Abstract
Single cell trapping increasingly serves as a key manipulation technique in single cell analysis for many cutting-edge cell studies. Due to their inherent advantages, microfluidic devices have been widely used to enable single cell immobilization. To further improve the single cell trapping efficiency, this paper reports on a passive hydrodynamic microfluidic device based on the “least flow resistance path” principle with geometry optimized in line with corresponding cell types. Different from serpentine structure, the core trapping structure of the micro-device consists of a series of concatenated T and inverse T junction pairs which function as bypassing channels and trapping constrictions. This new device enhances the single cell trapping efficiency from three aspects: (1) there is no need to deploy very long or complicated channels to adjust flow resistance, thus saving space for each trapping unit; (2) the trapping works in a “deterministic” manner, thus saving a great deal of cell samples; and (3) the compact configuration allows shorter flowing path of cells in multiple channels, thus increasing the speed and throughput of cell trapping. The mathematical model of the design was proposed and optimization of associated key geometric parameters was conducted based on computational fluid dynamics (CFD) simulation. As a proof demonstration, two types of PDMS microfluidic devices were fabricated to trap HeLa and HEK-293T cells with relatively significant differences in cell sizes. Experimental results showed 100% cell trapping and 90% single cell trapping over 4 × 100 trap sites for these two cell types, respectively. The space saving is estimated to be 2-fold and the cell trapping speed enhancement to be 3-fold compared to previously reported devices. This device can be used for trapping various types of cells and expanded to trap cells in the order of tens of thousands on 1-cm2 scale area, as a promising tool to pattern large-scale single cells on specific substrates and facilitate on-chip cellular assay at the single cell level.
INTRODUCTION
Traditional cell studies rely on large cell populations (i.e., 103–106 cells), where the averaged measurements over the whole population were used to represent cellular responses.1 These conventional approaches assume that cells are homogenous and uniform. However, recent research studies have evidenced that cells in one population, even under the same condition, are indeed heterogeneous2–5 in both phenotypes and genotypes.6 As one example, the Mathies group1 revealed that the siRNA knockdown of the GAPDH gene in individual Jurkat cells leads to two distinct subgroups: one with moderate (≈50%) and the other with complete (≈0%) gene silencing. This heterogeneity is bound to be masked by conventional bulk measurements. These gradually recognized facts give rise to increasing interests in single cell analysis, enabling researchers to deal with genomics, proteomics, transcriptomics, and metabolomics systematically at the single cell resolution.7
In the field of single cell analysis, one key issue is to obtain a large number of single cells for statistical analysis, featured by high throughput.8 Conventional high-throughput methods for single cell analysis include the well-established techniques such as flow cytometry, which, however, cannot simultaneously achieve the desired sensitivity, precision, throughput, and economy.9 Dielectrophoresis-based cell manipulation,10,11 though increasingly attractive to researchers, needs expensive electric apparatus with multiple channels to produce alternating-current fields. This approach requests complicated design and fabrication of electrodes when a large number of single cells are manipulated individually.
As an alternative, microfluidics can meet the needs of high-throughput single cell analysis with favorable capabilities of reagent volume controlling, cell handling, device automation, and multiple component integration, to name just a few.12 In the last decade, numerous microfluidic devices have been proposed for single cell analysis in the fields of cell culture,13 cell differentiation,14 cell sorting and immobilization,15 inter-cellular communication,16,17 and cell signaling and responses to external stimuli.18,19
In most of these techniques, single cell trapping is a key step without which the subsequent cell manipulation and assays cannot proceed. Thus, tremendous efforts have been made in the field of single cell capture, which can be categorized into two main types: the contact approach and the noncontact approach,20 depending on whether the cells are in touch with support-providing solid surfaces.
Among noncontact methods, two approaches based on stagnation point flows and microeddies, respectively, are the most widely studied and used. Using the stagnation point flow, Start et al.21 managed to immobilize nanotubes in highly viscous solutions, and Tanyeri et al.20 trapped 100 nm-1 μm particles in low viscous solutions. Although this approach is featured with high spatial resolution, it has been rarely applied to high-efficiency cell trapping, because the average diameters of eukaryocyte cells are generally much larger than 1 μm, exceeding the upper limit for an object captured based on this approach.
