Abstract
Protein–protein interactions are fundamental to the understanding of biological processes. Affinity purification coupled to mass spectrometry (AP-MS) is one of the most promising methods for their investigation. Previously, complexes were purified as much as possible, frequently followed by identification of individual gel bands. However, todays mass spectrometers are highly sensitive, and powerful quantitative proteomics strategies are available to distinguish true interactors from background binders. Here we describe a high performance affinity enrichment-mass spectrometry method for investigating protein–protein interactions, in which no attempt at purifying complexes to homogeneity is made. Instead, we developed analysis methods that take advantage of specific enrichment of interactors in the context of a large amount of unspecific background binders. We perform single-step affinity enrichment of endogenously expressed GFP-tagged proteins and their interactors in budding yeast, followed by single-run, intensity-based label-free quantitative LC-MS/MS analysis. Each pull-down contains around 2000 background binders, which are reinterpreted from troubling contaminants to crucial elements in a novel data analysis strategy. First the background serves for accurate normalization. Second, interacting proteins are not identified by comparison to a single untagged control strain, but instead to the other tagged strains. Third, potential interactors are further validated by their intensity profiles across all samples. We demonstrate the power of our AE-MS method using several well-known and challenging yeast complexes of various abundances. AE-MS is not only highly efficient and robust, but also cost effective, broadly applicable, and can be performed in any laboratory with access to high-resolution mass spectrometers.
Protein–protein interactions are key to protein-mediated biological processes and influence all aspects of life. Therefore, considerable efforts have been dedicated to the mapping of protein–protein interactions. A classical experimental approach consists of co-immunoprecipitation of protein complexes combined with SDS-PAGE followed by Western blotting to identify complex members. More recently, high-throughput techniques have been introduced; among these affinity purification-mass spectrometry (AP-MS)1 (1–3) and the yeast two-hybrid (Y2H) approach (4–6) are the most prominent. AP-MS, in particular, has great potential for detecting functional interactions under near-physiological conditions, and has already been employed for interactome mapping in several organisms (7–15). Various AP-MS approaches have evolved over time, that differ in expression, tagging, and affinity purification of the bait protein; fractionation, LC-MS measurement, and quantification of the sample; and in data analysis. Recent progress in the AP-MS field has been driven by two factors: A new generation of mass spectrometers (16) providing higher sequencing speed, sensitivity, and mass accuracy, and the development of quantitative MS strategies.
In the early days of AP-MS, tagged bait proteins were mostly overexpressed, enhancing their recovery in the pull-down. However, overexpression comes at the cost of obscuring the true situation in the cell, potentially leading to the detection of false interactions (17). Today, increased MS instrument power helps in the detection of bait proteins and interactors expressed at endogenous levels, augmenting the chances to detect functional interactions. In some simple organisms like yeast, genes of interest can directly be tagged in their genetic loci and expressed under their native promoter. In higher organisms, tagging proteins in their endogenous locus is more challenging, but also for mammalian cells, methods for close to endogenous expression are available. For instance, in controlled inducible expression systems, the concentration of the tagged bait protein can be titrated to close to endogenous levels (18). A very powerful approach is BAC transgenomics (19), as used in our QUBIC protocol (20), where a bacterial artificial chromosome (BAC) containing a tagged version of the gene of interest including all regulatory sequences and the natural promoter is stably transfected into a host cell line.
The affinity purification step has also been subject to substantial changes over time. Previously, AP has been combined with nonquantitative MS as the readout, meaning all proteins identified by MS were considered potential interactors. Therefore, to reduce co-purifying “contaminants,” stringent two-step AP protocols using dual affinity tags like the TAP-tag (21) had to be employed. However, such stringent and multistep protocols can result in the loss of weak or transient interactors (3), whereas laborious and partially subjective filtering still has to be applied to clean up the list of identified proteins. The introduction of quantitative mass spectrometry (22–25) to the interactomics field about ten years ago was a paradigm shift, as it offered a proper way of dealing with unspecific binding and true interactors could be directly distinguished from background binders (26, 27). Importantly, quantification enables the detection of true interactors even under low-stringent conditions (28). In turn, this allowed the return to single-step AP protocols, which are milder and faster, and hence more suitable for detecting weak and transient interactors.
Despite these advances, nonquantitative methods—often in combination with the TAP-tagging approach—are still popular and widely used, presumably because of reagent expenses and labeling protocols used in label-based approaches. However, there are ways to determine relative protein abundances in a label-free format. A simple, semiquantitative label-free way to estimate protein abundance is spectral counting (29). Another relative label-free quantification strategy is based on peptide intensities (30). In recent years high resolution MS has become much more widely accessible and there has been great progress in intensity-based label-free quantification (LFQ) approaches. Together with development of sophisticated LFQ algorithms, this has boosted obtainable accuracy. Intensity-based LFQ now offers a viable and cost-effective alternative to label-based methods in most applications (31). The potential of intensity-based LFQ approaches as tools for investigating protein–protein interactions has already been demonstrated by us (20, 32, 33) and others (34, 35). We have further refined intensity-based LFQ in the context of the MaxQuant framework (36) using sophisticated normalization algorithms, achieving excellent accuracy and robustness of the measured “MaxLFQ” intensities (37).
Another important advance in AP-MS, again enabled by increased MS instrument power, was the development of single-shot LC-MS methods with comprehensive coverage. Instead of extensive fractionation, which was previously needed to reduce sample complexity, nowadays even entire model proteomes can be measured in single LC-MS runs (38). The protein mixture resulting from pull-downs is naturally of lower complexity compared with the entire proteome. Therefore, modern MS obviates the need for gel-based (or other) fractionation and samples can be analyzed in single runs. Apart from avoiding selection of gel bands by visual examination, this has many advantages, including decreased sample preparation and measurement time, increased sensitivity, and higher quantitative accuracy in a label-free format.
In this work, we build on many of the recent advances in the field to establish a state of the art LFQ AE-MS method. Based on our previous QUBIC pipeline (20), we developed an approach for investigating protein–protein interactions, which we exemplify in Saccharomyces cerevisiae. We extended the data analysis pipeline to extract the wealth of information contained in the LFQ data, by establishing a novel concept that specifically makes use of the signature of background binders instead of eliminating them from the data set. The large amount of unspecific binders detected in our experiments rendered the use of a classic untagged control strain unnecessary and enabled comparing to a control group consisting of many unrelated pull-downs instead. Our protocol is generic, practical, and fast, uses low input amounts, and identifies interactors with high confidence. We propose that single-step pull-down experiments, especially when coupled to high-sensitivity MS, should now be regarded as affinity enrichment rather than affinity purification methods.
EXPERIMENTAL PROCEDURES
Yeast Strains
For all experiments GFP-tagged yeast strains originating from the Yeast-GFP Clone Collection were used, a library with 4156 GFP-tagged proteins representing about 63% of S. cerevisiae open reading frames (39). The haploid parental strain of this library, BY4741 (ATCC 201388), served as an initial control strain and to construct the strain pHis3-GFP-HIS3kMX6 (short name pHis3-GFP). To do so, we used the His3 locus in BY4741, which is nonfunctional because of a deletion of several amino acids in the middle of the coding sequence. We amplified a cassette containing a GFP gene without start codon and a His3 gene of Saccharomyces kluyveri under control of the TEF promoter and terminator out of the vector pFA6a-GFP(S65T)-HIS3kMX6. This cassette was integrated into the His3 locus of BY4741 directly after the original His3 promoter and start codon by homologous recombination, replacing the rest of the nonfunctional His3 sequence. As a result, our pHis3-GFP strain is able to synthesize histidine and expresses moderate amounts of cytosolic GFP just as the tagged library strains.
