Abstract
SRC homology 2 domain-containing leukocyte phosphoprotein of 76 kDa (SLP-76) is a cytosolic adaptor protein that plays an important role in the T-cell receptor–mediated T-cell signaling pathway. SLP-76 links proximal receptor stimulation to downstream effectors through interaction with many signaling proteins. Previous studies showed that mutation of three tyrosine residues, Tyr112, Tyr128, and Tyr145, in the N terminus of SLP-76 results in severely impaired phosphorylation and activation of Itk and PLCγ1, which leads to defective calcium mobilization, Erk activation, and NFAT activation. To expand our knowledge of the role of N-terminal phosphorylation of SLP-76 from these three tyrosine sites, we characterized nearly 1000 tyrosine phosphorylation sites via mass spectrometry in SLP-76 reconstituted wild-type cells and SLP-76 mutant cells in which three tyrosine residues were replaced with phenylalanines (Y3F mutant). Mutation of the three N-terminal tyrosine residues of SLP-76 phenocopied SLP-76-deficient cells for the majority of tyrosine phosphorylation sites observed, including feedback on proximal T-cell receptor signaling proteins. Meanwhile, reversed phosphorylation changes were observed on Tyr192 of Lck when we compared mutants to the complete removal of SLP-76. In addition, N-terminal tyrosine sites of SLP-76 also perturbed phosphorylation of Tyr440 of Fyn, Tyr702 of PLCγ1, Tyr204, Tyr397, and Tyr69 of ZAP-70, revealing new modes of regulation on these sites. All these findings confirmed the central role of N-terminal tyrosine sites of SLP-76 in the pathway and also shed light on novel signaling events that are uniquely regulated by SLP-76 N-terminal tyrosine residues.
Signaling events induced by the T-cell receptor (TCR)1 play an essential role in the adaptive immune response, important for T-cell proliferation, differentiation, and cytokine secretion. TCR engagement results in sequential activation of Src kinase Lck and Fyn, which phosphorylates the CD3ζ-chain immunoreceptor tyrosine-based activation motifs (ITAMs) (1). Phosphorylated ITAMs recruit and activate the Syk family protein kinase ZAP-70, which phosphorylates the transmembrane scaffold linker for activation of T cells (2), as well as SH2 domain-containing leukocyte protein of 76 kDa (SLP-76) (3), forming a signalosome complex essential for the assembly of downstream signaling proteins.
SLP-76, as an adaptor protein, lacks intrinsic enzymatic function but serves as an essential protein scaffold, recruiting other proteins for correct localization during T-cell signaling. Studies with SLP-76-deficient mice and SLP-76-deficient T-cell lines revealed a very profound role for SLP-76 in T-cell development and activation (4–7). In SLP-76-deficient Jurkat T cells, defects were observed in phosphorylation and activation of PLCγ1, calcium mobilization, Erk activation, and cytokine gene transcription following TCR ligation (6). SLP-76 consists of three domains: an N-terminal acidic region containing three tyrosine residues, Tyr112, Tyr128, and Tyr145; a central proline-rich region; and a C-terminal SH2 domain (7). Upon TCR activation, SLP-76 is recruited to the linker for activation of T cells signaling complex through binding with GADS (8), nucleating the interaction of signaling proteins, including PLCγ1, Itk, Vav, Nck, and adhesion and degranulation adaptor protein (9). PLCγ1 is recruited to the SLP-76 signaling complex through binding to both LAT and SLP-76. Phosphorylated Tyr145 of SLP-76 is recognized by the SH2 domain of the Tec family kinase Itk, which also binds to the proline-rich domain of SLP-76 (10). This interaction maintains Itk in an active conformation (7). The binding of PLCγ and active Itk to SLP-76 leads to the phosphorylation and activation of PLCγ1 and subsequent generation of the second messengers inositol 1,4,5-trisphosphate and diacylglcycerol (11). SLP-76 also regulates cytoskeletal rearrangement through the assembly of a tri-molecular signaling complex with Vav and Nck (12). In addition, the interaction between the tyrosine-phosphorylated adaptor protein and the SH2 domain of SLP-76 regulates integrin activation (13).
Besides its importance in regulating downstream signaling proteins, we recently revealed that SLP-76 plays an important role in mediating upstream signaling proteins (14). In a phosphoproteomic study examining cells deficient in SLP-76, SLP-76 was required for mediation of the phosphorylation of PAG (14), which transmits negative regulatory signals in complex with Csk (15). In addition, this earlier study revealed that the absence of SLP-76 perturbs the phosphorylation of Lck and, subsequently, a large number of Lck-regulated signaling molecules (i.e. CD3ε, -δ, -γ, and -ζ chains; ZAP-70) (14). These findings led to the hypothesis that SLP-76 mediates both PAG negative feedback and ERK positive feedback of Lck (14).
Phosphorylation of three N-terminal tyrosine residues is essential for the function of SLP-76 (16). Upon phosphorylation by ZAP-70, phosphorylated Tyr112 and Tyr128 bind to SH2 domains of Vav (17–20), Nck (12, 21), and the p85 subunit of phosphatidylinositol 3-kinase (22), whereas phosphorylated Tyr145 is recognized by the SH2 domain of Itk (10). N-terminal tyrosines of SLP-76 are required for the TCR-induced phosphorylation and activation of Itk and PLCγ1 (7).
However, the current understanding of N-terminal tyrosines of SLP-76 is incomplete, especially regarding their role in the newly discovered feedback regulation of the phosphorylation of upstream signaling proteins. For further elucidation of the function of SLP-76 N-terminal tyrosines in the regulation of the TCR signaling pathway, a wide-scale view of temporal changes in TCR signaling components is required. Quantitative mass-spectrometry-based phosphoproteomics is a powerful way to achieve this goal by enabling the system-wide identification of sites on proteins phosphorylated in the T cell, as well as the quantification of protein phosphorylation (14, 23–28). In this study, a wide-scale quantitative phosphoproteomic method was used to identify TCR-responsive tyrosine phosphorylation sites and to gain system-wide insight into the role of N-terminal tyrosine residues of SLP-76 in the TCR signaling pathway.
EXPERIMENTAL PROCEDURES
Cell Culture, SILAC Labeling, and T-cell Stimulation
The SLP-76 mutant cell line J14–2D1 (also known as Y3F mutant) and its wild type (WT) SLP-76 reconstituted derivative J14–76-11 (6) were provided by Deborah Yablonski at Israel Institute of Technology. All cells were initially maintained in RPMI 1640 medium (Hyclone, Logan, UT) supplemented with 10% heat-inactivated undialyzed FBS (Hyclone), 2 mm l-glutamine, 100 U/ml penicillin G, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA) in a humidified incubator with 5% CO2 at 37 °C. SILAC was performed as described (25). Briefly, after 5 days, all cell lines were washed twice with RPMI 1640 medium without arginine and lysine (Invitrogen) and reconstituted in RPMI 1640 medium containing either 12C6,14N4 arginine and 12C6,14N2 lysine (Sigma) or 13C6,15N4 arginine and 13C6,15N2 lysine (Cambridge Isotope Laboratories, Andover, MA) supplemented with 10% heat-inactivated dialyzed FBS (Sigma), 2 mM l-glutamine, 100 U/ml penicillin G, 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C for seven cell doublings. The concentrations of lysine and arginine used in SILAC labeling of Jurkat cells in experiments described here were 0.22 mM and 0.38 mM, respectively.
