Abstract
With the decrease in sequencing costs, personalized genome sequencing will eventually become common in medical practice. We therefore write this series of three reviews to help non-geneticist clinicians to jump into the fast-moving field of personalized medicine. In the first article of this series, we reviewed the fundamental concepts in molecular genetics. In this second article, we cover the key concepts and methods in genetic epidemiology including the classification of genetic disorders, study designs and their implementation, genetic marker selection, genotyping and sequencing technologies, gene identification strategies, data analyses and data interpretation. This review will help the reader critically appraise a genetic association study. In the next article, we will discuss the clinical applications of genetic epidemiology in the personalized medicine area.
Keywords: Genetic epidemiology, genome-wide association study, guidelines, heritability, modes of inheritance, next-generation sequencing, study design.
WHAT IS GENETIC EPIDEMIOLOGY?
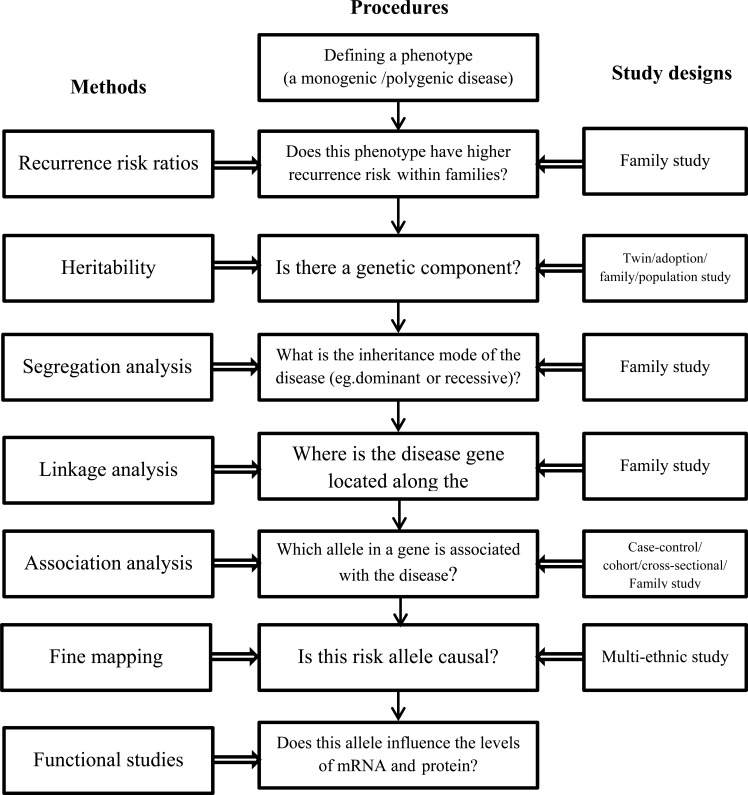
Genetic epidemiology emerged in the 1960s at the crossroads of multiple disciplines such as molecular genetics, epidemiology and biostatistics. Genetic epidemiology studies the role of genetic factors in determining health and disease in families and in populations, as well as the interplay of genetic determinants with specific environmental exposures. Morton elegantly defined genetic epidemiology as "a science which deals with the etiology, distribution, and control of disease in groups of relatives and with inherited causes of disease in populations" [1]. In this article, we aim to illustrate how to identify genetic variants associated with a disease including the relevant concepts, study designs and statistical analyses classically used in genetic epidemiology. Due to the complexity of the steps needed to explore genetic variation in common diseases, we provide a diagram which outlines how this paper is structured (Fig. 1). The questions illustrate the step by step procedures to conduct genetic epidemiology research; the methods show the parameters which are measured, and the third column lists the study designs most commonly used in genetic epidemiology.
Fig. (1).
Framework outlining the procedures, methods and study designs to identify the genetic determinants of common diseases.
PHENOTYPE
A phenotype represents the observable physical or biochemical characteristics of an individual or a group of organisms, as determined by both genetic make-up and environmental influences. In human genetics, phenotypes refer to traits as diverse as diseases, biochemical measurements or the levels of expression of a gene transcript. A phenotype can be binary (e.g. presence or absence of schizophrenia), categorical (e.g. personality disorders) or quantitative (e.g. hippocampal volume [2]). The ideal phenotype should be clinically and biologically relevant, not too rare, and inexpensive, thus allowing large-scale discovery and replication studies feasible. It should be well defined so that measurement errors, misclassification and heterogeneity can be minimized [3].
MODES OF INHERITANCE
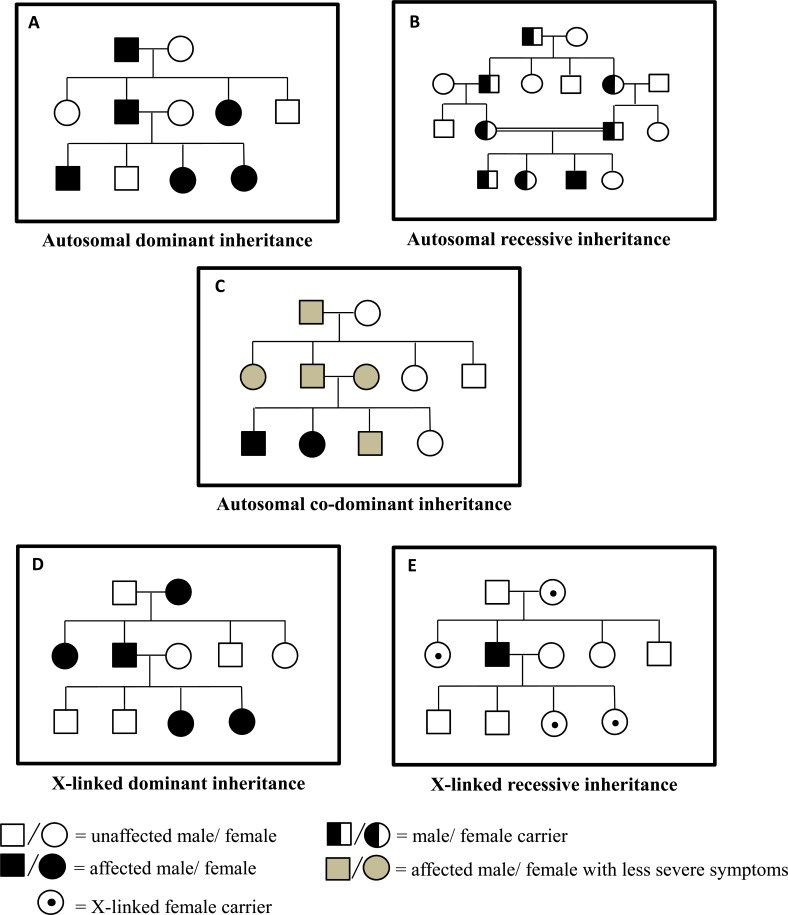
There are five basic patterns of Mendelian inheritances (Fig. 2). Punnett squares which are used to predict the chance of genetic disease in children for parents with an increased risk are presented in Fig. 3. First, autosomal dominant inheritance explains more than 50% of Mendelian diseases. One deleterious copy of the gene is sufficient to confer the disease. Both males and females have 50% risk of being affected and the disease occurs in every generation. Huntington’s disease follows an autosomal dominant mode of inheritance [4]. If each copy of the gene contributes to the trait and the heterozygote generates an intermediate phenotype, this is called co-dominant (e.g. ABO blood type) or additive inheritance (e.g. genetic effects from most risk alleles). Generally speaking, the concept of co-dominant includes additive models. If the trait is quantitative, when the heterozygotes have a mean level which is the average of two types of homozygotes means it is an additive model. An autosomal recessive disease only occurs when an individual harbors two deleterious copies at the locus. In most cases, both parents of the affected person are healthy heterozygous carriers of risk allele [5]. In accordance with Mendel’s Laws, every offspring has a 25% probability of developing the disease. Offspring of consanguineous marriages are more likely to develop autosomal recessive disorders because consanguinity increases the risk to inherit two identical mutations [5]. Sometimes, individuals develop autosomal recessive disorders in non-consanguineous pedigrees because they carry two mutant alleles for the same gene, but with those two alleles being different from each other (for example, two mutant alleles are at different loci). This phenomenon is called compound heterozygosity. Compound heterozygotes usually get ill later in life with less severe symptoms. Phenylketonuria, an inherited disorder that is characterized by seizures, delayed development, behavioral problems and psychiatric disorders, follows an autosomal recessive pattern of inheritance [6]. The fourth mode is X-linked recessive inheritance. A mutation in a gene located on the X chromosome causes a disease in males who are also called hemizygous (the gene mutation only occurs on the X chromosome) and in females who carry the mutant on each of the X chromosome. Thus, X-linked recessive diseases, such as X-linked mental retardation [7], affect more males than females. On the other hand, if only the father is affected, none of his sons will develop the disease, whereas all his daughters will carry the mutant allele. Fifth, X-linked dominant disorders are less common compared with X-linked recessive type. All the offspring of affected females have a 50% chance that they will inherit such a disease whereas all the daughters of an affected male will develop it. Usually, males are affected more severely than females as observed in Fragile X syndrome [5]. However, more female patients with X-linked dominant disorders are sometimes observed. In the Rett syndrome for instance, 50% of the males with the mutant allele miscarry before birth [8].
Fig. (2).
Modes of inheritance. Pedigrees with autosomal dominant inheritance (A), autosomal recessive inheritance (B), autosomal co-dominant inheritance (C), X-linked dominant inheritance (D), X-linked recessive inheritance (E).
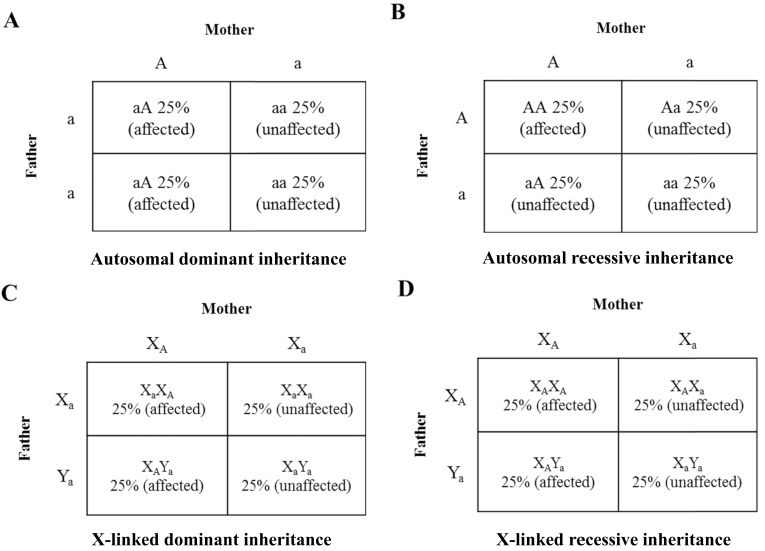
Fig. (3).