Culture of Yeast Strains and anti-GFP Immunoprecipitation
Tagged yeast strains, the parental strain BY4741 and the control strain pHis3-GFP were first grown on plates (YDP plates for BY4741, SC-His plates for all other strains) and then in YPD liquid medium at standard culture conditions. Cell growth was regularly examined by measuring OD600 nm. Yeast cells were grown until they reached an OD600 nm of around 1, followed by harvesting culture volumes equaling 50 ODs. For biochemical triplicates (experimental series 1 (ES1)), three times 50 ODs were harvested out of the same culture and from then on processed separately. For biological quadruplicates (experimental series 2 (ES2)), four different colonies were picked on different days and processed separately from the beginning. Yeast cell pellets were dissolved in 1.5 ml lysis buffer (150 mm NaCl, 50 mm Tris HCl pH 7.5, 1 mm MgCl2, 5% glycerol, 1% IGEPAL CA-630 (SIGMA-ALDRICH GmbH, Taufkirchen, Germany), Complete® protease inhibitors (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), and 1% benzonase (Merck KGgA, Darmstadt, Germany)), transferred into FastPrep® tubes (MP Biomedicals GmbH, Eschwege, Germany) containing 1 mm silica spheres (lysing matrix C, MP Biomedicals), frozen in liquid nitrogen and stored at −80 °C until lysis. The frozen samples were thawed and then lysed in a FastPrep24® instrument (MP Biomedicals) for 6 × 1 min at maximum speed. Lysates were cleared by a 10 min centrifugation step at 4 °C and 4000 × g; and 800 μl of the clear lysates were transferred into a deep-well plate for immunoprecipitation. IP of yeast protein complexes was essentially performed as described before for a mammalian cell culture system (20). IPs were performed on a Freedom EVO® 200 robot (Tecan Deutschland GmbH, Crailsheim, Germany) equipped with a MultiMACS™ M96 separation unit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) that contains a strong permanent magnet. (Miltenyi Biotec also supplies equipment for performing the same pull-downs in a manual fashion.) The basic steps of the IP protocol are as follows: First the lysates are mixed with 50 μl magnetic μMACS Anti-GFP MicroBeads (Miltenyi Biotec) and incubated for 15 min at 4 °C. Because of the favorable kinetics of the microbeads, tagged proteins are efficiently captured in only 15 min (40). Then the Multi-96 separation columns are equilibrated with 250 μl equilibration buffer (same as lysis buffer). After that, the lysates are added to the columns with the magnet turned on, retaining the magnetic MicroBeads on the column. Once all the liquid has passed through the columns, they are first washed with 3 × 800 μl ice cold wash buffer I (0.05% IGEPAL CA-630, 150 mm NaCl, 50 mm Tris HCl pH 7.5, and 5% glycerol), then with 2 × 500 μl of wash buffer II (150 mm NaCl, 50 mm Tris HCl pH 7.5, and 5% glycerol). Afterward 25 μl of elution buffer I (5 ng/μl trypsin, 2 m Urea, 50 mm Tris HCl pH 7.5, and 1 mm DTT) are added and the columns are incubated for 30 min at room temperature. In this “in-column digest,” the proteins are partially digested to allow elution from the columns, and reduced by DTT. Subsequently the resulting peptides are eluted and alkylated with 2 × 50 μl elution buffer II (2 m Urea, 50 mm Tris HCl pH 7.5, and 5 mm CAA), and collected in a 96-well plate.
The plate was incubated at room temperature overnight to ensure a complete tryptic digest. The next morning the digest was stopped by addition of 1 μl Trifluoroacetic acid (TFA) per well. The acidified peptides were loaded on StageTips (self-made pipette tips containing two layers of C18) to desalt and purify them according to the standard protocol (41). Every sample was divided onto two StageTips to give one “working” StageTip and one “backup” StageTip. The StageTips were stored at 4 °C until the day of LC-MS/MS measurement.
LC-MS/MS Measurement
Samples were eluted from StageTips with 2 × 20 μl buffer B (80% ACN and 0.5% acetic acid). The organic solvent was removed in a SpeedVac concentrator for 20 min, then the remaining 4 μl of peptide mixture were acidified with 1 μl of buffer A*(2% ACN and 0.1% TFA) resulting in 5 μl final sample size. 2 μl of each sample were analyzed by nanoflow liquid chromatography on an EASY-nLC system (Thermo Fisher Scientific, Bremen, Germany) that was on-line coupled to an LTQ Orbitrap classic (Thermo Fisher Scientific) through a nanoelectrospray ion source (Thermo Fisher Scientific). A 15 cm column with 75 μm inner diameter was used for the chromatography, in-house packed with 3 μm reversed-phase silica beads (ReproSil-Pur C18-AQ, Dr. Maisch GmbH, Germany). Peptides were separated and directly electrosprayed into the mass spectrometer using a linear gradient from 5.6% to 25.6% acetonitrile in 0.5% acetic acid over 100 min at a constant flow of 250 nl/min. The linear gradient was followed by a washout with up to 76% ACN to clean the column for the next run. The overall gradient length was 134 min. The LTQ Orbitrap was operated in a data-dependent mode, switching automatically between one full-scan and subsequent MS/MS scans of the five most abundant peaks (Top5 method). The instrument was controlled using Tune Plus 2.0 and Xcalibur 2.0. Full-scans (m/z 300–1650) were acquired in the Orbitrap analyzer with a resolution of 60,000 at 400 m/z. The five most intense ions were sequentially isolated with a target value of 1000 ions and an isolation width of 2 m/z and fragmented using CID in the linear ion trap with a normalized collision energy of 40. The activation Q was set to 0.25, the activation time to 30 ms. Maximum ion accumulation times were set to 500 ms for full scans and 1000 ms for MS/MS scans. Dynamic exclusion was enabled; with an exclusion list size of 500 and an exclusion duration of 180 s. Standard MS parameters were set as follows: 2.2 kV spray voltage; no sheath and auxiliary gas; 200 °C heated capillary temperature and 110 V tube lens voltage.
Raw Data Processing
All raw files were analyzed together using the in-house built software MaxQuant (36) (version 1.4.0.6). The derived peak list was searched with the built-in Andromeda search engine (42) against the reference yeast proteome downloaded from Uniprot (http://www.uniprot.org/) on 03–20-2013 (6651 sequences) and a file containing 247 frequently observed contaminants such as human keratins, bovine serum proteins, and proteases. Strict trypsin specificity was required with cleavage C-terminal after K or R, allowing up to two missed cleavages. The minimum required peptide length was set to seven amino acids. Carbamidomethylation of cysteine was set as a fixed modification (57.021464 Da) and N-acetylation of proteins N termini (42.010565 Da) and oxidation of methionine (15.994915 Da) were set as variable modifications. As no labeling was performed, multiplicity was set to 1. During the main search, parent masses were allowed an initial mass deviation of 4.5 ppm and fragment ions were allowed a mass deviation of 0.5 Da. PSM and protein identifications were filtered using a target-decoy approach at a false discovery rate (FDR) of 1%. The second peptide feature was enabled. The match between runs option was also enabled with a match time window of 0.5 min and an alignment time window of 20 min. Relative, label-free quantification of proteins was done using the MaxLFQ algorithm (37) integrated into MaxQuant. The parameters were as follows: Minimum ratio count was set to 1, the FastLFQ option was enabled, LFQ minimum number of neighbors was set to 3, and the LFQ average number of neighbors to 6, as per default. The “proteinGroups” output file from MaxQuant is available in the supplement (supplemental Table S1), as well as all spectra for single-peptide-based protein identifications (supplemental Spectra).
Data Analysis
Further analysis of the MaxQuant-processed data was performed using the in-house developed Perseus software (version 1.4.2.30). The “proteingroups.txt” file produced by MaxQuant was loaded into Perseus. First, hits to the reverse database, contaminants and proteins only identified with modified peptides were eliminated. Then the LFQ intensities were logarithmized, and the pull-downs were divided into ES1 and ES2 and from then on analyzed separately. Samples were first grouped in triplicates or quadruplicates and identifications were filtered for proteins having at least three or four valid values in at least one replicate group, respectively. For every bait a separate grouping was defined, and the data was individually filtered for proteins containing at least two (ES1) or three (ES2) valid values in the specific bait pull-downs. After this, missing values were imputed with values representing a normal distribution around the detection limit of the mass spectrometer. To that end, mean and standard deviation of the distribution of the real intensities were determined, then a new distribution with a downshift of 1.8 standard deviations and a width of 0.25 standard deviations was created. The total matrix was imputed using these values, enabling statistical analysis. Now a student's t-tests was performed comparing the bait pull-down (in replicates) to its individual bait specific control group (BSCG). This BSCG contained all other pull-downs in the data set except those of known complex members. This whole procedure of individual filtering, imputation and t test was repeated for every bait. The resulting differences between the logarithmized means of the two groups (“log2(bait/background”) and the negative logarithmized p values were plotted against each other using R (version 2.15.3) in “volcano plots.” We introduced two different cutoff lines with the function y = c/(x - x0), dividing enriched proteins into mildly and strongly enriched proteins (c = curvature, x0 = minimum fold change). The positions of the cutoff lines were defined for each experimental series separately by first plotting the distribution of all observed enrichment factors and deriving the standard deviation of this distribution. The x0 parameter for the inner curve and outer curve was then set to one and two standard deviations (rounded to one significant digit), respectively (supplemental Fig. S6B and S6F). The curvature parameters were obtained by overlaying all plots within one series, using only pull-downs of functional baits and rather small defined complexes (ES1: all but CDC73, PUP1, and PUP2; ES2: all but NUP84 and NUP145). The c parameter of the outer line was then adjusted to optimally separate true interactors from false positives (for more details see supplemental Fig.S6C, 6D, 6G, and 6H). The curvature of the inner line was then set to half of the curvature of the outer line. Cut-off parameters for ES1 were x0 = 0.9 and c = 4 for the inner curve, and x0 = 1.8 and c = 8 for the outer curve. Cutoff parameters for ES2 were x0 = 0.5 and c = 4 for the inner curve, and x0 = 1 and c = 8 for the outer curve. For all enriched proteins outside the inner cutoff line, we calculated the Pearson correlation of their LFQ intensity profile across all runs to the LFQ intensity profile of the corresponding bait. Enriched proteins were assigned to interactor confidence classes A, B, or C according to their position in the volcano plot and their correlation value. Cutoffs for the correlation scores were defined for both series individually by analyzing all correlations within one series using a quantile–quantile plot (Q–Q plot), which compares the real distribution of all correlation values to a theoretical normal distribution (supplemental Fig. S6E and 6F). The correlation cutoff was 0.55 for experimental series 1and 0.35 for experimental series 2. Note that these cutoff criteria do not represent absolute fixed values, but rather help to interpret the individual pull-down result.