Anti-CD3 and anti-CD4 (clones OKT3 and OKT4, eBioscience, San Diego, CA) stimulation was performed as described (28). Briefly, cells were washed once with 4 °C PBS and reconstituted at a concentration of 1 × 108 cells/ml in PBS. For each time point, 1 × 108 cells were treated with OKT3 and OKT4 antibodies at a concentration of 2.5 μg/ml of each antibody for 10 min at 4 °C. Cells were then cross-linked with 22 μg/ml goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) and incubated at 37 °C for 0, 1, 1.5, 2, 3, 5, 7, or 10 min. To halt the stimulation, the 1-ml uncentrifuged cell suspension was lysed with the addition of 5 ml of lysis buffer (9 m urea, 1 mM sodium orthovanadate, and 20 mM HEPES, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, pH 8.0), vortexed on a Vortex Mixer (Thermo Scientific) for 30 s, and then incubated for 20 min at 4 °C. Lysates were then sonicated at a 30-W output with two bursts of 30 s each and cleared at 12,000 × g for 15 min at 4 °C.
Protein Reduction, Alkylation, Digestion, and Peptide Immunoprecipitation
Protein concentrations were measured with the DC Protein Assay (Bio-Rad, Hercules, CA). Once protein concentrations were determined, a 5-mg equal portion of cell lysates from J14–2D1 and J14–76-11 was combined. The proteins in the lysate were reduced with 10 mM DTT for 20 min at 60 °C followed by alkylation with 100 mm iodoacetamide for 15 min at room temperature in the dark. Cell lysates were then diluted 4-fold with 20 mM HEPES buffer, pH 8.0, and digested with sequencing-grade modified trypsin (Promega, Madison, WI) in a 1:100 (w/w) trypsin:protein ratio overnight at room temperature. Tryptic peptides were acidified to pH 2.0 by the addition of 1/20 volume of 20% trifluoroacetic acid (TFA) for a final concentration of 1% TFA, cleared at 1800 × g for 5 min at room temperature, and desalted using C18 Sep-Pak plus cartridges (Waters, Milford, MA) as described (25), with the exception that TFA was used instead of acetic acid at the same required concentrations. Eluents containing peptides were lyophilized for 48 h to dryness.
Peptide immunoprecipitation was performed using p-Tyr-100 phosphotyrosine antibody beads (Cell Signaling Technology Danvers, MA). Dry peptides from each time point were reconstituted in ice-cold immunoaffinity purification buffer (5 mm MOPS, pH 7.2, 10 mm sodium phosphate, 50 mm NaCl) and further dissolved through gentle shaking for 30 min at room temperature and brief sonication in a sonicator water bath. Prior to peptide immunoprecipitation, a 10-pmol fraction of synthetic phosphopeptide LIEDAEpYTAK was added to each time-point sample as an exogenous quantitation standard. Peptide solutions were then cleared at 1800 × g for 5 min at room temperature, combined with p-Tyr-100 phosphotyrosine antibody beads, and incubated for 2 h at 4 °C. Beads were then washed three times with immunoaffinity purification buffer and twice with cold double-distilled H2O and eluted with 0.15% TFA. Eluted peptides were then desalted using C18 Zip Tip pipette tips (Millipore Corporation, Billerica, MA) as described (29).
All phosphoproteomic data represent the average of five total replicate analyses for each time point.
Automated Nano-LC/MS
LC/MS was performed as described previously (25). Tryptic peptides were analyzed via a fully automated phosphoproteomic technology platform (30, 31). Phosphopeptides were eluted into an LTQ/Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) through a PicoFrit analytical column (360-μm outer diameter, 75-μm inner diameter, fused-silica packed on a pressure bomb with 15 cm of 3-μm Monitor C18 particles; New Objective, Woburn, MA) with a reversed phase gradient (0% to 70% 0.1 m acetic acid in acetonitrile in 60 min, with a 90-min total method duration). An electrospray voltage of 1.8 kV was applied using a split flow configuration, as described previously (32). Spectra were collected in positive ion mode and in cycles of one full MS scan in the Orbitrap (m/z: 300–1700) followed by data-dependent MS/MS scans in the LTQ (∼0.3 s each) sequentially of the 10 most abundant ions in each MS scan with charge state screening for +1, +2, and +3 ions and a dynamic exclusion time of 30 s. The automatic gain control was 1,000,000 for the Orbitrap scan and 10,000 for the LTQ scans. The maximum ion time was 100 ms for the LTQ scan and 500 ms for the Orbitrap full scan. The Orbitrap resolution was set at 60,000.
Data Analysis
MS/MS spectra were searched against the non-redundant human UniProt complete proteome set database containing 72,078 forward (UniProt database release 2011.10.21) and an equal number of reversed decoy protein entries using Mascot algorithm version 2.2.07 from Matrix Science (Boston MA) (33). Peak lists were generated using extractMSn version 5, provided by Thermo Fisher, using a mass range of 600–4500. The Mascot database search was performed with the following parameters: trypsin enzyme cleavage specificity, two possible missed cleavages, 7-ppm mass tolerance for precursor ions, and 0.5-Da mass tolerance for fragment ions. The search parameters specified differential modification of phosphorylation (+79.9663 Da) on serine, threonine, and tyrosine residues; dynamic modification of methionine oxidationn (+15.9949 Da); and static modification of carbamidomethylation (+57.0215 Da) on cysteine. The search parameters also included differential modification for arginine (+10.00827 Da) and lysine (+8.01420 Da) amino acids for the SILAC labeling. To provide high-confidence phosphopeptide sequence assignments, we filtered Mascot results by Mowse score (>20) and precursor mass error (<2 ppm). The resulting unique peptide assignments were filtered down to a 1% false discovery rate (FDR) using a logistic spectral score filter (34). The FDR was estimated with the decoy database approach after final assembly of non-redundant data into heatmaps (35). To validate the position of the phosphorylation sites, we applied the Ascore algorithm (36) to all data, and the reported phosphorylation site position reflected the top Ascore prediction.
Quantitation of Relative Phosphopeptide Abundance
Relative quantitation of phosphopeptide abundance was performed via calculation of select ion chromatogram (SIC) peak areas for heavy and light SILAC-labeled phosphopeptides. For label-free comparison of phosphopeptide abundance in SLP-76 reconstituted Jurkat cells among different time points of TCR stimulation, individual SIC peak areas were normalized to the peak area of an exogeneously spiked standard phosphopeptide, LIEDAEpYTAK. The LIEDAEpYTAK phosphopeptide was added in the same amount to every LC/MS sample and accompanied cellular phosphopeptides through peptide immunoprecipitation, desalting, and reversed-phase elution into the mass spectrometer. Retention time alignment of individual replicate analyses was performed as described previously (37). Peak areas were calculated through inspection of SICs using in-house software programmed in Microsoft Visual Basic 6.0 and based on Xcalibur Development Kit 2.1 (Thermo Fisher Scientific). This approach used the ICIS algorithm available in the Xcalibur XDK with the following parameters: multiple resolutions of 8, noise tolerance of 0.1, noise window of 40, scans in baseline of 5, and inclusion of refexc peaks parameter value, which is false. SIC peak areas were determined for every phosphopeptide that was identified by MS/MS. In the case of a missing MS/MS spectrum for a particular peptide in a particular replicate, peak areas were calculated according to the peptide's isolated mass and the retention time calculated from retention time alignment. A minimum SIC peak area equivalent to the typical spectral noise level of 300 was required of all data reported for label-free quantitation.
