Abstract
Through unbiased metabolomics, we identified elevations of the metabolite 2-hydroxyglutarate (2HG) in renal cell carcinoma (RCC). 2HG can inhibit 2-oxoglutaratre (2-OG) dependent dioxygenases which mediate epigenetic events including DNA and histone demethylation. 2HG accumulation, specifically the D- enantiomer, can result from gain of function mutations of isocitrate dehydrogenase (IDH1, IDH2) found in several different tumors. In contrast, kidney tumors demonstrate elevations of the L enantiomer of 2HG (L-2HG). High 2HG tumors demonstrate reduced DNA levels of 5-hydroxymethylcytosine (5hmC) consistent with 2-HG mediated inhibition of TET (Ten Eleven Translocation) enzymes which convert 5-methylcystoine (5mC) to 5hmC. L-2HG elevation is mediated in part by reduced expression of L-2HG dehydrogenase (L2HGDH). L2HGDH reconstitution in RCC cells lowers L-2HG and promotes 5hmC accumulation. Additionally, L2HGDH expression in RCC cells reduces histone methylation and suppresses in vitro tumor phenotypes. Our report identifies L-2HG as an epigenetic modifier and putative oncometabolite in kidney cancer.
Keywords: renal cancer, 2-hydroxyglutarate, L2HGDH, 5-hydroxymethylcytosine, epigenetics
Introduction
One of the clearest examples of the role of metabolism in cancer is the recent identification of oncometabolites, small molecules with putative oncogenic properties. Mutations in tumors of fumarate hydratase (FH), succinate dehydrogenase (SDH), and isocitrate dehydrogenase 1 and 2 (IDH1/2) lead to elevated levels of fumarate, succinate, and 2-hydroxyglutarate (2HG), respectively(1, 2). In FH and SDH mutations, precursor metabolites (fumarate and succinate) accumulate due to loss of enzymatic activity. In the case of IDH1 and IDH2, highly conserved mutation “hot spots” in DNA of tumors result in the formation of a “neoenzyme” that leads to elevated 2HG (specifically the D- enantiomer) in gliomas and acute myeloid leukemias (AMLs) as well as other malignancies(2, 3). A unifying theme linking these three oncometabolites is their ability to inhibit a class of enzymes referred to as 2-oxoglutarate (2-OG) dependent dioxygenases (2OGDs). Members of this enzyme family include prolyl hydroxylases (PHDs), which are involved in the regulation of the transcription factor Hypoxia Inducible Factor-1α (HIF-1α), histone demethylases, and DNA hydroxylases(4). In the case of succinate accumulation, feedback inhibition is a proposed mechanism as succinate is a product of 2-OGD catalyzed reactions. In the case of fumarate and 2HG, competitive inhibition is the proposed mechanism due to structural similarity with the cofactor 2-OG. Collectively, these data suggest that metabolic perturbations in cancer can inhibit 2-OGD activities with potential effects on tumorigenesis.
Recent studies have focused on the ability of oncometabolites to inhibit DNA hydroxylation by the TET (Ten Eleven Translocation) enzymes (TETs 1-3) which convert 5-methylcystoine (5mC) to 5-hydroxymethylcytosine (5hmC). The oxidation of 5mc to 5hmC has been proposed to promote the demethylation of DNA, either via active or passive means(5). Alternatively, 5hmC has been proposed to be its own epigenetic mark with distinct effects on DNA architecture and gene expression(6). Despite these conflicting views, emerging data demonstrates reduced 5hmC levels in human cancer as well as animal tumor models indicating a role for 5hmC loss in carcinogenesis(7, 8). Further evidence for the role of 5hmC loss in malignancy is provided by the fact that TET2 is commonly mutated in human myeloid malignancies including acute myeloid leukemia (AML) as well as other myeloid disorders(9, 10). 2HG can inhibit TET enzymatic activity and promote loss of 5hmC(11). These studies have primarily examined the role of the D enantiomer of 2HG (D-2HG) which is markedly elevated in the setting of IDH mutations. Notably, both cell-free and in vitro studies demonstrate that the L enantiomer (L-2HG) is more potent at inhibiting 2OGDs including the TET enzymes(11, 12).
In this report, we demonstrate elevations of 2HG in clear cell renal cell carcinoma (ccRCC), the most common histological subtype of kidney cancer. In contrast to IDH mutant tumors, ccRCCs demonstrate elevations of L-2HG. In concordance with the ability of 2HG to inhibit TET enzymatic activity, tumors with elevations of 2HG demonstrated reduced levels of 5hmC in genomic DNA. We provide evidence that reduced mRNA and protein expression of L-2HG dehydrogenase (L2HGDH) in ccRCC promotes 2HG accumulation and 5hmC loss. Bioinformatic analysis demonstrates that copy number loss is associated with reduced L2HGDH expression in ccRCC. L2HGDH reconstitution in RCC cells lowers L-2HG, promotes 5hmC accumulation, and suppresses in vitro tumor phenotypes. Collectively, our data demonstrate a putative oncometabolite elevated in ccRCC with effects on the kidney cancer epigenome.