RESULTS
Establishing a High Performance AE-MS Method for Detecting Interactions in Yeast
First, we set out to develop a generic and robust, yet high performance affinity enrichment-mass spectrometry (AE-MS) method for investigating protein–protein interactions in yeast. This organism is amenable to genetic and biochemical approaches and has already served as a model in many of the classical interactome studies. We chose to work with a GFP-tag system, because this tag is well tolerated and highly specific antibodies have been generated. Furthermore, a library of GFP-tagged yeast strains is commercially available, covering about 4000 open reading frames, and also offering localization data (34). The GFP-tagged bait proteins in this library are expressed at endogenous levels, a great advantage for detecting functional interactions. We chose a subset of 36 strains from this library, containing tagged bait proteins that are members of characterized complexes from various cellular compartments and cover the entire abundance range of the yeast proteome (supplemental Fig. S1).
Next, we wished to construct a control strain that was as genetically similar to the strains of the library as possible. Because the parental strain of the GFP-library, BY4741, is histidine auxotroph and does not express GFP, we reintroduced the HIS3 selection marker gene and a GFP gene into the dysfunctional HIS3 locus of BY4741 (Experimental Procedures). The resulting control strain can be grown under the same conditions as the strains of the GFP library, expresses moderate amounts of cytosolic GFP and was termed pHIS3-GFP.
An overview of our AE-MS workflow is depicted in Fig. 1. We combined a mild detergent-based lysis buffer with extensive bead beating to efficiently extract yeast proteins without disrupting interactions. We investigated the needed input amounts, and found that a 50 ml yeast culture volume with an OD600 nm of 1.0 provided ample material for an IP experiment even with very low expressed baits. Starting from these initial 50 ODs of yeast cells allowed us to save material as backup at various stages of the sample preparation. The final amount injected into the mass spectrometer corresponded to only about 5.3 ODs; a very low amount of starting material, especially considering that baits were not overexpressed. The single-step affinity enrichment was performed with highly specific monoclonal anti-GFP antibodies coupled to magnetic microbeads in a flow-through column format using mild washing conditions to preserve weak or transient interactions (Experimental Procedures). The whole pull-down procedure was rather short, taking only about 2.5 h from lysis to elution. Proteins were eluted by in-column predigestion with trypsin, then digested to completion overnight. For all complexes tested, we found that the resulting peptides could be analyzed without any prefractionation in single-shot LC-MS/MS runs on Orbitrap instrumentation, which considerably shortens overall experiment time, provides greater reproducibility especially in a label-free format and higher sensitivity. All experiments were performed in several replicates; either biochemical triplicates (experimental series 1, ES1) or biological quadruplicates (experimental series 2, ES2).
Fig. 1.

Schematic representation of the AE-MS workflow. A, Endogenously expressed GFP-tagged proteins are extracted from yeast cells using mild, nondenaturing conditions. B = Bait, I = Interactor, U = Unspecific binder. B, Bait protein and specific interactors are enriched in a single-step immunoprecipitation using anti-GFP antibodies. Subsequently, bound proteins are digested into peptides. C, The peptide mixture is analyzed by single-shot liquid chromatography tandem mass spectrometry (LC-MS/MS) on an Orbitrap instrument. D, Raw data are processed with MaxQuant to identify and quantify proteins. The resulting label-free quantification (LFQ) intensity matrix is the basis for all downstream data analysis aimed at identifying interactors of the tagged bait proteins.
Raw data were analyzed using MaxQuant (36), providing ppm level mass accuracy, confident identification of proteins (False Discovery Rate of less than 1%), and accurate intensity-based label-free quantification, thanks to recently developed sophisticated normalization and matching algorithms (37). Remarkably, all our pull-downs resulted in the identification of thousands of unspecific binders in addition to the specific interactors, leading to quantification of about half of the yeast proteome in every single sample. On the one hand, this was because of the low stringent single-step protocol in which we attempt enrichment instead of proper purification of protein complexes. On the other hand, it resulted from the high instrument sensitivity of the LTQ Orbitrap instrument, and was also promoted by the “match between runs” algorithm in MaxQuant. Matching between runs transfers identifications from one MS run to another run, where the same peptide feature was present, but not selected for fragmentation and hence not identified. High confidence matching is enabled by the high mass precision of the Orbitrap and achieved using unique m/z and retention time information of the features, after the retention times of all runs have been aligned (43). Processing with matching between runs increased the number of available quantifications in the combined (ES1+ES2) unfiltered LFQ matrix of 196 samples times 2304 proteins from 45 to 80%. The very large number of proteins quantified per IP prompted us to establish novel data analysis strategies, exploiting the information-rich intensity-based LFQ data, as described in the following sections.
AE-MS Produces Internal Beadomes for Every Pull-down
Together, our pull-downs identified a large set of background binders specific for the affinity matrix and conditions used in our experiments. As these proteins are usually detected because they bind to the beads used in the purification, the totality of them has been called the “bead proteome” or “beadome” (44, 45). Instead of having to determine this beadome from separate control experiments, here we detect it as a byproduct in the specific pull-downs (“internal beadome”). In total, after standard filtering (Experimental Procedures) of the data we quantified 2245 different protein groups in the combined ES1 and ES2 experimental series (Fig. 2A). Per pull-down, we quantified on average 1860 proteins in ES1 and 1825 proteins in ES2. Only a tiny fraction of the detected proteins in each pull-down were actual interactors of the corresponding tagged protein. For example, using MCM2 as bait recovered the six MCM complex members along with 1891 unspecific background proteins on average. These six proteins constituted only 0.3% of all identified proteins and only 1.3% of the summed LFQ intensity in the corresponding pull-downs, although the bait was among the highest intense proteins.
Fig. 2.
The proteomic nature of the background in AE-MS. A, Heatmap of the LFQ intensities of all proteins identified in two experimental series (ES1 and ES2). Hierarchical row clustering was performed on the logarithmized LFQ intensities of more than 2000 quantified prey proteins in the 196 pull-downs, without data imputation. B, Histogram of the copy numbers of all proteins quantified in our pull-downs compared with the entire yeast proteome as in Kulak et al. C, The standard deviation of the LFQ intensity profile for each identified protein was calculated after imputing missing values. Proteins were then ranked according to the standard deviation of their profile. About 70% of detected proteins show a profile varying less than 1 log2 LFQ intensity unit and about 90% vary less than 1.5 log2 LFQ intensity units. D, Comparison of the control strain pHIS3-GFP with the two tagged strains SET1-GFP and PAF1-GFP; all measured in triplicates. The matrix of 36 correlation plots reveals very high correlations between LFQ intensities within triplicates (Pearson correlation coefficient > 0.977 for all strains). The correlation between different strains is always higher than 0.935. Average correlation of the corresponding nine comparisons were: SET1-GFP to PAF1-GFP 0.946, SET1-GFP to control strain 0.938, and PAF1-GFP to control strain 0.945. E, Zoom into the SET1-GFP_01 versus PAF1-GFP_01 correlation plot. The majority of proteins are detected at very similar LFQ intensities in both pull downs. The proteins that differ the most between the two strains are the members of the two targeted complexes highlighted in color.
