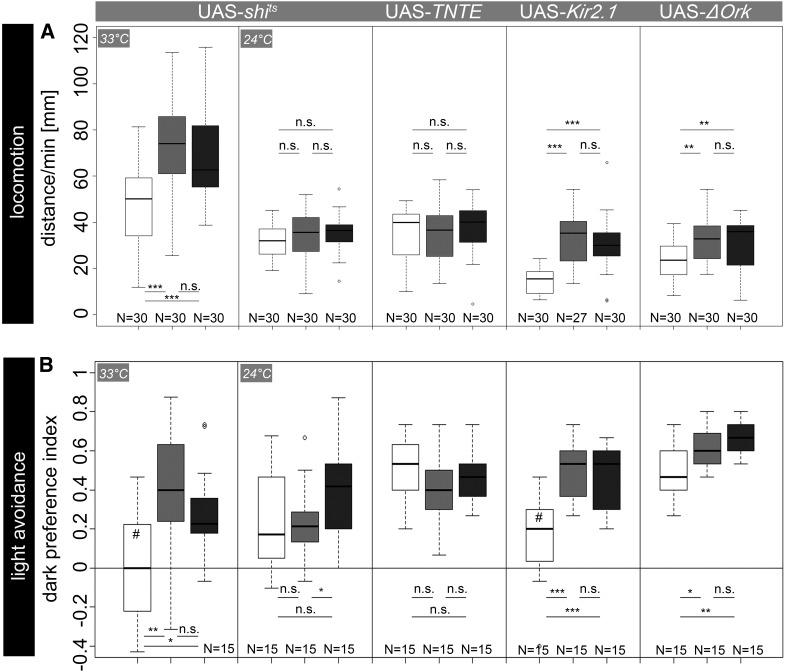
Figure 2.
(A) Locomotor behavior after neuronal silencing of OA/TA neurons via expression of UAS-shits, UAS-TNTE, UAS-Kir2.1, or UAS-ΔORK. Expression of UAS-shits reduced performance of experimental larvae compared to controls specifically at restrictive temperature. In contrast, no obvious effect appeared due to TNTE expression. After electrical silencing of OA/TA neurons with Kir2.1 or ΔOrk channels, experimental larvae crawled significantly less compared to controls. (B) lGMR-Gal4/UAS-shits larvae performed indistinguishably from chance level in rapid light avoidance at restrictive temperature. In contrast, performance of control larvae was significantly over chance level. Expression of TNTE in photoreceptor neurons had no effect on light avoidance. Expression of rectifier channels Kir2.1 and ΔOrk significantly reduced performances in experimental larvae compared to controls. Open box, experimental larvae; box with light shading, Gal4/+ larvae; box with dark shading, UAS/+ larvae. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05. #, not significantly different from chance level.
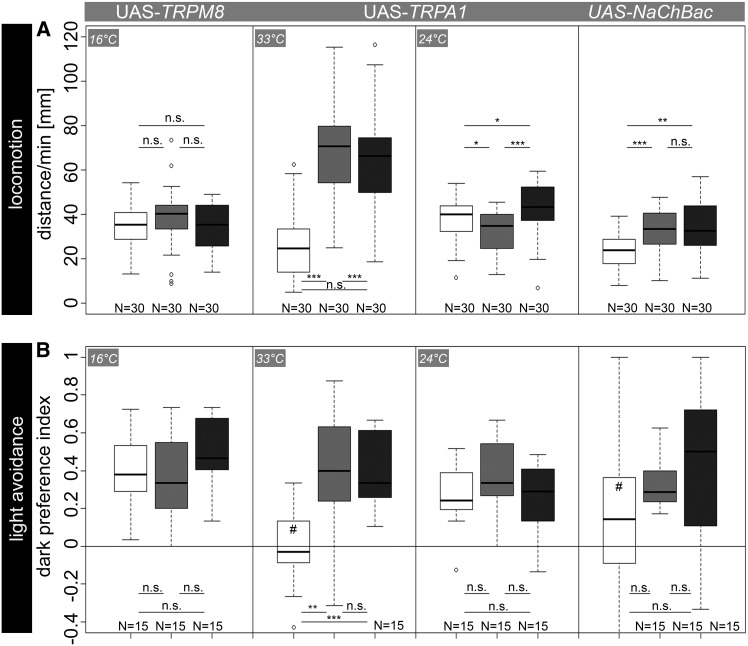
Figure 3.
(A) Locomotor behavior after artificial activation of OA/TA neurons using Tdc2-Gal4-directed expression of UAS-TRPM8, UAS-TRPA1, and UAS-NaChBac. Expression of UAS-TRPM8 did not affect larval locomotion at 16°. In contrast, thermogenetic activation of OA/TA neurons via UAS-TRPA1 reduced performance in experimental larvae compared to controls specifically at restrictive temperature. Additionally, expression of bacterial sodium channels via UAS-NaChBac reduced crawling distances significantly in Tdc2-Gal4/UAS-NaChBac larvae compared to controls. (B) Similar to the locomotion assay, rapid light avoidance was not affected in experimental larvae by TRPM8 expression. In contrast, dark preferences were abolished in experimental larvae after TRPA1 expression at restrictive temperature. Performance of control larvae was significantly different from chance level. In addition, lGMR-Gal4/UAS-NaChBac larvae showed no light avoidance behavior as performance scores were indistinguishable from chance level while controls performed over chance level. Here, experimental larvae performed not significantly different from controls. Open box, experimental larvae; box with light shading, Gal4/+ larvae; box with dark shading, UAS/+ larvae. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05. #, not significantly different from chance level.
