Abstract
O2 sensing in diverse protozoa depends on the prolyl 4 hydroxylation of Skp1 and modification of the resulting hydroxyproline with a series of five sugars. In yeast, plants, and animals, Skp1 is associated with F-box proteins. The Skp1–F-box protein heterodimer can, for many F-box proteins, dock onto cullin-1 en route to assembly of the Skp1–cullin-1–F-box protein–Rbx1 subcomplex of E3SCFUb ligases. E3SCFUb ligases conjugate Lys48-polyubiquitin chains onto targets bound to the substrate receptor domains of F-box proteins, preparing them for recognition by the 26S proteasome. In the social amoeba Dictyostelium, we found that O2 availability was rate-limiting for the hydroxylation of newly synthesized Skp1. To investigate the effect of reduced hydroxylation, we analyzed knockout mutants of the Skp1 prolyl hydroxylase and each of the Skp1 glycosyltransferases. Proteomic analysis of co-immunoprecipitates showed that wild-type cells able to fully glycosylate Skp1 had a greater abundance of an SCF complex containing the cullin-1 homolog CulE and FbxD, a newly described WD40-type F-box protein, than the complexes that predominate in cells defective in Skp1 hydroxylation or glycosylation. Similarly, the previously described FbxA–Skp1CulA complex was also more abundant in glycosylation-competent cells. The CulE interactome also included higher levels of proteasomal regulatory particles when Skp1 was glycosylated, suggesting increased activity consistent with greater association with F-box proteins. Finally, the interactome of FLAG-FbxD was modified when it harbored an F-box mutation that compromised Skp1 binding, consistent with an effect on the abundance of potential substrate proteins. We propose that O2-dependent posttranslational glycosylation of Skp1 promotes association with F-box proteins and their engagement in functional E3SCFUb ligases that regulate O2-dependent developmental progression.
Timely protein degradation is a cornerstone of cell cycling and the regulation of numerous physiological and developmental processes. Eukaryotes have evolved an extensive array of polyubiquitination enzymes to tag proteins on a protein-by-protein basis as a recognition marker for degradation in the 26S proteasome. The cullin-RING ubiquitin ligases (CRLs)1 are a prominent subgroup of these enzymes (1) and consist of an E3 architecture that includes a substrate receptor, an adaptor (in most cases), the cullin scaffold, the RING protein, and an exchangeable E2 ubiquitin donor that has been charged with ubiquitin (Ub) by an E1 enzyme. The first discovered and still prototypic example is the CRL1 class (2), also referred to as SCF on account of the names of its founding subunits, Skp1, cullin-1, and F-box proteins (FBPs). The CRL1 (or SCF) complexes utilize FBPs as substrate receptors, Skp1 as the adaptor linking the FBP to the N-terminal region of cullin-1 (Cul1), and Rbx1 as the RING protein that tethers the E2 Ub donor to the Cul1 C-terminal region (see Fig. 2B). CRL1s can be activated by neddylation of Cul1 by a Nedd8-specific E2, which mobilizes Rbx1 to afford rotational flexibility of the E2 and displaces the inhibitor Cand1, permitting docking of the Skp1–FBP heterodimer (3–5). Deneddylation mediated by the eight-subunit COP9 signalosome is required for in vivo activity, suggesting that Cand1 serves as a substrate exchange factor to allow for re-equilibration of SCF complexes from preexisting subunits. Each reaction cycle requires the exchange of a new E2-Ub and typically assembles a K48-linked polyUb chain that is recognized by the proteasome. Substrate specificity is conferred by FBPs, a gene family that numbers 69 in humans, 20 in budding yeast, 300 in Caenorhabditis elegans, and ∼800 in Arabidopsis. Some characterized FBPs can recognize perhaps a dozen or more substrates, and the coding of recognition and the meaning of their control by the same FBP is under intense investigation (6). Recognition is often activated by posttranslational modification of the substrate (often phosphorylation). Regulation of SCF Ub ligases has centered on the neddylation cycle, which potentially influences all seven known CRLs. Regulation of Skp1, investigated in this paper, would be specific to CRLs possessing Skp1, which include CRL1 and possibly the minor class CRL7 (7).
Fig. 2.
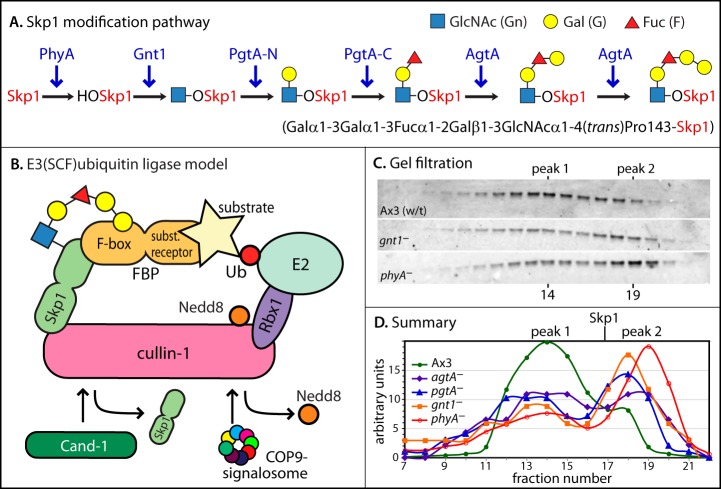
Skp1 modification pathway and global analysis of Skp1 interactions. A, Skp1 is sequentially modified by the indicated enzymes (in blue), resulting in the formation of a pentasaccharide at Pro143. B, model of the SCF complex in the context of the overall E3 Ub ligase, from studies in yeast, plants, and animals. Catalysis involves transfer of Ub from an exchangeable Ub-E2 conjugate to the substrate. Removal of Nedd8 by the COP9 signalosome facilitates binding of Cand1 to Cul1, which inhibits binding of Skp1 to Cul1. C, D, vegetative (growth stage) cells were filter-lysed, and a cytosolic fraction prepared via ultracentrifugation was chromatographed on a Superose 12 gel filtration column. Fractions were analyzed via Western blotting (representative examples are shown in C) followed by densitometry (D). The elution position of free Skp1 from a separate trial is indicated.
The basic SCF model is thought to be widespread among eukaryotes but has been extensively studied only in fungi/yeasts, plants, and animals. The broad phylogeny represented by protists includes many benign and pathogenic unicellular organisms of great economic, health, and environmental impact. Emerging evidence reveals that Skp1 in some of these groups is subject to a novel form of prolyl 4(trans)-hydroxylation and complex glycosylation (8). The roles of these Skp1 modifications have been most studied in the social amoeba Dictyostelium, which undergoes a starvation-induced developmental program during which individual amoebae chemotactically aggregate into an initial mound that then elongates into a migratory slug. Under appropriate conditions, the slug reorganizes to form a fruiting body consisting of a ball of spores supported by a vertical cellular stalk. The slug-to-fruit switch, referred to as culmination, and sporulation are regulated by checkpoints that are sensitive to multiple factors, including O2 (9–11). Functional studies of Dictyostelium Skp1 hydroxylation and glycosylation reveal roles in regulating the O2 dependence of culmination and sporulation (12–14). For example, wild-type (wt) cells require 7% to 10% O2 and phyA– requires 18% to 21% O2 in order to achieve 50% spore formation (a quantitative measure of fruiting body formation), whereas glycosylation mutants exhibit a complex pattern of intermediate requirements (13). In addition, at 21% O2, phyA– cells require an additional 3 to 4 h to complete development relative to their wt counterparts (14). In the apicomplexan Toxoplasma gondii, PhyA is also required for Skp1 glycosylation, and phyA– parasites are deficient in proliferation, especially at low O2 (15).
The idea that O2 availability is rate limiting for Skp1 modification was originally based on the observation that the Dictyostelium phyA– phenotype mimics that of wt cells in low O2 (9). However, the majority of Skp1 is hydroxylated and glycosylated in wt cells even at low O2 levels where culmination is blocked or delayed. Further analysis of a submerged development model, in which terminal development depended on an atmosphere of 70% to 100% O2 in order to overcome the diffusion barrier posed by the water layer, showed that at atmospheric O2 levels of 5% to 21% where sporulation was blocked, unmodified Skp1 accumulated to a higher level than at permissive O2 levels (10). As Skp1 modifications are thought to be irreversible, this likely resulted from slow hydroxylation of newly synthesized Skp1. To address this in a more physiological setting, we investigated nascent Skp1 directly using metabolic labeling with [35S]Met/Cys and verified that the rate of hydroxylation of newly synthesized Skp1 polypeptide was indeed inversely proportional to O2 levels, which makes PhyA-mediated hydroxylation of Skp1 an excellent candidate for the primary O2 sensor for culmination.
These modifications of Skp1 are of interest as a novel mechanism regulating the SCF ligase. Previously, we showed that hydroxylation and glycosylation of Dictyostelium Skp1 affect its conformation and promote binding to a soluble FBP, guinea pig Fbs1, in studies of purified proteins (16). Here we show that Dictyostelium Skp1 is indeed a subunit of a canonical SCF complex, as expected. The significance of undermodified Skp1 was examined via interactome analysis of Skp1 isoforms that accumulate in modification pathway mutants. Our findings revealed a lower abundance of SCF complexes than in wt cells, suggesting that Skp1 modification may promote SCF assembly and E3SCFUb ligase activities that control timely turnover of select proteins involved in developmental progression.
EXPERIMENTAL PROCEDURES
Metabolic Labeling and Isolation of Skp1
Cells (gnt1–) were grown axenically in HL-5 medium supplemented with 50 μg/ml streptomycin and 50 μg/ml ampicillin until they achieved a density of 2 × 106 cells per milliliter. Cells were recovered via centrifugation at 1000 × g for 1 min in a 15-ml conical polypropylene tube and resuspended at 107 cells per milliliter in 1 ml of fresh HL-5 pre-equilibrated overnight under an atmosphere of the indicated O2, with the balance made up of N2. Samples were shaken for 2.5 h under a flowing atmosphere of the same O2 percentage, at which time 0.71 μCi of [35S]Met/Cys (Tran35S-Label, >1000 Ci/mmol; MP Biomedicals, Santa Ana, CA) was introduced. Shaking incubation was continued until cells were recovered via centrifugation, resuspended in degassed PDF buffer (33.3 mm NaH2PO4, 10.6 mm Na2HPO4, 20.1 mm KCl, 5.8 mm MgSO4, pH 5.8) supplemented with 2 mm Na-dithionite, transferred to a 1.5-ml microcentrifuge tube, and centrifuged again. The cell pellet was snap frozen on dry ice and stored at −80 °C.