Punnett squares of inherited traits. Punnett squares are used to predict the chance of genetic disease in children for parents with an increased risk. The disease-causing mutation is denoted by A and the normal gene is denoted by a. A) Autosomal dominant inheritance: A mother with an autosomal dominant mutation has children with a father who is normal. They have 50% chance with each pregnancy of having a child (boy or girl) affected by the disease and a 50% chance having a child (boy or girl) unaffected. B) Autosomal recessive inheritance: A mother with an autosomal mutation has children with a father who also has the same autosomal mutation. They have 25% chance with each pregnancy of having a child (boy or girl) affected, a 50% of chance having a child unaffected but with the same mutation (carriers), and 25% chance having a child unaffected with normal genotypes. C) X-linked dominant inheritance: A mother with an X-linked mutation has children with a father who is normal. They have 25% chance with each pregnancy of having a girl affected by the disease and a 25% chance having a boy affected. The rest of the children are unaffected with normal genotypes. D) X-linked recessive inheritance: A mother with an X-linked mutation has children with a father who also has a copy of X-linked mutation. They have 25% chance with each pregnancy of having a girl affected by the disease and a 25% chance having a boy affected. The other half of the girls are unaffected but are the mutant carriers and the other half of the boys are unaffected with normal genotypes.
Departure from classical Mendelian patterns of inheritance often occurs and can be explained by different mechanisms that include incomplete penetrance, variable expressivity, genomic imprinting effects, mosaicism, mitochondrial inheritance, de novo mutations, overdominance or digenic inheritance. Incomplete penetrance refers to a situation in which the occurrence of the disease in individuals who harbour the same disease-causing allele is less than 100%. Although the mutant allele does not inevitably cause the disease, it is still passed to the offspring. On the other hand, individuals who inherit the same mutant allele may experience a different level of severity of the disease. This phenomenon is called variable expressivity. Incomplete penetrance and variable expressivity are commonly observed in autosomal dominant and X-linked recessive disorders and can be explained by the effect of modifying genes or by differential regulation of gene expression [9]. For instance, microdeletion of 15q13.3 shows incomplete penetrance of autism and a wide spectrum of mental retardation [10, 11]. Genomic imprinting is a phenomenon by which imprinted alleles are silenced such that the genes are expressed in a parent-of-origin-specific and mono-allelic manner [12]. In other words, the genes are expressed only from the non-imprinted allele inherited from the mother (maternal imprinting) or from the father (paternal imprinting). Imprinting is an epigenetic process that involves DNA methylation or histone methylation mechanisms with no alteration of the genetic sequence [12]. These epigenetic marks are established in the germline cells and are maintained throughout all somatic cells of an organism. Genomic imprinting has an important role in fetal and placental growth and development [13, 14]. Angelman or Prader–Willi syndromes are classical examples of genetic defects in genes submitted to parental imprinting [15]. When the paternal copy is imprinted and silenced, a deletion of 15q12 inherited from the mother causes Angelman syndrome. On the other contrary, if the maternal copy is imprinted and silenced, the deletion inherited from the father leads to Prader-Willi syndrome. Genomic DNA in every single cell of an individual is the same. But, if a mutation occurs during mitotic cell divisions of the developing fetus, it can give rise to mosaicism of at least two populations of cells (somatic or germline) that are genetically different. Mosaicism may explain a substantial fraction of unusual clinical observations, for example, mosaic structural variations are two-fold more frequent in schizophrenic cases than in controls [16]. A very small but functionally important portion of genomic DNA resides in the cytoplasm of mitochondria. Mitochondrial DNA can only be inherited from the mother, because mitochondria present in sperm are eliminated from the embryo. Another unique feature of mitochondrial DNA is that it is randomly distributed into daughter cells during mitosis and meiosis, leading to remarkably variable expressivity in mitochondrial diseases. Schizophrenia and bipolar disease have been reported to present excessive maternal inheritance, and mutations in mitochondrial DNA are also related to these disorders [17-19]. There is a probability of 10-6 to have a de novo mutation in any types of inheritance modes. The de novo mutations in autosomal recessive diseases are more frequent than autosomal dominant and X-linked disorders. The over-dominant mode of inheritance is rarely observed in humans [20]. In that model, the mean of the heterozygotes is higher than the mean of two types of homozygotes. Sometimes, a disease occurs only if two mutations in two different genes are present in the same individual which belongs to a digenic mode of inheritance [21]. Digenic inheritance has been reported in severe familial forms of insulin resistance [22]. Most of the time, non-Mendelian modes of inheritance observed in human diseases result from polygenic genetic architectures (see the section below).
FAMILIAL AGGREGATION, HERITABILITY AND SEGREGATION ANALYSES
Clinicians are used to collecting family history information related to a particular disease in order to assess whether a person is at risk of developing similar problems. A more frequent recurrence of a disease in a pedigree may be because of their shared environmental exposure (e.g. toxin), however, most of the time it indicates that the disease has a hereditary component. Familial aggregation analysis answers the question of whether the relatives of the affected person (proband) are more likely to suffer the same disease compared with the general population at a specific point of time. If the phenotype is qualitative, familial aggregation is measured by recurrence risk ratio in relatives λR (Table 1) [23]. A greater λ is expected in first degree than in second degree relatives of the affected person if genetic factors play a role in the occurrence of the disease [23]. A λR of 2 and above is a good indication that the causes of the underlying familial aggregation warrant further study [24]. Very high relative risk ratios λS for siblings have been observed for autism (λS=75), schizophrenia (λS=10) and bipolar disorder (λS=15) [25] in which shared genes greatly contribute to the familial recurrence of the diseases. If the phenotype is quantitative, familial aggregation is measured by intra-family correlation coefficients (ICC) which is the proportion of the total variance in the phenotype attributed to differences between families. The larger λR or ICC, the greater the familial component of the trait will be [23]. Neither λR nor ICC distinguishes genetic from environmental components, because family relatives share not only genes but also similar environment. For example, familial aggregation for depression could be due to either shared genes or similar environmental factors, such as socioeconomic status of the family.
Table 1.
Measurements of familial aggregation, heritability and linkage analysis.
| Measurements | Formula | Thresholds | |
|---|---|---|---|
| Familial Aggregation | recurrence risk ratio in relatives λR [23] | λR =prevalence of the disease in the relatives of the affected individual / prevalence of the disease in the general population [24] | 2 [24] |
| Heritability | the proportion of total phenotypic variability explained by genetic variance in a particular population [26] | Twin study: h2=2(rMZ-rDZ) Adoption study: h2=2rPO [27] Population-based: (narrow- sense) h2= variance of additive genetic effects/total variance of the observed phenotype [26] |
There is no consensus on the minimum threshold of heritability needed to follow-up with gene identification program. A heritability estimate of 30% maybe considered as the minimum [3]. |
| Linkage study | LOD: logarithm of the odds score [75] | LOD(θ)=log10[Likelihood(θˆ )/Likelihood(θ=0.5)] [75] | 3.3 [75] |
rMZ: correlation coefficient of the trait between monozygotic twins
rDZ: correlation coefficient of the trait between dizygotic twins
rPO: correlation coefficient of the trait between a biological parent and an adopted-away child
Θ is the probability of a recombination event (recombination fraction) between a genetic marker and the disease locus. Observed Θˆ can be obtained by counting recombinants and non-recombinants when the genotypes of individuals within a family are available.
Heritability reflects the proportion of total phenotypic variability explained by genetic variance in a particular population at a specific time. When only additive genetic effects are accounted for in the genetic variance, heritability is named narrow-sense heritability or just heritability (h2); when all genetic variance from additive, dominant and epistatic (gene × gene interaction) effects is accounted for, heritability is defined as broad-sense heritability (H2) [26]. Twin and adoption studies are ideal experimental designs to estimate heritability because of their natural separation of genetic and environmental components [26]. In twin studies, monozygotic (MZ) twins share 100% of their genome whereas dizygotic (DZ) twins share 50%. If genetic factors play a role in the phenotype, the correlation coefficient of the phenotype between MZs should be significantly higher than in DZs. The calculation of the heritability is listed in Table 1. These calculations are based on the assumption that MZ pairs and DZ pairs grow up in an identical environment [27]. There is a methodological concern that twins are not representatives of the general population [28]. In practice, the assumption of identical environment in twin studies may be difficult to hold. Twins may display difference in delivery process, special life events, and interactions with teachers or friends. In an alternative adoption study, a biological parent and an adopted-away offspring, or a full sibling and an adopted-away full sibling share 50% of genes that attribute to their resemblance in the trait. The heritability in this situation assumes they have different environmental exposures (Table 1). When the traits are binary, a liability scale model in which a disease arises when the determined probability exceeds a certain threshold, or the statistical models developed for quantitative traits may be applied [29, 30] Although the assumptions underlying the twin and adoption studies are not always met in practice, many important findings have been discovered from such designs [31]. Structural equation models have been used to estimate heritability with consideration of shared and non-shared environment effects by collecting diverse environmental variables [32]. Recently, Yang et al. has developed a GCTA model, a tool that estimates heritability using genome-wide association study (GWAS) data and unrelated individuals for both quantitative and binary traits [33, 34]. The phenotypic variance explained by this model is from all the SNPs (including imputed SNPs) rather than individual SNPs associated with this phenotype. It has been applied to estimate the heritability in intelligence and schizophrenia [35, 36]. Heritability is an important concept in genetics but is often misunderstood [26]. Heritability does not influence a trait in itself, but it can play a role in the variation of a trait. Therefore, heritability estimate cannot be used as an indicator of the individual risk. Heritability may vary in different populations and change over time. It is important to select a phenotype in a population with a substantial heritability to identify the genetic determinants underlying the trait [24]. Studies have shown that schizophrenia, bipolar disorder and autism are highly heritable traits with heritability greater than 80%, whereas drug dependence shows moderate heritability of 50-60% [37]. We do not encourage gene identification programs if traits show heritability estimates lower than 30%, as these programs may become a ‘geneticist’s nightmare’ [3, 38].