Results
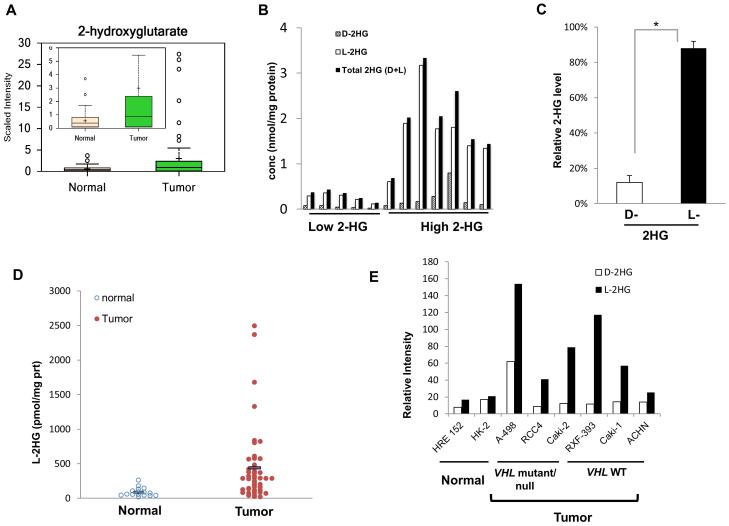
We analyzed 59 matched tumor/normal pairs utilizing an unbiased metabolomics profile (manuscript in preparation). This initial analysis identified statistically significant elevations of 2HG (greater than 5-fold) in ccRCC relative to normal renal parenchyma (Figure 1A). However, multiple tumors demonstrated elevations of 2HG more than 10-fold higher in normal tissue. Investigation for somatic mutations in RCC using both the cBioPortal for Cancer Genomics (to analyze TCGA data sets) and the Sanger COSMIC database did not demonstrate any evidence for IDH mutations in ccRCC (data not shown). 2HG is known to occur in two enantiomers, D(R) and L(S). We therefore analyzed metabolite extracts from both high and low 2HG tumors via liquid chromatography tandem mass spectrometry (LC-MS/MS) for levels of each 2HG enantiomer (Figure 1B). Of notable significance is that the predominant enantiomer present in these cells is the L enantiomer (Figure 1B). Analysis of high 2HG tumors demonstrated that the L enantiomer accounted for approximately 90% of the 2HG present in these tumors (Figure 1C). We validated our findings in a separate cohort (Figure 1D). Clinical information present on this cohort is provided in Supplemental Table 1. We therefore analyzed a panel of RCC lines for 2HG elevations in addition to non-transformed renal epithelial cells HK-2 and HRE 152. Consistent with our tissue analysis, several RCC lines demonstrated increases in 2HG with predominant contribution made by the L- enantiomer (Figure 1E). Notably, L-2HG elevation was found in cells both with and without alterations of VHL, the most commonly altered gene in ccRCC. Analysis of tumor samples confirmed L-2HG elevation in tumors both with and without VHL coding mutations (supplemental Table 2). Collectively, these demonstrate elevations of L-2HG in ccRCC.
Figure 1. L-2-hydroxyglutarate (L-2HG) is elevated in RCC tumors and cell lines.
Human kidney samples were obtained by surgical resection and metabolites were extracted for metabolite profiling analysis by GC/MS analysis. (A) 2HG was significantly increased in primary tumor (Tumor) compared to adjacent benign kidney tissue (Normal). Inset graph present with smaller scale. (B) High and low 2HG tumors were analyzed by tandem MS to resolve the enantiomeric distribution of 2HG within these tumors. (C) Relative ratio of D-2HG and L-2HG against total 2HG in high 2HG RCC samples. Error bars represent standard error of mean. (*) p <0.05. (D) L-2HG levels were measured in another cohort of samples from a separate biorepository. (E) Enantiomeric resolution of 2HG in a panel of nontransformed and transformed lines of renal origin.