The unspecific binders identified in our internal beadome cover the entire abundance range, with only a small bias toward more highly abundant proteins when compared with the yeast proteome as a whole (46) (Fig. 2B). GOBP and GOCC term analysis by category counting of the identified proteins did not indicate cellular functions or compartments that are strongly over-or underrepresented (supplemental Fig. S2A). However, the intensity at which we detect proteins in the beadome is dependent on two factors: their abundance in the proteome and their affinity to the beads. Whereas low abundant proteins are generally not found at high intensities in the beadome, the intensities of high abundant proteins can vary from high to low signals (supplemental Fig. S2B and 2C). Pearson correlation between beadome intensity and proteome copy numbers was 0.53 for both ES1 and ES2. Next, we performed 2D enrichment analysis (47), in which we compared protein annotations between beadome and proteome in an intensity-dependent fashion. The major protein classes that showed higher intensities in the beadome than what would be expected from their cellular abundance were RNA or DNA related (e.g. ribosome, spliceosome, nucleolus, and DNA recombination). This confirms former findings that ribosomal proteins have a high affinity to the beads. Interestingly, proteins in metabolic categories, which are ubiquitously present in pull-downs because of their high abundance, tended to be de-enriched (supplemental Fig. S2D and 2E). We conclude that the beadome is in essence a scaled down version of the proteome, albeit with some preferences related to general protein binding properties.
The reproducible identification of unspecific binders across all runs is of course correlated with their intensity; higher intense background binders are more likely to always be detected, whereas background binders that are close to the level of detection may only be identified in some of the runs. Therefore, the LFQ intensity matrix contains missing values among the lower intense proteins (marked gray in Fig. 2A). To enable statistical analysis, such missing values can be “imputed.” Therefore, after discarding proteins that are not reproducibly detected in at least one replicate group, we imputed the remaining missing values using a normal distribution around the detection limit of the mass spectrometer. These simulated low intensity values fit well into the profiles of the low abundant proteins, and because of its randomness, imputation does not create artifacts in t-tests or in intensity profile analyses. A comparison of the data set processed with and without matching identifications between runs, and the result of imputation are illustrated in supplemental Fig. S3.
Most of the background proteins are characterized by highly similar intensities in nearly all of the pull-downs within an experimental series, and we denote these as typical background binders. Both in ES1 and ES2 for about 90% of all detected proteins the standard deviation of their intensity profile was lower than 1.5 log2 LFQ intensity units; and for about 70% even lower than 1 (Fig. 2C). As expected, this analysis also confirms that proteins with higher intensity tend to have more stable background profiles. Next to the typical background binders, we also found a small number of proteins with irregular profiles. Those atypical background binders are usually among the lower intense proteins. Both types of unspecific binders can readily be distinguished from a specific interactor, whose profile ideally fluctuates mildly around an average background intensity and only deviates from that behavior in specific pull-downs, where it is detected reproducibly and at higher intensities. The relationship of mean LFQ intensity and standard deviation of the intensity profile as well as the profiles of some typical and atypical unspecific binders are further documented in supplemental Fig. S4. Again, there is a clear trend that the intensity profiles of higher intense proteins have a smaller standard deviation. Among the proteins with the highest standard deviation (>1.5 log2 LFQ intensity units) many bait proteins and interactors are found.
A closer look at the heat map in Fig. 2A reveals the background in ES1 and ES2 to be slightly different. Sample preparation was similar in both experiments; however, ES1 and ES2 were measured on two different LC-MS systems of the same type but at different time periods, which introduces noticeable variation of the corresponding background. The variation between pull-downs is lower in ES2 because samples were measured directly after each other in contrast to ES1 where samples were measured in blocks according to baits. Because of the slight variations in the background signature between ES1 and ES2, data analysis was performed separately for each experimental series. The differences between ES1 and ES2 allowed us to study the influence of these workflow parameters.
Exploiting the High Coverage Background for Identifying Protein Complexes
Evidently, the extremely large number of unspecific binders detected in addition to the specific interactors in AE-MS represents a completely different experiment readout than that of classic AP-MS protocols. This large background needs specialized data analysis, which is; however, not aimed at removing the unspecific binders, but instead exploits them for high confidence detection of interactors. We recognized four different ways in which the unspecific binders detected in our pull-downs can be used beneficially.
First, they form the basis for intensity-based LFQ in MaxQuant. To produce reliable and accurate quantification results, the normalization procedure performed in MaxQuant requires a background proteome that is assumed to be unchanging. This function is provided here by a large number of unspecific binders identified in all samples. Normalization can then correct for differences in sample loading and sample concentration, which is a prerequisite to making the pull-downs comparable at all and constitutes the basis for further data analysis.
Second, the unspecific binders can serve as a quality control. We observed that deviation of the detected background binders from the standard behavior can indicate insufficient quality of a specific pull-down, which easily became apparent by hierarchical clustering of the data matrix. As an example, see the vertical stripe close to the middle of ES2 in Fig. 2A, which is a replicate of a pHIS3-GFP pull-down. Close inspection of the raw data revealed generally low peptide intensities and polymer contamination in this sample. In another case, a difference in background signature was not because of sample quality, but seemed to be because of the nature of the tagged complex: All six proteasome pull-downs reproducibly featured a slightly but clearly different background than the other pull-downs. This can be explained by the fact that proteasome subunits have high cellular copy numbers and are part of a very large complex; together this alters conditions on the beads, “crowding out” some of the normally observed background binders.
Third, the high number of unspecific binders reproducibly quantified in all samples resulted in very high correlations between different pull-downs. In Fig. 2D, these correlations are plotted for two tagged strains, SET1-GFP and PAF1-GFP, and the control strain pHIS3-GFP. Within triplicate pull-downs, the average Pearson correlation coefficients were always greater than 0.977. Between the different strains, correlation was always higher than 0.935, indicating that the intensities of the background proteins in the three yeast strains are highly similar. In fact, the correlation of SET1-GFP to PAF1-GFP was even higher than the correlation of SET1-GFP to the control strain pHIS3-GFP (0.945 versus 0.937). The proteins most changing in intensity between the two pull-downs were the expected SET1 and PAF1 interactors (Fig. 2E). These findings led us to investigate the possibility of comparing pull-downs not to an untagged control strain as it is usually done, but instead to compare them to each other, which will be further explored in the next section.
Finally, we reasoned that next to the pair-wise correlation of samples across all protein intensities, pair-wise correlation of intensity profiles across all samples should contain meaningful information. Specifically, intensity profiles of true interactors across all pull-downs, when compared with the intensity profile of the corresponding bait, should be correlated. The characteristic profile of interactors compared with the unchanging profile of typical background binders or the random profile of atypical background binders could therefore be useful in verifying interactor candidates, as we will demonstrate later on.
Defining Interactors by Comparing Against Other Tagged Strains
To identify interactors of a specific bait protein in the presence of the large amount of background binders, we performed a student's t test comparing the LFQ intensities of all proteins identified in replicates of that bait with the LFQ intensities of all proteins identified in the control (Experimental Procedures). When the resulting differences between the log2 mean protein intensities between bait and control are plotted against the negative logarithmized p values in volcano plots, the unspecific background binders center around zero. The enriched interactors appear on the right side of the plot, whereas ideally no proteins should appear on the left side when comparing to an empty control, as these would represent proteins depleted by the bait, which is not expected to happen. The higher the difference between the group means (i.e. the enrichment) and the p value (i.e. the reproducibility), the more the interactors move to the top right corner of the plot, which is the area of highest confidence for a true interaction.
We started by comparing a specific pull-down to an empty control strain as it is usually done in AP-MS experiments. First we used BY4741, the parental strain of the GFP library, as control; however, cross-reactivity of the anti-GFP antibody could occur in the complete absence of GFP. Therefore, we had constructed pHIS3-GFP, a control strain highly similar to the strains of the GFP library, as it could be grown under the same selective conditions and expressed moderate amounts of cytosolic GFP (see above). When we compared the pHIS3-GFP control strain to its parental strain BY4741, we detected only one yeast protein to be enriched, which was imidazoleglycerol-phosphate dehydratase, the protein the HIS3 gene encodes for (Fig. 3A). This illustrates that GFP does not interact with any yeast protein, and furthermore demonstrates that our AE-MS workflow is sensitive to an extent that it picks up genetic differences between strains. This confirms the benefits of using a control strain as similar as possible to the actual bait strain, and supports our hypothesis that other tagged strains of the GFP-library could present an excellent control, as they are genetically identical except for the different tagged protein. When we tested this idea on the example of the SKI complex we indeed did not observe any differences in the identified interactors of the bait SKI2, whether we compared with pHis3-GFP or a tagged strain, e.g. SMC2-GFP (Fig. 3B and 3C). As the only side-effect the specific interactors of the other strain now appeared as de-enriched proteins. (We note that even this could be put to good use in certain cases, as it in principle enables detection of the interactors of two different bait proteins in only one comparison and without employing a control.)