On the other hand, Lin et al.22 demonstrated a Lorenz-force-driven oscillation flow which creates four stable eddies at orthogonal locations to retain particles and cells in the size range of 1–15 μm. Since the particles or cells captured in the micro-vortices are constantly circling, this method finds restricted use in cases where cell immobilization is desired. Recently, Hur et al.23 utilized inertially driven flow at sharp corners to create micro-vortices in order to selectively separate and trap (more specifically, enrich) larger cells of interest. Although this method works well for cell enrichment, it cannot be used for cell trapping and immobilization.
Due to the limitation of the “noncontact” method, most microfluidic single cell trapping devices fall into the contact type. Yang et al.24 proposed H-shaped structures which consist of two main parallel microfluidic channels connected by interconnecting microfluidic channels. Cells loaded from one of the two main channels are trapped at junction points of the H-shape structures. This idea is quite simple and straightforward. However, the device is incapable of separating trapped cells and thus it can be only treated as a quasi single cell capturing device.
Di Carlo et al.25 fabricated a Pachinko-style hydrodynamic trapping array consisting of trapping posts with recesses to capture single or low-number cells. This kind of design was reported with high trapping efficiency (>50% trap taken) and high robustness. Subsequently, this group extended the design to extract intracellular spatial-temporal information via fluorescent imaging, confirming its capability of high-throughput single cell analysis.26 Very recently, Zhang et al.27 improved the design by positioning cell trap sites on the side of the channel walls. Though it recorded success in trapping cells with predefined patterns, this approach is based on stochastic flow and cannot work with rare cell samples.
Several studies utilized the principle of “least flow resistance path” to direct single cells into the trap sites along the main channel sequentially.17,28–30 Among these methods, the most efficient demonstration to date was reported to achieve 99% single cell loading in 30 min.17 Although functioning in a robust way, this approach still needs further improvement. In order to realize the “least flow resistance path” principle, the main channel should be extended long enough to produce a higher flow resistance than the cell trapping channel, which inevitably increases the chip area and cell loading time. Therefore, a higher spatial and temporal efficiency remains to be a challenge.
Here, we proposed a passive hydrodynamic microfluidic device for high-efficiency single-cell capture based on the “least flow resistance path” principle. The trapping structure of the microfluidic device in this paper improves the previously reported serpentine geometry.28 In particular, the serpentine structure has been replaced by a series of concatenated T and inverse T junction pairs, and the geometry has been optimized and a compact form has been achieved to improve packing density and assay time. Compared with previous studies using the same principle, this design can achieve a higher spatial efficiency, i.e., capturing more cells per unit area. Under the same dimensions, the number of cell trap sites of the new design is twice that of the Takeuchi group28 as well as the West group17 and many times that of the Lutolf group.30 Furthermore, the trapping process of this design can be accomplished within 10 min to fill 400 trap sites, which is at least a 3-fold decrease in operation times compared to what was reported by the West group.17