Cell pellets were resuspended in 110 μl of 8 m urea, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl with protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin); incubated on ice for 15 min and then at 55 °C for 5 min; and clarified via centrifugation at 16,000 × g for 30 min at 4 °C. 50 μl was transferred to a 1.5-ml microfuge tube containing 450 μl of RIPA buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (w/v) Na-deoxycholate, 0.1% (w/v) SDS, with protease inhibitors as above). Samples were pre-cleared by the addition of 10 μl of 1 mg/ml normal mouse IgG or 10 μl of preimmune serum (UOK85 or UOK77) and incubated for 1 h on ice, after which we added 50 μl of a 25% slurry of pre-washed (in RIPA buffer) Protein A beads (UltraLink resin; Pierce) or 25 μl of Protein-G beads (UltraLink resin; Pierce) and rotated the samples at 4 °C for 1 h. Beads were sedimented for 10 s in a personal microcentrifuge, and the supernatant was transferred to a fresh tube. Total Skp1 was immunoprecipitated by incubation with 5 μg of mAb 4E1 followed by incubation with Protein G resin and centrifugation as described above. HO-Skp1 was similarly isolated via incubation with 5 μg of affinity-purified UOK85 rabbit IgG followed by incubation with Protein A beads and centrifugation as described above. Total Skp1 was alternatively isolated via the addition of 10 μl of beads covalently modified with 2 mg/ml of affinity-purified UOK77 anti-Skp1 rabbit IgG (described below), incubation at 4 °C for 2 h, and centrifugation. Affinity purification involved adsorption to Skp1 (UOK77) or HO-Skp1 (UOK85) covalently coupled to CNBr-activated Sepharose CL-4B, elution with 0.1 m glycine-HCl (pH 2.8), and immediate neutralization. Beads were washed in two cycles of resuspension in 1 ml of RIPA buffer, centrifugation, transfer of the beads to a fresh tube, and a final RIPA buffer wash, after which the samples were stored at −80 °C. The first supernatant was saved at −80 °C for assay of the total incorporation and protein content. Antibody and bead amounts and incubation times were the minimum required in order to recover maximal Skp1 (typically >75%).
Skp1 was released from the beads via the addition of 50 μl of 1× Laemmli sample buffer and boiling for 3 min, after which it was applied to a 7–20% SDS-PAGE gel. The gel was stained in Coomassie Blue, destained, incubated in H2O for 15 min and then 1 m Na salicylate for 60 min, and dried against a sheet of 3MM Whatman filter paper. The gel was autoradiographed against BioMax MR film (Kodak, New Haven, CT), using a Fisher Biotech l-Plus intensifying screen, at −80 °C. The film image was digitized and analyzed densitometrically using ImageJ (version 1.47, National Institutes of Health, Bethesda, MD).
Total protein was determined using a BCA assay (Pierce), and total 35S incorporation was determined via precipitation with trichloroacetic acid, recovery on a GF/C filter, transfer to Bio-Safe NA scintillation fluid, and quantitation using a Beckman scintillation counter.
Gel Filtration of Cytosolic Extracts
Vegetative (growth-stage) cells were harvested as described above; resuspended in 50 mm Tris-HCl (pH 7.4), 0.25 m sucrose, and protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin, added just before use); and lysed by means of forced filtration through a 5-μm pore diameter filter as described previously (17). A cytosolic extract was prepared via ultracentrifugation (100,000 × g for 60 min) at 4 °C and chromatographed on a Superose 12 gel filtration column at 22 °C pre-equilibrated in 50 mm Tris-HCl (pH 7.4), 50 mm NaCl, 1 mm DTT, and 10% glycerol. Fractions were analyzed via SDS-PAGE and Western blotting for Skp1 using mAb 4E1, and they were quantitated densitometrically as previously described (13, 16). phyA– cells were strain HW288, gnt1– cells were HW503, pgtA– cells were HW260, and agtA– cells were HW420.
CulE Tagging
The culE 5′ targeting sequence was amplified from Dictyostelium discoideum genomic DNA (Ax3 strain) via polymerase chain reaction (PCR) using primers 5′-AAGCGCGCCTGCAAGTGAGGATGCTGAAACTCTC and 5′-AGATCTGCTGCCGCTGCCAGCCATATATTCATATTTTCTTGC and ligated into pCR4-TOPO (Invitrogen). The culE 3′ targeting sequence was amplified and ligated in a similar manner using 5′-AAGCTTTGTGGTCTGCAGTGCAATCATTCAACCAG and 5′-CAGCTGCACCATGGCAATGGGTTAAC. The culE 5′-targeting sequence was excised via digestion with BssHII and BglII (italicized) and ligated into similarly digested pVS3 vector to yield pVSCulE5′. The pVS3 vector series (pVS3, pVS3C, pVS3E) was derived from pVS (18), pVSC (9), and pVSE (9), which differ in their discoidin 1γ, cotB, and ecmA promoters, respectively, through replacement of their myc tags with a sequence that encoded the following motifs: ATG/BclI/His6/BglII/3×FLAG/EcoRI/BirA/NcoI/TEV protease site/BamHI/TAA. We achieved this by cutting the pVS vectors with KpnI and BamHI and through replacement by ligating the following synthetic sequence annealed from complementary oligonucleotides and digested with the same restriction enzymes: GGTACCAAAAAAATGTCTGATCATCATCATCATCATCATAGATCTGATTATAAGGATGATGATGATAAGGATTATAAGGATGATGATGATAAGGATTATAAGGATGATGATGATAAGGAATTCGGTGGTGGTTTAAATGATATTTTTGAAGCACAAAAAATTGAATGGCATATCCATGGTGAAAACTTGTATTTCCAAGGTGGATCC. After site-directed mutagenesis to remove extraneous HindIII and PvuII sites in pVSCulE5′, we excised the culE 3′-targeting sequence via digestion with HindIII and PvuII (italicized) and ligated it into pVSCulE5′, generating pVSCulE5′3′. The floxed blasticidin S resistance cassette was excised from pLPBLP (19) using PstI and HindIII and ligated into the similarly digested pVSCulE5′3′ to yield pVSCulE5′3′-BsR. The culE-tagging construct, designed to engineer a carboxy-terminal 3×FLAG tag followed by a BirA recognition sequence at the endogenous locus of the culE gene, was then excised via digestion with PvuII and BssHII, briefly treated with BAL-31 exonuclease (18), and gel purified. Strain Ax3 cells were transformed via electroporation as previously described (20), selected in 10 μg/ml blasticidin S (MP Biomedicals) in HL-5+ medium, and cloned on Klebsiella aerogenes on SM-agar. Cell lysates from clones were initially analyzed for modification of the culE locus via Western blotting using the M2 anti-FLAG mAb (Sigma) and verified via PCR using primers BsRF and CE3FRev.
To disrupt phyA , the floxed blasticidin S resistance cassette was removed through transient expression of Cre recombinase as previously described (21). After confirmation of floxed blasticidin S resistance removal by PCR, the phyA locus was disrupted as previously described (22).
, the floxed blasticidin S resistance cassette was removed through transient expression of Cre recombinase as previously described (21). After confirmation of floxed blasticidin S resistance removal by PCR, the phyA locus was disrupted as previously described (22).
FbxD Overexpression
The FbxD coding region was amplified from D. discoideum genomic DNA via PCR using primers 5′-AAGGATCCTCATATGACTATAACTGTTGGGTTTGCTTT and 5′-AAGAGCTCTTAATCTAAATTAAAATTATATAATCTAACTAAATTATTGAAAAC and ligated into pCR4-TOPO (Invitrogen). The FbxD DNA was excised via digestion with BamHI and SacI (italicized in primer sequences) and ligated into a similarly digested pVSC vector (9), which inserted an amino-terminal FLAG tag and directs expression in prespore cells under control of the cotB promoter. Cells were transformed as described above in the presence of 10 μg/ml G-418 (Research Products International, Mount Prospect, IL) in HL-5+ medium, further selected with 20 or 120 μg/ml G-418 in HL-5 in gyrating flasks, and cloned as described above. For development, cells grown in HL-5 were washed via centrifugation, and 1.2 × 108 cells in PDF buffer were spread onto 10-cm non-nutrient agar plates. After 13 to 15 h, slugs were scraped off and analyzed for FLAG-FbxD expression via Western blotting using anti-FLAG mAb M2 (Sigma).
Co-immunoprecipitation
Aliquots of 3 × 107 slug cells, or 1.5 × 107 exponentially growing vegetative cells in order to match total protein content, were lysed in 250 μl of IP Buffer (0.2% or 1.0% Nonidet P-40 (as indicated), 50 mm HEPES-NaOH, pH 7.4, or 50 mm Tris-HCl, pH 8.0 (as indicated), 50 mm NaCl, 1 mm MgCl2, 0.1 mm EDTA, and the above-mentioned protease inhibitors) and allowed to incubate on ice for 10 min. Lysates were centrifuged at 16,000 × g for 15 min at 4 °C. For each IP, 50 μl of each S16 supernatant fraction was transferred to packed beads that were washed in IP Buffer. Co-IP of Skp1 was performed using 5 μl of packed pAb UOK77 conjugated to NHS-activated Sepharose beads (GE Healthcare) at 2 mg of protein per milliliter. Beads similarly prepared with total rabbit IgG were used in parallel. Co-IP of CulE-FLAG and FLAG-FbxD was performed using 2.5 μl of packed anti-FLAG M2-EZ view beads (Sigma). CulE-FLAG IPs contained 5 mm EDTA and 10 mm iodoacetamide to inhibit CulE deneddylation by cysteine peptidases. After rotation for 1 h at 4 °C, beads were collected by centrifugation at 2000 × g for 1 min and washed three times with 150 μl of IP buffer, with tubes exchanged after the first wash. Beads were boiled in SDS-PAGE sample buffer for 3 min. The equivalent of 6 × 106 slug cells or 3 × 106 vegetative cells was analyzed via SDS-PAGE and Western blotting using the M2 anti-FLAG mAb (Sigma) or the pan-specific Skp1 mAb 4E1, as well as Alexa-680-conjugated rabbit anti-mouse IgG. Fluorescence was detected using an Odyssey infrared scanner (Li-Cor, Lincoln, NE), and densitometric analysis was performed using ImageJ (version 1.47, National Institutes of Health).