Once twin studies, adoption studies, family studies or population-based studies of unrelated individuals have provided evidence that a trait has a genetic component, a segregation analysis with family data will answer the question of what is the best inheritance mode this trait follows [39]. It determines whether the transmission pattern of a trait in families is consistent with the expectation of one of the Mendelian inheritance modes we discussed above. Likelihood ratio test or chi-squared test is usually applied to examine whether a segregation ratio deviates from the expected under Mendelian laws, with no need for genetic marker information. For example, a dominant disease has a theoretical segregation ratio of 0.5. If the hypothesized Mendelian segregation ratio is true, it indicates the disease is determined by a single gene. Otherwise, the deviation may be an indication that the disease is determined by multiple genes, or caused by interplay between genetic and environmental factors, or the disease has an incomplete penetrance. Under these complicated circumstances, maximum likelihood tests are used to compare different inheritance models [40]. Therefore, segregation analysis seems appealing to typical Mendelian modes of inheritance. To a few notable exceptions (e.g. type 1 diabetes) [41] segregation analyses for complex diseases did not succeed in revealing the presence of a major gene and a clear pattern of inheritance [42].
SINGLE GENE DISORDERS VERSUS COMPLEX DISEASES
A single-gene disorder (also called a Mendelian or monogenic disorder) is caused by a single mutation in a single gene. It exhibits a familial pattern consistent with one of the Mendelian inheritance modes. According to the statistics of Mendelian Inheritance in Man (OMIM) (www.ncbi.nlm.nih.gov/omim), more than 5200 diseases follow a Mendelian inheritance pattern, and the underlying molecular basis of 66% of them has been elucidated. Sometimes, mutations in only one gene elucidate 100% of disease cases (e.g. Huntington’s disease). Sometimes, mutations in different genes lead to similar disease presentation. For instance, mutations in 15 different genes lead to the Bardet-Biedl syndrome [43, 44]. In that situation, the disease is referred as a heterogeneous monogenic disorder. The identification of genes responsible for single-gene diseases has made tremendous progress in the past 15 years and has greatly facilitated the understanding of disease-related molecular mechanisms. However, Mendelian segregation law which predicts discrete traits (like yellow/green, wrinkled/smooth peas in the original experiments) cannot explain many anthropometric features such as height and weight that show continuous variation. These quantitative traits do display familial clustering (e.g. relatives of the taller individuals tend to be taller than the general population), however, their transmission across generations does not follow clear Mendelian patterns of inheritance. In 1918, Ronald A Fisher, together with Sewall Wright and JBS Haldane, solved the dilemma by developing a polygenic inheritance theory using analysis of variance [45]. Multiple genes contribute to the continuous variation of a trait, each with allelic variation. Meanwhile, each allele follows Mendel’s segregation law and makes a small change in the total variance [45, 46]. Many common diseases (eg. cancers, diabetes, cardiovascular diseases, Alzheimer’s disease and schizophrenia) follow a polygenic model [47, 48]. Though the etiology of them is not completely understood, it is believed that they are caused by multiple genes and environmental factors and their interplay. The term complex disease is exchangeable with common disease and polygenic disease in the literature. It is important to pinpoint that monogenic genes exist in polygenic diseases, often initially identified in extreme end of the distribution of a trait. For example, more than sixty loci modestly contribute to the risk of obesity [49]. In addition, rare mutations or deletions at nine loci lead to monogenic forms of early-onset severe obesity and may explain 5-10% of obesity cases [49, 50].
Different models have been proposed to explain the genetic architecture of complex diseases. First, the common disease-common variant hypothesis (CDCV) states that risk variants are at relatively high frequency (>1%) in populations and modestly contribute to the risk of disease [51, 52]. The advent of GWAS has identified more than 2000 common loci modestly associated with complex traits and has given some credit to the CDCV hypothesis. However, the fact that common variants identified through large-scale GWAS consortium initiatives only explain a small proportion of heritability for most complex diseases excludes the possibility that CDCV hypothesis is the only relevant model [53, 54]. The second hypothesis, common disease-rare variant (CDRV), states that most of the common phenotypic variance are caused by rare variants (allele frequency <1%) with large effect sizes [55]. Recently, rare variants have been identified to play a role in several multifactorial disorders such as prostate cancer [56], inflammatory bowel disease [57] or type 2 diabetes [58]. Third, Dickson et al. recently proposed the synthetic association model in which the association of a common non-functional SNP with a disease may be the result of several disease-causing rare variants that have stronger effects and are tagged by the common SNPs [59]. Although the synthetic association hypothesis has been validated for specific SNPs associated with hearing loss, sickle cell anemia or Crohn’s disease [59, 60], it is unlikely to explain most of the associations between common variants and complex traits identified through GWAS [60, 61]. In fact, CDCV and CDRV models are complementary, and there is a growing consensus that multifactorial diseases may result from a combination of rare and common risk variants [62, 63].
IDENTIFICATION OF DISEASE PREDISPOSING GENETIC VARIANTS: STUDY DESIGNS
Different study designs can be used to identify disease-associated genetic variants in different contexts. Case-control and prospective cohort studies commonly used in classical epidemiology are also applied to genetic epidemiology. A case-control study recruits two groups of individuals who are diagnosed with (cases) or without (controls) a disease and determines the risk of being affected depending on different genotypes. This enables researchers to identify genes responsible for a disease (especially a less-common disease) in a time- and cost-efficient way, because adequate sample size is required to reach sufficient power to detect modest genetic effects. The major weaknesses of a case-control design are biases brought up by the retrospective recalls of exposures and misclassification of cases and controls [3, 64]. However, such biases are not a significant concern in a genetic association study because the genotypes (exposures) of individuals does not change with time [65]. However, when confounding factors of some exposures or gene-environment interactions are assessed, considerations to such biases are still relevant. Since genetic associations are sensitive to population stratification between cases and controls, individuals in both groups should come from the same population [66]. In some case-control studies, an enrichment sampling strategy may be applied to increase power to detect a novel genetic variant [67]. Such a strategy increases power but usually overestimates the relative risk. Therefore, it is necessary to replicate in a population-based sample or make a conclusion based on a specific group of people.
In a prospective cohort study, individuals without the disease at baseline are followed for a period of time and then the associations between genotypes and the incident disease status are assessed at the end of the study. Because the disease has not yet presented during sampling, it allows the researchers to control the potential selection bias and minimize the misclassification errors as well. This is why cohort studies are considered the gold standard for both classical and genetic epidemiology studies, but this is with the sacrifice of time and cost. For this reason, case-control studies are more popular in genetic epidemiology. An alternative study design, the nested case-control study, collects cases in a defined cohort and selects a specific number of controls among those who have not developed the disease yet at the time of assessment [68]. Such an approach shows its unique value in gene-environment interaction association studies because it increases the measurement accuracy of environmental exposures which is essential to increase statistical power to detect interactions [64, 69].
Population-based designs are desirable in genetic epidemiology but they require larger sample sizes than case-control designs to reach the same statistical power, the latter being enriched in a greater proportion of cases [70]. This limitation can become critical if expensive technologies are used (e.g. genome-wide DNA arrays, whole genome sequencing).
Family-based designs are also widely used in genetic epidemiology, which are ideal to assess parental imprinting effects or in haplotype studies (the reconstruction of the haplotype phase is improved by the availability of parental genotypes) [71]. A case-parent triad design which consists of one affected offspring and the two parents in each family is commonly used. Given the same power, type I error threshold and risk allele frequency, the number of trios in family-based study is the same as the pairs in a case-control study, signifying 50% more individuals and 50% increased genotyping or sequencing costs are needed. For example, if the power is 90%, using two-sided P-value of 0.001 and an allele frequency of 20% in the control group, 3731 trios will be requested to detect an odds ratio of 1.20 in family-based design and 3731 pairs of case and control in a case-control study, representing 50% more participants. Case-parent triad design is also used to confirm an association from a case-control study because it is robust to population stratification. However, it is not well-adapted to late-onset diseases due to the difficulty or unavailability of DNA collection in parents [72]. There are also other family-based matching designs and corresponding statistical methods [73]. The main limitations are the lack of power, especially if the effect sizes are small, difficulties in recruiting required number of samples and the generalization of the discoveries from family-based studies to general populations [74, 75].
As mentioned above, the choice of an appropriate control is critical to conduct a valid case-control study. The case-only study is one of the designs which have no controls involved. As well explained by Khoury et al., this design is especially efficient in the context of gene-environment interaction studies when the assumption that the tested genotype and environmental exposure are independent in a given population is met [76, 77]. Case-only studies can only examines the departure from a multiplicative interaction model rather than an additive interaction model, which is also less accepted by the scientific community. Although the case-only study design provides better estimation and needs a smaller sample size than traditional case-control design, it also may increase type I error if the assumption is not true [77, 78]. In addition to gene-environment interactions, it has also been used in gene-gene interaction and pharmacogenetic studies [79, 80]. Pharmacogenetic interaction is a special type of gene-environment interaction and is designed to identify genetic variants which predict response to treatment. When case-only study design is applied, the assumption that there is no correlation between genetic variants and treatment assignment must been examined. Thus, a case-only design nested in a randomized controlled trial (RCT) provides an ideal model for pharmacogenetic studies in which treatment assignment is random and unrelated to genotypes [80].
HOW DO WE GET THE GENETIC INFORMATION?
DNA Extraction
Adequate quantity and quality of DNA from a large number of individuals are prerequisites for a successful genetic epidemiology study, both of which depend on the samples collected and DNA extraction methods. The samples stored in the Biobank of study centres may be buffy coat (mainly blood leukocytes), saliva (mainly buccal cells) or tissue biopsies. The buffy coat is most commonly used, but saliva is getting more and more popular because of its non-invasive nature and stability at room temperature. Modern DNA extraction methods are fast, non-toxic and reach high yields. A general DNA extraction procedure consists of cell lysis by alkaline, protein removal by salt precipitation and DNA recovery by ethanol precipitation [81]. Extracted DNA is dissolved in appropriate buffer and stored in small aliquots at -70°C for long-term storage, but repeated freezing and thawing should be avoided.