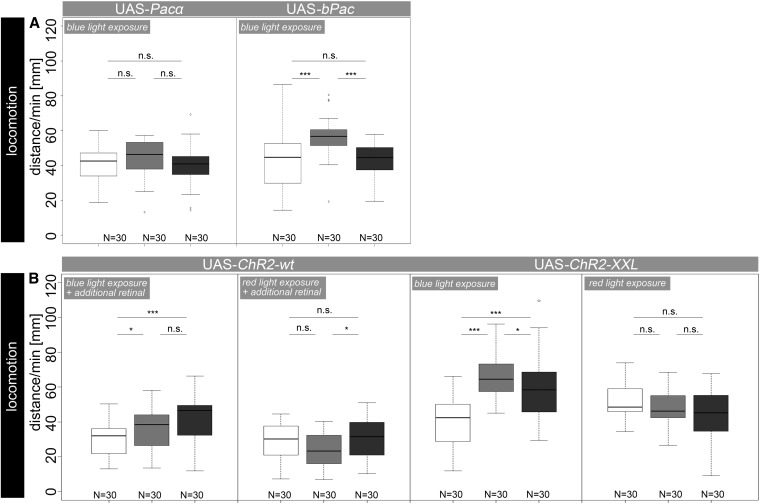
Figure 4.
Locomotor behavior after optogenetic activation of OA/TA neurons. Expression of UAS-Pacα and UAS-bPac did not affect larval locomotor behavior as experimental larvae performed indistinguishably from control larvae during illumination. In contrast, blue light exposure specifically affected locomotor behavior of Tdc2-Gal4/UAS-ChR2-wt and Tdc2-Gal4/UAS-ChR2-XXL as they crawled significantly shorter distances compared to both controls. Remarkably, under red light conditions experimental larvae performed not significantly different from controls. Open box, experimental larvae; box with light shading, Gal4/+ larvae; box with dark shading, UAS/+ larvae. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05.
Results
To evaluate the potencies of various effector genes to interfere with neurotransmission in the Drosophila larva, we expressed 15 different effector genes either in OA/TA neurons in the central nervous system (CNS) or in photoreceptor neurons in the BOs (Figure 1). Based on the expression pattern of Tdc2-Gal4 and lGMR-Gal4, we used larval locomotion and larval light avoidance as the behavioral readout. The results are summarized in Table 1. Information about the action of the tested effector genes in published behavioral studies on Drosophila larvae is compiled in Supporting Information, Table S1.
Effector genes inducing apoptotic cell death
The crudest way to interfere with neuronal transmission is to ablate neurons by the expression of different pro-apoptotic genes. Several studies suggest that Drosophila cell death is highly dependent on DIAP1 protein, a member of the inhibitor of apoptosis (IAP) family (Yoo et al. 2002). IAPs bind to active caspases and inhibit their proteolytic function by triggering caspase degradation. IAPs also bind to pro-apoptotic genes like grim, rpr (reaper), and hid (head involution defective), which prevents caspase binding and inhibition and therefore causes cell death (Kornbluth and White 2005). Although the distinct proteins may act on different targets within the apoptotic cascade, they also seem to functionally interact (Wing et al. 1998). For example, embryonic midline studies revealed that Grim alone is sufficient to ablate CNS midline cells in contrast to Hid and Rpr, indicating different apoptotic capabilities among the three gene products (Wing et al. 1998). In contrast, rpr expression alone is sufficient to ablate neurons (e.g., McNabb et al. 1997). Expression of the clostridian diphtheria toxin A (DTA) causes a general inhibition of protein synthesis and is therefore capable of inducing cell death (Bellen et al. 1992; Han et al. 2000). Toxicity is achieved by enzymatic inactivation of eukaryotic elongation factor-2 (Pappenheimer 1977). As DTA is lethal, an attenuated version I is used in Drosophila (DTI) (Bellen et al. 1992; Han et al. 2000).
Targeted expression of UAS-rpr, UAS-grim, or UAS-hid,rpr in OA/TA neurons significantly reduced the larval crawling distance per minute (Figure 1A). Although not immobile, experimental larvae showed impaired forward locomotion reduced to ∼50% of control levels. Tdc2-Gal4/UAS-grim larvae moved significantly less compared to Gal4/+ and UAS/+ larvae (P < 0.001). Similarly, the expression of UAS-rpr in Tdc2-Gal4-positive neurons led to reduced distances of the experimental larvae compared to controls (P < 0.001). Since different studies suggested that a combined expression of pro-apoptotic genes might have synergistic effects (Wing et al. 1998; Selcho et al. 2012), we expressed hid and rpr, using Tdc2-Gal4 to ablate OA/TA neurons. Tdc2-Gal4/UAS-hid,rpr larvae crawled significantly less than Tdc2-Gal4/+ and UAS-hid,rpr/+ larvae (P < 0.001), while the effect seemed to be at best just slightly stronger compared to the expression of grim and rpr alone. Inhibition of protein synthesis by expression of UAS-DTI significantly reduced larval crawling distances per minute compared to those of Tdc2-Gal4/+ and UAS-DTI/+ (P < 0.001; Figure 1A).
Targeted expression of pro-apoptotic genes in lGMR-Gal4-positive photoreceptor neurons led to results similar to those of the locomotion assay. lGMR-Gal4/UAS-grim larvae showed reduced rapid light avoidance indicated by higher numbers of larvae remaining in the illuminated quarters during the preference test (Figure 1B). Performance of experimental larvae was significantly reduced compared to that of lGMR-Gal4/+ and UAS-grim (P < 0.001) controls. Experimental lGMR-Gal4/UAS-rpr larvae showed reduced preference scores compared to both control groups (P < 0.001). Remarkably, expression of either grim or rpr alone did not lead to complete impaired light avoidance of experimental larvae (both P < 0.05 against chance level). In contrast, performance was indistinguishable from chance level after expression of combined hid and rpr (P > 0.05), whereas performance of control larvae was significantly over chance level (P < 0.001 for lGMR-Gal4/+ and UAS-hid,rpr/+). Similarly, inhibition of protein synthesis in photoreceptor neurons by expression of UAS-DTI completely abolished light avoidance behavior (P < 0.05 against chance level) while both control groups performed over chance level (P < 0.0001 compared to lGMR-Gal4/+ and UAS-DTI/+).