Mass Spectrometry Analysis
For interactome analysis, CulE-FLAG, FLAG-FbxD, or Skp1 was immunoprecipitated as described above, scaled up to 6 × 107 slug cells (∼50 pg of protein per cell) or 3 × 107 vegetative stage cells (∼100 pg of protein per cell). The method was similar to that described previously (23). The washed Ab beads were eluted with 8 m urea, 50 mm Tris-HCl, pH 8.5. Eluates were separated from their IgG resin pellets via centrifugation, and one-half of each was brought to 5 mm tris(2-carboxyethyl)phosphine, incubated for 30 min at room temperature, and then alkylated with 10 mm iodoacetamide for 20 min in the dark. After reduction/alkylation, samples were diluted with three volumes of Tris buffer and digested with 4 μg/ml sequencing-grade trypsin for 16 h at 37 °C. Reactions were stopped by the addition of TFA to 1%, salts were removed by means of solid phase extraction on 100-μl OMIX C18 tips following the manufacturer's instructions (Agilent, Santa Clara, CA), and peptide eluates were lightly dried by means of vacuum centrifugation. Samples were brought to 80 μl and analyzed in triplicate in three successive LC-MS/MS analyses on a Thermo Hybrid LTQ Orbitrap-XL mass spectrometer. For LC-MS/MS, 10 μl of peptides were injected onto a 5-cm trap column (3-μm Magic C18 AQ) in a vented column configuration and then subjected to analytical separation on a 40-cm Picofrit column (75-μm inner diameter; New Objective, Woburn, MA) packed with 3-μm Magic C18 AQ and terminated with an integral fused silica emitter. Peptides were eluted at 250 nL/min in a 1.6%–40% acetonitrile/0.1% formic acid gradient over 110 min, ionized via nanospray, and analyzed using a “Top-6” methodology consisting of one full-range Fourier transform MS scan (nominal resolution of 60,000 full width at half-maximum, 360 to 1400 m/z, using lock mass 445.1200) and up to six data-dependent MS/MS scans performed in the linear ion trap. MS/MS settings included a trigger threshold of 8000 counts, monoisotopic precursor selection, an isolation width of 2 m/z, a normalized collision energy of 35%, an activation Q of 0.25 for 30 ms, and rejection of parent ions that had unassigned charge states or that were previously identified as contaminants on blank gradient runs. Centroided ion masses were extracted using the extract_msn.exe utility in Bioworks 3.3.1 and were used for database searching with Mascot v2.2.04 (Matrix Science, Boston, MA). Searches against the D. discoideum protein list (from dictyBase, May 17, 2012; 12,430 protein sequence models) used the following search parameters: 5 ppm parent ion mass tolerance, 0.6 Da fragment ion tolerance, one missed tryptic cleavage, pyroglutamate cyclization of N-terminal Gln, oxidation of Met, formylation or acetylation of the protein N terminus, and iodoacetamide alkylation of Cys.
Peptide and protein identifications were validated using Scaffold v2.2.00 (Proteome Software, Portland, OR) and the PeptideProphet algorithm (21) using Scaffold's high-mass-accuracy scoring option. Protein acceptance thresholds were set at a 1% false discovery rate, based upon at least two peptides, each identified at a 1% false discovery rate. In addition, at least two spectral counts had to have been obtained for all three technical replicates of at least one individual analysis. At these settings, three, zero, or zero decoy proteins (i.e. reverse translated peptides) were identified from a population of 282, 527, or 406 total protein identifications from the pooled data shown in supplemental Tables S2–S4 and summarized in Figs. 4–6. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy principles of parsimony. Differences in spectral counts were assessed using Scaffold (unnormalized total spectrum counts, with the minimum spectrum count value set to zero). To ensure a high level of rigor with regard to the protein–protein interactions described, proteins reported in supplemental Tables S2–S4 were excluded from further analysis if their spectral counts from negative-control co-IPs were more than zero and >10% of those observed in the “bait” pulldowns, or if they were ribosomal subunits, chaperones (except for a subgroup in the FLAG-FbxD co-IPs), or intraorganellar or secretory proteins (most members of the latter category were already excluded by the previous criteria).
Fig. 4.
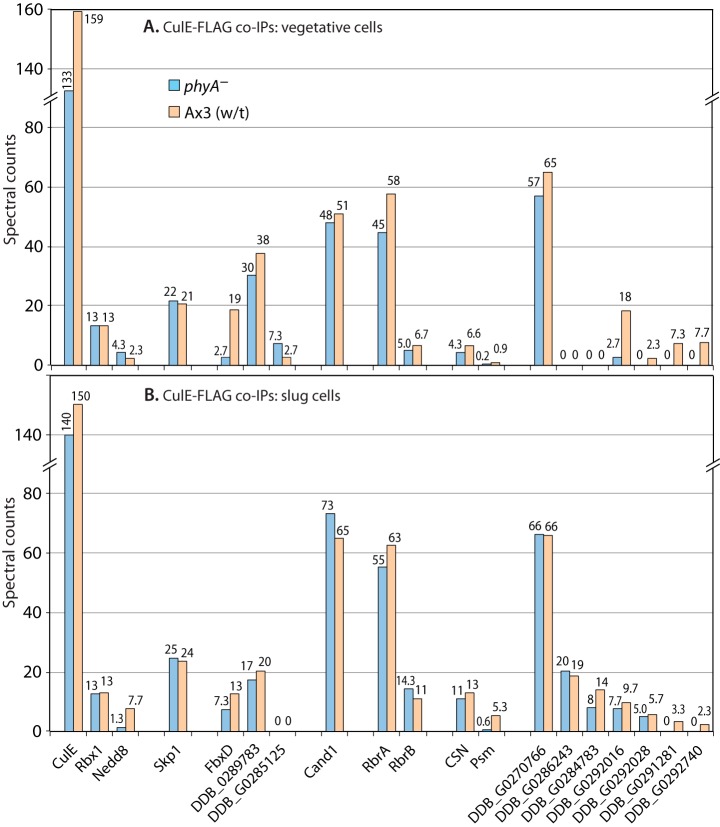
Proteomic analysis of CulE interactions. Vegetative (A) or slug-stage (B) wt or phyA– cells whose culE locus was FLAG-tagged were subjected to co-IP with anti-FLAG (mAb M2) beads, and potential interactors were identified based on mass spectrometric identification of tryptic peptides and exclusion criteria as described in “Experimental Procedures.” Average spectral counts of peptides detected for each protein in triplicate replicates of the single experiment for each panel are plotted in descending order (left to right) of total abundance in phyA– cells. For the protein complexes represented by CSN and PSM (proteasome), average values of detected polypeptides are reported; full data are in supplemental Fig. S1. Protein descriptors and RNA expression patterns are in supplemental Table S1. Full data are shown in supplemental Table S2.
Fig. 5.
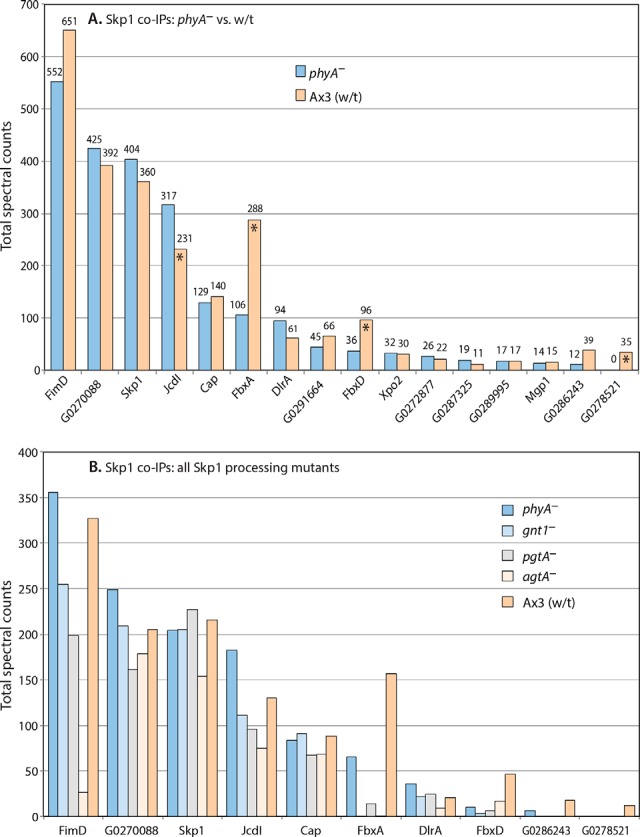
Proteomic analysis of Skp1 interactions. Slug-stage cells were subjected to co-IP with affinity-purified anti-Skp1 (UOK77)-coupled beads. Data from four co-IPs conducted under two conditions (pH 7.4 or pH 8.0; 0.2% or 1% Nonidet P-40), each conducted in triplicate, were pooled owing to the absence of systematic differences, and total spectral counts are reported. A, Ax3 and phyA– are compared. Asterisks denote those proteins for which the quantitative difference between wt and phyA– cells was observed in all trials in which the protein was detected. B, comparison of phyA–, gnt1–, pgtA–, and agtA– and wt (Ax3) cells. Protein descriptors and RNA expression patterns are in supplemental Table S1. Full data are shown in supplemental Table S3.
Fig. 6.