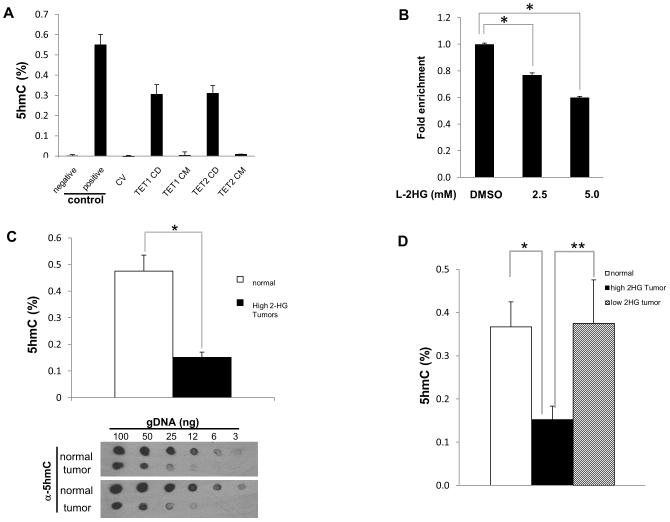
2HG elevations have been identified as inhibiting TET enzymatic activity thereby leading to reduced levels of 5hmC in the context of IDH mutation(11). We utilized an ELISA-based assay to quantitate absolute 5hmC levels. To validate the assay, we overexpressed the catalytic domains (CD) of TET1 and TET2 in HEK-293 cells in addition to enzymatically inactive mutants (CM) of TET1 and TET2. Consistent with previous data(11), cultured cells including HEK-293 cells express low levels of 5hmC (Figure 2A). However, transient expression of TET1 or TET2 CD was able to raise 5hmC levels, whereas CM forms of either TET1 or TET2 could not (Figure 2A). We therefore examined levels of 5hmC in the context of 2HG elevation. Consistent with prior data, L-2HG octyl-ester treatment reduced DNA 5hmC levels in HK-2 renal epithelial cells (Figure 2B). We confirmed increases in intracellular 2-HG levels following ester treatment (data not shown). Tumors with elevated 2HG levels demonstrate significantly reduced levels of 5hmC relative to matched normal tissue on ELISA analysis (Figure 2C-upper panel). Dot-blot assay with an antibody specific to 5hmC in DNA identified that high 2HG tumors demonstrated reduced levels of 5hmC relative to normal kidney (Figure 2C-lower panel). In contrast, tumors with low 2HG levels did not demonstrate reduced 5hmC levels (Figure 2D). Collectively, these data indicate that raised levels of 2HG in ccRCC are associated with 5hmC loss.
Figure 2. Increased L-2HG is associated with loss of 5-hmC in RCC tumors.
(A) Validation of ELISA for 5hmC. HEK293 cells were transiently transfected with plasmids expressing control vector (CV), TET1 wild-type catalytic domain (CD) and mutant catalytic domain (CM), TET2 wild-type catalytic domain (CD) and mutant catalytic domain (CM). Cells were harvested and genomic DNA was examined to determine 5hmC level. (B) HK-2 renal epithelial cells were treated with L-2HG octyl ester for 4 hours and assayed for 5hmC levels via ELISA. (C) 5hmC levels between normal and high L-2HG RCC tumor samples were analyzed by ELISA (upper panel) and dot blot assay (lower panel). (D) 5hmC levels in normal, low 2HG tumors, and high 2HG tumors were determined by ELISA. Error bars represent standard error of mean. (*) p <0.005, (**) p <0.05.
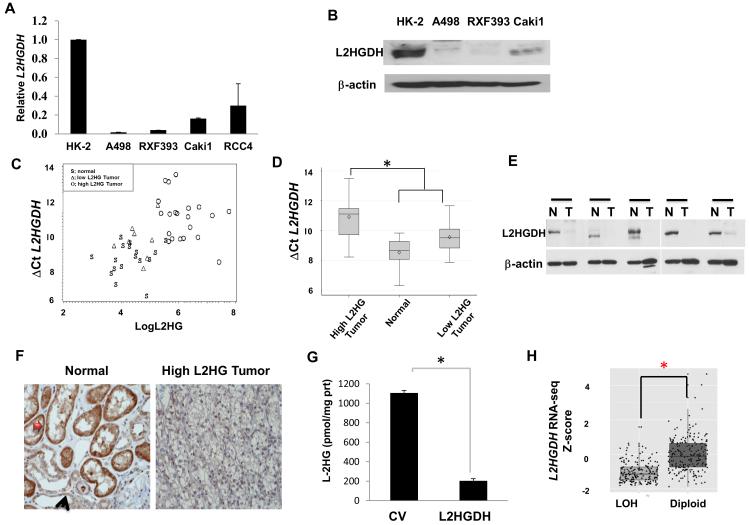
We next wanted to identify potential factors that promote L-2HG accumulation in ccRCC. L-2-hydroxyglutaric aciduria is an inborn error of metabolism linked to loss of function mutations of the gene encoding L-2HG dehydrogenase (L2HGDH)(13). Notably, several RCC lines demonstrated reduced mRNA and protein expression of L2HGDH relative to nontransformed renal epithelial cells (Figures 3A and 3B). We found that A498 and RXF-393 demonstrate elevated L-2HG levels with concomitant reduced L2HGDH mRNA/protein expression (Figures 1E, 3A, 3B). These data prompted us to examine the relationship between L-2HG levels and L2HGDH expression in patient samples. We analyzed L2HGDH mRNA expression in normal kidney, low L-2HG tumors, and high L-2HG tumors. Elevated L-2HG tumors had higher ΔCt L2HGDH (Ct L2HGDH- Ct RPL0) indicating lower L2HGDH expression when compared with low L-2HG tumors and normal kidney (Figures 3C and 3D). Immunoblot analysis confirmed mRNA findings as primary tumors with elevated 2HG had reduced protein levels of L2HGDH relative to normal tissue (Figure 3E). Immunohistochemical analysis also demonstrated reduced L2HGDH expression in high L-2HG tumor relative to normal kidney. Notably, proximal tubule cells, the likely cell of origin for ccRCCs, have prominent L2HGDH expression relative to distal tubule cells (Figure 3F). We next examined if re-expression of