Genotyping
Single nucleotide polymorphisms (SNPs) represent more than 90% of the entire genomic variants. SNPs have been initially detected by direct sequencing and genotyping of 270 individuals in the context of the Human Genome Project and HapMap Project and more recently through the 1000 Genomes Project. There are over 38 million validated human SNPs in the dbSNP database (dbSNP Build 137) (https://www.ncbi.nlm.nih.gov/SNP/). In the past two decades, many genotyping methods have been developed, with most of them assuming a bi-allelic feature of most SNPs in human. The commonly used approaches include restriction fragment length polymorphism (RFLP), differential hybridization (TaqMan), allele-specific primer extension (SNaPshot, SNPstream, pyrosequencing), allele-specific oligonucleotide ligation (Applied Biosystems SNPlex), allele-specific extension (Illumina Omni Whole-Genome Arrays) and single-base extension (Affymetrix 6.0) which can be detected by mass spectrometry (Sequenom MassArray), fluorescent light (TaqMan, Applied Biosystems), bioluminescent light, electrophoresis or high-resolution melting curves (Roche Applied Sciences LightTyper) [82, 83]. Generally speaking, all these methods are performed in two different formats: homogeneous reactions (in solution) and heterogeneous reactions (in solution and a solid phase such as a microtiter well plate, latex beads, a glass slide, or a silicon chip). The former has limited capability of multiplexing which is to examine more than one SNP at a time; while the latter one is flexible in multiplexing ranging from a few to a hundred to several million SNPs. Because of their intrinsic characteristics, each genotyping method has unique applications and multiplexing capability. For examples, TaqMan SNP Genotyping Assays (Applied Biosystems) identify the genotypes of single SNP at a time with great precision and is widely used in candidate-gene association and replication studies even with large sample size [84]. The Sequenom MassArray uses a single-base primer extension genotyping method followed by distinguishing DNA base by molecular weight. It has high resolution but moderate multiplexing, and it is appropriate for small number of SNPs [85]. The more recent genome-wide genotyping arrays can accommodate up to 4.8 million genetic markers, including single nucleotide polymorphisms (SNPs) and probes for the detection of copy number variations (CNV). Therefore, some platforms work better for single SNPs or a few targeted SNPs in many individuals, some are suitable for small to moderate number (a few hundred to a few thousand) of SNPs on a few subjects at one time, and others are the best choice for several million SNPs on one subject at one time, depending on the aim and design of a particular study. Customized design may also be applied to genotyping on a single SNP or moderate number of SNPs. More than 4.5 million predesigned probes are available to customized uses with TaqMan genotyping [83]. Table 2 gives a simple guideline on how to choose an appropriate genotyping platform, and the updated capacity of each platform is always available on the commercial websites.
Table 2.
Genotyping methods and study designs.
| Number of SNPs to be Genotyped | Study Designs | Genotyping Methods |
|---|---|---|
| 1-10 | Candidate gene studies Replication studies |
TaqMan LightTyper Pyrosequencing |
| 1-500 | Replication studies Linkage studies Fine-mapping studies |
SNaPshot SNPlex Sequenom MassARRAY Illumina Golden Gate with BeadXpress readout |
| 384-3,072 | Linkage studies Fine-mapping studies Disease-specific SNPs Pathway-specific SNPs |
Illumina Golden Gate with iScan readout |
| 6,000-70,000 | Linkage studies Fine-mapping studies Disease-specific SNPs Pathway-specific SNPs |
Illumina Infinium iSelec Custom Beadchip |
| >500,000 Up to 4.8 million |
GWAS (SNPs, CNVs) | Illumina Omni Whole-Genome Array Affymetrix 6.0 Array |
Sequencing
Sequencing is a method to determine the exact sequence of nucleotides from a fragment of DNA or the whole genome. It not only examines the presence of the bi-allelic variants reported in databases, but also provides information on all possible polymorphisms (including those with 3 or 4 alleles). Sequencing is the ideal method to characterize the sequence of a new genome or to identify rare genetic variants not reported in SNP databases. Due to its current cost, sequencing has not yet been an efficient and economical way to genotype SNPs. Sanger sequencing (the first generation sequencing method), which was described in 1977 [86], experienced many technical revolutions and eventually developed into today’s automated Sanger sequencing [87, 88]. The completion of the Human Genome Project led to tremendous improvements in the Sanger sequencing method, including the development of whole-genome shotgun sequencing and a parallel sequencing initiative of the human genome by the company Celera Genomics [89]. However, Sanger sequencing is still expensive and laborious, and faster and more affordable methods to sequence DNA have been in great demand from broad research interests such as variant association studies, comparative genomics, population evolution and clinical diagnostics. High-throughput next generation sequencing (NGS), first launched in 2005, involves “massively parallel” sequencing and offers to sequence up to hundreds of millions of DNA fragments in a single platform. It cost $2.7 billion and 12 years to complete the Human Genome Project with Sanger sequencing, but it is now possible to obtain a personal whole-genome sequence at a cost of $1,000 [90].
Currently the DNA polymerase-dependent sequencing strategies are widespread on the market and can be classified as single nucleotide addition (SNA), cyclic reversible termination (CRT) and real-time sequencing. Here we will introduce three major platforms which are commercially available, in combination with their unique sequencing principles (Table 3). Roche/454 was the first developed NGS, using “pyrosequencing” technique of DNA [91, 92]. The current Roche/454 GS FLX+ Sequencer is able to produce 700 Mb of sequence with 99.997% accuracy for single reads of 1,000 bases in length (http://454.com/products/gs-flx-system/index.asp).
Table 3.
Characteristics of sequencing platforms.
| Platform | Sequencing Technology | Sequencing Reaction | Capacity | Efficiency (bp/Read) |
|---|---|---|---|---|
| Roche/454 | Single nucleotide addition (pyrosequencing) | Synthesis | 700 MB | 1,000 |
| Illumina/Solexa Genome Analyzer | Cyclic reversible termination | Synthesis | 600Gb | 100 |
| Applied Biosystems/ (SOLiD) | Real-time sequencing | Ligation | 320Gb | 50-75 |
| Applied Biosystems/ION Torrent | Semiconductor | Synthesis | 2Gb | 200-400 |
The second NGS approach is the Illumina/Solexa Genome Analyzer which uses cyclic reversible termination sequencing method and currently dominates the market. The capacity of the newest model generates up to 600 Gb of bases per run with a read length of about 100 bases (http://www.illumina.com/technology/solexatechnology. ilmn). This is less than Roche/454 due to less efficient incorporation of modified nucleotides.
Another NGS system is Applied Biosystems Supported Oligonucleotide Ligation and Detection (SOLiD) sequencer based on sequencing by ligation [93]. The complicated process is well illustrated in Metzker’s paper [92]. SOLiD systems have two independent flow cells and allow two completely different experiments to be run at the same time. The updated SOLiD system can yield 320 Gb of sequence per run with a 99.99% accuracy and a read length of 50-75 bases (http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Sequencing/Next-Generation-Sequencing/).
Recently, the novel sequencing technology ION Torrent arose on the market. It does not need any modified nucleotides. Its chemistry rationale is very simple. During the process of DNA synthesis, the incorporation of each dNTP causes the release of a hydrogen ion. The hydrogen ion changes pH in the solution, which can be detected by an ion-sensitive field-effect transistor (ISFET) detector [94]. This method enables a fast, accurate, inexpensive, and simple massively parallel sequencing. Ion Personal Genome Machine (PGM) and Ion Proton sequencers load amplified DNA fragments into micro wells of a high-density Ion chip to perform sequencing. The changed pH can be detected by an ion sensitive layer beneath the wells and converted into voltage changes. The change in voltage is proportional to the type and number of nucleotides incorporated and recorded. These smaller and cheaper sequencers can produce up to 2 Gb output per run with a read length of 200-400 bases.
In addition to the strategies discussed above, many other technologies are under development and all the methods will continue to compete and improve [88]. Currently, it is not easy to predict which approach will be the winner of the future sequencing market. NGS is certainly another ground-breaking revolution in biology and medicine after the completion of the Human Genome Project, making personal whole-genome studies more than just a dream. The 1000 Genomes Project has used Illumina/Solexa and Roche/454 platforms to sequence whole genomes and has validated up to 38 million SNPs, 1.4 million short insertions and deletions, and more than 14,000 larger deletions [95]. Whole-genome sequencing plays a unique role in facilitating a deeper and broader understanding of the spectrum of genetic variants and their pathogenesis in complex diseases, clinical diagnosis and personalized health decision-making. It will eventually come into daily practice in the near future, however, current cost and analytical challenges limit its applicability [90, 96]. An alternative solution to this may be to apply NGS to target specific sequences of interest, for example, whole-exome sequencing which sequences the entire protein-coding genes. In spite of constituting approximately 1% of the human genome, protein-coding regions include 85% of mutations associated with Mendelian diseases [97]. Meanwhile, non-synonymous variants predict with a high likelihood a functional change [98]. As such, the whole-exome is a relevant subset of the genome to search for genetic variants with large effect sizes and has been used to dissect the genetic architectures of Mendelian and complex disorders [99, 100]. Exome sequencing by NGS, in conjunction with developed strategies in study design and analytic methods, has had a great success in identifying causal alleles for several dozen Mendelian disorders [99]. Although more challenging, whole-exome sequencing has also been an effective strategy in identifying coding variants associated with complex diseases such as autism spectrum disorders and schizophrenia [101-103]. Compared to whole-genome sequencing, whole-exome sequencing is currently a more widely accepted strategy to search for rare variants because of its cost-effectiveness, the simpler data analysis and interpretation.
GENE IDENTIFICATION STRATEGIES
The identification of genes responsible for Mendelian and complex diseases may enable a better understanding of their pathology, provide efficient molecular targets for innovative therapeutic drugs, and help to better predict disease risk in populations for targeted prevention. In the past decade, a remarkable progress has been made in the journey of discovering disease-causing genes. However, more than 30% of the underlying genes leading to Mendelian disorders are still unknown, and the identified genetic variants to complex diseases account for only a small portion of heritability. In order to pursue gene identification efforts, traditional and novel gene identification strategies are introduced below.