A label-free data heatmap was generated for the comparison of phosphopeptides in SLP-76 reconstituted cells through a time course of receptor stimulation as previously described (25). The magnitude of change of the heatmap color was calculated based on the natural log of the ratio of the fold change of each individual phosphopeptide peak area compared with the geometric mean for that phosphopeptide across all time points, as described previously (30). In the heatmap representation, the geometric mean of a given phosphopeptide across all time points was set to the color black. A blue color represented below-average abundance and yellow represented above-average abundance for each unique phosphopeptide. Blanks in the heatmap indicated that a clearly defined SIC peak was not observed for that phosphopeptide in any of the replicate analyses for that time point. The heatmap colors were generated from the average of the LIEDAEpYTAK standard phosphopeptide normalized SICs in the five replicate experiments. The coefficient of variation (cv) was calculated for each heatmap square. Label-free p values were calculated from the replicate data for each time point compared with the time point with the minimum average peak area for that phosphopeptide. Q values for multiple hypothesis tests were also calculated for each time point based on the determined p values using the R package QVALUE as previously described (38, 39). A white dot on a label-free heatmap square indicated that a significant difference (Q value < 0.05) was detected for that phosphopeptide and time point relative to the time point with the minimal value.
In the second type of heatmap, SILAC ratios corresponding to phosphopeptide abundance differences between J14–2D1 and J14–76-11 cells across the time course of receptor stimulation were represented. For the SILAC heatmap, a black color represented a ratio of 1 between J14–2D1 and J14–76-11 for the peak area of a given phosphopeptide at that time point. A red color represented less abundance and green represented higher abundance of the given phosphopeptide in J14–2D1 relative to J14–76-11 cells. The magnitude of change of the heatmap color was calculated as described (25). Q values were also calculated between the J14–2D1 and J14–76-11 cell replicate measurements for each phosphopeptide and time point. A white dot on a heatmap square indicated that a significant change (Q value < 0.05) was observed between the replicate data from the J14–2D1 and J14–76-11 samples for that time point and phosphopeptide.
Western Blot Analysis
Total cellular protein from 9 m urea cell lysates was diluted 1:1 with a 2× sample loading buffer (4% SDS, 125 mm Tris-HCl, pH 6.8, 20% v/v glycerol, 5% 2-mercaptoethanol, 0.01% bromphenol blue) for each proteomic sample. Equal amounts of protein, as measured by the DC Protein Assay, were separated via 4–20% SDS-PAGE on Precise Tris-HEPES gel (Thermo Fisher Scientific) and electroblotted onto an Immobilon membrane (Millipore). The membrane was blocked for 30 min in Odyssey blocking buffer at room temperature (Li-Cor, Lincoln, NE) and then incubated with the primary antibody diluted 1:1000 overnight at 4 °C. Primary antibodies used in this study were mouse monoclonal ANTI-FLAG® M2 antibody F3165 (Sigma-Aldrich), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit antibody (Cell Signaling Technology), p44/42 MAPK (Erk1/2) mouse antibody (Cell Signaling Technology), phospho-PLCγ1 (Tyr783) rabbit antibody (Cell Signaling Technology), and PLCγ1 rabbit antibody (Cell Signaling Technology). The membrane was washed four times for 5 min at room temperature in PBS/0.1% Tween-20. The membrane was then incubated with anti-mouse IgG (Li-Cor) or anti-rabbit IgG (Li-Cor) for 45 min in Odyssey blocking buffer at room temperature and washed three times for 5 min with PBS/0.1% Tween-20. Bands were visualized using an Odyssey Imaging System (Li-Cor).
RESULTS
A quantitative phosphoproteomic analysis of TCR signaling was used to compare SLP-76 reconstituted Jurkat T cell line J14–76-11 and SLP-76 mutant cell line J14–2D1, in which three N-terminal tyrosine residues were replaced with phenylalanines (Y3F mutant). For each cell line, a receptor stimulation time course experiment including eight time points and five replicates was performed. The resulting phosphoproteomic data represent a wide-scale view of the temporal changes of tyrosine phosphorylation events following TCR stimulation in the presence of SLP-76 Y3F. In all, 934 unique tyrosine phosphorylation sites residing on 658 proteins were identified at a 1% FDR. The expression level of SLP-76 in the SLP-76 Y3F cells and reconstituted cells was tested via Western blot with an anti-Flag M2 antibody. Analysis of nine replicates showed no significant difference in the SLP-76 expression level in J14–76-11 (SLP-76 WT reconstituted) and J14–2D1 (SLP-76 Y3F mutant) (supplemental Fig. S1A).
Phosphoproteomic Sample Quantitation and Statistical Analysis
After processing of phosphoproteomic samples and generation of raw MS phosphopeptide identifications, high-quality sequence assignments were determined using stringent criteria as described in “Experimental Procedures.” Relative quantitation of phosphopeptide abundance via calculation of SIC peak areas was performed for each phosphopeptide at each time point. Ratios were also calculated through comparison of the SIC peak area of a phosphopeptide from SLP-76 Y3F to SLP-76 WT cells at each time point. A total of five replicate experiments were performed, and SIC peak areas and heatmaps were generated from the average values. For each sequenced phosphopeptide, two different visual representations of quantitative data in the form of heatmaps were generated to reflect either label-free or SILAC ratio data. For label-free heatmaps, the fold change in phosphorylation for each identified phosphopeptide was compared across TCR stimulation time points in SLP-76 WT cells. For SILAC heatmaps, the fold change for each phosphopeptide peak area between SLP-76 Y3F and SLP-76 WT cells was compared. A complete list of sequence and phosphorylation site assignments of all identified phosphopeptides with corresponding SIC peak areas calculated either from MS/MS or accurate mass/aligned retention time and statistics can be found in supplemental Table S1. A complete list of tyrosine phosphorylated peptides with MOWSE scores > 20 and mass error < 2 ppm including reversed database hits from every replicate and time point of TCR stimulation are provided in supplemental Table S2. A summary of the number of unique tyrosine phosphorylated peptides and phosphorylation sites identified through the MS/MS database search at a 1% FDR in each replicate and time point can be found in supplemental Table S3. Phosphopeptide replicate SIC peak areas calculated directly from MS/MS-identified peptides or from peptides identified via accurate mass and retention time alignment showed a high degree of correlation (J14–76-11: r = 0.872 ± 0.006 from MS/MS, r = 0.794 ± 0.011 from retention time alignment; J14–2D1: r = 0.880 ± 0.006 from MS/MS, r = 0.791 ± 0.012 from retention time alignment) (supplemental Table S4). Supplemental Fig. S2 shows a pairwise comparison of replicate SIC peak areas from J14–76-11 at 0 min calculated using MS/MS retention times or retention times based on spectral alignment. Moreover, scatter plots comparing average SIC peak areas calculated from MS/MS retention times or retention time spectral alignment showed a high degree of correlation (r = 0.914) (supplemental Fig. S3). In all, a total of 2295 unique peptides containing 862 unique tyrosine phosphorylation sites on 659 proteins were identified at a 1% FDR after filtering and assembly, of which 1940 unique peptides containing 745 unique tyrosine phosphorylation sites on 555 proteins showed statistically significant changes between SLP-76 Y3F and SLP-76 WT cells (Q value < 0.05) and 1646 unique peptides containing 665 unique tyrosine phosphorylation sites on 482 proteins showed statistically significant TCR-responsive changes in SLP-76 reconstituted cells (Q value < 0.05). The mass spectrometry proteomics raw data and annoted MS/MS spectra for all post-translational modification containing peptides have been deposited to the ProteomeXchange (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (40) with the dataset identifier PXD001094. Annotated MS/MS spectra for all post-translational modification containing peptides from this study are also provided in supplemental Fig. S4.