L2HGDH in RCC cells could lower L-2HG levels. Using lentivirus, we stably expressed L2HGDH cDNA in RCC cells and confirmed expression by immunoblotting (Supplemental Figure 1S). L2HGDH expression significantly reduced intracellular L-2HG levels in A498 (figure 3G). The L2HGDH locus is located on chromosome 14q, a region commonly lost in ccRCC(14). We therefore examined the relationship between copy number loss and gene expression alterations with Level 3 RNAseq data from TCGA. ccRCC tumors with loss of a single copy (i.e. loss of heterozygosity) were associated with significantly reduced mRNA expression of L2HGDH compared with diploid tumors (Figure 3H). Collectively these data demonstrate that reduced expression of L2HGDH promotes L-2HG accumulation in ccRCC.
Figure 3. L2HGDH is reduced in RCC tumors and cell lines.
(A) The mRNA expression of L2HGDH in RCC lines relative to nontransformed renal epithelial cells (HK-2). (B) The protein expression of L2HGDH in RCC lines relative to nontransformed renal epithelial cells. (C and D) L2HGDH mRNA levels measured by real time RT-PCR in normal, low L-2HG tumor, and high L-2HG tumor and plotted as a function of L-2HG levels and graphically displayed. *p< 0.001. (E) High 2HG tumors and matched normal tissue were analyzed for L2HGDH protein levels by immunoblotting (T, RCC tumor; N, benign tissue). (F) Immunohistochemistry for L2HGDH in normal kidney and high L-2HG tumor. Red and black arrows denote proximal and distal tubular epithelial cells, respectively. (G) L-2HG levels were measured in A498 cells stable transduced with control vector and L2HGDH cDNA. (H) Analysis of TCGA data assessing the effects of copy loss on mRNA gene expression of L2HGDH. LOH, loss of heterozygosity. (*) p < 0.0001.
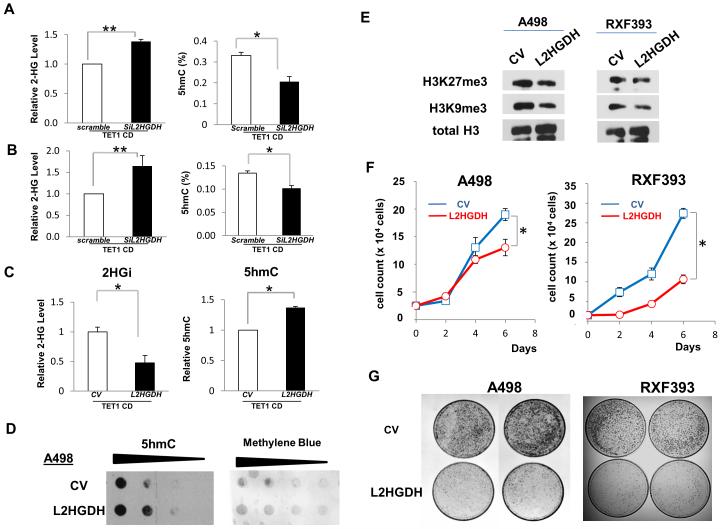
We next determined whether modulation of L2HGDH expression could impact levels of 2HG as well as 5hmC levels. We therefore cotransfected HEK-293 cells with TET1 CD (to raise 5hmC levels to a detectable range) and either siRNA to L2HGDH or control siRNA. Immunoblotting confirmed TET1 CD expression and knockdown of L2HGDH (Supplemental Figure 2S). Consistent with its role in L2HG metabolism, L2HGDH knockdown led to raised levels of cellular 2HG (figure 4A – left panel). Concomitantly, we identified decreased levels of 5hmC with L2HGDH knockdown consistent with the ability of L2HG to inhibit TET enzymatic activity (Figure 4A- right panel). Similar results were obtained in HK-2 renal epithelial cells (figure 4B- and Supplemental Figure 2S). As a complement to our knockdown approach, we overexpressed TET1 CD and either control vector or L2HGDH cDNA in HEK-293 cells. Immunoblotting confirmed transgene expression (Supplemental Figure 3S). Under basal culture conditions, L2HGDH overexpression had little impact on cellular 2HG levels consistent with the fact that HEK-293 cells express high levels of L2HGDH mRNA and protein levels (data not shown). We therefore challenged cells with esterified L-2HG (octyl-2HG) to raise intracellular levels of 2HG. Following octyl-2HG challenge, L2HGDH cDNA overexpression in HEK-293 cells led to reduced cellular 2HG levels relative to control cells consistent with the role of L2HGDH in the metabolism of L-2HG (figure 4C-left panel). Consistent with reduced 2HG levels, L2HGDH overexpressing cells demonstrate increased 5hmC levels compared to vector control transfected cells (figure 4C-right panel). Consistent with these data, reexpression of L2HGDH in A498 cells led to an increase in 5hmC levels as compared with control vector (figure 4D). As noted, prior studies have demonstrated that L-2HG can inhibit histone demethylases and hence promote histone methylation. We therefore examined the impact