Genetic Linkage Studies
Linkage analysis aims to map the location of a disease-causing loci by looking for genetic markers that co-segregate with the disease within pedigrees, though the disease causing allele has not to be directly genotyped [75]. Linkage is based on the facts that recombination occurs between homologous chromosomes during meiosis and recombination likelihood increases with the distance between two loci, a random probability from zero to 0.5. When a marker allele is inherited along with the disease in pedigrees, it strongly suggests that the disease-causing locus is located in the vicinity of the genetic marker on the chromosome. A set of 400 highly-informative microsatellite markers (repeated sequences of DNA fragments less than 10 bp [104]) equally distributed across the genome is generally selected in a whole-genome linkage analysis. More recently, a set of 6,000-10,000 markers have been proposed by different companies to perform linkage analysis.
Different linkage approaches are chosen depending on the type of disease (monogenic or polygenic) or trait (dichotomous or quantitative). Parametric or model-based linkage analysis is used if the disease follows one of the typical Mendelian inheritance modes. Results of linkage analysis are often reported as logarithm of the odds (LOD) score which is a function of the parameter θ. θ is the probability of a recombination event (recombination fraction) between a genetic marker and the disease locus [75]. LOD score analysis is equivalent to likelihood ratio test, assessing the null hypothesis H0 of θ=0.5 (absence of linkage) versus alternative hypothesis H1 of θ<0.5 (presence of linkage). In the simplest scenario with a known inheritance model, complete penetrance, no de novo mutations and no phenocopies (different environmental exposures and genetic variants lead to the same disease), θ is estimated by the maximum likelihood method, thus giving rise to a maximum LOD score (Table 2). The higher the LOD score is, the stronger the evidence of linkage will be. Historically, a rule of thumb states that a LOD score above 3 is sufficient to claim a significant linkage, based on the critical value from Morton [105]. An even higher LOD score of 3.3 is required to ensure the genome-wide type I error of 0.05. Other complicated model-based cases with incomplete penetrance, phenocopies and mutations, and more relaxed LOD score thresholds are discussed in detail by Ziegler and Konig [30]. Linkage analysis has successfully mapped genes responsible for Mendelian disorders such as the Wolfram syndrome on the short arm of chromosome 4 [106, 107].
Little is known about loci predisposing to complex diseases, and attributing a clear Mendelian pattern of inheritance within families for such a locus is impossible. As a result, model-based linkage analyses do not apply to complex trait linkage analyses and model-free linkage analyses have been developed. The fundamental rationale underlying model-free linkage analysis is that the genetic resemblance in the affected sibling pairs is more similar in certain regions of the genome if the disease is heritable. Therefore, the statistical tests assess whether the observed degree of genotypic similarity exceeds the expected value. Instead of measuring recombinant fraction of θ, genotypic similarity is measured by the identical by descent (IBD) value which refers to the number of alleles inherited from the same common ancestor in a pair of relatives. The IBD values can be 0, 1, or 2. If the distribution of IBD values is determined, model-free linkage analysis examines whether allele sharing in affected siblings is different from the expected distribution. More generally, it tests whether the mean number of IBD shared alleles departs from the expected value of 1 in sibling pairs [108]. Excess of IBD sharing can also be tested by other methods such as the maximum non-parametric LOD score test and Wald test [30] which successfully identified the HLA region associated with type I diabetes [109].
Linkage studies also apply to quantitative traits such as cholesterol or glucose level. The approaches for model-free linkage analysis of quantitative traits include the Haseman-Elston, variance component methods among others [30]. A region between markers D9S925 and D9S741 on chromosome 9p associated with high-density lipoprotein-cholesterol concentration in Mexican Americans was initially identified with variance component analysis [110]. However, true linkage has been hard to find in complex trait studies, likely due to the modest effect sizes of genetic variants, allelic heterogeneity, or gene by environment interactions in complex diseases [25, 111].
Homozygosity Mapping
Homozygosity mapping is a powerful tool to map genes responsible for recessive Mendelian disorders in consanguineous pedigrees [112]. With this approach less than a dozen of affected individuals are needed and more importantly no additional family members are required to identity a disease-causing locus. These advantages render it possible to map disease loci of many rare recessive disorders when it is impossible to collect adequate number of families as linkage analysis usually requires. The principle underlying this approach is that if the offspring of a consanguineous marriage (for example sibling, first-cousin, and second-cousin) is affected with a recessive inherited disease, a large region spanning the disease locus is homozygous by decent [112]. For instance, a child of a consanguineous couple has a coefficient of inbreeding F of 1/4, 1/16, 1/64 for sibling, first-cousin, and second cousin, respectively. Assuming the frequency of the disease allele in this population is q, the probability of homozygosity by decent at the disease locus is α= F*q/[F*q+(1-F)*q2]. If q is far smaller than F, α is close to 1, indicating the greatest chance to be homozygous. The comparison of homozygous regions in several affected family members, along with traditional linkage analysis and a sufficiently dense genetic map, can narrow down the location of a gene underlying a recessive disease. Low-density restriction fragment length polymorphism (RFLP), microsatellite linkage maps, and more recently high-density SNP arrays have been used in homozygosity mapping gene identification. For instance, the use of a high-density GeneChip containing 57,244 SNPs identified the linked region for autosomal recessive Bardet-Biedl syndrome which was initially missed by linkage studies with 400 highly informative microsatellites in a small Israeli Bedouin consanguineous pedigree [113].
Candidate Gene Studies
This approach is hypothesis-driven and has been widely used in genetic association studies before the advent of GWAS. Candidate genes are selected based on prior knowledge of their potential role on the trait of interest from in vivo, in vitro or in silico studies in animals or humans [114, 115]. One important advantage of the candidate gene approach is to restrict the number of hypotheses tested and to relax the multiple testing correction thresholds in comparison with genome-wide approaches. One limitation of the candidate gene approach is its dependence on the level of current knowledge of a specific gene. The success rate of candidate gene studies has been low, in part due to the limited understanding of the molecular and genetic mechanisms in complex diseases [66]. Selecting strong candidate genes on the basis of converging arguments from different research disciplines has been more successful, as illustrated by the identification of SNPs in APOE4 associated with Alzheimer disease (AD) [116]. APOE4 gene was indeed located on the proximal long arm of chromosome 19, in a region of linkage for late-onset AD [117]. In addition, apolipoprotein E (apoE) was a key protein related to AD [116].
Genome-Wide Association Studies
Hypothesis-free GWAS exhaustively test the genotype-phenotype associations across up to 4.8 million genetic markers and represent to date the most efficient way to identify common variants (MAF> 1%) associated with complex diseases [118]. Along with the advanced high-throughput technology, more and more SNPs and copy number variants (CNVs) are validated by the 1000 Genomes Project, which enable the current genotyping arrays to include rare variants and CNVs in addition to common variants. GWAS have identified several risk variants associated with bipolar disorder [119] or schizophrenia [120]. However, there are two major limitations of GWAS. First, a very stringent level of significance is required to adjust for multiple testing. Second, most of the statistically significant associations lack a biological support [121-123].
Whole-Genome/Whole-Exome Sequencing
Whole-genome/whole-exome sequencing strategies are currently efficiently applied to identify rare variants associated with Mendelian or complex traits. Whole-genome/whole-exome sequencing is not just an alternative way for genotyping as it also detects novel mutations not catalogued in SNP databases and additional alleles beyond bi-alleles. The biggest challenge in whole-genome/whole-exome sequencing experiments is how to analyze a huge sequencing dataset to identify the novel causal genes for either Mendelian or complex diseases [124]. Usually, 20,000 to 30,000 variants are found through each whole-exome run. Unreliable variants are first removed by data quality control procedures (e.g. read coverage less than five, inconsistency among the reads). If the investigators focus their attention on potentially deleterious rare coding variants, variants located outside the coding regions and synonymous coding variants are filtered out. Then the most important step with substantial reduction of the number of variants is to exclude known polymorphisms in human population based on appropriate databases [125]. At this step, approximately 150-500 non-synonymous or splicing variants remain to be potentially causal variants. Additional filtering methods may include in silico functional evaluation of mutations, candidate gene, linkage, homozygosity mapping, de novo and overlap strategies [124]. Becker et al. have successfully used homozygosity mapping, in combination with an exome sequencing strategy, to elucidate the genetic basis of osteogenesis imperfecta [126]. They found 318 non-synonymous variants after several filtering strategies. Among them, 17 were autosomal homozygous, but only three were in the regions with the larger stretch of homozygous loci. In combination with overlap strategy and functional testing, truncating mutations in gene SERPINF1 were identified as causal loci leading to autosomal-recessive osteogenesis imperfecta [126].
HOW TO INTERPRET GENETIC ASSOCIATIONS IN COMPLEX DISEASE?
Power of a Study
In genetic epidemiology, most genetic variants confer small to modest effect sizes with an odds ratio (OR) lower than 1.5, indicating that a large sample size is needed in a population-based association study. For example, if the risk allele frequency in controls is 20%, 1763 cases and 1763 controls are needed to detect an OR of 1.3 at a type I error level of 0.001 (two-sided) and power of 90% [3]. The requirement for such large sample sizes can be difficult to achieve by single teams and as a result researchers have to pool samples in large-scale international consortium initiatives to reach an adequate power. These power estimations also imply that many previously published case-control studies were underpowered. This may explain why many promising associations were never replicated [127]. Replication of an association study in an independent sample is recommended. The sample size for the replication study should take into account of the risk of overestimation of the true effect in the initial sample (a phenomenon called the Winner’s curse effect) [128, 129]. Statistical power may be even more a concern in genetic association studies involving rare variants, and the desired number of individuals may not be feasible in practice [130]. To deal with these issues, researchers select designs where additional copies of the variant of interest can be sampled (perhaps in large pedigrees or in a founder population). They also pool together variants likely to have an impact on the function of a specific gene and compare the global distribution of these variants in case control designs [131].