Phosphorylation of Canonical TCR Signaling Proteins
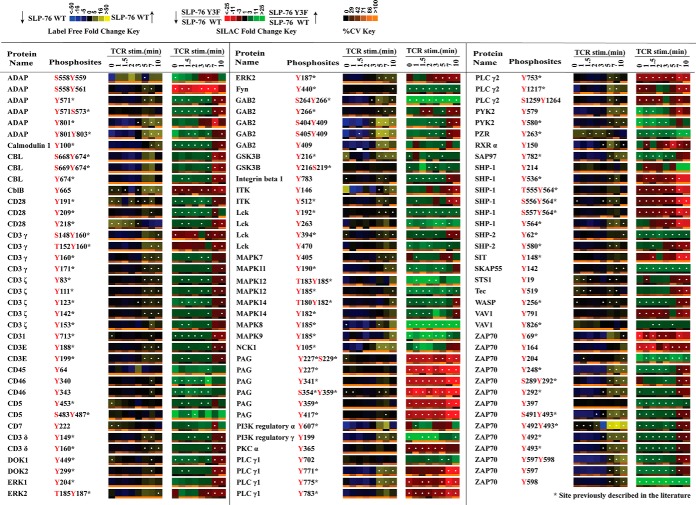
Of the confidently identified phosphopeptides in the analysis, 140 tyrosine phosphorylation sites on 58 proteins were found within the subset of proteins annotated in KEGG as TCR signaling pathway proteins, of which 113 phosphorylation sites on 49 proteins showed a statistically significant change in relative abundance (Q value < 0.05) between SLP-76 Y3F and SLP-76 WT cells (Fig. 1 and Fig. 2). The SLP-76 Y3F mutant cells had constitutive decreases in phosphorylation on PLCγ1, PLCγ2, PAG, and SHP-1. Significantly decreased phosphorylation in the SLP-76 Y3F cells was observed at later time points on Erk1 at Tyr204 and Erk2 at Tyr187, whereas the changes at early time points were not statistically significant. The quantitative phosphoproteomic data for a selection of sites were confirmed by Western blot analysis (supplemental Figs. S1B and S1C). Phosphorylation was elevated at early time points and decreased at later time points in SLP-76 Y3F cells on Lck, TCR proteins (CD3ε, -δ, -γ, and -ζ chains; ZAP-70), and other proteins (Itk, ADAP, DOK1, DOK2, PYK2). Furthermore, the temporal pattern of phosphorylation in Y3F relative to WT for a subset of sites diverged from the typical pattern of phosphorylation in other neighboring sites within the same protein. For example, Lck Tyr192, Fyn Tyr440, PLCγ1 Tyr702, ZAP-70 Tyr204, and Tyr397 were all identified with elevated constitutive phosphorylation in SLP-76 Y3F cells, whereas ZAP-70 Tyr69 was identified with constitutively decreased phosphorylation.
Fig. 1.
Quantitative phosphoproteomic analysis of known TCR signaling proteins. Listed is a portion of the data collected representing proteins annotated in KEGG as T-cell receptor signaling proteins. Heatmaps were calculated from five replicate experiments. The label-free heatmap represents the temporal change in the phosphorylation of proteins from wild-type SLP-76 reconstituted cells (J14–76-11) through a time course of TCR stimulation. In the label-free heatmaps, black squares represent a phosphopeptide abundance equal to the geometric mean for that phosphopeptide across all time points. Yellow represents levels of phosphorylation above the average, and blue corresponds to less than average abundance (as indicated in the color legend). Within the label-free heatmap, white dots indicate a statistically significant difference (Q value < 0.05) in the fold change in phosphopeptide abundance for that time point in the SLP-76 wild-type reconstituted cells. In the second SILAC heatmap, SILAC ratios between Y3F mutant cells (J14–2D1) and SLP-76 reconstituted cells (J14–76-11) are represented for each phosphopeptide at each time point according to the SILAC heatmap color key. Black signifies no change. Red represents reduced phosphorylation in Y3F mutant cells, and green represents elevated phosphorylation in Y3F mutant cells. White dots on SILAC heatmap squares indicate a statistically significant difference (q value < 0.05) in the comparison between Y3F mutant and SLP-76 reconstituted cell SILAC ratios for that time point. Below each heatmap time point is a separate heatmap representing the coefficient of variation (cv) for that time point. According to the cv color key, black represents 0% cv, and more orange shading represents a greater cv. Blanks in the heatmaps indicate that a clearly defined SIC peak was not observed for that phosphopeptide at that time point.
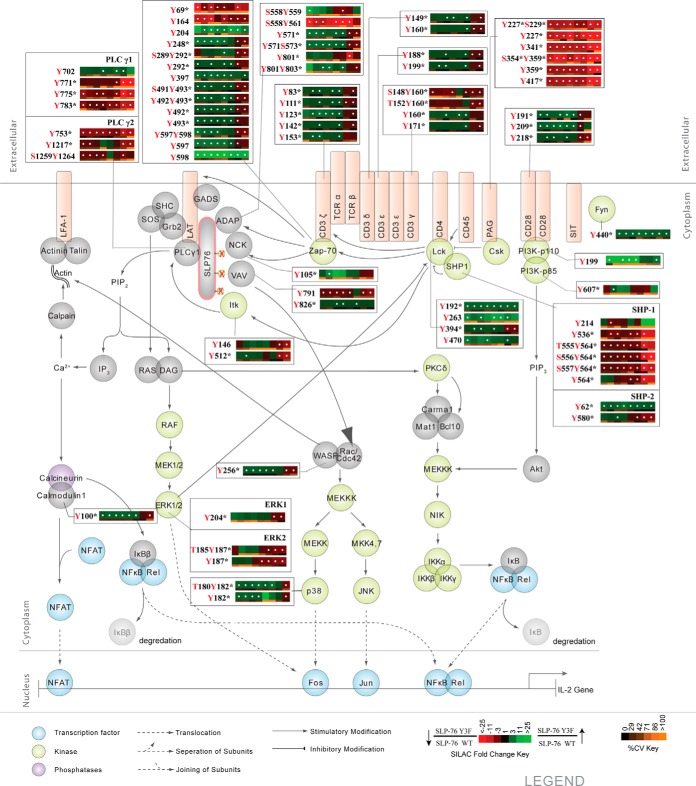
Fig. 2.
Effects of N-terminal tyrosine residues of SLP-76 on the canonical TCR signaling pathway. The canonical T-cell signaling pathway is represented with SILAC heatmap quantitation and the corresponding identified phosphorylation sites. Heatmaps were calculated from the average of five replicate experiments. White dots within a heatmap square indicate a statistically significant difference (Q value < 0.05) in the comparison between Y3F and SLP-76 reconstituted cells. SILAC heatmaps are described in detail in Fig. 1.