of L2HGDH effects on histone methylation patterns. Stable expression of L2HGDH in both A498 and RXF393 cells led to reduced H3K27me3 and H3K9me3 levels consistent with reduced L-2HG levels and ensuing activation of histone demethylase activity (Figure 4E). Collectively, these demonstrate that modulation of L2HGDH expression can impart effects on both DNA and histone modifications. Given the emerging role of epigenetic alterations in RCC, we examined the effects on L2HGDH expression on in vitro phenotypes. Notably, L2HGDH expression reduced proliferation and colony formation in A498 and RXF393 RCC cells (Figure 4F and 4G).
Figure 4. Knockdown or ectopic expression of L2HGDH is associated with changes of intracellular L-2HG concentration and DNA 5hmC level.
siRNA to L2HGDH was co-transfected with TET1 catalytic domain (CD) in HK-2 cells (A) and HEK293 cells (B). Noncoding scramble was used as control siRNA (siControl). Mass spectrometry confirmed raised 2HG with L2HGDH knockdown (left panels). Genomic DNA was also isolated to determine 5hmC level by ELISA (right panels). (C) HEK293 cells were transiently co-transfected with TET1 CD and either L2HGDH cDNA or control vector (CV). Cells were subsequently challenged for 4hrs with 1mM L-2HG octyl ester. Metabolites were extracted for measurement of intracellular total 2HG level and analyzed by LC-MS (left panel). Genomic DNA was also isolated to determine 5hmC level by ELISA (right panel). Error bars indicate standard deviation from at least two independent experiments. (**) p < 0.005, (*) p < 0.05 (D) 5hmc dot blot assay in A498 cells +/- L2HGDH cDNA. Methylene blue blot is included for loading control. (E)Histone immunoblotting in A498 and RXF393 cells +/- L2HGDH cDNA. (F and G) Proliferation and colony formation assays in RCC cells +/- L2HGDH cDNA.
Discussion
Here, we demonstrate elevations of L-2HG in clear cell renal cell carcinoma, the most common histology. We also demonstrate a novel mechanism for L-2HG elevations via the reduced mRNA and protein expression of L2HGDH in part due to loss of the L2HGDH gene. Previous studies demonstrate that elevated cellular levels of L-2HG are present in an inborn error of metabolism that results from L-2HG dehydrogenase (L2HGDH) deficiency(13). Although rare, a significant proportion of patients develop tumors suggesting a role for L-2HG in carcinogenesis. In particular, L2HGDH deficiency has been linked to brain tumors and Wilm’s tumor(15, 16). To the best of our knowledge, this is the first report to demonstrate elevations primarily of the L- enantiomer in cancer. This is in contrast to elevations of D-2HG that have been identified in a growing list of cancers with IDH mutations. Notably, a recent study reported 2HG increases in breast cancers in the absence of IDH mutations(17). However, the specific enantiomer was not reported.
Our data do not exclude alternative mechanisms for L-2HG accumulation in ccRCC. Prior studies have demonstrated that “off-target” activity of malate dehydrogenase (MDH1/MDH2) can convert 2-OG to L-2HG(18). Substrates for this reaction include glucose and glutamic acid(19). It is well established that glutamine can be converted to glutamate (via glutaminases), which can be eventually converted to 2-OG. This is particularly relevant given recent studies by Wise et al. demonstrating that HIF can promote modest increases in 2HG synthesis via IDH mediated reductive carboxylation of glutamine-derived 2-OG(20). However, enantiomeric resolution of 2-HG was not performed in this study. Nevertheless, given recent studies on reductive glutamine metabolism in VHL deficient RCC cells (which demonstrate HIF stabilization)(21, 22), these alternative mechanisms warrant further investigation.