Data Quality Control (QC)
Genotyping errors cause genotype misclassification and have the potential risk of decreased power, leading to false associations [132]. The procedures to remove the uncertain individuals and DNA markers are critical steps before statistical analysis of associations. It is recommended to conduct QC on the individuals before QC on the DNA markers [133]. Individuals with discordant sex information, inaccurate phenotypic data, or a conflicting ethnicity between self-reported and genetically determined should be identified and removed. Individuals with low DNA quality (e.g. displaying >10% missing genotypes in a genotyping array) should also be taken out. At the genetic marker level, the genotyping method should be reliable and the laboratory protocols should be standard. The concordance rate of duplicated samples must be higher than 99% (usually > 10% of the entire sample are re-genotyped with the same or a different genotyping method). SNPs with a genotyping call rate (percentage of successfully genotyped individuals)<95%, a significant deviation from a Hardy-Weinberg equilibrium (HWE) test [134] (P HWE < 0.005 in the control group), a significant difference in the missing genotype rates between cases and controls, or a very low allele frequency should be filtered out. In a family-based study, an additional check of Mendelian inconsistencies should be conducted [30].
According to the workflow of NGS, standard protocols for QC should be developed and implemented at each step including DNA extraction, targeted gene enrichment, library preparation and sequencing. Current NGS technologies have higher raw per-base error rates than Sanger sequencing [135]. However, this shortcoming can be compensated to some extent by increasing the coverage depth of sequencing, checking the presence of a mutation in related individuals or validating the findings by Sanger sequencing [136, 137]. False-positive association may also result from a difference of coverage depth between cases and control groups [138].
Statistical Analysis
A genetic model (i.e., dominant, additive, recessive) needs to be defined prior to any genotype-phenotype association study. If the underlying genetic model is unknown, an additive model is frequently assumed, but testing the three models is more informative. Given two alleles A and B (B is risk allele) and three genotypes AA, AB and BB at a locus, AA is coded as 0, AB as 1 and BB as 2, and a 2×3 contingency table is created under an additive model as illustrated in the Table 4. In the simplest scenario in which cases and controls are matched for confounding factors (e.g. age, sex), the Cochran-Armitage test is used to test the association between the allele B and a trait, which is similar to Peason’s χ2 test but taking into account the order of risk of the three genotypes (AA<AB<BB) [139]. Meanwhile, the odds ratios (OR) are often calculated to provide a measure of the strength of the associations. If individuals have one risk allele B, the risk of having the disease is OR1=(b/a)/(e/d)=bd/ae times higher than those who has no risk allele B; and if individuals have two copies of B, the risk of being affected is OR2=(c/a)/(f/d)=cd/af times higher than those who has no B. If the outcome is binary (presence or absence of the disease), a simple logistic regression can also be applied. The exponential of the regression coefficient equals to the increased OR with per additional B. If the outcome is a continuous (or quantitative) variable, a linear regression model will be used. The beta coefficient from a linear regression analysis means how much increase in the outcome for each additional risk allele B. Compared to Cochran-Armitage and Peason’s χ2 tests, the advantage of using a linear or logistic regression is that they allow for the adjustment for the confounding factors such as age, sex and including of gene × gene and gene × environment interaction terms into the model [140]. When the outcome is a count/rate or a time-to-events, a Poisson regression model or a Cox proportional hazard model will be chosen, respectively. As a result, relative risk (RR) or hazard ratio (HR) will be estimated [141]. Sophisticated methods such as the kernel association test have been recently developed to assess the association of groups of rare variants with a disease or a quantitative trait [142, 143].
Table 4.
A 2x3 contingency table in an additive model.
| AA | AB | BB | |
|---|---|---|---|
| Case | a | b | c |
| Control | d | e | f |
a, b, c are the counts of individuals with genotypes of AA, AB, BB respectively in cases, and d, e, f are the counts of individuals with genotypes of AA, AB, BB respectively in controls.
From the perspective of statistics, GWAS analysis is just an extension of the single-SNP analysis and covariates can also be adjusted in linear or logistic regression models. One issue is that most of the significant associations at the nominal level (P < 0.05) are likely to be spurious in the context of the many tests performed in GWAS [144]. There is no universal standard to obtain a critical value for adjustment; nevertheless, the Bonferroni correction, Bayesian procedures and false-discovery rate (FDR) are widely used to define an appropriate threshold of significance level accounting for multiple testing. The Bonferroni correction considers a simple setting in which the type I error α level is 0.05 and n independent SNPs are tested, the adjusted significance level α’ should meet α=1-(1-α’)n and then α’≈α/n. If 1 million SNPs are independently tested whether they are associated with a trait in a GWAS context, the Bonferroni-adjusted threshold will be 0.05 / 1,000,000 = 5 ×10-8, which is a genome-wide significance level frequently reported in the GWAS literature [145]. The Bonferroni correction is overly conservative because many SNPs being tested are in linkage disequilibrium and tightly correlated each other. The Bayesian approach is based on the prior probability of true positive association from previous evidence [146]. As a result, the P-values are far less stringent and the thresholds are different from study to study and from researcher to researcher. The false-discovery rate (FDR) method measures the false rate of the rejected null hypotheses (detected associations) rather than focusing on the presence of at least one error, resulting in an increase in power [147, 148]. A FDR of 0.05 is usually adapted and indicates that 5% of the detected associations are random results. However, in GWAS, because the majority of the null hypotheses are true, FDR does not provide a substantial advantage in comparison with the Bonferroni correction.
Multiple testing presents new challenges in whole-exome and whole-genome sequencing experiments due to the massive amount of genetic data generated by these methods. Because there are many rare variants which are expected to have larger effect sizes and more severe functional impacts, it is not practical to use the same threshold across all the variants. Several recommendations are proposed and different analytic packages are in implementation [131, 149]. Some authors suggest gene-based or pathway-based tests [131], while others recommend different thresholds would be generated according to cut-offs derived from different allele frequencies. Probably, a permutation-based approach is more accurate to handle multiple testing by naturally taking into account allele frequency and correlated alleles [100].
Most genetic association studies focus on the main effects of variants contributing to the development of a disease. However, predisposing SNPs identified to date only explain a small portion of the heritability of many complex diseases. Gene by gene (G×G) and gene by environment (G×E) interactions are critical components of the architecture of complex traits and have been proposed to explain at least a fraction of the “missing heritability” [47, 54]. May a variant missed by a classical GWAS have an increased effect in presence of another genetic variant or in a specific environment? This hypothesis can be tested by incorporating interaction terms into a SNP-based linear or logistic regression model [65]. When a systematic search for G×G epistatic interactions is undertaken, the power dramatically decreases due to the numerous combinations of any two SNP tests. If two SNPs interactions are systematically investigated in a first generation GWAS (e.g. 300,000 SNPs), 100 billion epistasis tests will be performed, resulting in an exceptionally stringent Bonferroni-corrected significance threshold of 5 × 10-13 [150]. As a result, the few epistasis studies using GWAS data published up to date failed to identify G×G interactions significant after multiple testing correction [151, 152]. Compared to an epistatic study, a G×E interaction study is more feasible in the context of GWAS, although an empirical rule states that the samples needed are four times larger than those needed for studying the main effect [153]. Currently, three classical methods are used to identify G×E interactions [154]. The first tests G×E interactions using biologic candidate genes and/or GWAS validated loci. This is currently the more commonly used approach in literature. The second approach is the hypothesis-free Genome-Environment Wide Interaction Study (GEWIS), which systematically tests G×E interactions across the genome. Multiple testing decreases the statistical power in GEWIS. The third method of variance prioritization (VP) prioritizes SNPs on the basis of heterogeneity in the variance of a quantitative trait among three genotypes of a bi-allelic SNP [155]. It selects a subset of SNPs for G×E interaction tests, thus increasing the chance to detect potential associations missed by GEWIS.
All the commonly used statistical software (such as SAS, SPSS or STAT et al.) can be used to analyze genetic data. PLINK [156] is a free and very efficient tool to deal with genetic quality control and data analysis, especially for GWAS data. R software is more and more used in genetic epidemiology as many packages with specific genetic functions are programmed and it is free online.
Meta-Analysis
An individual linkage or association study is rarely conclusive in genetic epidemiology; therefore replication studies are always required. Following the same rules as in traditional clinical epidemiology, meta-analysis is also applied to genetic epidemiology. Meta-analysis combines relevant but independent studies and increases the power of the analysis and the precision of the effect size by increasing sample size, thus providing more precise evidence of association [157]. Usually, more weight is assigned in the meta-analysis to studies displaying a larger sample size or a greater event rate. Both the sample size and event rate can be reflected in the variance estimate. Therefore, a usual way to assign a weight to individual studies in a meta-analysis is to use inverse variance, even though alternative methods exist (e.g. Mantel-Haenszel test). The estimation of the degree of between-study heterogeneity is important in the inter-pretation of meta-analyses [158]. Between-study heterogeneity is measured by I2 which is a modified Cochran’s Q statistic [159]. Because this test has a low power, a p value of less than 0.1 is considered as significant heterogeneity. Usually, I2 values of 25%, 50% and 75% represent low, moderate and high levels of between-study heterogeneity, respectively. If heterogeneity exists, subgroup or sensitivity analysis may further be performed to assess the causes of such heterogeneity (e.g. study ascertainment). New global fixed-effect (FE) and random-effects (RE) meta-analytic methods have been recently proposed to deal with heterogeneity between studies [160]. The recent emergence of international consortia and the conduct of large-scale meta-analyses of genetic association studies have revolutionized the field and have led to an important yield of novel disease-predisposing loci. For instance, a recent meta-analysis of the 5, 10-methylenetetrahydrofolate reductase (MTHFR) gene variant C677T in 29,502 subjects has confirmed its associations with schizophrenia, bipolar disorder and unipolar depressive disorder and suggests a shared genetic susceptibility among distinct psychiatric disorders [161]. Numbers matter but do not always lead to success. Recently, the psychiatric GWAS consortium conducted a mega-analysis for major depressive disorder in 18,759 subjects followed by a replication in 57,478 samples. They did not find genome-wide significant association signal and concluded that the sample was still underpowered to identify common variants associated with major depression [162].