To confirm the efficiency of pro-apoptotic genes and DTI we visualized Tdc2-positive cells after expression of pro-apoptotic genes and DTI (Figure S1, A–C, E–G, and I–K) by anti-Tdc2 immunostaining (Pech et al. 2013) that labels OA/TA neurons within the brain and ventral nerve cord. In addition, we expressed 10×UAS-myr::GFP (Pfeiffer et al. 2010) to analyze the efficiency of cell ablation and protein synthesis inhibition with the Gal4 expression pattern as anti-Tdc2 antibody labels neurons not included in the expression pattern of Tdc2-Gal4 (Figure S1, D, H, and L). Expression of pro-apoptotic genes grim, rpr, and combined hid and rpr in OA/TA neurons led to a similar strong reduction in cell number, showing the efficiency of the effectors. Nevertheless, in all cases a small, varying number of Tdc2-Gal4-positive neurons escaped apoptosis. The number of GFP-expressing cells seemed to be slightly lower in larvae expressing hid,rpr (6.6 ± 1.2 surviving cells) compared to larvae expressing either grim (9.4 ± 0.4 surviving cells) or rpr (10 ± 0.4 surviving cells). In detail, ventral paired median (VPM) neurons in thoracic neuromeres t1–t3 and abdominal neuromere a1 as well as one dorsal unpaired neuron in abdominal neuromere a9 seemed to consistently survive apoptosis, while ventral unpaired median (VUM) neurons in thoracic and abdominal neuromeres seemed to consistently die (Figure S1). Furthermore, labeling of OA/TA neurons via anti-Tdc2 and anti-GFP, respectively, indicated weak but consistent expression of Tdc2 or GFP after expression of UAS-DTI (Figure S1, M and P; 34.6 ± 0.24 cells survived in comparison to 42 ± 0.57 cells in control CNS). In contrast, expression of DTI as well as grim, rpr, and hid,rpr in photoreceptor neurons using lGMR-Gal4 completely ablated all photoreceptor neurons based on anti-Elav labeling (Figure S2, C, F, I, and L).
Effector genes inducing neuronal silencing
Impairment of neuronal transmission by genetical cell ablation is a crude manipulation and may cause various side effects. To avoid adaptations of the corresponding neuronal networks due to ablated cells, inhibition of synaptic transmission is probably a more cautious alternative to interfere with neuronal communication. Synaptic transmission relies on Ca2+-dependent neurotransmitter release from synaptic vesicles. At the presynapse, a multitude of proteins including SNARE proteins located in the vesicle (v-SNARE) and presynaptic membrane (t-SNARE) are required for proper fusion events. One of the key v-SNARE proteins for targeted vesicle fusion is neuronal Synaptobrevin (n-Syb). n-Syb is targeted by clostridial tetanus toxin that efficiently inhibits chemical transmission. Tetanus toxin consists of a heavy polypeptide chain required for proper binding to its neuronal target, plus a light chain. Expression of the light chain (UAS-TNT or UAS-TeTxLc) intracellularly cleaves n-Syb and thereby diminishes synaptic transmission (Sweeney et al. 1995).
Another way to interfere with neurotransmission is the expression of UAS-shits (a temperature-sensitive dominant-negative form of Dynamin) as this conditional effector can bypass developmental effects or synaptic compensation of constitutively silenced neurons (Kitamoto 2001). Dynamin encodes a GTPase required for normal endocytosis and is crucial for vesicle recycling and neuronal functionality. Ectopic expression of UAS-shits blocks neuronal transmission only at restrictive temperature (>29°). Since neuronal inhibition by Shits is achieved only at restrictive temperature, this tool allows a rapid and reversible inhibition in a spatially and temporally controlled manner (Kitamoto 2001). A limitation of this effector gene is the necessity to increase temperature during the experiment, which might cause side effects.
A third way to induce neuronal silencing is via overexpression of permanently open K+ channels (Baines et al. 2001; Nitabach et al. 2002). Kir2.1 is a human inward rectifying potassium channel. Neuronal overexpression of Kir2.1 hyperpolarizes neurons and reduces the probability of evoked action potential generation and neurotransmitter release at the presynapse, while spontaneous release of neurotransmitters seems to be unaffected (Baines et al. 2001). Similar to Kir2.1 channels, the Drosophila ΔOrk outward rectifying potassium channels hyperpolarize neurons and inhibit normal synaptic transmission (Nitabach et al. 2002). ΔOrk channels act like a K+-selective hole in the cell membrane without any voltage or time dependence of the open state inducing currents similar to natural leak conductance. The native function of this channel in Drosophila is unknown (Goldstein et al. 1996).
In our experiments, Shits appeared to be the most potent transgenic tool to block neurotransmission (Kitamoto 2001). At 24°, both larval locomotion and rapid light avoidance were unaffected in Shits-expressing larvae (Figure 2, A and B; P > 0.05). In contrast, at 33° Tdc2-Gal4/UAS-shits larvae moved significantly less than Tdc2-Gal4/+ and UAS-shits/+ (P < 0.001) controls. Although experimental larvae moved less than controls, mean distances for all groups were higher at 33° than in any other experiment at 24°, indicating a generally increased locomotor activity at higher temperatures. In some behavioral paradigms, this might strongly affect behavioral outcomes. In these cases, transgenic tools other than Shits might be better suited to silence synaptic transmission. We therefore tested Tdc2-Gal4/UAS-TNTE larvae that performed indistinguishably from both Tdc2-Gal4/+ and UAS-TNTE/+ (P > 0.05) controls, indicating no efficient block of neuronal activity in OA/TA neurons. As both Shits and TNT affect chemical transmission and thus likely leave electrical synapses unaffected, we next tested the ion channels Kir2.1 and ΔOrk that alter the membrane potential. Tdc2-Gal4/UAS-Kir2.1 larvae showed significantly reduced distance scores (P < 0.001). Along the same line, expression of UAS-ΔOrk significantly reduced the crawling distances of experimental larvae compared to controls (P < 0.01). Notably, by trend the expression of UAS-Kir2.1 seemed to be more efficient than the expression of UAS-ΔOrk (see Figure 2A).