2HG is part of a growling list of small molecules referred to as oncometabolites, which refers to small molecules with putative transforming properties. Recent studies have linked D-2HG to the promotion of leukemogenesis(23). In particular, D-2HG promotes cytokine independence and blocks differentiation in hematopoietic cells. Mutations in tumors of fumarate hydratase (FH) and succinate dehydrogenase (SDH) lead to elevated levels of fumarate and succinate, respectively, due to loss of enzymatic activity. Germline mutations of FH predispose individuals to the development of renal cancer as well as cutaneous and uterine leiomyomas(24). Germline SDHB mutations have also been linked to renal cancer as well pheochromocytomas and paragangliomas(25, 26). Hence, a notable finding is that all 3 metabolites are linked to RCC.
A unifying theme amongst these three oncometabolites is their ability to inhibit 2-oxoglutarate (2-OG) dependent dioxygenases (2OGDs). Members include prolyl hydroxylases (PHDs), 5mC DNA hydroxylases (TETs 1-3), and histone demethylases. Multiple studies have demonstrated that fumarate, succinate, and 2HG can inhibit TET enzymatic activity(11, 27, 28). Consistent with these data, we identified reduced 5hmC levels in tumors with elevations of 2HG. The fold changes of 2HG we have identified in RCC are more modest relative to the 2HG elevations noted in the context of IDH mutations. A prior study by Choi et al. analyzed 2-HG (both D and L) in IDH mutant tumors by the same methodology as our study(29). The reported range of D-2HG in these tumors ranged from ≈20-200 nmol/mg protein. We identified multiple tumors with L-2HG levels within 10-fold of IDH mutant tumors. Notably, prior studies have demonstrated that the L enantiomer of 2HG is a far more potent inhibitor of 2OGDs (including TETs) compared to the D enantiomer(11, 12, 27).
Given the multiple 2OGDs, it may be difficult to discern which of these are significant from a tumorigenic standpoint. Several lines of evidence suggest the importance of 2-OGDs relating to histone and DNA biology in kidney cancer. Recurrent mutations of UTX which encodes a H3K27 demethylase that requires 2-OG have been found in RCC(30). In addition, recent studies demonstrate increased expression of the catalytic subunit of the polycomb repressive complex 2(PRC2), enhancer of zeste homologue 2 (EZH2), in ccRCC(31, 32). The PRC2 promotes H3K27 methylation. Our findings demonstrate that reexpression of L2HGDH in RCC cells reduces H3K27 methylation. Collectively, these data strongly suggest the importance of this histone mark in renal carcinogenesis. A notable finding from two independent studies in AML is that TET2 mutations are mutually exclusive from IDH mutations(10, 33). The common link between these two seemingly unrelated mutations is their effect on TET enzymatic activity- either through loss of function mutations (TET2 mutations) or, in the case of IDH mutation, 2HG mediated inhibition of TET enzymatic activity through competition with the cofactor 2-OG. As noted, mutations of TET2 have been identified in myeloid disorders such as AML and glioma. Notably, recent deep sequencing efforts on RCC has identified TET2 mutations in RCC(34). However, whether these mutants result in loss of function with effects of 5hmC levels has not been determined. Additionally, reduced expression of TET enzymes has been demonstrated in a variety of tumors(7, 8). Correspondingly, reduced levels of 5hmC have been identified in cancer as well(7, 8). Given these data, L-2HG like has multiple biologically relevant 2-OGD targets.
As noted previously, the TETs promote the conversion of 5mC to 5hmC which is currently thought to promote DNA demethylation. Hence, it might be expected that loss of TET activity, either due to mutation or inhibition, would result in DNA hypermethylation. However, the reported effects on global DNA methylation in the context of TET2 mutations in myeloid disorders has been conflicting(10, 35). Alternatively, 5hmC may represent its own epigenetic mark with specific effects on gene expression, perhaps through the recruitment of specific transcriptional regulators. These data point to the complexity of 5hmC biology and the need for further studies clarifying the mechanisms by which this DNA modification impacts gene methylation, regulation and expression in the setting of elevated L-2HG.
In summary, our data demonstrate elevated levels of 2HG in RCC, specifically the L-enantiomer. We demonstrate that elevations of 2HG are associated with loss of 5hmC levels in RCC samples and that these changes are mediated by the reduced expression of L2HGDH. Specifically, we demonstrate the first putative oncometabolite in the most common form of kidney cancer and its effects on the epigenetic landscape. Our data add to the growing body of evidence demonstrating the interplay between intermediary metabolism and nucleic acid biology in cancer.