Fig. 3.
Comparing to unrelated tagged strains. All pull-downs in this figure were measured in quadruplicates. Cut-off lines were those of ES2 (see Experimental Procedures). Red dots represent members of the SKI complex and blue dots represent members of the condensin complex. A, Comparison of the control strain pHIS3-GFP against its parental strain BY4741. B, Classic comparison of a tagged strain against an untagged control strain, in this case SKI2-GFP against pHIS3-GFP. C, SKI2-GFP compared with an unrelated tagged strain, SMC2-GFP. D, SKI2-GFP compared with 8 × pHIS3-GFP in quadruplicate (= 32 control pull-downs). E, SKI2-GFP compared with eight unrelated tagged strains in quadruplicate (APC1-GFP, CAF1-GFP, CCR4-GFP, PAF1-GFP, PEP5-GFP, SMC1-GFP, SMC2-GFP, and SNF4-GFP = 32 control pull-downs). F, SKI2-GFP compared with its bait specific control group (BSCG) consisting of all other pull-downs in the data set except for the SKI3 quadruplicate (= 116 control pull-downs).
A larger control group consisting of many control pull-downs should help to better identify interactors; and we next tested whether this holds true for our pull-downs. Comparing a specific pull-down to eight pHis3-GFP pull-downs, consisting of four biological replicates each, clearly led to better separation of interactors from the background cloud than just comparing to one pHIS3-GFP pull-down (compare Fig. 3D to Fig. 3B). The larger control group provided a less error-prone average background intensity of every protein, which in turn resulted in higher p values of the enriched true interacting proteins. This is particularly beneficial to separate weaker or transient interactors, which by their nature tend to only be mildly enriched, from the background cloud, as long as their low enrichment is highly reproducible. The more control pull-downs are included into the control group, the better the results should become. However, performing a large number of empty control experiments consumes considerable resources. In a human interactome study in 2007 for example, the authors conducted 202 control experiments (12). We reasoned that if we are able to compare tagged strains to each other, we would naturally obtain a large control group without any additional efforts. To test this concept, we first compared the SKI complex pull-downs to eight unrelated tagged strains. This resulted in the same or better statistical improvement of the interactors as we had obtained when using the same number of control strains (Fig. 3E and 3D). We chose the tagged strains serving as the control group to be unrelated to the specific bait of interest, in the sense that their tagged proteins do not reside in a known complex with this bait. To obtain the largest possible control group, we selected all unrelated pull-downs in the data set and termed this the “bait specific control group” (BSCG). If interacting proteins are included in the BSCG, they can increase the calculated average background intensity of interactors and therefore artificially decrease the t test result. For large control groups; however, wrong assignment would generally not dramatically change results, as demonstrated by comparing the SKI2 pull-downs against all other pull-downs in the data set (supplemental Fig. S5). Although we here constructed the BSCG from prior knowledge, it could also be constructed in an iterative way. In the case of SKI2, the BSCG included all pull-downs except the replicates of SKI3, resulting in 116 controls. This led by far to the best separation, placing the SKI complex into the far upper right corner of the volcano plot (Fig. 3F). Therefore, we concluded that other pull-downs can serve as excellent controls and in the following determined interactors by comparing each specific pull-down to its BSCG.
Combining Enrichment Over Background with Intensity Profile Analysis Leads to High Quality Interaction Data
To classify a protein as an interactor, we needed to introduce a cutoff that separates enriched proteins from the unchanged cloud of background binders centered around zero in the volcano plots. The position of this cutoff is crucial: A stringent cutoff leads to a low false positive rate, but may miss weaker or more transient interactors, whereas a permissive cutoff would include these, but at the cost of increasing false positives. To preserve information about weak or transient interactors, we decided to use a two cutoff strategy, which divides interactor candidates into mildly and strongly enriched proteins (Fig. 4A). To define the position of the two cutoff lines, we plotted the distribution of all enrichment factors within one series and placed two minimum fold change cutoffs at one and two standard deviations, respectively. Interestingly, in the case of ES2, the series with biological quadruplicates that had been measured in one block, the standard deviation was much lower than for ES1. The cutoff lines were placed once for all pull-downs within an experimental series with curvature parameters that best separate the outliers from the cumulative background cloud (for more details see Experimental Procedures and supplemental Fig. S6A–6H).
Fig. 4.
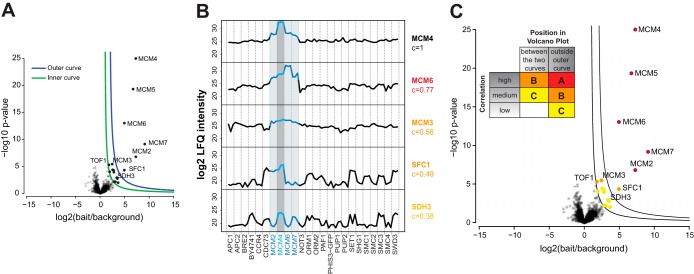
Classification of interactors. Proteins are classified as interactors according to their position in the volcano plot and according to their correlation to the corresponding bait protein. A, Volcano Plot. Potential interactors are preclassified according to their position in the volcano plot into “mildly enriched” (between the two curves) and “strongly enriched” (outside the blue curve) proteins B, Intensity profile analysis of some enriched proteins from the volcano plot in A. From top to bottom: intensity profile of MCM4 (the bait protein), MCM6, and MCM3 (true interactors), and SFC1 and SDH3 (false positives) with the according calculated correlation to the profile of MCM4. C, Same volcano plot as in A, but with classification of interactors. Insert: Enrichment, reproducibility and correlation are combined to score interactors into interactor confidence classes A, B and C. Proteins between the cutoff curves with a low correlation (lower than 0.1) were not considered at all. Both proteins between the cutoff curves with a medium correlation (between 0.1 and the series-specific correlation cutoff) and proteins outside the outer cutoff curve with a low correlation (lower than 0.1) were assigned to class C (noninteractors). Proteins between the cutoff curves with a high correlation (higher than the series-specific correlation cutoff) as well as proteins outside the outer cutoff curve with a medium correlation were assigned to class B (lower confidence interactors). Proteins outside the outer cutoff curve with a high correlation were assigned to class A (high confidence interactors).
We then introduced a new criterion to deal with the false positives among the mildly enriched interactors close to the cutoff lines. This criterion makes use of the above mentioned tendency of intensity profiles of true interactors of a bait protein to be correlated, because interacting proteins should be enriched whenever one of the complex members is tagged. Moreover, slight variations across samples because of background binding should be followed by all complex subunits. This concept requires a complete LFQ intensity matrix, produced by imputing missing values from a suitably chosen random distribution, to not artificially increase or decrease the correlation (Experimental Procedures). To evaluate the similarity of a given profile to the profile of the bait, we calculated the Pearson correlation of the two profiles; and this was repeated for every enriched protein (Fig. 4B). Although strongly enriched proteins generally show medium to high correlations, mildly enriched proteins generally show lower correlations, but with a much higher variation from high to even negative values (supplemental Fig. S7). This indicates that true interactors exist among those borderline interactors that can be detected with the help of the correlation analysis. For the example of the MCM4 pull-down in Fig. 4, five out of the six complex members were highly enriched, but one (MCM3) only scored a mild enrichment and moderate p value, but a high correlation (0.56), which led to its correct identification as an interactor of MCM4. In this exemplary pull-down, the detected true interactors showed an average correlation of 0.68 to the bait, whereas the detected unspecific binders showed an average correlation of 0.42. In ES2, the average correlation of detected unspecific binders was generally even lower. We determined a series specific correlation cutoff for ES1 and ES2 by evaluating the correlation of all proteins detected in all pull-downs in a Q-Q-plot, which visualizes the real distribution of all correlation values compared with a theoretical normal distribution (supplemental Fig. S6I and 6J). The point, where actual and theoretical distribution sharply deviated was chosen as the correlation cutoff. Correlation analysis worked particularly well with our data set, as it contains at least two entry points for every complex.
We then proceeded to group enriched proteins into interactor confidence classes A–C by their enrichment, p value and correlation to the bait as summarized in Fig. 4C. Class C proteins are proteins between the two cutoff lines with low or medium correlation to the bait and are not regarded as interactors. Class B proteins are proteins between the cutoff lines with high correlation or proteins outside the outer cutoff line with medium correlation, and represent lower confidence interactors. Finally, class A proteins are proteins outside the outer cutoff line with high correlation and are considered high confidence interactors. The result of the classification is shown for the MCM complex in Fig. 4C, and the same color scheme is used in all volcano plots throughout the supplemental Material ES1/ES2. Although we found the above classification scheme to be very efficient, it should not be seen as absolute, but rather as a help in interpreting the pull-downs results.