Similar results for silencing synaptic transmission were obtained in the light avoidance assay. No significant dark preference was shown after conditional silencing of photoreceptor neurons, using UAS-shits (P = 0.91; Figure 2B). In contrast, dark preferences of control larvae were significantly higher compared to those of lGMR-Gal4/UAS-shits larvae (P < 0.01 for lGMR-Gal4/+ and P < 0.05 for UAS-shits/+) and over chance level (both P > 0.05). Similar to larval locomotion, expression of UAS-TNTE did not lead to any significant changes in light avoidance (P > 0.05). However, lethality after the ubiquitous expression of UAS-TNTE driven by actin-Gal4 confirmed the general potency of this effector gene (data not shown). Furthermore, we used anti-TNT labeling to underline the inefficiency of UAS-TNTE in OA/TA neurons or photoreceptor neurons, respectively. While immunolabeling after expression driven by Tdc2-Gal4 (Figure S1Q) indicated presence of TNT in OA/TA neurons, no labeling was found in photoreceptors after expression driven by lGMR-Gal4. Thus, unaffected light avoidance in lGMR-Gal4/UAS-TNTE larvae might be due to a lack of TNT expression in photoreceptor neurons. Electrical silencing using either UAS-Kir2.1 or UAS-ΔOrk led to significantly reduced light avoidance of experimental larvae. The dark preference of lGMR-Gal4/UAS-Kir2.1 larvae was significantly reduced (P < 0.001). Although the dark preference of lGMR-Gal4/UAS-ΔOrk larvae was also reduced compared to that of controls (P < 0.05 for lGMR-Gal4/+ and P < 0.01 for UAS-ΔOrk/+), ΔOrk seemed to be again—by trend—less potent than Kir2.1 as lGMR-Gal4/UAS-Kir2.1 larvae performed not significantly different from chance level. Since both constructs are GFP tagged, we used anti-GFP labeling to confirm effector gene expression and to investigate whether the expression of permanently open potassium channels throughout development changes neuronal morphology. Neither expression of ΔOrk nor that of Kir2.1 obviously altered the morphology or arborization pattern of OA/TA neurons within the ventral nerve cord (data not shown).
Effector genes increasing neuronal excitability or intracellular signaling
While effector genes suppressing neuronal activity help to identify the necessity of defined neurons for a certain behavior, activators may also identify modulatory effects of these neurons onto the regarding circuit. To specifically activate neuronal activity or intracellular signaling in defined cells, we used effector genes encoding different cation channels, as neuronal activation can be achieved by influx of sodium or calcium or the decrease of potassium conductance. Two widely used activator genes are TRPA1 and TRPM8, members of the transient receptor potential (TRP) cation channel superfamily that are sensitive to different temperatures (Rosenzweig et al. 2005). TRPA1 channels activate in response to warm temperatures (Viswanath et al. 2003). In contrast, rat TRPM8 is responsible for sensing mild cold temperatures (Colburn et al. 2007; Peabody et al. 2009). UAS-Channelrhodopsin2 (UAS-ChR2-wt and UAS-ChR2-XXL), UAS-Pacα, and UAS-bPac expression was successfully used for optogenetic cell manipulation (see Table S1). ChR2 is a light-activatable cation channel from the flagellate Chlamydomonas reinhardtii with seven transmembrane domains and an all-trans chromophore, which responds to blue light stimulation (∼480 nm) by opening the internal channel (Nagel et al. 2003; Zhang et al. 2007; Dawydow et al. 2014). The open state of these channels allows Na+ and to a lower extent Ca2+ to enter the cell, which leads to membrane depolarization. Wild-type ChR2 (ChR2-wt) was shown to stimulate neuronal activity when additional all-trans-retinal is added to the food media to compensate for the limited cellular retinal availability. Low light transmission through the cuticle is known to be the bottleneck for optogenetic approaches in adult flies (Dawydow et al. 2014). ChR2-variant ChR2-XXL was recently shown to bypass this limitation as photosensitivity is 10,000 times higher than in ChR2-wt. In addition, ChR2-XXL efficiently stimulates neuronal activity without retinal supplementation (Dawydow et al. 2014). PACα is a subunit of a light-activatable adenylyl cyclase from the flagellate Euglena gracilis. The photoactivated adenylyl cyclase (PAC) is composed of two subunits: PACα and PACβ (Schröder-Lang et al. 2007). Expression of PACα in Drosophila allows cell manipulation by blue light stimulation due to increasing intracellular cAMP levels in a spatiotemporal manner. Additionally, a further Pac from sulfide-oxidizing Beggiatoa bacteria (bPac) was introduced to Drosophila for optogenetic approaches (Stierl et al. 2011). bPac carries a blue light-sensitive domain linked to a type III adenylyl cyclase, allowing cell manipulation similar to that in UAS-Pacα. bPac cyclase activity seems to be 3–4 times higher than Pacα activity. bPac thus needs ∼1000 times less light to induce similar cAMP changes in neurons (Stierl et al. 2011). Finally, expression of UAS-NaChBac, a bacterial voltage-gated sodium channel, can be used to permanently increase neuronal excitability (Ren et al. 2001; Nitabach et al. 2006). Expressed in oocytes, NaChBac was able to conduct Na+ inward currents with a lower activation threshold than endogenous Drosophila voltage-gated Na+ channels (Nitabach et al. 2006).
At 16°, Tdc2-Gal4/UAS-TRPM8 larvae crawled similar distances per minute to Tdc2-Gal4/+ and UAS-TRPM8/+ (P > 0.05) controls. In contrast, high temperature-induced opening of TRPA1 channels in OA/TA neurons led to significantly reduced crawling distances in experimental larvae compared to both control groups (P < 0.001; Figure 3A). Similar to the results achieved during the UAS-shits experiments, larval crawling was generally increased by high temperature (33°; Figure 2 and Figure 3). To test for the temperature specificity of TRPA1, we performed the experiment at room temperature (24°). As expected, there was no effect between experimental larvae and control larvae. Tdc2-Gal4/UAS-TRPA1 larvae crawled significantly longer distances per minute than Tdc2-Gal4/+ larvae (P < 0.05), but significantly shorter distances compared to UAS-TRPA1/+ larvae (P < 0.05). While TRP channels allow conditional activation of neurons specifically during an experiment, expression of bacterial sodium channels via UAS-NaChBac constitutively activates neurons, which might cause unspecific side effects. Nevertheless, Tdc2-Gal4/UAS-NaChBac larvae showed significantly reduced crawling distances compared to both controls (P < 0.001 for Tdc2-Gal4/+ and P <0.01 for UAS-NaChBac/+).