CONCLUSIONS
Genetic epidemiology is a relatively recent but fascinating research field in which expertise from different disciplines converge to elucidate genetic factors responsible for Mendelian and complex diseases. We comprehensively reviewed the key concepts and methods in genetic epidemiology including single gene disorders and complex diseases, study design implementation, genotyping and sequencing strategies, gene identification strategies, data analysis and data interpretation. We hope this review will help non-geneticist clinicians critically appraise a genetic association study and understand what makes a good genetic association study. With the decrease in sequencing costs, personalized genome sequencing will eventually become an instrument of common medical practice. In the next paper, we will review the past, current and coming applications of genetic knowledge in medical practice, and we will appreciate how far we are from the personalized medicine revolution.
ACKNOWLEDGEMENTS
We thank Jackie Hudson, Alexandra Mayhew and Hudson Reddon for editing of the manuscript, and the reviewers for their helpful comments. David Meyre is supported by a Tier 2 Canada Research Chair. Aihua Li is supported by a Queen Elizabeth II Graduate Scholarship in Science and Technology.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Morton NE. The future of genetic epidemiology. Ann Med. 1992;24:557–62. doi: 10.3109/07853899209167010. [DOI] [PubMed] [Google Scholar]
- 2.Erk S, Meyer-Lindenberg A, Schmierer P , et al. Functional impact of a recently identified quantitative trait locus for hippocampal volume with genome-wide support. Transl Psychiatry. 2013;3:e287. doi: 10.1038/tp.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li A, Meyre D. Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. Int J Obes (Lond) 2013;37:559–67. doi: 10.1038/ijo.2012.82. [DOI] [PubMed] [Google Scholar]
- 4.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.Sudbery P, Sudbery I, editors. Third edition ed: Pearson Education Limited; 2009. Human molecular genetics. [Google Scholar]
- 6.Scriver CR. The PAH gene. phenylketonria.and a paradigm shift. Hum Mutat. 2007; 28:831–45. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- 7.Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 8.Strachan T, Read A. Human molecular genetics . Garland Science. (4th ed.) 2011 [Google Scholar]
- 9.Raj A, Van Oudenaarden A. Nature nurture or chance stochastic gene expression and its consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Shachar S, Lanpher B, German JRetal. Microdeletion 15q13.: A locus with incomplete penetrance for autism mental retardation and psychiatric disorders. J Med Genet. 2009;46:382–8. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Bon BW, Mefford HC, Menten Betal. Further delineation of the 15q13 microdeletion and duplication syndromes A clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–23. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 13.Miozzo M, Simoni G. The role of imprinted genes in fetal growth. Biol Neonate. 2002;81:217–28. doi: 10.1159/000056752. [DOI] [PubMed] [Google Scholar]
- 14.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls RD. The impact of genomic imprinting for neurobehavioral and developmental disorders. J Clin Invest. 2000;105:413–8. doi: 10.1172/JCI9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruderfer DM, Chambert K, Moran J , et al. Mosaic copy number variation in schizophrenia. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2012.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon FJ, Stine OC, Meyers DA, Simpson SG, DePaulo JR. Patterns of maternal transmission in bipolar affective disorder. Am J Hum Genet. 1995;56:1277–86. [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein JM, Faraone SV, Chen WJ, Tsuang MT. Gender and the familial risk for schizophrenia.Disentangling confounding factors. Schizophr Res. 1992;7:135–40. doi: 10.1016/0920-9964(92)90043-5. [DOI] [PubMed] [Google Scholar]
- 19.Rollins B, Martin MV, Sequeira PA , et al. Mitochondrial variants in schizophrenia. bipolar disoder.and major depressive disorder. . PLoS One. 2009;4:e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wermter AK, Scherag A, Meyre D , et al. Preferential reciprocal transfer of paternal/maternal DLK1 alleles to obese children: First evidence of polar overdominance in humans. Eur J Hum Genet. 2008;16:1126–34. doi: 10.1038/ejhg.2008.64. [DOI] [PubMed] [Google Scholar]
- 21.Burghes AH, Vaessin HE, De La Chapelle A. Genetics.The land between Mendelian and multifactorial inheritance. Science. 2001;293:2213–4. doi: 10.1126/science.1065930. [DOI] [PubMed] [Google Scholar]
- 22.Savage DB, Agostini M, Barroso I , et al. Digenic inheritance of severe insulin resistance in a human pedigree. Nat Genet. 2002;31:379–84. doi: 10.1038/ng926. [DOI] [PubMed] [Google Scholar]
- 23.Risch N. Linkage strategies for genetically complex traits.I. Multilocus models. Am J Hum Genet. 1990;46:222–8. [PMC free article] [PubMed] [Google Scholar]
- 24.Burton PR, Tobin MD, Hopper JL. Key concepts in genetic epidemiology. Lancet. 2005;366:941–51. doi: 10.1016/S0140-6736(05)67322-9. [DOI] [PubMed] [Google Scholar]
- 25.Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M. Genomewide scans of complex human diseases: True linkage is hard to find. Am J Hum Genet. 2001;69:936–50. doi: 10.1086/324069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet. 2008;9:255–66. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 27.Urbanoski KA, Kelly JF. Understanding genetic risk for substance use and addiction: A guide for non-geneticists. Clin Psychol Rev. 2012;32:60–70. doi: 10.1016/j.cpr.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Kamin LJ, Goldberger AS. Twin studies in behavioral research: A skeptical view. Theor Popul Biol. 2002;61:83–95. doi: 10.1006/tpbi.2001.1555. [DOI] [PubMed] [Google Scholar]
- 29.Falconer DS. The inheritance of liability to diseases with variable age of onset. with particular reference to diabetes mellitus. Ann Hum Genet. 1967;31:1–20. doi: 10.1111/j.1469-1809.1967.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler A, Konig IR, editors. 2nd ed: Wiley-VCH Verlag GmbH & Co.; 2010. A statistical approach to genetic epidemiology. [Google Scholar]
- 31.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–82. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 32.Carlsson S, Ahlbom A, Lichtenstein P, Andersson T. Shared genetic influence of BMI. physical activity and type 2 diabetes: A twin study. Diabetologia. 2013;56:1031–5. doi: 10.1007/s00125-013-2859-3. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Lee SH, Goddard ME, Visscher PM, Yang , et al. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visscher PM, Yang J, Goddard ME. A commentary on 'common SNPs explain a large proportion of the heritability for human height' by (2010). Twin Res Hum Genet. 2010;13: 517–24. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- 35.Davies G, Tenesa A, Payton A , et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell SM, Wray NR, Stone JL , et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dick DM, Riley B, Kendler KS. Nature and nurture in neuropsychiatric genetics: where do we stand?. Dialogues Clin Neurosci. 2010;12:7–23. doi: 10.31887/DCNS.2010.12.1/ddick. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neel JV. Diabetes mellitus: A "thrifty" genotype rendered detrimental by "progress"?. Am J Hum Genet. 1962;14:353–62. [PMC free article] [PubMed] [Google Scholar]
- 39.Elston RC. Segregation analysis. AdHum Genet. 1981;11:63–120. doi: 10.1007/978-1-4615-8303-5_2. [DOI] [PubMed] [Google Scholar]
- 40.Moll PP, Burns TL, Lauer RM. The genetic and environmental sources of body mass index variability: The Muscatine Ponderosity Family Study. Am J Hum Genet. 1991;49:1243–55. [PMC free article] [PubMed] [Google Scholar]
- 41.Barrai I, Cann HM. Segregation Analysis of Juvenile Diabetes Mellitus. J Med Genet. 1965;2:8–11. doi: 10.1136/jmg.2.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez M. [Genetic markers and risk factors in diseases with complex etiology: psychiatric diseases]. Rev Epidemiol Sante Publique. 1993;41:306–14. [PubMed] [Google Scholar]
- 43.Leitch CC, Zaghloul NA, Davis EE , et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–8. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 44.Stoetzel C, Muller J, Laurier V , et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–56. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 46.Farrall M. Quantitative genetic variation A post-modern view. Hum Mol Genet. 2004;13:R1–7. doi: 10.1093/hmg/ddh084. [DOI] [PubMed] [Google Scholar]
- 47.Eichler EE, Flint J, Gibson G , et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schork NJ, Greenwood TA, Braff DL. Statistical genetics concepts and approaches in schizophrenia and related neuropsychiatric research. Schizophr Bull. 2007;33:95–104. doi: 10.1093/schbul/sbl045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choquet H, Meyre D. Molecular basis of obesity: Current status and future prospects. Curr Genomics. 2011;12:154–68. doi: 10.2174/138920211795677921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choquet H, Meyre D. Genetics of Obesity: What have we Learned?. Curr Genomics. 2011;12:169–79. doi: 10.2174/138920211795677895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES. The new genomics: Global views of biology. Science. 1996;274:536–9. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 52.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–10. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 53.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 54.Manolio TA, Collins FS, Cox NJ , et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–25. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 56.Gudmundsson J, Sulem P, Gudbjartsson DF , et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–9. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivas MA, Beaudoin M, Gardet A , et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonnefond A, Clement N, Fawcett K , et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson CA, Soranzo N, Zeggini E, Barrett JC. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunt KA, Mistry V, Bockett NA , et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–5. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstein DB, Chikhi L. Human migrations and population structure: What we know and why it matters. Annu Rev Genomics Hum Genet. 2002;3:129–52. doi: 10.1146/annurev.genom.3.022502.103200. [DOI] [PubMed] [Google Scholar]
- 63.Gibson G. Rare and common variants: Twenty arguments. Nat Rev Genet. 2011;13:135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manolio TA, Bailey-Wilson JE, Collins FS. Genes. environment and the value of prospective cohort studies. Nat Rev Genet. 2006;7:812–20. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 65.Suarez E, Sariol CA, Burguete A, McLachlan G. A tutorial in genetic epidemiology and some considerations in statistical modeling. P R Health Sci J. 2007;26:401–21. [PubMed] [Google Scholar]
- 66.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 67.Meyre D, Delplanque J, Chevre JC , et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 68.Thomas D. Gene--environment-wide association studies: Emerging approaches. Nat Rev Genet. 2010;11:259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moffitt TE, Caspi A, Rutter M. Strategy for investigating interactions between measured genes and measured environments. Arch Gen Psychiatry. 2005;62:473–81. doi: 10.1001/archpsyc.62.5.473. [DOI] [PubMed] [Google Scholar]
- 70.Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–7. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 71.Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations. gene-gene and gene-environment interactons.and parent-of-origin effects. Genet Epidemiol. 2004;26:167–85. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- 72.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 73.Cordell HJ, Clayton DG. Genetic association studies. Lancet. 2005;366:1121–31. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- 74.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 75.Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005;366:1036–44. doi: 10.1016/S0140-6736(05)67382-5. [DOI] [PubMed] [Google Scholar]