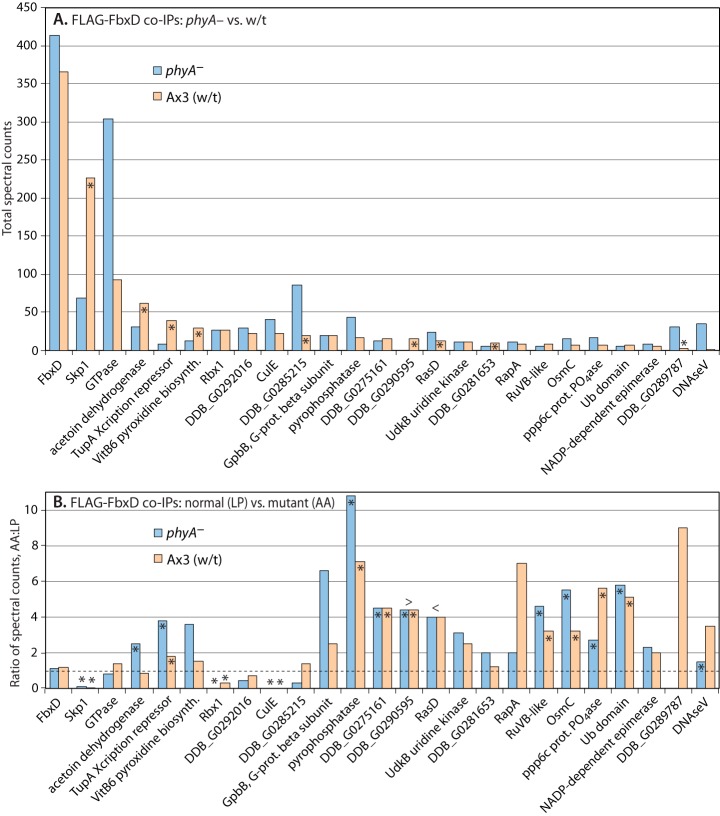
Proteomic analysis of FbxD interactions. Slugs expressing ectopic FLAG-FbxD or FLAG-FbxD(LP-AA), in the presence of endogenous FbxD, under control of the prespore cell-specific cotB promoter were harvested and subjected to co-IP with anti-FLAG (M2) beads as described in “Experimental Procedures.” Potential interactors were identified based on their presence in FLAG pulldowns, absence or near absence from control IPs conducted in non-FLAG-tagged strains, and other criteria as in Fig. 4. A, Ax3 and phyA– cells expressing FLAG-FbxD are compared. Data from three co-IPs conducted under two conditions (pH 7.4, 1% Nonidet P-40; pH 8.0, 0.2% Nonidet P-40), each conducted in triplicate, were pooled owing to a lack of systematic differences, and total spectral counts are plotted. Asterisks signify differences between wt (Ax3) and phyA– cells that were consistent in all trials. B, comparison of FLAG-FbxD and FLAG-FbxD(LP-AA) co-IPs from the two trials shown in A that included mutant FbxD. Ratio of total spectral counts (mutant:normal FbxD) are plotted. The dashed line indicates the ratio value corresponding to no change in cells expressing mutant versus normal FLAG-FbxD. Asterisks signify differences between mutant and normal FLAG-FbxD that were consistent in both trials. > or < refer to values that are greater than bar values immediately to the right or left, respectively (resulting from a denominator value of zero). Protein descriptors and RNA expression patterns are in supplemental Table S1. Full data are shown in supplemental Table S4.
RESULTS
O2 Level Is Limiting for Skp1 Hydroxylation in Cells
To compare the rates of Skp1 polypeptide synthesis and its prolyl hydroxylation, growth-stage cells were pulse-labeled with [35S]Met/Cys, split in half for separate immunoprecipitations for all Skp1 (pAb UOK77 or mAb 4E1) or the reaction product HO-Skp1 (pAb UOK85), and subjected to autoradiography of the [35S]Skp1 bands. Mutant gnt1– cells were used to prevent glycosylation of HO-Skp1, which blocks recognition by pAb UOK85. pAb UOK85 is highly specific for HO-Skp1 based on Western blotting (13, 16) and ELISA assays (24), and no [35S]Skp1 was detected using nonspecific antibodies (13). The experimental design (Fig. 1A) involved a 2.5-h pre-equilibration of gnt1– cells in 2.5%, 5%, 7.5%, or 21% O2 followed by metabolic pulse-labeling for up to 4 h and isolation of Skp1 isoforms.
Fig. 1.
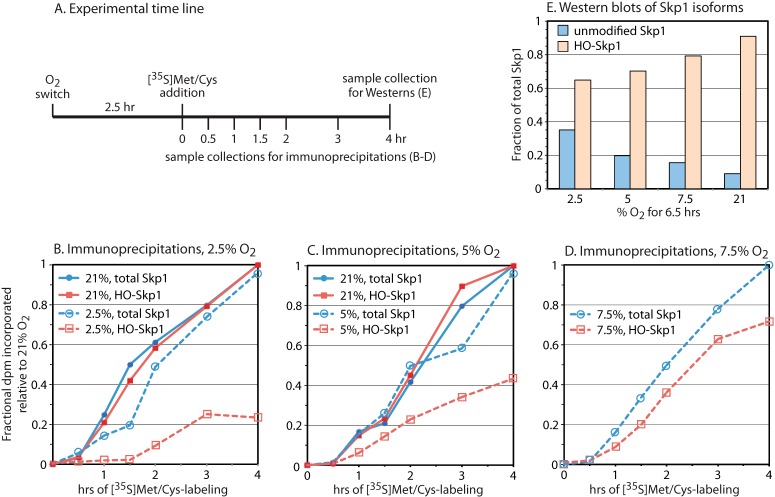
Rate of Skp1 hydroxylation in cells. A, gnt1– cells were transferred to fresh medium and shaken under an atmosphere of the indicated O2 percentage. Metabolic labeling with [35S]Met/Cys commenced at 2.5 h, and cells were harvested at the indicated times and processed for immunoprecipitation using antibodies specific for HO-Skp1 (pAb UOK85) or both HO-Skp1 and Skp1 (pAb UOK77 or mAb 4E1). B–D, time-dependent incorporation of 35S into HO-Skp1 and total Skp1 in cells held at the indicated O2 levels. Data in the same panel are from parallel experiments. Maximal incorporation values into total Skp1 and HO-Skp1 pools after 4 h at 21% O2 were adjusted to 1 to account for variations in antibody pulldown efficiency, and the factor was applied to the parallel low-O2 samples, except for the data in D, which employed the average of factors used for B and C. E, a parallel 4-h sample was subjected to whole cell Western blot analysis using HO-Skp1 (UOK85) and pan-specific (4E1) antibodies to estimate the fraction of Skp1 that was hydroxylated after 6.5 h of exposure to the indicated O2 level. The fraction of Skp1 that was unmodified at steady state in 21% O2 was set at 10% based on the ratio of unmodified to glycosylated Skp1 bands in the wild-type strain Ax3. The unmodified fraction at other O2 levels was derived from the modified fraction via subtraction.
At 21% O2, Skp1 polypeptide synthesis was observed to occur at a nearly linear rate after a lag of 30 min (solid blue lines in Figs. 1B and 1C). This is consistent with the estimated Skp1 half-life of 14 to 16 h based on pulse-chase labeling (13), as turnover of labeled Skp1 would be minimal at 4 h. The rate of HO-Skp1 synthesis was also linear, and similar to that of total Skp1 (solid red lines). Thus, in ambient atmosphere, prolyl hydroxylation of Skp1 occurred without a detectable delay, though a small delay could have been missed owing to different recovery efficiencies of the different antibodies.
Skp1 polypeptide synthesis was similar at 2.5% and 21% O2, despite an accompanying global reduction of 35S-dpm in 2.5% O2 into total cellular protein (65% less total [35S]protein after 4 h based on liquid scintillation counting of TCA precipitates). Thus Skp1 synthesis was preserved by selective regulation. In contrast, Skp1 hydroxylation was delayed with a lag phase of 1.5 h and subsequently occurred at about a 30% rate (dashed red line in Fig. 1B). De-hydroxylation of proline is not known to occur at the protein level, so it is likely that the delayed accumulation of HO-Skp1 reflects a reduced rate of prolyl hydroxylation by PhyA owing to limited availability of the O2 substrate and, potentially, the co-substrate α-ketoglutarate, whose levels may also be diminished at low O2 levels. In cells maintained at 5% O2, where total protein synthesis was >90% of that occurring at 21% O2, hydroxylation was initiated without a lag phase relative to polypeptide synthesis, but the rate was only 45% that of the Skp1 polypeptide (Fig. 1C). At 7.5% O2, the rate of hydroxylation rose to about 80% that in the polypeptide synthesis. Thus increasing O2 levels led to proportionately increased rates of Skp1 hydroxylation in cells.
The hydroxylation status of the total Skp1 pool after 6.5 h of exposure to different O2 levels was evaluated via Western blotting (Fig. 1E). The fraction of hydroxylated Skp1 increased proportionately with increasing O2 level, rising from 65% at 2.5% O2 to 70% at 5% O2, 79% at 7.5% O2, and 90% at 21% O2. Considering the 8-h doubling time of cells under these conditions and the ∼16-h Skp1 half-life reported at 21% O2 (13), ∼23% unmodified Skp1 is expected at 5% O2 ([40% due to growth replacement + 20% for protein turnover replacement − 8% for overlap] × 45% hydroxylation efficiency), and is close to the 20% value inferred from the Western blot analysis (Fig. 1E). The properties of nascent Skp1, which is slow to be hydroxylated at limiting O2 levels, were then investigated through comparison of the interactomes of modified and unmodified Skp1s at 21% O2 in cells whose Skp1 modifications are controlled genetically.
Global Assessment of Skp1 Interactions
To address whether O2 and other factors that control Skp1 processing might affect its interactions with other macromolecules, we analyzed a series of Skp1 modification pathway mutant strains that result in the accumulation of Skp1 isoforms at the processing step immediately prior to the enzymatic deficiency (Fig. 2A). Fresh S100 soluble fractions of vegetative (growth stage) cells were prepared and immediately fractionated via gel filtration. Based on Western blot analysis of the fractions (Fig. 2C), Skp1 eluted in two broad peaks centered at fractions 14 (peak 1) and 19 (peak 2). Whereas unmodified Skp1 from phyA– cells eluted mostly in peak 2, fully modified Skp1 from wt (Ax3) cells eluted mainly in peak 1. Increasing modification increased the peak 1:peak 2 ratio (Fig. 2D), as if each modification step contributed incrementally to the formation of larger complexes represented by the earlier eluting peak 1. Peak 2 might represent a simpler complex because of its later elution, but other unknown factors affected elution position, because free Skp1, which is a dimer, eluted at an intermediate position on this column.
Characterization of SCF Complexes from Dictyostelium Cells
Skp1 is best known as a subunit of the SCF complex, and the D. discoideum genome encodes many known subunits of the traditional CRL1/SCF complex as defined in yeasts, animals, and plants, as well as other proteins known to regulate CRL1 enzymes (supplemental Table S1). Of the five predicted cullins, three (CulA, CulE, and CulB) are most similar to conventional Cul1s based on sequence analysis using classifications described in Ref. 7 (supplemental Fig. S2). CulE was chosen for further analysis because of its detection in pilot interactome studies of FLAG-FbxD (see below). To minimize effects on CulE expression level, the culE chromosomal locus was tagged with a C-terminal 3×FLAG and BirA recognition sequence to allow for potential biotinylation. A similar approach used with Schizosaccharomyces pombe (5) and C. elegans (25) allowed for a description of the interactome of Cul1 via co-IP.