Although successfully used by Peabody et al. (2009), there was no effect in rapid light avoidance at 16° after expression of UAS-TRPM8 in photoreceptor neurons (P > 0.05; Figure 3B). To make sure that our temperature decrement is generally sufficient to activate TRPM8 channels, we expressed UAS-TRPM8 in motor neurons via OK6-Gal4. After short cold exposure at 16°, larvae were immobile, indicating this temperature decrement is sufficient to activate TRPM8 channels and thus induce cell activity (data not shown). In contrast, at 33° expression of UAS-TRPA1 using lGMR-Gal4 reduced dark preference scores of experimental larvae to zero (P > 0.05). Both control groups performed over chance levels and preferred dark quarters significantly more compared to lGMR-Gal4/UAS-TRPA1 (P < 0.01 for lGMR-Gal4/+ and P < 0.001 for UAS-TRPA1/+). In accordance with the results obtained for larval locomotion, there was no effect at room temperature (24°), indicating the temperature specificity of TRPA1 action (P > 0.05; Figure 3B).
Next, we expressed UAS-NaChBac in photoreceptor neurons. In contrast to the results obtained in the locomotion assay, lGMR-Gal4/UAS-NaChBac larvae performed not significantly different from controls (P > 0.05 for lGMR-Gal4/+ and UAS-NaChBac/+). The lack of significance, however, may rely on higher variance within this data set (Figure 3B): while control larvae performed over chance levels (both P < 0.01), experimental larvae showed no significant preference for darkness (P > 0.05), suggesting an increased excitability of photoreceptor neurons after expression of NaChBac channels.
Finally, we tested the optogenetic effectors UAS-Pacα, UAS-bPac, and two different versions of UAS-ChR2 to activate OA/TA neurons in larval locomotion. The expression of these effector genes in photoreceptor neurons was omitted, as it seems counterproductive to activate light-sensitive neurons via blue light exposure. Expression of UAS-Pacα in OA/TA neurons did not lead to any significant effect between experimental and control larvae (all P > 0.05; Figure 4A), indicating UAS-Pacα to be ineffective, possibly since an increase of intracellular cAMP in Tdc2-positive neurons does not affect larval locomotion. Similarly, cellular manipulation using expression of UAS-bPac did not lead to significant changes at lit conditions since experimental larvae performed not significantly different from UAS control larvae (P > 0.05; Figure 4A).
Expression of UAS-ChR2-wt using Tdc2-Gal4 significantly reduced larval crawling distances compared to those of Tdc2-Gal4/+ (P < 0.001) and UAS-ChR2/+ (P < 0.05) larvae at blue light exposure (Figure 4B). Also UAS-ChR2-XXL profoundly reduced larval locomotion after expression in OA/TA neurons, yet without retinal supplementation (P < 0.001) (Dawydow et al. 2014). To exclude unspecific effects induced by ChR2-acitvating blue light, we repeated the experiments under red light conditions. As expected, experimental larvae carrying either one of the two ChR2 variants performed indistinguishably from both control groups (all P > 0.05).
Discussion
Comparison of various effector genes to manipulate neuronal activity
Relatively simple neuronal circuits in Drosophila flies and especially larvae facilitate neurogenetic manipulations (see, e.g., Vogelstein et al. 2014) to investigate how the brain organizes behavior based on changing environmental information and innate needs. However, it is crucial to choose transgenic tools, which reliably and robustly manipulate the neuron type and behavior of interest. Here we compared the efficiency of 15 different effector genes in larval aminergic and cholinergic neurons that were previously shown to affect neuronal activity and corresponding behavior in Drosophila (Venken et al. 2011).
First, expression of different pro-apoptotic genes appeared to be highly efficient to affect both OA/TA neurons and photoreceptor neurons and thus larval locomotor and light avoidance behavior. In light avoidance, the combined expression of hid and rpr showed the highest efficiency as larval dark preference dropped to zero. Although expression of grim and rpr alone also strongly affected light avoidance in experimental larvae, they still performed over chance level, suggesting that the combined expression of two pro-apoptotic genes enhances the efficiency to induce cell death. This is in line with studies on adipokinetic hormone (AKH)-producing neurosecretory cells. Expression of UAS-hid,rpr was sufficient to consistently eliminate AKH cells whereas few cells survived after expression of UAS-hid or UAS-rpr alone (Isabel et al. 2005). GFP labeling after expression of hid and rpr in OA/TA neurons revealed a slightly stronger reduction in cell number compared to larvae expressing rpr or grim, while in all cases a small varying number of Tdc2-Gal4 positive neurons escaped apoptosis, possibly due to the low Gal4 expression in these cells. However, it seems difficult to assume that the—at best—slightly stronger reduction in larval distances per minute in larvae expressing hid,rpr compared to larvae expressing rpr or grim might rely on the higher number of ablated cells. This is in line with previous findings showing that adult paralysis is less efficiently induced by cell ablation than by neuronal silencing (Thum et al. 2006). Interestingly, in the larval locomotion assay not a single larva turned out to be fully immobile after cell ablation, neuronal silencing, or activation, respectively. This suggests that either OA/TA neurons modulate locomotion rather than command its initiation or functional redundancy is in place to compensate for impaired OA/TA signaling.
Expression of Shits, ΔOrk, and Kir2.1 appeared to efficiently silence neuronal transmission in OA/TA neurons and photoreceptor neurons. UAS-shits strongly reduced crawling distances in the locomotion assay and similar to the combined expression of hid and rpr reduced larval dark preference to zero. Silencing by ectopic potassium channel expression was efficient to reduce larval locomotion and larval dark preferences, with Kir2.1 having a slightly stronger effect than ΔOrk. In contrast, TNTE expression was insufficient to manipulate either larval locomotion or rapid light avoidance. Similarly, TNT was shown to fail in silencing adult photoreceptor neurons (Rister and Heisenberg 2006) and mushroom body neurons (Thum et al. 2006), indicating the presence of TNT-resistant neurons.