Comparison of Effect in TCR Signaling Pathway between SLP-76-deficient (J14) and SLP-76 Y3F Cells
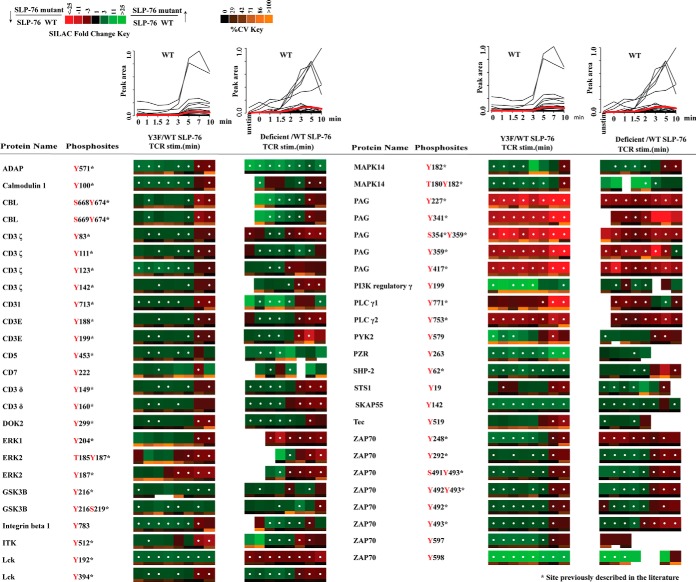
To better understand the role of the N-terminal tyrosine sites of SLP-76 in the TCR signaling pathway, we compared phosphorylation changes of other signaling molecules between cells lacking SLP-76 protein and SLP-76 Y3F cells. A quantitative phosphoproteomic study of SLP-76-deficient and SLP-76 WT cells was performed in our lab and published recently. In that study, 74 tyrosine phosphorylation sites on 35 proteins within the KEGG T-cell signaling pathway were identified with significant changes in relative abundance between SLP-76-deficient and SLP-76 WT cells (14). Among these sites, 49 tyrosine phosphorylation sites on 28 proteins were also identified with significant changes in relative abundance (Q value < 0.05) between SLP-76 Y3F and SLP-76 WT cells in the present study. SLP-76 WT (J14–76-11) cells were used as the control cell line in both studies. Therefore, site-by-site comparison of SILAC heatmaps comparing the phosphorylation changes between SLP-76 mutant cells and SLP-76 WT cells will aid exploration of the function of SLP-76, especially the typical N-terminal tyrosine domain. The phosphorylation changes of these 49 tyrosine phosphorylation sites between the two studies were compared (Fig. 3). On top of each heatmap, the abundance of phosphopeptides from J14–76-11 cells (SLP-76 WT reconstituted) during the time course of T-cell stimulation is compared. The plots show some delay in the initiation of TCR signaling in the SLP-76 Y3F study relative to the SLP-76-deficient study. Despite this slight experimental variation in the timing of receptor stimulation, the phosphorylation of the majority of signaling proteins showed the same phosphorylation change pattern when we compared the SLP-76-deficient and SLP-76 Y3F cells to the SLP-76 WT cells (Fig. 3). For example, a constitutive decrease in phosphorylation was observed on PLCγ1 Tyr771, PLCγ2 Tyr753 and PAG Tyr227, Tyr341, Ser354Tyr359, Tyr359, Tyr417. Also, a characteristic pattern with elevated phosphorylation at early time points and decreased phosphorylation at later time points was observed on Lck activation tyrosine residue Tyr394 and many Lck-regulated proteins (CD3ε, -δ, -γ, and -ζ chains; ZAP-70), as well as several other signaling proteins in both SLP-76 Y3F and SLP-76-deficient datasets. However, a divergent pattern was observed on Lck Tyr192 between these two studies.
Fig. 3.
Quantitative phosphoproteomic analysis of known TCR signaling proteins identified in both this Y3F experiment and the previous SLP-76-deficient datasets. SLP-76 reconstituted cells (WT) were used as a control in both studies. Line plots on top of the table represent the abundance of phosphopeptides observed from J14–76-11 (SLP-76 wild-type reconstituted cells) in either the Y3F study or the SLP-76-deficient cell study. Each black line represents one phosphopeptide, and the red line represents the average peak area of all the phosphopeptides in Fig. 3 at that time point. Heatmaps for Y3F experiments were calculated from the average of five replicate experiments, and heatmaps for J14 experiments were calculated from three replicate experiments. Green represents elevated phosphorylation in Y3F mutant cells or J14 cells relative to wild-type SLP-76 reconstituted cells, and red represents decreased phosphorylation. White dots within a heatmap square indicate a statistically significant difference (Q value < 0.05) in the comparison between Y3F or SLP-76-deficient (J14) and SLP-76 reconstituted cells. The SILAC heatmaps are described in detail in Fig. 1.
DISCUSSION
Tyrosine phosphorylation of three N-terminal tyrosine residues of SLP-76 is crucial for the function of SLP-76 (16). In this study, a quantitative mass-spectrometry-based phosphoproteomic strategy was utilized to study the systems-wide effects of phosphorylation of three N-terminal tyrosine residues of SLP-76 within TCR signaling.
Itk May Phosphorylate PLCγ1 and PLCγ2 at Novel Sites
Blocking the N-terminal phosphorylation of SLP-76 led to statistically significant decreases in the phosphorylation of Tyr771, Tyr775, and Tyr783 of PLCγ1 and Tyr753, Tyr1217, and Tyr1264 of PLCγ2 (Q value < 0.05) (supplemental Figs. S5B and S5C). It is well established that Itk mediates the phosphorylation of PLCγ1 at Tyr775 and Tyr783 (7), two phosphorylation sites that are required for the activation of PLCγ1 (41). The binding of the SH2–SH3 domain of Itk with SLP-76 maintains Itk in an active formation (11). SLP-76, particularly its N-terminal tyrosine residues, is required for the activation of Itk as well as the phosphorylation of both PLCγ1 activation sites (7). Thus, the constitutively decreased phosphorylation of Tyr775 and Tyr783 on PLCγ1 is consistent with previous studies. The similar SILAC pattern of constitutively decreased phosphorylation on Tyr771 suggests that this site might also be a substrate for Itk. PLCγ2 plays a crucial role in BCR-dependent calcium mobilization, and Tec-family kinases were demonstrated to phosphorylate PLCγ2 at Tyr753 and Tyr759, leading to PLCγ2-mediated calcium signaling (42). However, the study of the role of PLCγ2 in T cells is more limited. The constitutively decreased phosphorylation of PLCγ2 in Y3F cells suggests that Itk in T cells could also regulate PLCγ2 phosphorylation at Tyr753, Tyr1217, and Tyr1264.
Impaired activation of PLCγ1 would lead to reduced production of the second messenger molecules inositol 1,4,5-trisphosphate and diacylglcycerol. Diacylglcycerol serves as a direct activator for Ras and PKC, leading to the activation of the Ras–Erk–AP1 and NF-κB pathways, respectively (43). Erk2 activation was previously observed to be partially reduced in J14 (SLP-76 deficient cells) (6). Decreased phosphorylation of Tyr204 of Erk1 and Tyr187 of Erk2 was observed in SLP-76 Y3F cells relative to SLP-76 WT cells, as expected.