The interaction of CulE-FLAG with Skp1 was initially examined via Western blot analysis of co-IPs from slug cells. Co-IP with anti-FLAG (mAb M2) beads was optimized to recover all CulE-FLAG based on the absence of detectable CulE-FLAG remaining in the supernatant (not shown). CulE migrated as two bands corresponding to the apoprotein and the neddylated version as observed in other organisms (26–30), and similar levels were observed in phyA– and phyA+ (wt Ax3) cells (Fig. 3A). As >95% of CulE-FLAG is neddylated when cells are directly analyzed via Western blotting (not shown), the presence of non-neddylated CulE in these co-IP trials was evidently due to post-lysis deneddylation.
Fig. 3.
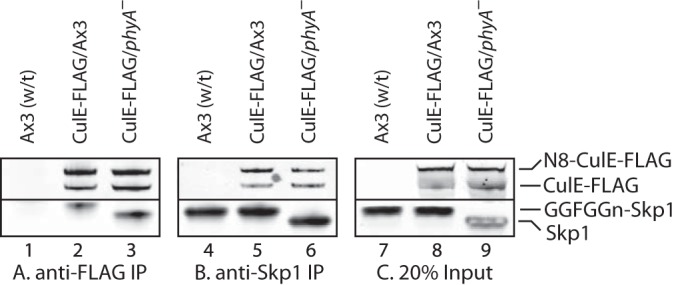
Western blot analysis of interaction of CulE with Skp1. Slug cells whose culE locus was tagged with a C-terminal 3×FLAG tag (CulE-FLAG) in a wt (Ax3) or phyA– background, or cells whose culE locus was not tagged, were solubilized in 1% Nonidet P-40/pH 7.4 IP buffer and immunoprecipitated with anti-FLAG (M2) (A) or anti-Skp1 (UOK77) (B). The presence of CulE-FLAG and Skp1 was probed via Western blotting with the same antibodies. CulE occurred as the apoprotein and a slower migrating species corresponding to the conjugate with Nedd8 (N8). Skp1 from wt cells migrated more slowly because of glycosylation. C, 20% of the input material. Note that a co-migrating band in phyA– cells interfered with the detection of Skp1 in lane 9.
Skp1 was also present in the co-IP, and its level was not reproducibly affected by the absence of PhyA, which caused Skp1 to migrate more rapidly owing to the absence of its glycosylation. Comparisons with input levels (Fig. 3C) and with Western blots of the co-IP supernatants (not shown) suggested that <20% Skp1 was associated with CulE under these conditions. This was likely due to partial dissociation associated with post-lysis deneddylation and association of a Skp1 subpopulation with CulA (see below). Co-IP with affinity-purified UOK77 beads recovered similar levels of Skp1 from both strains (Fig. 3B), with about 50% Skp1 captured based on comparison with the unbound fraction and the input (Fig. 3C). These co-IPs also contained CulE-FLAG, with no reproducible differences in the capture of neddylated or deneddylated CulE. Thus a fraction of Skp1 is normally associated with CulE, and the glycosylation status of Skp1 does not appear to influence the extent of association or the activation state of CulE.
The composition of the CulE-FLAG co-IPs was further examined using a proteomics strategy. The relative recoveries of CulE subunits in control versus bait IPs and in wt versus phyA– cells were compared by spectral counting (31). After filtering to remove contaminants from the CulE-associated proteome (see “Experimental Procedures”), CulE-FLAG was the most abundant protein detected in wt cells based on total spectral counts, and it appeared at similar levels in co-IPs from the same numbers of slug-stage (Fig. 4B) and growth-stage (vegetative) cells (Fig. 4A). Recovery of CulE-FLAG was similar in phyA– and wt cells. Many additional proteins were identified, including expected interactors (see Fig. 2B): the RING protein Rbx1, the Cul1-modifying polypeptide Nedd8, the inhibitory protein Cand1, all eight subunits of the COP9 signalosome (average value shown; see supplemental Fig. S1A for all data), and Skp1. Each of these proteins was similarly represented in mutant and wt vegetative and slug cells, with the Skp1 spectrum counting data confirming the Western blot analyses (Fig. 3). Both Skp1A and Skp1B, which differ by a single amino acid and are expressed at similar levels (32), were similarly represented (not shown). Differences in Nedd8 spectral counts between wt and phyA– cells were not reproducible (compare with Fig. 3), probably because variable deneddylation occurred in some trials (see above). Subunits of the 19S regulatory particle of proteasomes (PSM) were detected, and substantially higher levels were detected in slug versus vegetative cells (>5-fold) and in wt versus phyA– cells (>5-fold), as depicted by the values averaged over all detected proteasomal subunits (Fig. 4). Similar differences were observed for each of the subunits (supplemental Fig. S1B). All six AAA-ATPases and both scaffold proteins of the base region, as well as most subunits of the lid region (33), were detected, though only the base region was detected in growth-stage cells. RbrA and RbrB (Ring-between-Ring, most related to Ariadne-like ubiquitin ligases) were abundant CulE interactors, consistent with their presence in complexes with mammalian Cul1 (34). In addition, a number of proteins with unknown functions were detected. This group included three potential F-box proteins, based on sequence prediction algorithms: DDB_G0292312 (referred to as FbxD), DDB_G0281783, and DDB_G0285125 (protein I.D.s at dictyBase). Of these, FbxD was 2- to 5-fold more abundant in wt than in phyA– cells. Seven other proteins, ranging in levels from abundant to rare, were detected at one or both stages, which correlated approximately with levels of expression based on transcriptomic analyses (35) as summarized in supplemental Table S1. The lower abundance species tended to be more prevalent in wt than in phyA– pulldowns. Thus Dictyostelium expresses SCF-type Ub ligases involving CulE, which are related to previously described SCF-type Ub ligases containing the related CulA (36).
The effect of the phyA– background that blocks Skp1 modifications was mainly confined to an increased interaction of CulE with the predicted FBP FbxD and the 19S-regulatory particle of the proteasome. We investigated these effects further by examining the interactomes of Skp1 and FbxD.
The Skp1 Interactome
Because the interaction of FbxD with CulE is expected to be mediated by Skp1, we analyzed the interactome of Skp1 directly using an affinity-purified anti-Skp1 rabbit antiserum (UOK77) coupled to beads. Pilot studies conducted using Ax3 and phyA– strains expressing FbxD-FLAG (see below) indicated that an interaction with Skp1 was detected but inhibited by standard RIPA buffer, higher concentrations of Nonidet P-40, pH extremes, and time. Therefore, samples were not precleared, Nonidet P-40 was varied from 0.2% (most permissive) to 1.0% (lower background), pH ranged from 7.4 to 8.0, and bead washing was minimized. Four independent comparisons of Ax3 and phyA– slug cells were performed, and proteins were identified and filtered as described above. The same exclusion criteria were applied, but an additional requirement was that reported proteins appeared in at least two trials. As no systematic differences were seen across buffer conditions, spectral counts from the four independent co-IPs were pooled together (supplemental Table S2B). Skp1 was an abundant protein in these co-IPs, with the third most spectral counts (Fig. 5A), although it was 19th on the spectral counts list before exclusions were applied. Skp1 was on average 20% more abundant in the co-IPs of phyA– than in those of wt cells, as also observed in Western blots. Analysis of the supernatant fractions indicated that this was due to the greater capture efficiency of Skp1 in phyA– extracts (not shown), rather than to different levels of Skp1, which is consistent with evidence for altered complexes of Skp1 in wt cells (Figs. 2B and 2C).
Fifteen proteins were found to reproducibly associate with Skp1 in a specific fashion (Fig. 5A). Each was less abundant than Skp1, based on the assumption that proteins with higher Mr generate proportionately more spectral counts. The abundance of four proteins was reproducibly different between the wt and phyA– strains (Fig. 5A, asterisks). The three most abundant are predicted to contain an F-box sequence. The first, FbxD, was more abundant in Skp1 co-IPs from wt cells, consistent with its greater association with CulE in wt relative to phyA– cells (Fig. 4). A comparison with mutants that accumulate intermediate glycoforms indicated that full glycosylation is important for increased interaction with FbxD (Fig. 5B). FbxA was, like FbxD, also more enriched in Skp1 co-IPs from wt cells, and this also depended on full modification of Skp1. The third FBP-like protein, JcdI, was, in contrast, more enriched in Skp1 co-IPs from phyA– cells. A fourth potential FBP, DlrA, was not differentially enriched. Thus, of the 15 Skp1 interactors reported, 4 of the 8 most abundant are predicted to be FBPs and, of these, 3 are differentially enriched according to Skp1's modification status. The other four proteins were not differentially enriched. Of the seven less abundant proteins, only one, a predicted Tyr-kinase with ankyrin repeats (DDB-G0278521), was differentially enriched according to Skp1 modification status. CulA, previously observed to associate with the FBP FbxA (36), was also detected, but its abundance was too low to meet our reportability criteria, possibly because of competitive binding with pAb UOK77. CulE and Rbx1 were also detected as expected (Figs. 3 and 4), at similarly low levels.
In an attempt to complement these studies, we examined the interactome of FLAG-Skp1 overexpressed in Ax3 cells using anti-FLAG beads. However, no significant proteins interactors were detected, despite the utility of this construct in yeast (37). Interestingly, we previously noted that FLAG-Skp1 remains unmodified when expressed in Dictyostelium (14), indicating that perturbation of its N terminus interferes with not only its ability to enter SCF complexes, but also its hydroxylation and glycosylation.
The FbxD Interactome
FbxD possesses an F-box-like sequence and seven C-terminal WD40 repeats predicted to serve as a substrate receptor. To test the hypothesis that the F-box-like sequence mediates binding to Skp1, we changed the Leu-Pro motif near the N terminus of the F-box to Ala-Ala; this substitution interferes with FBP binding to Skp1 from Dictyostelium and other organisms (38–40). FbxD and FbxD(LP-AA) were N-terminally modified with a 3×FLAG tag and ectopically expressed under control of the prespore-specific cotB promoter, using an expression DNA that typically inserts chromosomally in tandem arrays (20). Similar FbxD-FLAG levels were expressed in all genetic backgrounds.
Cell fractionation studies performed in the absence of detergents showed that the great majority of FLAG-FbxD and the majority of FLAG-FbxD(LP-AA) were present in the soluble cytosolic fraction after centrifugation at 100,000 × g for 1 h (data not shown). Thus, this mutation did not grossly compromise FbxD expression or subcellular localization, consistent with previous studies showing that such mutations do not unduly distort the tertiary structure of FBP.