Neuronal activation was achieved by UAS-TRPA1 and UAS-NaChBac. Additionally, larval locomotion was affected in response to optogenetic activation using two variants of UAS-ChR2. Here, the newly developed ChR2-XXL seemed to be more efficient compared to the wild-type ChR2-wt, which is in line with the reported extended open-state lifetime, elevated cellular expression, and reduced dependence on retinal supplementation of ChR2-XXL (Dawydow et al. 2014). In this study we did not test light-inducible channels to activate photoreceptor neurons as photoactivation likely interferes with the light-sensing pathways. In contrast to the effector genes discussed above, UAS-TRPM8, UAS-bPac, and UAS-Pacα failed to alter the behaviors used in this study. Pacα and bPac expression was not sufficient to affect larval locomotion after blue light stimulation. Basal activity of endogenous adenylyl cyclases in response to certain physiological states of the animal and thus higher levels of cAMP before induced Pacα or bPac stimulation by light exposure may underlie this inefficiency. This is in particular likely for the OA/TA neurons, which are known to signal stress (Roeder 2005). Thus, stress-stimulated elevated cAMP levels might have been induced by handling during or prior to the experiment. An alternative explanation is that elevated intracellular cAMP levels in OA/TA neurons simply do not affect larval locomotor behavior. Furthermore, it is noteworthy to mention that Pacα and bPac change intracellular signaling by altering cAMP levels rather that change neuronal excitability. Thus, the general potency of light-inducible adenylyl cyclase (and probably for many more effector genes) seems to be highly dependent on the properties of the target neurons.
UAS-TRPM8 was expressed pan-neuronally, using elav-Gal4 to test its functionality to different temperature decrements (Peabody et al. 2009). Here 100% of flies fell down after temperature shift from 24° to ∼15° within 2.5 min, using one or three copies of UAS-TRPM8, respectively. The mildest shift to 18° was already sufficient to induce immobility in 60% of male flies, but not in female flies (Peabody et al. 2009). In our study we used a temperature shift from ∼23° to ∼16°, which was not sufficient to affect either larval locomotion or larval light avoidance behavior. Moreover, UAS-TRPA1 and UAS-TRPM8 were used to screen and identify neurons controlling motor output in adult flies (Flood et al. 2013). Surprisingly, both transgenic lines induced contrasting results driven by the same Gal4 lines, most probably based on differences in action potential frequency (Hamada et al. 2008; Peabody et al. 2009).
Benefits and drawbacks of effector gene use in larval Drosophila
In summary, UAS-hid,rpr, UAS-shits, and UAS-Kir2.1 seemed to be the most potent effector lines to impair neuronal transmission, since with these effector genes dark preferences of tested larvae were indistinguishable from chance levels. UAS-TRPA1 turned out to be most efficient in activating photoreceptor neurons as also here dark preferences dropped to zero, while the efficiency of ChR2 variants to manipulate photoreceptor neurons was not tested in this study. In the locomotion assay, UAS-TRPA1 and UAS-ChR2-XXL seemed to be equally capable to induce neuronal activation.
While cell ablation is the crudest way to manipulate neuronal signaling, it comes with the plus that the efficiency of cell ablations can easily be assessed by antibody staining. Electrical synapses can be modulated by the expression of the rectifier potassium channels UAS-Kir2.1 and UAS-ΔOrk. Both lines are available as a GFP-tagged version (Baines et al. 2001; Nitabach et al. 2002) (Figure S1 and Figure S2), allowing fluorescent detection to verify the ectopic expression of rectifier potassium channels and direct labeling of the manipulated neurons. In addition, UAS-ΔOrk is also available in a nonconducting version (Nitabach et al. 2002) to serve as a suitable genetic control. Both UAS-Kir2.1 and UAS-ΔOrk affected—albeit to a slightly different extent—larval locomotion and rapid light avoidance, indicating that both are a suitable choice. The largest benefit of UAS-shits is the possibility to induce fast and reversible conditional synaptic block by high temperatures. This eliminates possible developmental and adaptation effects. The same benefit applies for the usage of UAS-TRPA1 and UAS-TRPM8 (if functional in a given experiment). Also UAS-ChR2 variants and UAS-Pacα or UAS-bPac (if functional in a given experiment) can be expressed to specifically activate neurons or neuron populations only during the experiment and thereby omit developmental side effects.
While all light- and temperature-inducible effector genes share the benefit of spatiotemporal conditional usage within the experiment, the larval locomotion experiment revealed disadvantages of these thermo- and optogenetic tools. Light and especially higher temperature during the experiment changed naive behaviors in larvae. In addition, heat-inducible effector genes are less suitable to investigate temperature-dependent behaviors in Drosophila. The same is true for the combination of light-dependent behaviors and light-inducible effector genes such as UAS-ChR2 variants and UAS-Pacα or UAS-bPac. It should also be kept in mind that all effector genes studied here share the caveat that the behavioral readout after their usage gives no indication about the identity of the transmitter underlying the observed effects. For example, biogenic amines and neuropeptides are often coexpressed with classical neurotransmitters like acetylcholine or GABA (Nässel and Homberg 2006; Nässel 2009). In these cases, neuronal manipulation will likely affect the release of both biogenic amines/neuropeptides and classical transmitters. On the other hand, there is also evidence for differences between the molecular release mechanisms of amine/peptide-containing dense core vesicles and small transmitter-containing synaptic vesicles (e.g., Renden et al. 2001; Park et al. 2014), and biogenic amine and neuropeptide release may not be restricted to active synaptic zones (e.g., Karsai et al. 2013). This may explain the inefficiency of TNTE in OA/TA neurons (Sweeney et al. 1995). Thus, to fully understand the function of defined neurons within a neuronal network, it is essential to identify the functional signaling substance for a certain behavior.
Acknowledgments
The authors thank Mareike Selcho, Konrad Öchsner, and Georg Nagel for technical assistance and/or comments on the article and Andreas Thum, Martin Schwärzel, Robert Kittel, and Georg Nagel for providing flies. The authors declare no competing interests. D.P. and A.v.E. conceived and designed the experiments; D.P., L.v.G., R.L., and A.v.E. performed the experiments; D.P., A.v.E., L.v.G., R.L, R.R., C.W., and S.G.S. analyzed the results; and D.P., S.G.S. and C.W. wrote the article. This work was supported by a grant from the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg (to D.P.), by grants from the Swiss National Science Foundation (CRSII3_136307) and the European Research Council (ERC-2012-StG 309832-PhotoNaviNet) (to S.G.S.), and by the German Research Foundation (Deutsche Forschungsgemeinschaft), collaborative research center SFB 1047 “Insect timing,” project B2 (to C.W.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.172023/-/DC1.