How the intensity profile analysis can recognize false-positives is illustrated by the profiles of SFC1 and SDH3 in Fig. 4B. They represent atypical background binders (see above) fluctuating from low to high intensities across pull-downs. Because they appeared by chance in all of the replicates of the specific pull-down they scored both a good enrichment factor and p value. However, because of the fluctuations in their profiles, the correlation to the bait intensity profile is poor, which reclassifies SFC1 as lower confidence interactor and SDH3 as noninteractor. Without the correlation analysis, SFC1 would have been considered a high confidence interactor. Conversely, proteins that are only minimally but reproducibly enriched are likely to still be true interactors if they show good correlation (See MCM3 in Fig. 4B). Using the data set-dependent cutoff definition, the average complex coverage per pull-down (calculated as true positives/(true positives + false negatives), with true complex members derived from UniProt) was 74% for ES1 and even 83% for ES2. Among the 82 and 79 class A interactors, the false-positive rates (calculated as false positives/(true positives + false positives) were only 6 and 0% for ES1 and ES2 respectively. Among the 32 class B interactors in ES1, the false-positive rate was 53%; however, 15 out of these 17 false positives were downgraded from class A and therefore rightfully classified as lower confidence interactors. Among the 15 class B interactors in ES2, the false positive rate was 20%. False-negative rates in class C (calculated as true complex members falsely classified as class C/all proteins in class C) were very low with 3% (4 out of 133) for ES1 and 6% (2 out of 35) for ES2. For all the aforementioned calculations, the two large complexes (NPC and proteasome) as well as the complexes were no classification could be performed (APC2, CDC73, and TEF1) were excluded.
Defining Complexes of Various Sizes, Abundances, and Cellular Localizations
The bait proteins in our study had been selected to represent a wide range of cellular abundances (supplemental Fig. S1), localizations (e.g. cytosolic, nuclear, and membrane bound), and functions (e.g. cell cycle, transcription, translation-elongation, and transport). For each of the pull-downs, the volcano plot containing the results of our analysis is depicted in supplemental Material ES1 and/or supplemental Material ES2. All bait proteins and the page number of the corresponding volcano plot within the supplemental Material ES1/ES2 are summarized in a table on the first page of both files. Given the diversity of these complexes, they serve to illustrate different aspects of our method.
When we used very low abundant proteins as baits, we were still able to identify interactors with a surprisingly high complex coverage, especially considering that our system uses endogenous expression and relatively little input material. For instance the members of the anaphase promoting complex, which has a key regulatory role in the cell cycle, are expressed at an estimated average of about 70 copies per cell in unsynchronized yeast cells (46). Using APC1 (about 30 copies/cell) as the entry point to the APC, our standard pull-down protocol already identified 11 out of 13 APC members. The two missing complex members (APC9 and APC11) are potentially even lower abundant in unsynchronized cells as they were also not detected in a deep yeast proteome (46). Similarly, pull-down of the SET1/COMPASS histone methyltransferase complex by its SET1 (135 copies/cell) and SWD3 (74 copies/cell) subunits revealed all eight complex members as clear outliers in the volcano plots.
Conversely, we were also able to detect interactors of very high abundant proteins. Here the challenge is that these proteins often have very high background intensities – ranging in our workflow to a log2 intensity of up to about 36 – over which they can hardly be further enriched. For the elongation factor CAM1 (49,500 copies/cell, average log2 background intensity 29.9) we identified CAM1 itself and its direct interactor EFB1 with a moderate but clear enrichment but an extremely significant p value (p < 10−25). However, TEF1 (630,000 copies/cell, average log2 background intensity of 34.8), another elongation factor 1 complex member, did not register as an interactor as its background intensity is so high that it cannot be significantly further enriched. Even when we tagged TEF1, this bait was not an outlier, although all three interactors CAM1, EFB1, and TEF4 were significantly enriched. We also targeted another very high abundant complex, the ribosome-associated complex (RAC) through its components SSZ1 (59,450 copies/cell, average log2 background intensity of 32.2) and ZUO1 (45,188 copies/cell, average log2 background intensity of 31.4). Although SSZ1 only retrieved itself as outlier, when we tagged ZUO, we could indeed detect SSZ1 with mild enrichment but with a very good p value (p < 10−22).
Although the above examples serve as positive controls, illustrating aspects of our affinity enrichment workflow, we also we detected some interactors that are not part of the stable, known core complexes. The MCM complex presents the core of the replicative DNA helicase in yeast and forms a double hexameric ring around the DNA (48). We identified TOF1 (Topoisomerase 1-associated factor 1) which is not part of the core helicase but which has been shown to interact and regulate it (49). TOF1 is an example of an interactor that was promoted to likely interactor status (class B), because of its high correlation with complex members.
The yeast proteasome consists of a 20S core particle composed of 28 α and β-subunits assembled into four rings, and a 19S regulatory particle on both sides of the core composed of 19 proteins. As the proteasome is a highly dynamic holocomplex, its purification is not trivial (50). Using two 20S members, PUP1 (β subunit) and PUP2 (α subunit), retrieved the complete 20S complex and most of the 19S members. Additionally, we found a number of transient interactors, such as the proteasome activator BLM10, the proteasome stabilizing component ECM29, the proteasome chaperone PBA1 and the uncharacterized protein YCR076C. The latter has already been reported to interact with proteasome core particle subunits (51), an association that we now confirm. Other enriched proteins found in the PUP1/PUP2 pull-downs that are not reported to interact with the complex could be proteasome substrates.
The nuclear pore complex (NPC) represents an example of a very large complex (about 30 different proteins in multiple copies) that is embedded in the nuclear membrane (52). Performing pull-downs with two of the subunits (NUP84 and NUP145), we found many components of the NPC (19 and 16 respectively), which, remarkably, is more than what was identified for these two baits in a dedicated membrane interactome (53). Additionally, we identified proteins that are not only components of the NPC but also of the spindle pole body (SPB), namely CDC31 (54, 55) and NDC1 (56). Consequently, other components of the SPB including SPC110 and SPC42 were among the outliers. We also identified the inner nuclear membrane protein HEH2, which has been proposed to be important for a proper distribution of nuclear pores across the nuclear envelope (57).
Two further examples are PAF1 (RNA polymerase II-associated protein 1), pull-down of which resulted in all five core complex members as well as RPO21. This protein is a subunit of the RNA polymerase II. Likewise pull-down of PEP5, a member of the HOPS complex, retrieved all its members, and furthermore VPS8, a component of the CORVET complex sharing four subunits (PEP3,PEP5,VPS16, and VPS33) with the HOPS complex (58).
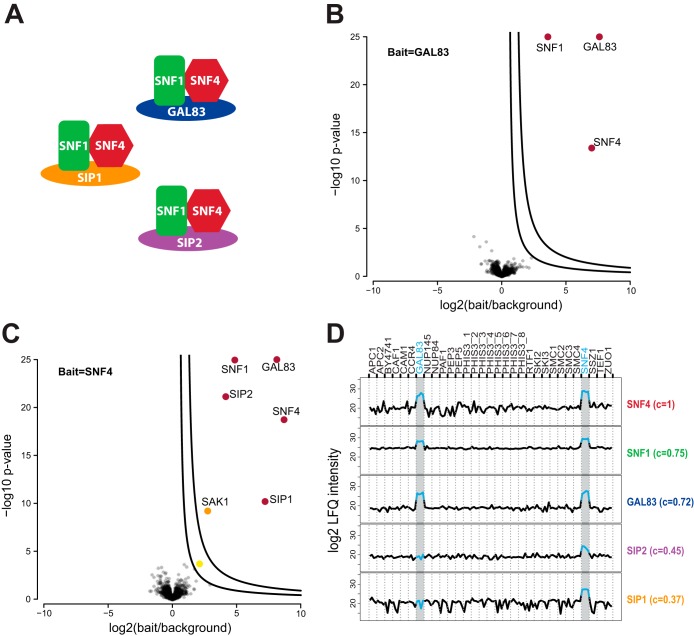
Apart from core and transient, proteins can also be mutually exclusive complex members. As an example, the SNF1 protein kinase complex is a hetero-trimeric complex consisting of the alpha subunit SNF1, the gamma subunit SNF4, and one of three alternative beta subunits SIP1, SIP2, or GAL83 (Fig. 5A) (59). This complex proved to be a good case to investigate the effects of mutually exclusive complex members on the intensity profile analysis. We used SNF4 and GAL83 as baits, hence SIP1 and SIP2 were only identified in the SNF4 pull-down, as expected (Fig. 5B and 5C). Nevertheless they showed a correlation of 0.37 and 0.45, respectively, to the bait SNF4 (Fig. 5D), which was higher than the correlation cutoff (0.35 for ES2). This demonstrates the usefulness of correlation analysis for associating even alternative members with the core complex. This complex also illustrates the need for several entry points per complex to recapitulate more complicated complex arrangements such as alternative cellular subcomplexes. Using SNF4 as bait, we additionally identified the protein SAK1, which is an upstream kinase that activates SNF1 (60).