In proteomic surveys of the co-IPs, similar levels of FbxD-FLAG were obtained from wt and phyA– cells (Fig. 6A). In contrast, Skp1 recovery with FbxD was 3-fold reduced in phyA– cells, which was consistent with the glycosylation dependence of the reciprocal co-IP (Fig. 5). Co-IPs of FLAG-FbxD(LP-AA) recovered little Skp1 from either strain (AA:LP ratio ≪1 in Fig. 6B), confirming the F-box dependence of the interaction. A putative GTPase (DDB_G0283979) bound to FLAG-FbxD in a pattern complementary to that of Skp1, but the small increase in binding to mutant FLAG-FbxD(LP-AA) (Fig. 6B) indicates that the GTPase does not compete with Skp1 binding, unless the GTPase also depends on an intact F-box. CulE and the associated Rbx1 were observed in the FbxD complexes, as expected from the reciprocal co-IP (Fig. 4); the absence of a reproducible difference between wt and phyA– cells might be due to the lower spectral count level from these co-IPs. Notably, CulA and CulB were not detected, suggesting that FbxD is selective for CulE. Aside from predicted chaperone proteins, 25 additional FbxD-associated proteins were identified, and the 20 most abundant are included in Fig. 6. Interestingly, five reproducibly favored binding to FLAG-FbxD when Skp1 was glycosylated, whereas three favored binding when Skp1 was unmodified (Fig. 6A). The latter three are candidates for substrates that are expected to accumulate in association with FbxD in the absence of binding to Skp1, although at this point differential abundance in the extract cannot be excluded. Nevertheless, the ≥2-fold increase observed for 12 proteins in co-IPs from cells expressing FLAG-FbxD(LP-AA) relative to FLAG-FbxD regardless of Skp1 modification status (Fig. 6B) reinforces their candidacy as substrates to be explored in the future.
The FLAG-FbxD co-IPs also contained members of the Hsp20 (HspG, HspL, HspM), Hsp70 (HspH), and Hsp90 (HspD) classes of heat-shock chaperones, and all members of the Cct/TriC class of chaperonins (supplemental Fig. S3). Unlike other Hsp-like proteins, including HspA (Hsp60-like), HspB (Hsc70-like), and HspE-1 (Hsc70-like), these chaperones were not observed in control co-IPs. These chaperones were also not detected in the co-IPs targeting Skp1 or CulE-FLAG, except for the Ccts, which were also observed in the control Skp1 co-IPs and thus excluded. The spectral counts for individual chaperones ranged from very low to up to 153 for the experimental series shown in Fig. 6, and thus represent a major FLAG-FbxD interactor if summed over the 13 Hsps and eight Ccts detected. In addition, the levels of co-immunoprecipitated Hsps were reproducibly increased by 2-fold to greater than 10-fold for mutant FLAG-FbxD(LP-AA) (supplemental Fig. S3). The Cct subunits were also increased, but mostly by ≤2-fold. The association of Hsps and Ccts might have been due at least in part to the unnaturally high level of expression of FLAG-FbxD in these strains, but the increased association with mutant FLAG-FbxD(LP-AA) is consistent with assisting in maintaining its solubility when the hydrophobic F-box domain is unable to interact with Skp1. Hsp70 and the Cct complex were previously implicated in the assembly of the VHL-type CRL2 complex (41), which is related to the SCF-type CRL1 complex analyzed here.
To confirm the glycosylation-dependent interaction of FbxD with Skp1, we analyzed three additional biological replicates via Western blotting. For this study we examined the matched set of Skp1 modification mutants that each express FLAG-FbxD at approximately similar levels. As illustrated by the example shown in Fig. 7A using Skp1 as the bait, FLAG-FbxD exhibited the greatest association with fully modified Skp1, partial association with the trisaccharide form FGGn-Skp1, and less association with less modified isoforms. Thus increased binding of glycosylated Skp1 to FbxD observed in the native co-IPs (Fig. 5) was not trivially due to different levels of FbxD available for binding. For confirmation, anti-FLAG was used in reciprocal co-IPs. Unlike the Skp1 co-IPs, the anti-FLAG co-IPs captured essentially all their bait (data not shown). The same pattern of interaction was observed (Fig. 7B), confirming that a greater fraction of FbxD was associated with Skp1 in cells that could fully glycosylate Skp1. In contrast, minimal FLAG-FbxD(LP-AA) associated with Skp1 (Fig. 7C), confirming its F-box dependence. Thus, formation of the trisaccharide promotes the Skp1–FbxD interaction in cells, which is further enhanced by completion of the pentasaccharide.
Fig. 7.
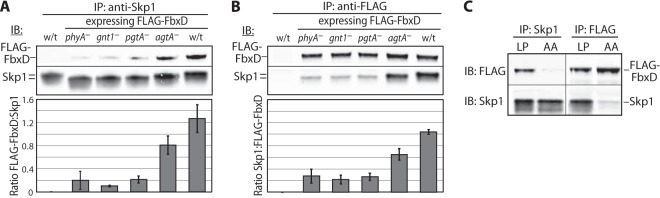
Glycosylation dependence of the FbxD interaction with Skp1. Slug extracts (1% Nonidet P-40, pH 7.4) of pathway knockout strains expressing FLAG-FbxD (as in Fig. 6) were subjected to co-IP using anti-Skp1 (A) or anti-FLAG (B) antibodies. Representative Western blots probed with mAb 4E1 (pan-specific toward Skp1 isoforms) or anti-FLAG (M2) are shown. The migration of Skp1 decreased with increasing modification. Bar graphs represent densitometric analysis of Western blots from three independent trials (average values ± S.E.). Intensities were normalized to the maximum signal within each experiment. C, similar analysis of FLAG-FbxD(LP-AA) expressed in wt slug cells.
FLAG-FbxD Delays Development in an F-box- and Glycosylation-dependent Fashion
Previous studies showed that disruption of phyA delays developmental progression, mainly at the culmination stage (transition from slug to fruiting body), by ∼3 h at 21% O2 (14). Our novel findings suggested that the underlying mechanism might be an O2- and prolyl-hydroxylation-regulated interaction with FbxD and FbxA. To address this, we analyzed the developmental timing of the FLAG-FbxD overexpression strains. All strains achieved the slug stage of development at about the same time (11 h under the conditions employed here) (Fig. 8). FLAG-FbxD overexpression delayed culmination by about 3 h in wt cells, almost as much as the 4-h delay caused by blocking Skp1 modification. We suggest that overexpression of this FBP upsets the proteostatic network in cells via an as yet uncertain mechanism. An intermediate effect was rendered by the F-box mutant FLAG-FbxD(LP-AA), indicating that inhibition is partially dependent on Skp1 interaction. In contrast, FLAG-FbxD overexpression had no effect on phyA– cells, which is consistent with their acting in the same pathway where Skp1 interaction was already reduced when Skp1 was not modified.
Fig. 8.
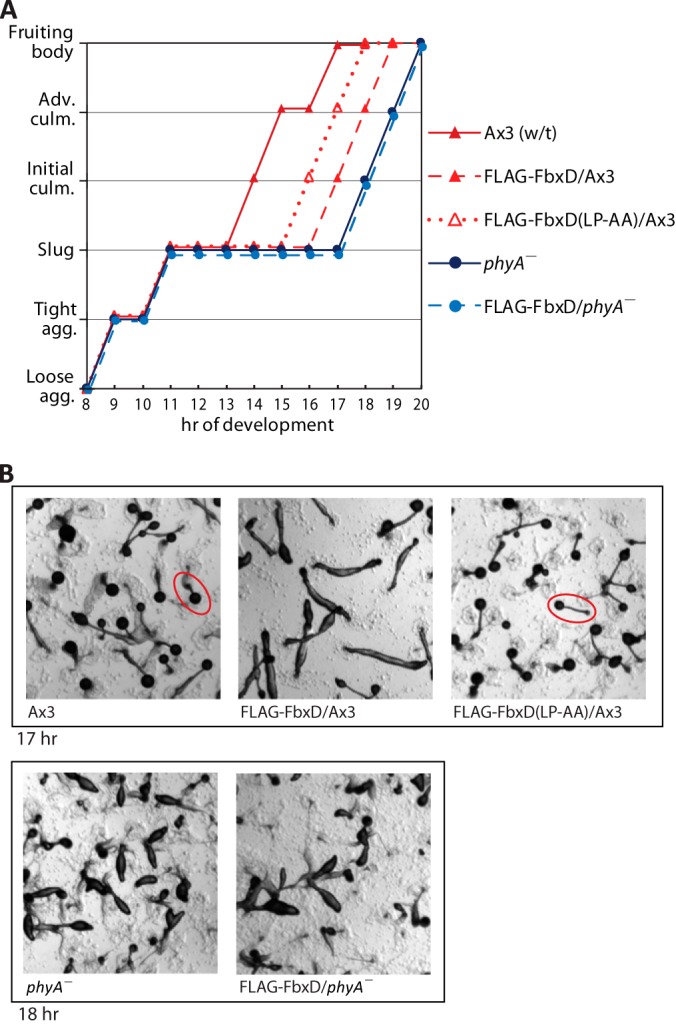
FbxD overexpression inhibits development. A, cells were uniformly dispensed onto 10-cm non-nutrient agar plates at 21% O2 to induce development and imaged at hourly intervals. Morphological status is plotted as a function of time. Typical fruiting bodies (1 to 2 mm tall) are encircled in red. Data are from one of two independent trials, each with similar outcomes. B, representative images of strains at the indicated times of development.
DISCUSSION
Dictyostelium SCF Complexes
Results presented here confirm bioinformatic findings and limited experimental data that predict that Dictyostelium assembles a canonical family of conventional SCF complexes and corresponding E3SCFUb ligases (Fig. 9). The Dictyostelium SCF repertoire appears to be expanded, however, by the existence of multiple Cul1s, including the previously described CulA, CulE identified here, and possibly CulB (42). Each possesses typical Cul1-like sequence motifs (supplemental Fig. S2). In this work, CulE was associated with the Dictyostelium homolog of Rbx1, with relative spectral counts suggesting a stoichiometric relationship. CulE was mostly neddylated, implying its constitutive activation. Skp1 was a prominent interactor of CulE. Three new potential F-box proteins were detected in addition to FbxA (36). One, FbxD, was validated by site-directed mutagenesis and, of the three, preferentially associated with CulE. As CulE was presumably expressed at native levels, these interactions may represent what occurs in the cell. Candidate E2 sequences were not detected, but their association is expected to be labile.