Communicating editor: N. Perrimon
Literature Cited
- Baines R. A., Uhler J. P., Thompson A., Sweeney S. T., Bate M., 2001. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21: 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., D’Evelyn D., Harvey M., Elledge S. J., 1992. Isolation of temperature-sensitive diphtheria toxins in yeast and their effects on Drosophila cells. Development 114: 787–796. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Cobb M., Scott K., Pankratz M., 2009. Gustation in Drosophila melanogaster. SEB Exp. Biol. Ser. 63: 1–38. [PubMed] [Google Scholar]
- Colburn R. W., Lubin M. L., Stone D. J., Jr, Wang Y., Lawrence D., et al. , 2007. Attenuated cold sensitivity in TRPM8 null mice. Neuron 54: 379–386. [DOI] [PubMed] [Google Scholar]
- Cole S. H., Carney G. E., McClung C. A., Willard S. S., Taylor B. J., et al. , 2005. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 280: 14948–14955. [DOI] [PubMed] [Google Scholar]
- Dawydow, A., R. Gueta, D. Ljaschenko, S. Ullrich, M. Hermann et al., 2014 Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. USA 111: 13972–13977. [DOI] [PMC free article] [PubMed]
- Flood, T. F., M. Gorczyca, B. H. White, K. Ito, and M. Yoshihara, 2013 A large-scale behavioral screen to identify neurons controlling motor programs in the Drosophila brain. G3 3: 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B., Stocker R. F., Tanimura T., Thum A. S., 2009. Smelling, tasting, learning: Drosophila as a study case. Results Probl. Cell Differ. 47: 139–185. [DOI] [PubMed] [Google Scholar]
- Goldstein S. A., Price L. A., Rosenthal D. N., Pausch M. H., 1996. ORK1, a potassium-selective leak channel with two pore domains cloned from Drosophila melanogaster by expression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 13256–13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N., Rosenzweig M., Kang K., Pulver S. R., Ghezzi A., et al. , 2008. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454: 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. D., Stein D., Stevens L. M., 2000. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development 127: 573–583. [DOI] [PubMed] [Google Scholar]
- Heckscher E. S., Lockery S. R., Doe C. Q., 2012. Characterization of Drosophila larval crawling at the level of organism, segment, and somatic body wall musculature. J. Neurosci. 32: 12460–12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C., Edwards T., Yasuyama K., Wisotzki B., Schneuwly S., et al. , 2002. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J. Neurosci. 22: 9255–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel G., Martin J. R., Chidami S., Veenstra J. A., Rosay P., 2005. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288: R531–R538. [DOI] [PubMed] [Google Scholar]
- Kane E. A., Gershow M., Afonso B., Larderet I., Klein M., et al. , 2013. Sensorimotor structure of Drosophila larva phototaxis. Proc. Natl. Acad. Sci. USA 110: E3868–E3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai G., Pollák E., Wacker M., Vömel M., Selcho M., et al. , 2013. Diverse in- and output polarities and high complexity of local synaptic and non-synaptic signaling within a chemically defined class of peptidergic Drosophila neurons. Front. Neural Circuits 7: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene A. C., Sprecher S. G., 2012. Seeing the light: photobehavior in fruit fly larvae. Trends Neurosci. 35: 104–110. [DOI] [PubMed] [Google Scholar]
- Keene A. C., Mazzoni E. O., Zhen J., Younger M. A., Yamaguchi S., et al. , 2011. Distinct visual pathways mediate Drosophila larval light avoidance and circadian clock entrainment. J. Neurosci. 31: 6527–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T., 2001. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 47: 81–92. [DOI] [PubMed] [Google Scholar]
- Kong E. C., Woo K., Li H., Lebestky T., Mayer N., et al. , 2010. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS ONE 5: e9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S., White K., 2005. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm). J. Cell Sci. 118: 1779–1787. [DOI] [PubMed] [Google Scholar]
- Kutsukake M., Komatsu A., Yamamoto D., Ishiwa-Chigusa S., 2000. A tyramine receptor gene mutation causes a defective olfactory behavior in Drosophila melanogaster. Gene 245: 31–42. [DOI] [PubMed] [Google Scholar]
- McNabb S. L., Baker J. D., Agapite J., Steller H., Riddiford L. M., et al. , 1997. Disruption of a behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron 19: 813–823. [DOI] [PubMed] [Google Scholar]
- Moses K., Rubin G. M., 1991. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 5: 583–593. [DOI] [PubMed] [Google Scholar]
- Nagaya Y., Kutsukake M., Chigusa S. I., Komatsu A., 2002. A trace amine, tyramine, functions as a neuromodulator in Drosophila melanogaster. Neurosci. Lett. 329: 324–328. [DOI] [PubMed] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., et al. , 2003. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100: 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel D. R., 2009. Neuropeptide signaling near and far: How localized and timed is the action of neuropeptides in brain circuits? Invert. Neurosci. 9: 57–75. [DOI] [PubMed] [Google Scholar]
- Nassel D. R., Homberg U., 2006. Neuropeptides in interneurons of the insect brain. Cell Tissue Res. 326: 1–24. [DOI] [PubMed] [Google Scholar]
- Nitabach M. N., Blau J., Holmes T. C., 2002. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495. [DOI] [PubMed] [Google Scholar]
- Nitabach M. N., Wu Y., Sheeba V., Lemon W. C., Strumbos J., et al. , 2006. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J. Neurosci. 26: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer A. M., Jr, 1977. Diphtheria toxin. Annu. Rev. Biochem. 46: 69–94. [DOI] [PubMed] [Google Scholar]