MATERIALS AND METHODS
Conceptual design
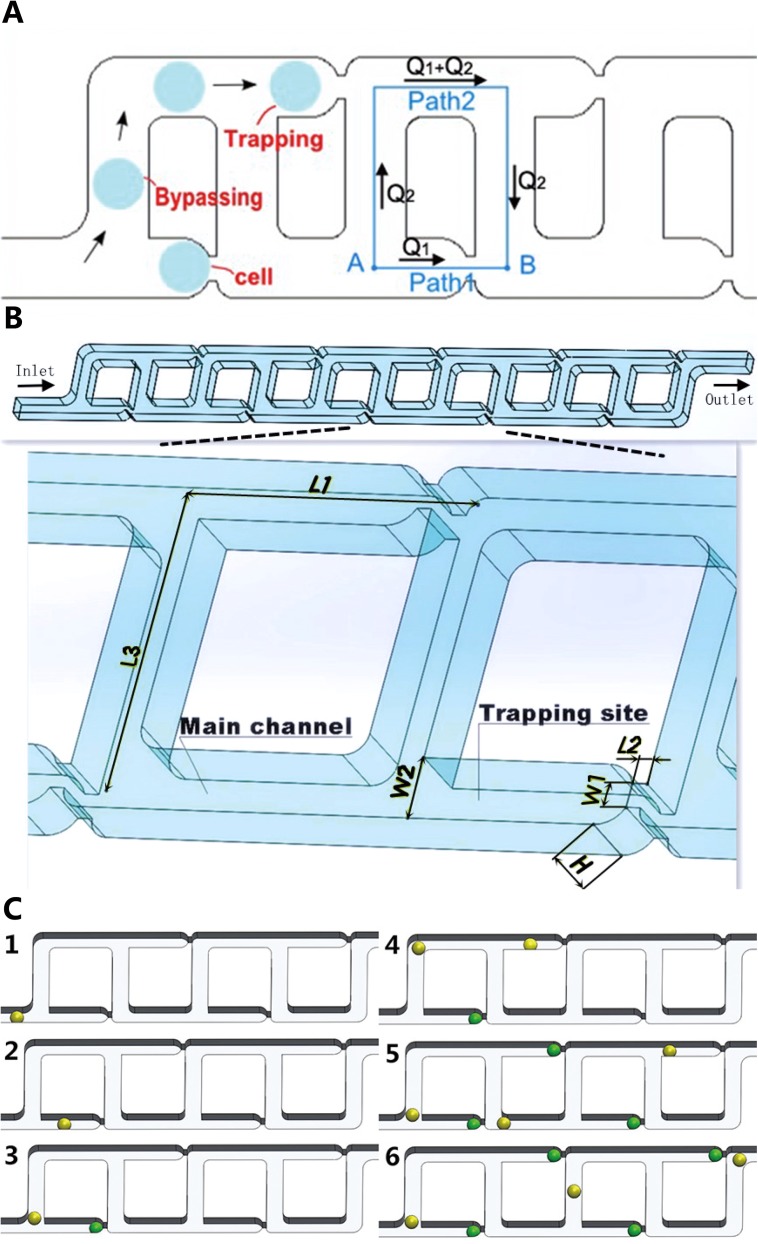
Fig. 1 shows the schematic view for the proposed microfluidic device. Resembling a ladder, each channel of the micro-device is composed of a set of connecting T and inverse T junction pairs. In each junction pair, the fluid carrying cells has two paths to flow. For example, in Fig. 1(a), from point A to B, path 1 represents a shortcut for the fluid straightforward through a narrower constriction while path 2 is a detour channel which is much longer than path 1. In this design, the main channel has a fairly constant geometry used for cell medium bypassing while the constrictions serve as trap sites. The main channel transits to the constrictions via arc contours for smooth fluid transition.
FIG. 1.
Microfluidic devices for single cell trapping. (a) Schematic view of the microfluidic channels for hydrodynamic trapping using the “least flow resistance path” principle. (b) 3-D view of the ten-trapping-site example model selected for analysis. The geometric parameters affecting the trapping efficiency are labeled. (c) Illustration of the single cell trapping principle. (1) one cell flows into the main channel, (2) hydrodynamic forces guide the cell into the first trap, (3–4) the next cell flows in and moves towards the next available trap, (5–6) the process is repeated till all the traps are filled with cells, while the hydrodynamic forces keep cells securely docked in the course of experiments. For visualization purposes, green stands for trapped cells and yellow stands for cells to be trapped.
According to the “least flow resistance path” principle, the micro-device was designed to guarantee that the volume flow rate Q1 in path 1 is higher than Q2 in path 2. In another word, Q1/Q2 (denoted as Q ratio below) should exceed 1. In this way, the majority of cell medium can be directed to pass through path 1 rather than path 2, carrying the cell in the flow into the trap.28,30 To this end, the flow resistance of path 1 should be less than that of path 2. Here, in this design, although path 1 includes a much narrower constriction which can increase the flow resistance significantly, the path 2 was carefully designed to be long enough, eventually producing a higher flow resistance than path 1.
When individual cells are transported by medium into the trap sites, they act as plugs, increasing the flow resistance of path 1 dramatically to produce a smaller-than-one Q ratio value. Under this condition, the cell medium prefers to travel in the main channel, which allows cells to travel around the filled trap site, and to be captured in the next empty trap sites in the downstream. Therefore, the device works in the following manner. When cell medium flows constantly inside the channel, all cells to be trapped line up autonomously in a queue, which moves forward as a whole along the channel, driven by the hydrodynamic forces. Every time, the first cell in the queue is trapped by the first empty trap site in the downstream of the flow. Simultaneously, the second cell in the queue moves (does NOT need to stop or wait) and gets closer to the second trap site. They become the first cell and first empty trap site at the end of each cycle. In such a repetitive and sequential manner, all sites in each channel capture single cells, as illustrated in Fig. 1(c). Note that as a proof-of-concept demonstration, the design proposed in this study has four parallel channels which work simultaneously and independently (i.e., without cross-talk). Actually, the number of parallel channels can be easily scaled up.