SLP-76 Y3F Is Sufficient to Replicate the Majority of Phosphoproteome Changes Observed in SLP-76-deficient Cells
With the advancement of instruments and methods, the number of tyrosine sites identified with statistically significant phosphorylation changes was improved almost 2-fold in this SLP-76 Y3F study relative to the previous SLP-76-deficient study. Nevertheless, among 47 tyrosine sites identified in both studies, the majority of them, especially 25 important tyrosine sites in the TCR signaling pathway, showed the same phosphorylation change pattern (Fig. 3). For example, decreased phosphorylation among the entire time course was observed in downstream tyrosine sites such as PLCγ1/2 and Erk 1/2 in both studies.
Constitutively decreased phosphorylation on Tyr227, Tyr341, Tyr359, and Tyr417 of PAG was observed in the SLP-76 Y3F study (supplemental Fig. S5A) and the previous SLP-76-deficient study. PAG is known to negatively regulate TCR signaling via recruitment of C-terminal Src kinase (Csk). Active Csk phosphorylates the C-terminal inhibitory tyrosine residues of Src-family kinases such as Lck and Fyn, down-regulating their kinase activities (15). In fact, the PAG–Csk complex represents a critical component of the negative homeostatic regulatory feedback mechanism restraining Lck activity (15). However, the regulation of phosphorylation of PAG is not well understood. Previous studies indicated that Fyn, but not Lck, is predominantly responsible for the phosphorylation of PAG in resting peripheral T cells (44). Interestingly, although one study suggested that CD45 phosphatase could act on Tyr371 of PAG (45), another failed to confirm this finding (15). Our observation suggested that phosphorylation of the three N-terminal tyrosine residues of SLP-76 is required for the phosphorylation of PAG. One possible explanation for this observation is that phosphorylation of N-terminal tyrosine resides of SLP-76 is important in mediating the recruitment of Fyn to PAG. Another possibility is that phosphorylation of SLP-76 is required for the activity of Fyn or CD45.
Elevated phosphorylation at early time points and decreased phosphorylation at later time points was observed in the activation loop of Lck at Tyr394 in the present SLP-76 Y3F study (supplemental Fig. S6A) and the previous SLP-76-deficient study. These findings reveal a previously undescribed regulatory network in which N-terminal tyrosine residues on SLP-76 are involved in both positive and negative feedback pathways that regulate the phosphorylation of the activation loop of Tyr394 on Lck (14). Constitutively reduced phosphorylation on a variety of tyrosine residues on PAG in mutant cells would be expected to lead to a reduction in recruitment of the negative feedback regulator Csk, resulting in constitutively increased phosphorylation of Lck within its activation loop. Phosphorylation of Erk was increased with 5 to 10 min of TCR stimulation in cells reconstituted with WT SLP-76 (Fig. 1, label-free heatmap). We recently showed in Jurkat T cells (46), as others have shown in primary T cells (47), that inhibition of Erk activation leads to inhibition of positive feedback pathways through decreased phosphorylation of Ser59 on Lck (48, 49). Our data support the hypothesis that N-terminal tyrosine residues of SLP-76 regulate competing negative feedback through Csk and positive feedback through Erk. According to this hypothesis, at early time points, Y3F SLP-76 mutant inhibition of PAG negative feedback outcompetes inhibition of Erk positive feedback because Erk is not robustly phosphorylated and activated. But at later time points when the phosphorylation and activation of Erk are much higher, inhibition of Erk positive feedback overcomes the inhibition of Csk negative feedback, leading to decreased phosphorylation at Lck Tyr394 in the SLP-76 Y3F mutant.
Elevated phosphorylation at early time points and decreased phosphorylation in later time points was also observed on CD3ε, -δ, -γ, and -ζ chains (supplemental Fig. S7) and ZAP-70 (supplemental Fig. S6B) in SLP-76 Y3F mutant cells. Phosphorylation of ITAM domains on the ε, δ, γ, and ζ CD3 subunits by Lck is key to the initiation of signaling cascades that characterize T-cell activation (50–55). In Y3F mutant cells, elevated phosphorylation at early time points and decreased phosphorylation at later time points were observed on Tyr149 and Tyr160 of the CD3δ chain (Q value < 0.05). A similar trend of phosphorylation change was also observed on CD3ε; Tyr188 and Tyr199, two sites known to be regulated by Lck (56); and Tyr83, Tyr111, Tyr123, Tyr142 of the CD3ζ chain. Besides the TCR ITAMs, a similar phosphorylation change pattern was also observed on Zap-70 Tyr292, Tyr492, and Tyr493. It is well established that Lck mediates the phosphorylation of ZAP-70 at Tyr493 to increase kinase activity of ZAP-70 in stimulated T cells (57). Autophosphorylation of both Tyr292 and Tyr492 on ZAP-70 is dependent on the initial phosphorylation of Tyr493 (58).
Early increased and late decreased phosphorylation in the SLP-76 Y3F mutant was also observed on Tyr512 of Itk (supplemental Fig. S6C). Itk is recruited to the cell membrane through the interaction of its pleckstrin homology domain with the membrane phosphatidylinositol 3,4,5-trisphosphate, where it becomes phosphorylated at Tyr512 within its activation loop by Lck (59, 60). The TCR-induced global tyrosine phosphorylation of ITK was decreased in SLP-76-deficient T cells over the entire time course by immunoprecipitation of Itk from cell lysates followed by probe with pan-specific anti-phosphotyrosine (7). Nevertheless, this Western blot result did not measure phosphorylation at the specific site Tyr512 of Itk, as the blot experiment was not site specific. In addition, our observation is consistent with the hypothesis of N-terminal tyrosine SLP-76-mediated competing positive and negative feedback pathways, as Itk is a known substrate of Lck (60). Taken together, these data suggest that SLP-76, especially its N-terminal tyrosine sites, regulates competing positive and negative feedback pathways that regulate not only Lck, but also its substrates (TCR ITAMs, ZAP-70, and Itk).
Proline-rich Domain of SLP-76 Plays a Key Role in Phosphorylation of Lck Tyr192
Although the majority of sites responded similarly to the mutation of N-terminal tyrosine residues and to the complete removal of SLP-76, Lck Tyr192 responded differently. Constitutively increased phosphorylation was observed in SLP-76 Y3F cells (supplemental Fig. S6A), compared with constitutively decreased phosphorylation in SLP-76-deficient cells. The difference in Lck Tyr192 phosphorylation observed between mutant Y3F SLP-76 and SLP-76-deficient cells suggests that a domain of SLP-76 outside of the N-terminal tyrosine domain is regulating interaction between Lck and SLP-76. A previous study suggested that the interaction between Lck and SLP-76 is regulated by the interaction between the Lck SH3 domain and a proline-rich domain of SLP-76 outside of the N-terminal tyrosine domain (61). Our data, in combination with these previous data, support a new hypothesis that the regulation of Lck Tyr192 but not Tyr394 phosphorylation is independent of N-terminal tyrosine phosphorylation of SLP-76 and may be regulated by the interaction between Lck and the SLP-76 proline-rich domain.
Supplementary Material
Acknowledgments
We thank Dr. Deborah Yablonski at the Technician-Israel Institute of Technology for generously providing us with the Jurkat clones J14–2D1 (Y3F mutant cells) and J14-76-11 (SLP-76 reconstituted cells), and for the insightful discussions.
Footnotes
Author contributions: Q.J. and A.R.S. designed research; Q.J. and Y.D. performed research; Q.J. analyzed data; Q.J. and A.R.S. wrote the paper.