Fig. 5.
Correlation analysis and mutually exclusive binding. A, Schematic representation of the three alternate SNF1 protein kinase complexes. B, Volcano plot of GAL83 compared with its bait-specific control group (BSCG). C, Volcano plot of SNF4 compared with its BSCG. D, Intensity profiles of the gamma subunit SNF4, the alpha subunit SNF1, and the three alternate beta subunits GAL83, SIP1, and SIP2 as well as their correlation to the bait SNF4.
DISCUSSION
For about two decades, AP-MS techniques have been used as tools for investigating protein complexes, and they have been improved greatly during this time. Previously, protein complexes were extensively purified, to reduce the amount of copurifying unspecific binders as much as possible. However, such stringent purification becomes unnecessary as soon as AP is coupled to high resolution, quantitative MS. Quantification can distinguish the true interactors from contaminants. Therefore, protocols can be less stringent, preserving weaker interactions, while resulting in a higher background. In this work, we have taken this concept to its logical conclusion by employing low stringent single-step enrichment of protein complexes followed by label-free quantitative MS analysis in which we co-purify a very large number of unspecific binders representing about half of the yeast proteome. Complexes can still be confidently identified because of their enrichment in specific bait pull-downs versus all other pull-downs. As we do not aim to purify but only to enrich, we suggest terming such methods AE-MS. Our methodology is solely based on intensity-based label-free quantification, which has advanced considerably and for pull-downs is now comparable with label-based quantification approaches like SILAC (20, 33).
Identification of a large number of background binders is unavoidable with modern MS instrumentation. Perhaps counterintuitively, our results demonstrate that these unspecific proteins can actually be beneficial, elevating them from a nuisance to an essential part of the analysis. Apart from their essential use in normalization, they are indicators of the reproducibility within a specific workflow and serving as quality control. As unchanging background binders greatly outnumber changing interactors, pull-downs are highly similar to each other, which in turn obviates the need for a dedicated control strain. Finally, we have shown that reproducible detection of unspecific binders allows further characterization of interactor candidates by correlating their intensity profiles to the profile of the bait. Using our pipeline, we identified interactors of a diverse set of endogenously expressed bait proteins with high confidence, starting from minimal input amounts of unlabeled yeast, and requiring modest measuring times despite replicate analysis. In medium or large-scale projects, our workflow automatically provides a large control group, without actually performing any control pull-downs. However, as illustrated with the SKI complex, using only one tagged strain as control (or an empty stain) already correctly identified all complex members, demonstrating the feasibility of AE-MS also for small scale projects.
Although a large improvement, our AE-MS workflow does not solve all issues in MS-based interaction studies. Membrane complexes always present a challenge because of their hydrophobic nature. However, our protocol yielded excellent results for the HOPS vacuolar membrane complex and the nuclear pore complex without adapting it in any way. For the SPOTS complex, we only retrieved two out of the six complex members. Adapting the type of detergent or the detergent concentration in the lysis buffer may help to better identify membrane complexes (53). To further verify interactors, we have introduced intensity profile analysis, which proved to be very helpful for upgrading weaker interactors and uncovering false positives. As this method relies on correlation to the bait profile, it could; however, not be used in three cases where we did not detect the bait as an outlier (in ES1: APC2 and CDC73; in ES2: TEF1). In the case of CDC73, the bait was incorrectly tagged in the strain we used, as we subsequently found by a control PCR. For APC2 the very low copy number was presumably the reason, as even in ES2 where we found APC2, it was only identified with two peptides. Finally, as already mentioned, for TEF1 the background intensity was so high that it did not form a useful profile. However, the intensity profiling only serves as additional information, and in all these cases the correct interacting proteins were still identified through their enrichment. A final potential caveat for the intensity profile analysis are newly identified proteins interacting with several baits, which decreases their correlation score. However, provided their enrichment is high, they would still be considered (class B) interactors. Examination of the actual intensity profile of such promiscuous interactors could also help in judging whether weak correlation to the bait is caused by strong fluctuation between all samples, making the protein a false positive, or caused by strong fluctuation between several replicate groups, making it a potential link between several complexes.
The two largest yeast interactomes published in 2006 by Gavin et al. and Krogan et al. both employed TAP-tagging coupled to nonquantitative MS and among other frequency filtering of detected proteins to remove unspecific binders (9, 10). This can be problematic in the case of atypical background binders that appear spontaneously at high intensity in only some pull-downs. In our AE-MS approach, pull-downs are performed in replicates, hence such proteins are rarely scored as interactors. Even if an atypical background binder is by chance detected in all replicates, the intensity profile analysis can still uncover it. With very few exceptions, all of the proteins listed as contaminant in the above studies were also found in our data set. However, they did not appear as interactors in any of our pull-downs other than where expected. The data sets of Gavin et al. and Krogan et al. only share about one quarter of detected interactions (61) and did not contain 1/3 or 1/2 of the baits that we had tagged here, respectively. For each of the pull-downs that we could compare between all three studies (APC2, BRE2, CCR4, NUP84, NUP145, POP2, RTF1, SET1, SKI2, SMC1, SSZ1, and SWD3) the complex coverage was equal or better using the AE-MS method. In one case, we only retrieved EFB1 as interactors of CAM1 whereas Gavin et al. also found TEF1 and TEF2. Although these proteins were also found in a mock TAP-tag purification and therefore included in the contaminant list, we reason that more stringent purification could be helpful for detecting interactors of extremely high expressed proteins such as CAM1.
Recent interaction proteomics efforts typically at least employ semiquantitative approaches; however, removal of contaminants can still be problematic. There is an ongoing collaborative effort to establish a “contaminant repository for affinity purification,” the “CRAPome,” containing control pull-downs from various laboratories performed under various experimental conditions (62). In the case of yeast 17 control pull-downs are currently available, of which 12 have been performed using GFP-tagged proteins and nano-magnetic beads. However, a larger number of controls may be necessary to comprehensively cover all nonspecific binders and thereby avoid incorrectly classifying a nonspecific binder as an interactor. Our AE-MS method sidesteps this problem, as the samples themselves are the controls. The minor but clear differences between our two experimental series (Fig. 2A) demonstrate that minor changes in the workflow like using a different machine of the same type can already alter the detected low abundant background binders, making the notion of a universal CRAPome problematic.
From the differences between the two experimental series we also conclude that for the most optimal output, AE-MS experiments should be executed in a reproducible manner from sample preparation to MS measurement, which should ideally be conducted on one machine and in one batch as in ES2. However, the MaxLFQ normalization algorithm successfully corrected for most of the variability in the ES1 series in general and in the proteasome pull-downs in particular, resulting in excellent results even for ES1.
To perform the AE-MS workflow described here, only three elements were needed: tagged proteins of interest, a high resolution LC-MS system, and sophisticated software to quantify proteins and analyze the data. Here we used the LTQ Orbitrap classic, which—although not being the latest Orbitrap technology—proved to be sufficient for identifying even very low abundant protein complexes. Such technology is now widely accessible, as is the MaxQuant software for performing accurate intensity-based label-free quantification and the Perseus program for statistical analysis of the data. Our AE-MS protocol is equally suited to investigate a small, medium or large number of samples. For a smaller set of samples, SILAC labeling could easily be implemented, which might provide even more accurate ratios in the case of borderline enrichment. More and more AP-MS workflows already use single-step protocols and employ high resolution MS, and therefore rather represent AE-MS methods. The shift in the conceptual framework from AP-MS to AE-MS and the development of sophisticated analysis tools for AE-MS experiments should contribute to higher quality interaction data, thereby making studies more comparable, and helping to solve open challenges in the interactomics field.
Supplementary Material
Acknowledgments
We thank Nils A. Kulak for input regarding yeast culture, Jürgen Cox for advice regarding data analysis and Roland Wedlich-Söldner for providing the strains of the yeast-GFP clone collection.
Footnotes
Author contributions: E.C.K. and M.M. designed research; E.C.K. performed research; M.Y.H. contributed new reagents or analytic tools; E.C.K., M.Y.H., and M.M. analyzed data; E.C.K. and M.M. wrote the paper.