Fig. 9.
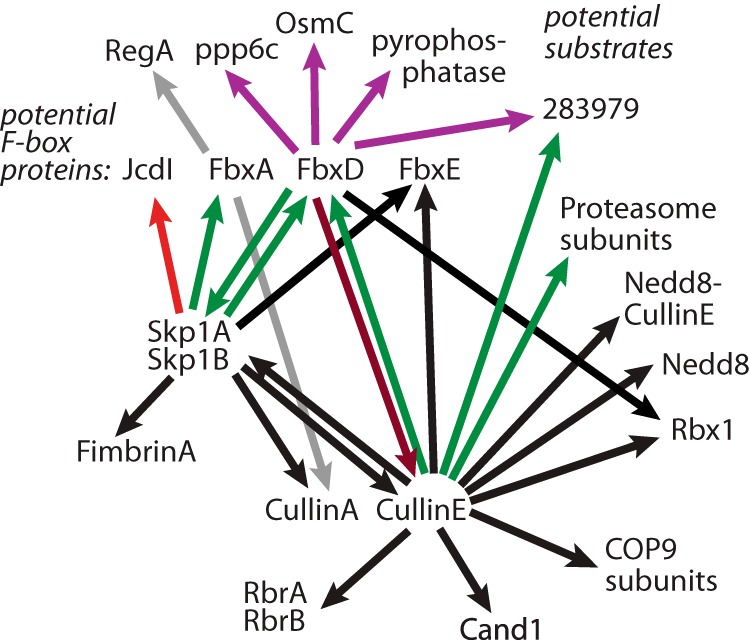
Summary of Skp1 interactome. Arrows depict interactions detected in the present or previous studies. The tail denotes the bait, and the tip the prey. Black indicates an interaction not affected by the modification status of Skp1. Green indicates an interaction that was greater in wt than in phyA– cells, and red denotes an interaction that was less. Gray indicates a previously described interaction (36) whose dependence on Skp1 modification has not been examined.
The CulE interactome also included additional proteins that could be classified as (i) regulatory, (ii) function related, (iii) previously observed but unknown, and (iv) unknown. Known regulators included Cand1 and the COP9 signalosome (CSN). All eight subunits of the previously characterized Dictyostelium CSN (43), expected to mediate CulE deneddylation, were found in association with CulE (Fig. 3). CSN5 and CSN2 are essential for Dictyostelium proliferation, but because the CSN may also affect other CRLs, a specific role for CRL1s is not certain. As expected, Dictyostelium expresses other genes expected for CulE neddylation (supplemental Table S1). In other organisms, Cand1 associates only with deneddylated Cul1, suggesting that the CulE–Cand1 interaction might have occurred following post-lysis deneddylation. Based on evidence that yeast and mammalian Cand1 is an exchange factor for Skp1–FBP complexes (3–5), Cand1 binding to CulE is expected to be competitive with Skp1 (see Fig. 2B).
Other CulE interactors were the Ariadne-related Ring-between-Ring protein RbrA and its homolog RbrB. RbrA was previously observed in association with mammalian Cul1, but the significance of this interaction is poorly understood (34). Disruption of the Dictyostelium RbrA gene results in compromised fruiting body formation (44) in a way that resembles effects of Skp1 modification mutants, consistent with a possible functional interaction. Seven additional interacting candidates were identified (Fig. 3), but the significance of these unknown proteins is unclear.
Finally, all 10 subunits of the 19S-proteasome regulatory particle, but none from the 20S-core particle, were detected (supplemental Fig. S1B). Association of the SCF complex with the proteasome has been previously observed (45, 46) and might reflect functional coordination as the polyubiquitinated substrate is transferred for proteolytic degradation.
Role of Skp1 Glycosylation
In contrast to the majority of the CulE interactome, which is largely unaffected by Skp1's O2-regulated glycosylation, FbxD and the 19S-proteasome regulatory particle amounts were elevated by Skp1 glycosylation (Fig. 9). FbxD was enriched 2- to 5-fold in wt versus phyA– cells, and the 19S regulatory particle was enriched 5- to 9-fold. As glycosylation appears to occur only on the Skp1 component of SCF complexes (8, 13, 14), this posttranslational modification might promote the assembly of or stabilize the SCFFbxD complex. Analysis of the Skp1 interactome confirmed increased association of FbxD with fully glycosylated Skp1 (2.6-fold) and, to a lesser extent, with FGGn-Skp1, relative to the less modified isoforms (Figs. 5 and 6). A similar effect was observed for another documented FBP, FbxA. FbxA was not observed in the CulE pulldown, but given that FbxA is known to interact with CulA (36), FBP interactions might be Cul1-subtype specific, consistent with direct FBP–Cul1 contacts for some FBPs (47). Increased interaction of glycosylated Skp1 with FbxD was observed even when FLAG-FbxD was ectopically overexpressed in prespore cells, and reciprocal co-IPs confirmed the greater interaction of FbxD with glycosylated Skp1s (Figs. 6 and 7). General promotion of SCF assembly by glycosylation was suggested by gel filtration analysis of cytosolic extracts (Fig. 2). This contrasts with a proposed role for phosphorylation of yeast Skp1, which might inhibit FBP interactions (48). Stimulation of SCF assembly might explain the increased 19S-proteasomal regulatory particle interaction detected in the CulE co-IPs. Proteasomal subunits were not detected in the Skp1 pulldowns, but notably, not all Skp1 was accessible to the antibody, potentially because of steric interference in some complexes.
The mechanism by which FbxD and FbxA interact more efficiently with glycosylated Skp1 remains to be determined. We recently showed that a model soluble FBP, guinea pig Fbs1, binds preferentially to GGFGGn-Skp1 and Gn-Skp1 relative to HO-Skp1 and Skp1 in vitro in the absence of other proteins (16). This suggests that glycosylation might affect Skp1 directly to promote its interactions with FBPs. However, the pattern differs from that of FbxD and FbxA, where GGFGGn-Skp1 binds substantially better than FGGn-Skp1 and Gn-Skp1. Possibly, peripheral sugars of the glycan interact with certain Dictyostelium F-box domains to stabilize the interaction. Alternatively, an additional factor that recognizes the glycan (or is blocked by the glycan) contributes to differential recognition. To this point, 11 other proteins were less abundantly but reproducibly detected in the Skp1 interactome. One was reproducibly differentially affected by Skp1 glycosylation, and thus might also contribute to selective interactions according to Skp1 glycosylation status. Further studies are needed to distinguish whether increased FBP interactions reflect differential affinity or rates of assembly, or selective stability following cell disruption. The decreased interaction of fully glycosylated Skp1 with JcdI (Fig. 5) raises the possibility of differential effects of glycosylation on substrate receptor and non-receptor subclasses of FBPs, such that glycosylation might affect FBP selectivity in the competitive environment occurring in the cell. FbxD, like almost all known FBPs (except guinea pig Fbs1), is insoluble when expressed in Escherichia coli in the absence of Skp1 (data not shown), making direct biochemical analysis challenging.
FBPs are typically insoluble in the absence of Skp1, and both FLAG-FbxD expressed in phyA– cells and mutant FLAG-FbxD(LP-AA) are soluble in extracts, so how does this occur if they are not binding Skp1? Two of the detected FLAG-FbxD interactors shown in Fig. 6 match the predicted pattern of increased binding to FLAG-FbxD in phyA– cells and increased binding to mutant FLAG-FbxD(LP-AA) in either background (RasD and a predicted pyrophosphatase). Chaperones are also candidates to maintain FbxD and other FBPs in solution until Skp1 becomes available, as suggested by the increased association of Hsp20, Hsp70, Hsp90, and Cct/TriC proteins with mutant relative to native FLAG-FbxD (supplemental Fig. S3). Although we did not observe an increased association of these chaperones with FLAG-FbxD in phyA– relative to wt cells, Hsp70 and Cct/TriC proteins have been implicated in the assembly of the related CRL2 E3 Ub ligases (41). In mammalian cells, some FBPs can bind nuclear exportin-1 in place of Skp1 (49). Although the related Dictyostelium protein Xpo2 was detected, it was not enriched in the Skp1 co-IPs from phyA– versus wt cells.
With its C-terminal WD40 repeats, FbxD is likely to bind multiple substrates like other FbxW-type FBPs such as mammalian βTrCP and FbxW7 and yeast Grr1 (6). Many of the FLAG-FbxD interactors are potential substrates owing to their increased association when the F-box was mutated, and a subset of these was increased in phyA– relative to wt cells (Fig. 6). Prediction of substrates is, however, confounded by potentially different levels of the interacting protein available for binding and potential targeting by other polyubiquitin ligases. Poor evolutionary conservation of FBP sequences precludes prediction of FbxD and other FBP substrates, which are under investigation using orthogonal approaches.
Role of Skp1 Modification in O2 Sensing
We propose a novel model in which the activity of certain FBPs such as FbxD and FbxA is regulated by cellular nutritional status. In this model, hydroxylation of newly synthesized Skp1 is controlled by the level of ambient O2 (Fig. 1). Skp1 might also respond to other metabolic factors such as the co-substrate α-ketoglutarate and another measure of the nutritional or metabolic status of the cells (i.e. sugar-nucleotide concentrations to support glycosylation). We propose that O2 levels low enough to inhibit culmination trap newly synthesized Skp1 in its unglycosylated form, and this isoform persists for a period of time inversely proportional to the available O2. Hypoglycosylation of Skp1 is predicted to inhibit its association with developmentally regulated and newly synthesized substrate receptor FBPs such as FbxD and FbxA. Thus, limitations in O2 and other metabolic substrates could regulate the assembly of key E3SCFUb-ligase complexes, which in turn would regulate the polyubiquitination and degradation of proteins that control the progression of Dictyostelium development. This model is consistent with (i) data from a submerged O2-restricted model of development, where accumulation of unmodified Skp1 correlated with inability to sporulate (10); (ii) data showing that wt cells require 7% to 10% O2 for culmination, whereas phyA– mutant cells require 18% to 21% (9); (iii) data showing that the O2 requirement for culmination is affected by the Skp1 concentration in a way that depends on the Pro143 modification target (14); and (iv) new data suggesting that FLAG-FbxD overexpression inhibits development via a mechanism that involves Skp1 interaction, but not in phyA– mutant cells that have a reduced capacity to bind FLAG-FbxD (Fig. 8). The model has parallels with O2 sensing in animals, where the O2-dependent PhyA homolog PHD2 influences the proteome through prolyl hydroxylation of the transcriptional co-factor hypoxia inducible factor-α, which leads to recognition by the E3VHLUb ligase (homologous to the E3SCFUb ligase) and consequent degradation by the 26S proteasome (50). In contrast, hydroxylation and glycosylation of Dictyostelium Skp1 do not affect its stability (13, 14), but perhaps they affect the activity of E3SCFUb ligases, resulting in, by inference, control of the proteome via its degradation arm. Overall, these studies implicate conditional modifications of Skp1 in the assembly of nascent Dictyostelium SCF complexes as novel mechanisms for SCF regulation of development in Dictyostelium and cell cycling in Toxoplasma.