* This research is based in part upon work conducted using the Rhode Island NSF/EPSCoR Proteomics Share Resource Facility, which is supported in part by National Science Foundation EPSCoR Grant No. 1004057, National Institutes of Health Grant No. 1S10RR020923, a Rhode Island Science and Technology Advisory Council grant, and the Division of Biology and Medicine, Brown University. We acknowledge financial support from National Institutes of Health Grant No. R01AI083636.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TCR
- T-cell receptor
- Csk
- C-terminal Src kinase
- Erk
- extracellular signal-regulated kinase
- FDR
- false discovery rate
- Fyn
- proto-oncogene tyrosine-protein kinase Fyn
- ITAM
- immunoreceptor tyrosine-based activation motif
- Itk
- interleukin 2–inducible T-cell kinase
- Lck
- lymphocyte-specific protein tyrosine kinase
- PAG
- phosphoprotein-associated glycolipid-enriched membrane protein
- PLCγ
- phospholipase C γ
- SIC
- select ion chromatogram
- SILAC
- stable isotope labeling of amino acids in cell culture
- SLP-76
- SH2 domain-containing leukocyte protein of 76 kDa
- WT
- wild type
- ZAP-70
- zeta-chain-associated protein kinase 70.
REFERENCES
- 1. Zamoyska R., Basson A., Filby A., Legname G., Lovatt M., Seddon B. (2003) The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol. Rev. 191, 107–118 [DOI] [PubMed] [Google Scholar]
- 2. Zhang W., Sloan-Lancaster J., Kitchen J., Trible R. P., Samelson L. E. (1998) LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell 92, 83–92 [DOI] [PubMed] [Google Scholar]
- 3. Bubeck Wardenburg J., Fu C., Jackman J. K., Flotow H., Wilkinson S. E., Williams D. H., Johnson R., Kong G., Chan A. C., Findell P. R. (1996) Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem. 271, 19641–19644 [DOI] [PubMed] [Google Scholar]
- 4. Pivniouk V., Tsitsikov E., Swinton P., Rathbun G., Alt F. W., Geha R. S. (1998) Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell 94, 229–238 [DOI] [PubMed] [Google Scholar]
- 5. Clements J. L., Yang B., Ross-Barta S. E., Eliason S. L., Hrstka R. F., Williamson R. A., Koretzky G. A. (1998) Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science 281, 416–419 [DOI] [PubMed] [Google Scholar]
- 6. Yablonski D., Kuhne M. R., Kadlecek T., Weiss A. (1998) Uncoupling of nonreceptor tyrosine kinases from PLC-gamma 1 in an SLP-76-deficient T cell. Science 281, 413–416 [DOI] [PubMed] [Google Scholar]
- 7. Bogin Y., Ainey C., Beach D., Yablonski D. (2007) SLP-76 mediates and maintains activation of the Tec family kinase ITK via the T cell antigen receptor-induced association between SLP-76 and ITK. Proc. Natl. Acad. Sci. U.S.A. 104, 6638–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singer A. L., Bunnell S. C., Obstfeld A. E., Jordan M. S., Wu J. N., Myung P. S., Samelson L. E., Koretzky G. A. (2004) Roles of the proline-rich domain in SLP-76 subcellular localization and T cell function. J. Biol. Chem. 279, 15481–15490 [DOI] [PubMed] [Google Scholar]
- 9. Yablonski D., Weiss A. (2001) Mechanisms of signaling by the hematopoietic-specific adaptor proteins, SLP-76 and LAT and their B cell counterpart, BLNK/SLP-65. Adv. Immunol. 79, 93–128 [DOI] [PubMed] [Google Scholar]
- 10. Su Y. W., Zhang Y., Schweikert J., Koretzky G. A., Reth M., Wienands J. (1999) Interaction of SLP adaptors with the SH2 domain of Tec family kinases. Eur. J. Immunol. 29, 3702–3711 [DOI] [PubMed] [Google Scholar]
- 11. Qi Q., August A. (2007) Keeping the (kinase) party going: SLP-76 and ITK dance to the beat. Sci. STKE 2007, pe39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bubeck Wardenburg J., Pappu R., Bu J. Y., Mayer B., Chernoff J., Straus D., Chan A. C. (1998) Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity 9, 607–616 [DOI] [PubMed] [Google Scholar]
- 13. Musci M. A., Hendricks-Taylor L. R., Motto D. G., Paskind M., Kamens J., Turck C. W., Koretzky G. A. (1997) Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J. Biol. Chem. 272, 11674–11677 [DOI] [PubMed] [Google Scholar]
- 14. Cao L., Ding Y., Hung N., Yu K., Ritz A., Raphael B. J., Salomon A. R. (2012) Quantitative phosphoproteomics reveals SLP-76 dependent regulation of PAG and Src family kinases in T cells. PLoS One 7, e46725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brdicka T., Pavlistova D., Leo A., Bruyns E., Korinek V., Angelisova P., Scherer J., Shevchenko A., Hilgert I., Cerny J., Drbal K., Kuramitsu Y., Kornacker B., Horejsi V., Schraven B. (2000) Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J. Exp. Med. 191, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang N., Motto D. G., Ross S. E., Koretzky G. A. (1996) Tyrosines 113, 128, and 145 of SLP-76 are required for optimal augmentation of NFAT promoter activity. J. Immunol. 157, 3769–3773 [PubMed] [Google Scholar]
- 17. Raab M., daSilva A. J., Findell P. R., Rudd C. E. (1997) Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR zeta/CD3 induction of interleukin-2. Immunity 6, 155–164 [DOI] [PubMed] [Google Scholar]
- 18. Onodera H., Motto D. G., Koretzky G. A., Rothstein D. M. (1996) Differential regulation of activation-induced tyrosine phosphorylation and recruitment of SLP-76 to Vav by distinct isoforms of the CD45 protein-tyrosine phosphatase. J. Biol. Chem. 271, 22225–22230 [DOI] [PubMed] [Google Scholar]
- 19. Tuosto L., Michel F., Acuto O. (1996) p95vav associates with tyrosine-phosphorylated SLP-76 in antigen-stimulated T cells. J. Exp. Med. 184, 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu J., Motto D. G., Koretzky G. A., Weiss A. (1996) Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity 4, 593–602 [DOI] [PubMed] [Google Scholar]
- 21. Wunderlich L., Farago A., Downward J., Buday L. (1999) Association of Nck with tyrosine-phosphorylated SLP-76 in activated T lymphocytes. Eur. J. Immunol. 29, 1068–1075 [DOI] [PubMed] [Google Scholar]
- 22. Shim E. K., Moon C. S., Lee G. Y., Ha Y. J., Chae S. K., Lee J. R. (2004) Association of the Src homology 2 domain-containing leukocyte phosphoprotein of 76 kD (SLP-76) with the p85 subunit of phosphoinositide 3-kinase. FEBS Lett. 575, 35–40 [DOI] [PubMed] [Google Scholar]
- 23. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L. F., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2 p. ra46. [DOI] [PubMed] [Google Scholar]
- 24. Matsumoto M., Oyamada K., Takahashi H., Sato T., Hatakeyama S., Nakayama K. I. (2009) Large-scale proteomic analysis of tyrosine-phosphorylation induced by T-cell receptor or B-cell receptor activation reveals new signaling pathways. Proteomics 9, 3549–3563 [DOI] [PubMed] [Google Scholar]
- 25. Nguyen V., Cao L. L., Lin J. T., Hung N., Ritz A., Yu K. B., Jianu R., Ulin S. P., Raphael B. J., Laidlaw D. H., Brossay L., Salomon A. R. (2009) A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell. Proteomics 8, 2418–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. E., White F. M. (2006) Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J. Immunol. 176, 2833–2843 [DOI] [PubMed] [Google Scholar]