- 76.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: Case-control studies with no controls. Am J Epidemiol. 1996;144:207–13. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- 77.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–62. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 78.Kazma R, Dizier MH, Guilloud-Bataille M, Bonaiti-Pellie C, Genin E. Power comparison of different methods to detect genetic effects and gene-environment interactions. BMC Proc. 2007;1(Suppl 1 ):S74. doi: 10.1186/1753-6561-1-s1-s74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10:167–70. [PubMed] [Google Scholar]
- 80.Pierce BL, Ahsan H. Case-only genome-wide interaction study of disease risk. prognosis and treatment. Genet Epidemiol. 2010;34:7–15. doi: 10.1002/gepi.20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Visvikis S, Schlenck A, Maurice M. DNA extraction and stability for epidemiological studies. Clin Chem Lab Med. 1998;36:551–5. doi: 10.1515/CCLM.1998.094. [DOI] [PubMed] [Google Scholar]
- 82.Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet. 2001;2:235–58. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 83.Edenberg HJ, Liu Y. Laboratory methods for high-throughput genotyping. Cold Spring Harb Protoc. 2009;2009:pdb top62. doi: 10.1101/pdb.top62. [DOI] [PubMed] [Google Scholar]
- 84.Song Y, You NC, Hsu YH , et al. FTO polymorphisms are associated with obesity but not diabetes risk in postmenopausal women. Obesity (Silver Spring) 2008;16:2472–80. doi: 10.1038/oby.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jurinke C, Van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2002;187:179–92. doi: 10.1385/1-59259-273-2:179. [DOI] [PubMed] [Google Scholar]
- 86.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hutchison CA., 3rd DNA sequencing: Bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–37. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–76. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 89.Venter JC. Genome-sequencing anniversary.The human genome at 10: Successes and challenges. Science. 2011;331:546–7. doi: 10.1126/science.1202812. [DOI] [PubMed] [Google Scholar]
- 90.Mardis ER. The $1.000 geome.the $100.,000 analysis?. Genome Med. 2010;2:84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–9. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 92.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 93.Shendure J, Porreca GJ, Reppas NB , et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–32. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 94.Rothberg JM, Hinz W, Rearick TM , et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 95.Abecasis GR, Auton A, Brooks LD , et al. An integrated map of genetic variation from 1.092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease. future approaches for complex disease. Nat Genet. 2003;33 Suppl:228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 98.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–39. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bamshad MJ, Ng SB, Bigham AW , et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 100.Kiezun A, Garimella K, Do Retal. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–30. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O'Roak BJ, Vives L, Girirajan Setal. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanders SJ, Murtha MT, Gupta AR , et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Girard SL, Gauthier J, Noreau Aetal. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–3. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 104.Krebs JE, Goldstein ES, Kilpatrick ST, editors. 10th ed. Jones and Bartlet Publishers.; 2011. Lewin's Gene X. [Google Scholar]
- 105.Morton NE. Sequential tests for the detection of linkage. Am J Hum Genet. 1955;7:277–318. [PMC free article] [PubMed] [Google Scholar]
- 106.Polymeropoulos MH, Swift RG, Swift M. Linkage of the gene for Wolfram syndrome to markers on the short arm of chromosome 4. Nat Genet. 1994;8:95–7. doi: 10.1038/ng0994-95. [DOI] [PubMed] [Google Scholar]
- 107.Inoue H, Tanizawa Y, Wasson J , et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet. 1998;20:143–8. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- 108.Blackwelder WC, Elston RC. A comparison of sib-pair linkage tests for disease susceptibility loci. Genet Epidemiol. 1985;2:85–97. doi: 10.1002/gepi.1370020109. [DOI] [PubMed] [Google Scholar]
- 109.Davies JL, Kawaguchi Y, Bennett ST , et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371:130–6. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 110.Arya R, Duggirala R, Almasy L , et al. Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat Genet. 2002;30:102–5. doi: 10.1038/ng810. [DOI] [PubMed] [Google Scholar]
- 111.Saunders CL, Chiodini BD, Sham P , et al. Meta-analysis of genome-wide linkage studies in BMI and obesity. Obesity (Silver Spring) 2007;15:2263–75. doi: 10.1038/oby.2007.269. [DOI] [PubMed] [Google Scholar]
- 112.Lander ES, Botstein D. Homozygosity mapping: A way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–70. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 113.Chiang AP, Beck JS, Yen HJ , et al. Homozygosity mapping with SNP arrays identifies TRIM32. an E3 ubiquitin liase.as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A. 2006; 103:6287–92. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu M, Zhao S. Candidate gene identification approach: Progress and challenges. Int J Biol Sci. 2007;3:420–7. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tranchevent LC, Capdevila FB, Nitsch D, De Moor B, De Causmaecker P, Moreau Y. A guide to web tools to prioritize candidate genes. Brief Bioinform. 2011;12:22–32. doi: 10.1093/bib/bbq007. [DOI] [PubMed] [Google Scholar]
- 116.Saunders AM, Strittmatter WJ, Schmechel D , et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–72. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 117.Pericak-Vance MA, Bebout JL, Gaskell PC, Jr, et al. Linkage studies in familial Alzheimer disease: Evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48:1034–50. [PMC free article] [PubMed] [Google Scholar]
- 118.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381:1654–62. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 120.Ripke S, Sanders AR, Kendler KS , et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–7. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 122.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 123.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 124.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–7. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abecasis GR, Altshuler D, Auton A , et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker J, Semler O, Gilissen C , et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88:362–71. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zollner S, Pritchard JK. Overcoming the winner's curse: Estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhong H, Prentice RL. Correcting "winner's curse" in odds ratios from genomewide association findings for major complex human diseases. Genet Epidemiol. 2010;34:78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fawcett KA, Wheeler E, Morris AP , et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes. 2010;59:741–6. doi: 10.2337/db09-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: Practical aspects of rare variant association studies. Hum Mol Genet. 2012;21:R1–9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: Causes. consequences and solutions. Nat Rev Genet. 2005;6:847–59. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- 133.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–73. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–86. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chan EY. Next-generation sequencing methods: Impact of sequencing accuracy on SNP discovery. Methods Mol Biol. 2009;578:95–111. doi: 10.1007/978-1-60327-411-1_5. [DOI] [PubMed] [Google Scholar]
- 136.Stitziel NO, Kiezun A, Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ku CS, Cooper DN, Polychronakos C, Naidoo N, Wu M, Soong R. Exome sequencing: Dual role as a discovery and diagnostic tool. Ann Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 138.Garner C. Confounded by sequencing depth in association studies of rare alleles. Genet Epidemiol. 2011;35(4):2618. doi: 10.1002/gepi.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Slager SL, Schaid DJ. Case-control studies of genetic markers: Power and sample size approximations for Armitage's test for trend. Hum Hered. 2001;52:149–53. doi: 10.1159/000053370. [DOI] [PubMed] [Google Scholar]
- 140.Saito YA, Talley NJ, De Andrade M, Petersen GM. Case-control genetic association studies in gastrointestinal disease: Review and recommendations. Am J Gastroenterol. 2006;101:1379–89. doi: 10.1111/j.1572-0241.2006.00587.x. [DOI] [PubMed] [Google Scholar]
- 141.Burton PR, Scurrah KJ, Tobin MD, Palmer LJ. Covariance components models for longitudinal family data. Int J Epidemiol. 2005;34:1063–77. doi: 10.1093/ije/dyi069. [DOI] [PubMed] [Google Scholar]
- 142.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rice TK, Schork NJ, Rao DC. Methods for handling multiple testing. Adv Genet. 2008;60:293–308. doi: 10.1016/S0065-2660(07)00412-9. [DOI] [PubMed] [Google Scholar]
- 145.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–72. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 147.Bretz F, Landgrebe J, Brunner E. Multiplicity issues in microarray experiments. Methods Inf Med. 2005;44:431–7. [PubMed] [Google Scholar]
- 148.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goldstein DB, Allen A, Keebler J , et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet. 2013;14:460–70. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 151.Tao S, Feng J, Webster T , et al. Genome-wide two-locus epistasis scans in prostate cancer using two European populations. Hum Genet. 2012;131:1225–34. doi: 10.1007/s00439-012-1148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Greliche N, Germain M, Lambert JC , et al. A genome-wide search for common SNP x SNP interactions on the risk of venous thrombosis. BMC Med Genet. 2013;14:36. doi: 10.1186/1471-2350-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 154.Franks PW. Gene x environment interactions in type 2 diabetes. Curr Diab Rep. 2011;11:552–61. doi: 10.1007/s11892-011-0224-9. [DOI] [PubMed] [Google Scholar]
- 155.Pare G, Cook NR, Ridker PM, Chasman DI. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: A report from the Women's Genome Health Study. PLoS Genet. 2010;6:e1000981. doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Purcell S, Neale B, Todd-Brown K , et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Normand SL. Meta-analysis: Formulating. evaluaing.combining., and reporting. Stat Med . 1999; 18:321–59. doi: 10.1002/(sici)1097-0258(19990215)18:3<321::aid-sim28>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 158.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The quality of meta-analyses of genetic association studies: A review with recommendations. Am J Epidemiol. 2009;170:1333–43. doi: 10.1093/aje/kwp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Neupane B, Loeb M, Anand SS, Beyene J. Meta-analysis of genetic association studies under heterogeneity. Eur J Hum Genet. 2012;20:1174–81. doi: 10.1038/ejhg.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Peerbooms OL, Van Os J, Drukker M , et al. Meta-analysis of MTHFR gene variants in schizophrenia. bipolar disorder and unipolar depressive disorder: Evidence for a common genetic vulnerability?. Brain Behav Immun. 2011;25:1530–43. doi: 10.1016/j.bbi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 162.Ripke S, Wray NR, Lewis CM , et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]