Supplementary Material
Acknowledgments
We thank Chris Schafer for providing the pVS3 vectors.
Footnotes
Author contributions: M.O.S. and C.M.W. designed research; M.O.S., Y.X., H.v., P.W., and S.D.H. performed research; S.D.H. contributed new reagents or analytic tools; M.O.S., Y.X., H.v., and C.M.W. analyzed data; M.O.S. and C.M.W. wrote the paper.
* This research was supported by Grant No. R01 GM037539 from the National Institutes of Health and by funds from the OUHSC Provost for the Summer Undergraduate Research Experience.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CRL
- cullin-RING ubiquitin ligase
- Ub
- ubiquitin
- FBP
- F-box protein
- Cul1
- cullin-1
- wt
- wild type
- IP
- immunoprecipitation
- co-IP
- co-immunoprecipitation
- CSN
- COP9 signalosome
- SCF
- Skp1–Cul1–FBP complex.
REFERENCES
- 1. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 2. Willems A. R., Schwab M., Tyers M. (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695, 133–170 [DOI] [PubMed] [Google Scholar]
- 3. Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., Larimore E. A., Rome M., Zheng N., Clurman B. E., Hess S., Shan S. O., Deshaies R. J. (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N. T., Rabut G., Kurz T. (2013) CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 4, 1641. [DOI] [PubMed] [Google Scholar]
- 5. Wu S., Zhu W., Nhan T., Toth J. I., Petroski M. D., Wolf D. A. (2013) CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 4, 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skaar J. R., Pagan J. K., Pagano M. (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. West C. M., Wang Z. A., van der Wel H. (2010) A cytoplasmic prolyl hydroxylation and glycosylation pathway modifies Skp1 and regulates O2-dependent development in Dictyostelium. Biochim. Biophys. Acta 1800, 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. West C. M., van der Wel H., Wang Z. A. (2007) Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development 134, 3349–3358 [DOI] [PubMed] [Google Scholar]
- 10. Xu Y., Wang Z. A., Green R. S., West C. M. (2012) Role of the Skp1 prolyl-hydroxylation/glycosylation pathway in oxygen dependent submerged development of Dictyostelium. BMC Dev. Biol. 12, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonner J. T., Lamont D. S. (2005) Behavior of cellular slime molds in the soil. Mycologia 97, 178–184 [DOI] [PubMed] [Google Scholar]
- 12. Wang Z. A., van der Wel H., Vohra Y., Buskas T., Boons G. J., West C. M. (2009) Role of a cytoplasmic dual-function glycosyltransferase in O2 regulation of development in Dictyostelium. J. Biol. Chem. 284, 28896–28904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang D., van der Wel H., Johnson J. M., West C. M. (2012) Skp1 Prolyl 4-hydroxylase of Dictyostelium mediates glycosylation-independent and -dependent responses to O2 without affecting Skp1 stability. J. Biol. Chem. 287, 2006–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z. A., Singh D., van der Wel H., West C. M. (2011) Prolyl hydroxylation- and glycosylation-dependent functions of Skp1 in O2-regulated development of Dictyostelium. Dev. Biol. 349, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu Y., Brown K. M., Wang Z. A., van der Wel H., Teygong C., Zhang D., Blader I. J., West C. M. (2012) The Skp1 protein from Toxoplasma is modified by a cytoplasmic prolyl 4-hydroxylase associated with oxygen sensing in the social amoeba Dictyostelium. J. Biol. Chem. 287, 25098–25110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheikh M. O., Schafer C. M., Powell J. T., Rodgers K. K., Mooers B. H., West C. M. (2014) Glycosylation of Skp1 affects its conformation and promotes binding to a model F-Box protein. Biochemistry 53, 1657–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ketcham C., Wang F., Fisher S. Z., Ercan A., van der Wel H., Locke R. D., Sirajud-Doulah K., Matta K. L., West C. M. (2004) Specificity of a soluble UDP-galactose:fucoside α1,3galactosyltransferase that modifies the cytoplasmic glycoprotein Skp1 in Dictyostelium. J. Biol. Chem. 279, 29050–29059 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y., Zhang P., West C. M. (1999) A linking function for the cellulose-binding protein SP85 in the spore coat of Dictyostelium discoideum. J. Cell Sci. 112 (Pt 23), 4367–4377 [DOI] [PubMed] [Google Scholar]
- 19. Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A. R. (2004) A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32, e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pang K. M., Lynes M. A., Knecht D. A. (1999) Variables controlling the expression level of exogenous genes in Dictyostelium. Plasmid 41, 187–197 [DOI] [PubMed] [Google Scholar]
- 21. Linkner J., Nordholz B., Junemann A., Winterhoff M., Faix J. (2012) Highly effective removal of floxed Blasticidin S resistance cassettes from Dictyostelium discoideum mutants by extrachromosomal expression of Cre. Eur. J. Cell Biol. 91, 156–160 [DOI] [PubMed] [Google Scholar]
- 22. van der Wel H., Ercan A., West C. M. (2005) The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-α class of animal prolyl 4-hydroxylases. J. Biol. Chem. 280, 14645–14655 [DOI] [PubMed] [Google Scholar]
- 23. Voruganti S., Lacroix J. C., Rogers C. N., Rogers J., Matts R. L., Hartson S. D. (2013) The anticancer drug AUY922 generates a proteomics fingerprint that is highly conserved among structurally diverse Hsp90 inhibitors. J. Proteome Res. 12, 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. West C. M., van der Wel H., Chinoy Z., Boons G.-J., Gauthier T. J., Taylor C. M., Xu Y. (2014) Generating isoform-specific antibodies: lessons from the nucleocytoplasmic glycoprotein Skp1. In Glycoscience: Biology and Medicine (Taniguchi N., Endo T., Hart G., Seeberger P., Wong C.-H., eds), Springer, New York, in press. [Google Scholar]
- 25. Bosu D. R., Feng H., Min K., Kim Y., Wallenfang M. R., Kipreos E. T. (2010) C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Dev. Biol. 346, 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Read M. A., Brownell J. E., Gladysheva T. B., Hottelet M., Parent L. A., Coggins M. B., Pierce J. W., Podust V. N., Luo R. S., Chau V., Palombella V. J. (2000) Nedd8 modification of Cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol. Cell. Biol. 20, 2326–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohh M., Kim W. Y., Moslehi J. J., Chen Y., Chau V., Read M. A., Kaelin W. G., Jr. (2002) An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 3, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liakopoulos D., Doenges G., Matuschewski K., Jentsch S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. del Pozo J. C., Estelle M. (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. U.S.A. 96, 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pintard L., Kurz T., Glaser S., Willis J. H., Peter M., Bowerman B. (2003) Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13, 911–921 [DOI] [PubMed] [Google Scholar]
- 31. Lundgren D. H., Hwang S. I., Wu L., Han D. K. (2010) Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 [DOI] [PubMed] [Google Scholar]
- 32. West C. M., Kozarov E., Teng-umnuay P. (1997) The cytosolic glycoprotein FP21 of Dictyostelium discoideum is encoded by two genes resulting in a polymorphism at a single amino acid position. Gene 200, 1–10 [DOI] [PubMed] [Google Scholar]
- 33. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelsall I. R., Duda D. M., Olszewski J. L., Hofmann K., Knebel A., Langevin F., Wood N., Wightman M., Schulman B. A., Alpi A. F. (2013) TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 32, 2848–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parikh A., Miranda E. R., Katoh-Kurasawa M., Fuller D., Rot G., Zagar L., Curk T., Sucgang R., Chen R., Zupan B., Loomis W. F., Kuspa A., Shaulsky G. (2010) Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 11, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mohanty S., Lee S., Yadava N., Dealy M. J., Johnson R. S., Firtel R. A. (2001) Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seol J. H., Shevchenko A., Shevchenko A., Deshaies R. J. (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3, 384–391 [DOI] [PubMed] [Google Scholar]
- 38. Kondo-Okamoto N., Ohkuni K., Kitagawa K., McCaffery J. M., Shaw J. M., Okamoto K. (2006) The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol. Biol. Cell 17, 3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Price C. T., Al-Quadan T., Santic M., Rosenshine I., Abu Kwaik Y. (2011) Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 40. Russell I. D., Grancell A. S., Sorger P. K. (1999) The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145, 933–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melville M. W., McClellan A. J., Meyer A. S., Darveau A., Frydman J. (2003) The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23, 3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang B., Kuspa A. (2002) CulB, a putative ubiquitin ligase subunit, regulates prestalk cell differentiation and morphogenesis in Dictyostelium spp. Eukaryot. Cell 1, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosel D., Kimmel A. R. (2006) The COP9 signalosome regulates cell proliferation of Dictyostelium discoideum. Eur. J. Cell Biol. 85, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 44. Whitney N., Pearson L. J., Lunsford R., McGill L., Gomer R. H., Lindsey D. F. (2006) A putative Ariadne-like ubiquitin ligase is required for Dictyostelium discoideum development. Eukaryot. Cell 5, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Farras R., Ferrando A., Jasik J., Kleinow T., Okresz L., Tiburcio A., Salchert K., del Pozo C., Schell J., Koncz C. (2001) SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Voloshin O., Bakhrat A., Herrmann S., Raveh D. (2012) Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS One 7, e39210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schlman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 48. Beltrao P., Albanese V., Kenner L. R., Swaney D. L., Burlingame A., Villen J., Lim W. A., Fraser J. S., Frydman J., Krogan N. J. (2012) Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nelson D. E., Laman H. (2011) A competitive binding mechanism between Skp1 and Exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J. Biol. Chem. 286, 19804–19815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ratcliffe P. J. (2013) Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J. Physiol. 591 (Pt 8), 2027–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.