Materials and Methods
Plasmids
Expression plasmids of FLAG-tagged TET1 CD, TET1 CM, TET2 CD, TET2 CM were kindly provided by Dr. Yi Zhang through Addgene. The L2HGDH expression plasmid pcDNA3.1-L2HGDH was constructed by cloning the full-length human L2HGDH cDNA amplified from human kidney epithelial HK-2 cells into pcDNA3.1 using primers 5’-TTTGAATTCATGGTGCCAGCGCTGCGTTAT-3' and 5'- TTTGGTACCTTATAATTCAAATCTTTGTTGTACTTCATCTGCAATC-3'.
Cell Culture and Transfection
All lines were acquired from ATCC except RXF-393 (NCI), HRE152 (J.A. Copland, Mayo Jacksonville) and RCC4 (P. Ratcliffe, Oxford). Cell lines were periodically tested for mycoplasma. No other authentication was performed. For transient transfection in HEK293, cells were transfected using Lipofectamine 2000 (Invitrogen) and calcium phosphate methods for RNA interference and cDNA expression, respectably. For L2HGDH knockdown in HK-2 cells, cells were transfected with pooled siRNA reagent (Thermo Fisher) using Amaxa 4D Nucleofector 4D nucleofector (Lonza) system according to the manufacturer’s protocol. A nontargeting scramble siRNA pool was used as a negative control (Thermo Fisher).
RNA extraction and Quantitative RT-PCR
Total RNA was extracted with Trizol (Invitrogen) from RCC patient samples and RCC cell lines. cDNA was generated reverse using VILO RT kit (Invitrogen) and then used as template for L2HGDH Taqman expression assay probe (Applied Biosystems). Ribosomal protein (RPL0) was used to normalize data in tissue samples. For cell line analysis, L2HGDH mRNA expression level was quantified by using 2-ΔΔCT method normalized to GAPDH. For tissues, the ΔCt L2HGDH (Ct L2HGDH- Ct RPL0) was measured for normal tissue, low L-2HG, and high L-2HG tumor. Multivariate analyses of variance (MANOVA) followed by Analysis of Variance (ANOVA) were conducted in SAS Version 9.3 to compare the expression level (ΔCt L2HGDH) between the three groups. We used SAS GLM contrast to compare the High group to the others (Normal and Low). L-2HG values were log-transformed to meet the assumptions for parametric tests.
Quantitative 5-hydroxymethylation (5-hmC) analysis
For DNA dot blotting, genomic DNA was denatured, serially diluted in NaOH/EDTA solution and spotted on positively charged Nylon membranes (Roche Applied Science). The membrane was crosslinked (UVP) and then blocked with 5% milk in TBST for 30 min, followed by the incubation with the anti-5-hmC antibody (Active Motif) overnight at 4° C and HRP-conjugated anti-rabbit IgG secondary antibody for 1 hr at room temperature. After washing three times with TBST, the membrane was treated with ECL and scanned. For quantitation of 5hmC, Quest 5-hmC DNA ELISA kits (Zymo Research) were used followed the manufacturer’s protocol. Briefly, anti-5-hydroxymethylcytosine polyclonal antibody was coated to the bottom of well and denatured 100 ng genomic DNA was added. To detect DNA bound to anti-5-hmC pAb, anti-DNA HRP antibody and HRP developer was applied. Greenish-blue color was analyzed in the wells by a plate reader at 405-450nm detection.
Metabolite extraction and Chromatography/Mass Spectrometry of 2-Hydroxyglutarate (2HG)
For GC/MS and LC/MS analysis of tissues, human kidney samples including normal and RCC were extracted and prepared for analysis as described in the Supplemental Methods and as previously described(36).
For 2HG enantiomer analysis (i.e. D- and L-2HG quantification), samples were analyzed as previously described(37). Briefly, extracts were derivatized with DATAN (diacetyl-L-tartaric acid), which permits enantiomeric analysis, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis and normalized to protein levels.
For total 2HG (D-2HG+L-2HG) measurement of samples from in vitro studies, cell pellets were washed in 1x PBS three times, extracted with 10% cold trichloroacetic acid (TCA) and precipitate was removed by centrifugation. TCA in the supernatant was removed by vortexing with 4 volumes of 1,1,2-trichlorotrifluoroethane (FREON)-trioctylamine (Sigma) mixture and the upper aqueous layer was collected for analysis after centrifugation. Samples were analyzed by ion chromatography coupled with negative electrospray mass spectrometry (RFIC-MS) (Dionex) and 2HG was determined by selected ion monitoring (SIM 147.1). 2HG in cell extracts was quantified by using a calibration curve of 2HG and normalized to protein content. Unless otherwise noted, 2HG refers to total levels (D- + L-).