* This work was supported by the Bundesministerium für Bildung und Forschung (grant number FKZ01GS0861, DiGtoP consortium) and the European Commission's 7th Framework Program PROSPECTS (HEALTH-F4-2008-201648).
 This article contains supplemental Figs. S1 to S7, Table S1, Spectra, and Experimental Series S1 and S2.
This article contains supplemental Figs. S1 to S7, Table S1, Spectra, and Experimental Series S1 and S2.
DATA AVAILABILITY: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (63). (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (64) with the data set identifier PXD000955.
1 The abbreviations used are:
- AP-MS
- Affinity purification mass spectrometry
- AE-MS
- Affinity enrichment mass spectrometry
- GFP
- Green fluorescent protein
- (Co-)IP
- (Co-) Immunoprecipitation
- Y2H
- Yeast two-hybrid
- BAC
- Bacterial artificial chromosome
- QUBIC
- Quantitative BAC green fluorescent protein interactomics
- TAP
- Tandem affinity purification
- LFQ
- Label-free quantification
- MaxLFQ
- MaxQuant Label-free quantification
- CAA
- Chloroacetamide
- ES
- Experimental series
- FDR
- False discovery rate
- SC
- Synthetic complete
- YPD
- Yeast extract peptone dextrose
- BSCG
- Bait specific control group
- NPC
- Nuclear pore complex
- SPB
- Spindle pole body.
REFERENCES
- 1. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 2. Oeffinger M. (2012) Two steps forward–one step back: advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics 12, 1591–1608 [DOI] [PubMed] [Google Scholar]
- 3. Gavin A. C., Maeda K., Kuhner S. (2011) Recent advances in charting protein–protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 [DOI] [PubMed] [Google Scholar]
- 4. Fields S., Song O. (1989) A novel genetic system to detect protein–protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 5. Rajagopala S. V., Sikorski P., Caufield J. H., Tovchigrechko A., Uetz P. (2012) Studying protein complexes by the yeast two-hybrid system. Methods 58, 392–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parrish J. R., Gulyas K. D., Finley R. L., Jr. (2006) Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17, 387–393 [DOI] [PubMed] [Google Scholar]
- 7. Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 8. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 9. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dumpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 10. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 11. Butland G., Peregrin-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 12. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart II, Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuhner S., van Noort V., Betts M. J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., Castano-Diez D., Chen W. H., Devos D., Guell M., Norambuena T., Racke I., Rybin V., Schmidt A., Yus E., Aebersold R., Herrmann R., Bottcher B., Frangakis A. S., Russell R. B., Serrano L., Bork P., Gavin A. C. (2009) Proteome organization in a genome-reduced bacterium. Science 326, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 14. Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D. Y., Cenaj O., McKillip E., Shah S., Stapleton M., Wan K. H., Yu C., Parsa B., Carlson J. W., Chen X., Kapadia B., VijayRaghavan K., Gygi S. P., Celniker S. E., Obar R. A., Artavanis-Tsakonas S. (2011) A protein complex network of Drosophila melanogaster. Cell 147, 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., Qin J. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Makarov A. (2000) Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 17. Gibson T. J., Seiler M., Veitia R. A. (2013) The transience of transient overexpression. Nat. Methods 10, 715–721 [DOI] [PubMed] [Google Scholar]
- 18. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poser I., Sarov M., Hutchins J. R., Heriche J. K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A. W., Pelletier L., Kittler R., Hua S., Naumann R., Augsburg M., Sykora M. M., Hofemeister H., Zhang Y., Nasmyth K., White K. P., Dietzel S., Mechtler K., Durbin R., Stewart A. F., Peters J. M., Buchholz F., Hyman A. A. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 22. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 23. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 24. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 25. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 26. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 27. Ranish J. A., Yi E. C., Leslie D. M., Purvine S. O., Goodlett D. R., Eng J., Aebersold R. (2003) The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33, 349–355 [DOI] [PubMed] [Google Scholar]
- 28. Paul F. E., Hosp F., Selbach M. (2011) Analyzing protein–protein interactions by quantitative mass spectrometry. Methods 54, 387–395 [DOI] [PubMed] [Google Scholar]
- 29. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 30. Bondarenko P. V., Chelius D., Shaler T. A. (2002) Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 74, 4741–4749 [DOI] [PubMed] [Google Scholar]
- 31. Nahnsen S., Bielow C., Reinert K., Kohlbacher O. (2013) Tools for label-free peptide quantification. Mol. Cell Proteomics 12, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
- 33. Eberl H. C., Spruijt C. G., Kelstrup C. D., Vermeulen M., Mann M. (2013) A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 49, 368–378 [DOI] [PubMed] [Google Scholar]
- 34. Choi H., Glatter T., Gstaiger M., Nesvizhskii A. I. (2012) SAINT-MS1: protein–protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J. Proteome Res. 11, 2619–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poulsen J. W., Madsen C. T., Young C., Poulsen F. M., Nielsen M. L. (2013) Using guanidine-hydrochloride for fast and efficient protein digestion and single-step affinity-purification mass spectrometry. J. Proteome Res. 12, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 36. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 37. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagaraj N., Kulak N. A., Cox J., Neuhauser N., Mayr K., Hoerning O., Vorm O., Mann M. (2012) System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteomics 11, M111 013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 40. Hubner N. C., Mann M. (2011) Extracting gene function from protein–protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods 53, 453–459 [DOI] [PubMed] [Google Scholar]
- 41. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, prefractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 42. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 43. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111 014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rees J. S., Lowe N., Armean I. M., Roote J., Johnson G., Drummond E., Spriggs H., Ryder E., Russell S., St Johnston D., Lilley K. S. (2011) In vivo analysis of proteomes and interactomes using Parallel Affinity Capture (iPAC) coupled to mass spectrometry. Mol. Cell. Proteomics 10, M110 002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 47. Cox J., Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nedelcheva M. N., Roguev A., Dolapchiev L. B., Shevchenko A., Taskov H. B., Shevchenko A., Stewart A. F., Stoynov S. S. (2005) Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J. Mol. Biol. 347, 509–521 [DOI] [PubMed] [Google Scholar]
- 50. Forster F., Unverdorben P., Sledz P., Baumeister W. (2013) Unveiling the long-held secrets of the 26S proteasome. Structure 21, 1551–1562 [DOI] [PubMed] [Google Scholar]
- 51. Hatanaka A., Chen B., Sun J. Q., Mano Y., Funakoshi M., Kobayashi H., Ju Y., Mizutani T., Shinmyozu K., Nakayama J., Miyamoto K., Uchida H., Oki M. (2011) Fub1p, a novel protein isolated by boundary screening, binds the proteasome complex. Genes Genet. Syst. 86, 305–314 [DOI] [PubMed] [Google Scholar]
- 52. Fernandez-Martinez J., Rout M. P. (2012) A jumbo problem: mapping the structure and functions of the nuclear pore complex. Curr. Opin. Cell Biol. 24, 92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Babu M., Vlasblom J., Pu S., Guo X., Graham C., Bean B. D., Burston H. E., Vizeacoumar F. J., Snider J., Phanse S., Fong V., Tam Y. Y., Davey M., Hnatshak O., Bajaj N., Chandran S., Punna T., Christopolous C., Wong V., Yu A., Zhong G., Li J., Stagljar I., Conibear E., Wodak S. J., Emili A., Greenblatt J. F. (2012) Interaction landscape of membrane–protein complexes in Saccharomyces cerevisiae. Nature 489, 585–589 [DOI] [PubMed] [Google Scholar]
- 54. Spang A., Courtney I., Fackler U., Matzner M., Schiebel E. (1993) The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chial H. J., Rout M. P., Giddings T. H., Winey M. (1998) Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143, 1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yewdell W. T., Colombi P., Makhnevych T., Lusk C. P. (2011) Lumenal interactions in nuclear pore complex assembly and stability. Mol. Biol. Cell 22, 1375–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Balderhaar H. J., Ungermann C. (2013) CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126, 1307–1316 [DOI] [PubMed] [Google Scholar]
- 59. Nath N., McCartney R. R., Schmidt M. C. (2002) Purification and characterization of Snf1 kinase complexes containing a defined Beta subunit composition. J. Biol. Chem. 277, 50403–50408 [DOI] [PubMed] [Google Scholar]
- 60. Hedbacker K., Carlson M. (2008) SNF1/AMPK pathways in yeast. Front. Biosci. 13, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kiemer L., Costa S., Ueffing M., Cesareni G. (2007) WI-PHI: a weighted yeast interactome enriched for direct physical interactions. Proteomics 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 62. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.