- Park D., Li P., Dani A., Taghert P. H., 2014. Peptidergic cell-specific synaptotagmins in Drosophila: localization to dense-core granules and regulation by the bHLH protein DIMMED. J. Neurosci. 34: 13195–13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls D., Selcho M., Gendre N., Stocker R. F., Thum A. S., 2010. Drosophila larvae establish appetitive olfactory memories via mushroom body neurons of embryonic origin. J. Neurosci. 30: 10655–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody N. C., Pohl J. B., Diao F., Vreede A. P., Sandstrom D. J., et al. , 2009. Characterization of the decision network for wing expansion in Drosophila using targeted expression of the TRPM8 channel. J. Neurosci. 29: 3343–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech U., Pooryasin A., Birman S., Fiala A., 2013. Localization of the contacts between Kenyon cells and aminergic neurons in the Drosophila melanogaster brain using SplitGFP reconstitution. J. Comp. Neurol. 521: 3992–4026. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, B.D., T.T. Bgo, K.L. Hibbard, C. Murphy, A. Jenett et al., 2010 Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. [DOI] [PMC free article] [PubMed]
- Ren D., Navarro B., Xu H., Yue L., Shi Q., et al. , 2001. A prokaryotic voltage-gated sodium channel. Science 294: 2372–2375. [DOI] [PubMed] [Google Scholar]
- Renden R., Berwin B., Davis W., Ann K., Chin C.-T., et al. , 2001. Drosophila CAPS is an essential gene that regulates dense-core vesicle release and synaptic vesicle fusion. Neuron 31: 421–437. [DOI] [PubMed] [Google Scholar]
- Rister J., Heisenberg M., 2006. Distinct functions of neuronal synaptobrevin in developing and mature fly photoreceptors. J. Neurobiol. 66: 1271–1284. [DOI] [PubMed] [Google Scholar]
- Roeder T., 2005. Tyramine and octopamine: ruling behavior and metabolism. Annu. Rev. Entomol. 50: 447–477. [DOI] [PubMed] [Google Scholar]
- Rosenzweig M., Brennan K. M., Tayler T. D., Phelps P. O., Patapoutian A., et al. , 2005. The Drosophila ortholog of vertebrate TRPA1 regulates thermotaxis. Genes Dev. 19: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswati S., Fox L. E., Soll D. R., Wu C. F., 2004. Tyramine and octopamine have opposite effects on the locomotion of Drosophila larvae. J. Neurobiol. 58: 425–441. [DOI] [PubMed] [Google Scholar]
- Schroder-Lang S., Schwarzel M., Seifert R., Strunker T., Kateriya S., et al. , 2007. Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods 4: 39–42. [DOI] [PubMed] [Google Scholar]
- Schroll C., Riemensperger T., Bucher D., Ehmer J., Voller T., et al. , 2006. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol. 16: 1741–1747. [DOI] [PubMed] [Google Scholar]
- Selcho M., Pauls D., Han K. A., Stocker R. F., Thum A. S., 2009. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS ONE 4: e5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcho M., Pauls D., El Jundi B., Stocker R. F., Thum A. S., 2012. The role of octopamine and tyramine in Drosophila larval locomotion. J. Comp. Neurol. 520: 3764–3785. [DOI] [PubMed] [Google Scholar]
- Selcho M., Pauls D., Huser A., Stocker R. F., Thum A. S., 2014. Characterization of the octopaminergic and tyraminergic neurons in the central brain of Drosophila larvae. J. Comp. Neurol. 522: 3485–3500. [DOI] [PubMed] [Google Scholar]
- Sprecher S. G., Desplan C., 2008. Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature 454: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher S. G., Pichaud F., Desplan C., 2007. Adult and larval photoreceptors use different mechanisms to specify the same Rhodopsin fates. Genes Dev. 21: 2182–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher S. G., Cardona A., Hartenstein V., 2011. The Drosophila larval visual system: high-resolution analysis of a simple visual neuropil. Dev. Biol. 358: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M., Stumpf P., Udwari D., Gueta R., Hagedorn R., et al. , 2011. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286: 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R. F., 2008. Design of the larval chemosensory system. Adv. Exp. Med. Biol. 628: 69–81. [DOI] [PubMed] [Google Scholar]
- Sweeney S. T., Broadie K., Keane J., Niemann H., O’Kane C. J., 1995. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14: 341–351. [DOI] [PubMed] [Google Scholar]
- Thum A. S., Knapek S., Rister J., Dierichs-Schmitt E., Heisenberg M., et al. , 2006. Differential potencies of effector genes in adult Drosophila. J. Comp. Neurol. 498: 194–203. [DOI] [PubMed] [Google Scholar]
- Tripodi M., Evers J. F., Mauss A., Bate M., Landgraf M., 2008. Structural homeostasis: compensatory adjustments of dendritic arbor geometry in response to variations of synaptic input. PLoS Biol. 6: e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Gueta R., Nagel G., 2013. Degradation of channelopsin-2 in the absence of retinal and degradation resistance in certain mutants. Biol. Chem. 394: 271–280. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Simpson J. H., Bellen H. J., 2011. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72: 202–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath V., Story G. M., Peier A. M., Petrus M. J., Lee V. M., et al. , 2003. Opposite thermosensor in fruitfly and mouse. Nature 423: 822–823. [DOI] [PubMed] [Google Scholar]
- Vogelstein J. T., Park Y., Ohyama T., Kerr R. A., Truman J. W., et al. , 2014. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344: 386–392. [DOI] [PubMed] [Google Scholar]
- von Essen A. M., Pauls D., Thum A. S., Sprecher S. G., 2011. Capacity of visual classical conditioning in Drosophila larvae. Behav. Neurosci. 125: 921–929. [DOI] [PubMed] [Google Scholar]
- Vosshall L. B., Stocker R. F., 2007. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30: 505–533. [DOI] [PubMed] [Google Scholar]
- Wang Y., Pu Y., Shen P., 2013. Neuropeptide-gated perception of appetitive olfactory inputs in Drosophila larvae. Cell Rep. 3: 820–830. [DOI] [PubMed] [Google Scholar]
- Wing J. P., Zhou L., Schwartz L. M., Nambu J. R., 1998. Distinct cell killing properties of the Drosophila reaper, head involution defective, and grim genes. Cell Death Differ. 5: 930–939. [DOI] [PubMed] [Google Scholar]
- Yoo S. J., Huh J. R., Muro I., Yu H., Wang L., et al. , 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4: 416–424. [DOI] [PubMed] [Google Scholar]
- Zhang W., Ge W., Wang Z., 2007. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. Eur. J. Neurosci. 26: 2405–2416. [DOI] [PubMed] [Google Scholar]
- Zhou L., Schnitzler A., Agapite J., Schwartz L. M., Steller H., et al. , 1997. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. USA 94(10): 5131–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]