Statistical Analysis
Unless otherwise noted, statistical analyses were carried out using the program “R” and Microsoft excel software. Comparisons between groups for statistical significance were performed with a two-tailed paired t-test and ANOVA. A P-value of <0.05 was considered statistically significant in all cases.
Supplementary Material
Significance.
Here, we report elevations of the putative oncometabolite L-2HG in the most common subtype of kidney cancer and describe a novel mechanism for the regulation of DNA 5hmC levels. Our findings provide new insight into the metabolic basis for the epigenetic landscape of renal cancer.
Acknowledgements
The authors gratefully acknowledge Marcia Grayson, April Mitchem, and Lee Whitworth for expert technical assistance. Research reported in this publication was supported by the NIH (P30 CA013148). The authors also acknowledge the assistance of the UTHSCSA Genomic Resource Core Laboratory and the UTHSCSA Nucleic Acids Core Facility which are supported by UTHSCSA and NIH-NCI P30CA054174 (CTRC of UTHSCSA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial Support:
NIH K08 CA138774 (SS)
Urology Care Foundation and Astellas Pharma Rising Star Award (SS)
America Cancer Society RSG-12-127-01 CNE (SS)
NIH R01 NCI CA131272 (KB)
CTSA/IIMS UL1RR025767 (SS,CL)
Cancer Prevention Research Institute of Texas RP120190 (CL,KB)
Footnotes
Conflicts Of Interest: None
References
- 1.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Human molecular genetics. 2005;14:2231–9. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 2.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG., Jr Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–45. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–52. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics & chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–5. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–9. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 10.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rzem R, Veiga-da-Cunha M, Noel G, Goffette S, Nassogne MC, Tabarki B, et al. A gene encoding a putative FAD-dependent L-2-hydroxyglutarate dehydrogenase is mutated in L-2-hydroxyglutaric aciduria. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16849–54. doi: 10.1073/pnas.0404840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer research. 2009;69:4674–81. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroni I, Bugiani M, D'Incerti L, Maccagnano C, Rimoldi M, Bissola L, et al. L-2-hydroxyglutaric aciduria and brain malignant tumors: a predisposing condition? Neurology. 2004;62:1882–4. doi: 10.1212/01.wnl.0000125335.21381.87. [DOI] [PubMed] [Google Scholar]
- 16.Rogers RE, Deberardinis RJ, Klesse LJ, Boriack RL, Margraf LR, Rakheja D. Wilms tumor in a child with L-2-hydroxyglutaric aciduria. Pediatr Dev Pathol. 2010;13:408–11. doi: 10.2350/09-12-0768-CR.1. [DOI] [PubMed] [Google Scholar]
- 17.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rzem R, Vincent MF, Van Schaftingen E, Veiga-da-Cunha M. L-2-hydroxyglutaric aciduria, a defect of metabolite repair. J Inherit Metab Dis. 2007;30:681–9. doi: 10.1007/s10545-007-0487-0. [DOI] [PubMed] [Google Scholar]
- 19.Struys EA, Gibson KM, Jakobs C. Novel insights into L-2-hydroxyglutaric aciduria: mass isotopomer studies reveal 2-oxoglutaric acid as the metabolic precursor of L-2-hydroxyglutaric acid. J Inherit Metab Dis. 2007;30:690–3. doi: 10.1007/s10545-007-0697-5. [DOI] [PubMed] [Google Scholar]
- 20.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nature genetics. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 25.Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. American journal of human genetics. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, et al. Germline SDHB mutations and familial renal cell carcinoma. Journal of the National Cancer Institute. 2008;100:1260–2. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 27.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development. 2012;26:1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nature medicine. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinz S, Weikert S, Magheli A, Hoffmann M, Engers R, Miller K, et al. Expression profile of the polycomb group protein enhancer of Zeste homologue 2 and its prognostic relevance in renal cell carcinoma. J Urol. 2009;182:2920–5. doi: 10.1016/j.juro.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Wagener N, Macher-Goeppinger S, Pritsch M, Husing J, Hoppe-Seyler K, Schirmacher P, et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC cancer. 2010;10:524. doi: 10.1186/1471-2407-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaidzik VI, Paschka P, Spath D, Habdank M, Kohne CH, Germing U, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30:1350–7. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 34.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature genetics. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 35.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 37.Rakheja D, Boriack RL, Mitui M, Khokhar S, Holt SA, Kapur P. Papillary thyroid carcinoma shows elevated levels of 2-hydroxyglutarate. Tumour Biol. 2011;32:325–33. doi: 10.1007/s13277-010-0125-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.