- 27. Navarro M. N., Goebel J., Feijoo-Carnero C., Morrice N., Cantrell D. A. (2011) Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat. Immunol. 12, 352–U111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salomon A. R., Ficarro S. B., Brill L. M., Brinker A. (2003) Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 100, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 30. Yu K. B., Salomon A. R. (2009) PeptideDepot: flexible relational database for visual analysis of quantitative proteomic data and integration of existing protein information. Proteomics 9, 5350–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu K. B., Salomon A. R. (2010) HTAPP: high-throughput autonomous proteomic pipeline. Proteomics 10, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ficarro S. B., Salomon A. R., Brill L. M., Mason D. E., Stettler-Gill M., Brock A., Peters E. C. (2005) Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 19, 57–71 [DOI] [PubMed] [Google Scholar]
- 33. Perkins D. N., Pappin D. J. C., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 34. Yu K. B., Sabelli A., DeKeukelaere L., Park R., Sindi S., Gatsonis C. A., Salomon A. (2009) Integrated platform for manual and high-throughput statistical validation of tandem mass spectra. Proteomics 9, 3115–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 36. Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 37. Demirkan G., Yu K., Boylan J. M., Salomon A. R., Gruppuso P. A. (2011) Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS One 6, e21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Storey J. D. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 [Google Scholar]
- 39. Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Serrano C. J., Graham L., DeBell K., Rawat R., Veri M. C., Bonvini E., Rellahan B. L., Reischl I. G. (2005) A new tyrosine phosphorylation site in PLC gamma 1: the role of tyrosine 775 in immune receptor signaling. J. Immunol. 174, 6233–6237 [DOI] [PubMed] [Google Scholar]
- 42. Humphries L. A., Dangelmaier C., Sommer K., Kipp K., Kato R. M., Griffith N., Bakman I., Turk C. W., Daniel J. L., Rawlings D. J. (2004) Tec kinases mediate sustained calcium influx via site-specific tyrosine phosphorylation of the phospholipase C gamma Src homology 2-Src homology 3 linker. J. Biol. Chem. 279, 37651–37661 [DOI] [PubMed] [Google Scholar]
- 43. Hickman S. P., Yang J., Thomas R. M., Wells A. D., Turka L. A. (2006) Defective activation of protein kinase C and Ras-ERK pathways limits IL-2 production and proliferation by CD4(+)CD25(+) regulatory T cells. J. Immunol. 177, 2186–2194 [DOI] [PubMed] [Google Scholar]
- 44. Yasuda K., Nagafuku M., Shima T., Okada M., Yagi T., Yamada T., Minaki Y., Kato A., Tani-Ichi S., Hamaoka T., Kosugi A. (2002) Cutting edge: Fyn is essential for tyrosine phosphorylation of Csk-binding protein/phosphoprotein associated with glycolipid-enriched microdomains in lipid rafts in resting T cells. J. Immunol. 169, 2813–2817 [DOI] [PubMed] [Google Scholar]
- 45. Davidson D., Bakinowski M., Thomas M. L., Horejsi V., Veillette A. (2003) Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell. Biol. 23, 2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Helou Y. A., Nguyen V., Beik S. P., Salomon A. R. (2013) ERK positive feedback regulates a widespread network of tyrosine phosphorylation sites across canonical T cell signaling and actin cytoskeletal proteins in Jurkat T cells. PLoS One 8, e69641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stefanova I., Hemmer B., Vergelli M., Martin R., Biddison W. E., Germain R. N. (2003) TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 4, 248–254 [DOI] [PubMed] [Google Scholar]
- 48. Winkler D. G., Park I., Kim T., Payne N. S., Walsh C. T., Strominger J. L., Shin J. (1993) Phosphorylation of Ser-42 and Ser-59 in the N-terminal region of the tyrosine kinase p56lck. Proc. Natl. Acad. Sci. U.S.A. 90, 5176–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joung I., Kim T., Stolz L. A., Payne G., Winkler D. G., Walsh C. T., Strominger J. L., Shin J. (1995) Modification of Ser59 in the unique N-terminal region of tyrosine kinase p56lck regulates specificity of its Src homology 2 domain. Proc. Natl. Acad. Sci. U.S.A. 92, 5778–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nika K., Tautz L., Arimura Y., Vang T., Williams S., Mustelin T. (2007) A weak Lck tail bite is necessary for Lck function in T cell antigen receptor signaling. J. Biol. Chem. 282, 36000–36009 [DOI] [PubMed] [Google Scholar]
- 51. Mustelin T., Abraham R. T., Rudd C. E., Alonso A., Merlo J. J. (2002) Protein tyrosine phosphorylation in T cell signaling. Front. Biosci. 7, d918–d969 [DOI] [PubMed] [Google Scholar]
- 52. Straus D. B., Weiss A. (1992) Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell 70, 585–593 [DOI] [PubMed] [Google Scholar]
- 53. Samelson L. E., Patel M. D., Weissman A. M., Harford J. B., Klausner R. D. (1986) Antigen activation of murine T-cells induces tyrosine phosphorylation of a polypeptide associated with the T-cell antigen receptor. Cell 46, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 54. Baniyash M., Garcia-Morales P., Luong E., Samelson L. E., Klausner R. D. (1988) The T cell antigen receptor zeta chain is tyrosine phosphorylated upon activation. J. Biol. Chem. 263, 18225–18230 [PubMed] [Google Scholar]
- 55. Mustelin T., Altman A. (1989) Do Cd4 and Cd8 control T-cell activation via a specific tyrosine protein-kinase. Immunol. Today 10, 189–192 [DOI] [PubMed] [Google Scholar]
- 56. Guirado M., de Aos I., Orta T., Rivas L., Terhorst C., Zubiaur M., Sancho J. (2002) Phosphorylation of the N-terminal and C-terminal CD3-epsilon-ITAM tyrosines is differentially regulated in T cells. Biochem. Biophys. Res. Commun. 291, 574–581 [DOI] [PubMed] [Google Scholar]
- 57. Chan A. C., Dalton M., Johnson R., Kong G. H., Wang T., Thoma R., Kurosaki T. (1995) Activation of Zap-70 kinase-activity by phosphorylation of tyrosine-493 is required for lymphocyte antigen receptor function. EMBO J. 14, 2499–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong G. H., Dalton M., Wardenburg J. B., Straus D., Kurosaki T., Chan A. C. (1996) Distinct tyrosine phosphorylation sites in ZAP-70 mediate activation and negative regulation of antigen receptor function. Mol. Cell. Biol. 16, 5026–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. August A., Sadra A., Dupont B., Hanafusa H. (1997) Src-induced activation of inducible T cell kinase (ITK) requires phosphatidylinositol 3-kinase activity and the pleckstrin homology domain of inducible T cell kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 11227–11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heyeck S. D., Wilcox H. M., Bunnell S. C., Berg L. J. (1997) Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. J. Biol. Chem. 272, 25401–25408 [DOI] [PubMed] [Google Scholar]
- 61. Sanzenbacher R., Kabelitz D., Janssen O. (1999) SLP-76 binding to p56lck: a role for SLP-76 in CD4-induced desensitization of the TCR/CD3 signaling complex. J. Immunol. 163, 3143–3152 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.