The microfluidic devices proposed in this study have several advantages that enhance the trapping efficiency compared to previously reported devices. First, the microfluidic device has a small footprint in each trapping unit. Due to the compact configuration of concatenated T and inverse T junction pairs through the whole channel, there is no need to deploy very long bypassing channels which take much space in a device.
Second, the “deterministic” trapping scheme saves a great deal of cell samples. In this device, the cell medium flows through everywhere of the channel and the cells in the medium are trapped one by one. The first cell in the cell queue lined up in the main channel is captured by the first empty constriction in the downstream in a repetitive way. Therefore, in principle, only the same number of cells as that of the trap sites is needed to fill all the sites. This brings a huge benefit when rare cells (e.g., stem cells or circulating tumor cells) are under manipulation.
The third advantage is high throughput. Due to the first advantage featured with a small footprint for the trap sites, cell medium flowing a certain distance can traverse more trap sites. This indicates a faster speed of cell trapping. Furthermore, a large number of sites (e.g., M) in one channel and multiple parallel channels (e.g., N) in one microfluidic device can be deployed, producing a trapping of M by N cells in each batch.
Theoretical modeling
To model this design mathematically, the Darcy-Weisbach equation is used to determine the pressure drop or pressure difference in the micro channels and the Hagen-Poiseuille flow problem solution is applied in this situation.28 The pressure difference can be expressed by the following equation:31
| (1) |
where ƒ is the Darcy friction factor, L is the length of the channel, ρ is the fluid density, V is the average velocity of the fluid, D is the hydraulic diameter. represents the sum of losses due to the liquid contraction, convergence, scattering, and the resulting vortices caused by complicated geometry. It has neither explicit and compact mathematical forms nor empirical values in literature for precise modeling in this paper and will be therefore treated by the equivalent substitution method.
In case of a typical geometry, the maximum Reynolds number calculated at experimental flow speeds is less than 10, indicating laminar characteristics of the flow system. D can be further expressed as 4A/P for a rectangular channel, and V as Q/A, where A and P are the cross-sectional area and the perimeter of the channel, and Q is the volume flow rate. The Darcy friction factor ƒ is related to the aspect ratio, α, and Reynolds number, , where μ is the fluid viscosity. The aspect ratio is defined as either height/width or width/height such that 0 ≤ α ≤ 1.
For fully developed laminar flow in rectangular channels, the following expression32 is used to predict the friction constant C(α):
| (2) |
In order to form a fully developed laminar flow, L/D > 300 is the minimum requirement.33 Regarding the device proposed in this study, since L/D is less than 10, the micro channels in the device are not long enough for the flow to become fully developed under laminar flow conditions. In such a case, the following expression32 for friction factors accounts for both the developing and fully developed laminar flow regions in the channels:
| (3) |
where is calculated as in Eq. (2) and the entrance length is defined as
| (4) |
To shorten the terms in mathematical derivation, we ignore the losses in Eq. (1) and by substituting Eqs. (2)–(4) to Eq. (1), the expression is obtained as follows:
| (5) |
Through a series of deductions and calculations shown in the Appendix, it is found that when Q2 < 600 nL/min, the Q ratio can be explicitly expressed by the following simplified equation:
| (6) |
where W1, W2, L1, L2, L3, and H are geometric parameters shown in Fig. 1, , , , .
Equation (6) has not yet incorporated the fluidic losses of Eq. (1). By taking the equivalent substitution method, all the fluidic losses could be treated collectively in a simplified form as K times of any term in the algebraic fraction of Eq. (6). Because the narrow segment of path 1 has the most significant (or non-negligible) losses due to the vortices caused by contracting in one end and scattering in the other end, we only put the factor K to the middle term of the denominator and have
| (7) |
where K is a constant to be determined. Since in the current design, the fluidic volume flow rate used in experiments (typically 3.75 nL/min) is at least 2-order of magnitude lower than 600 nL/min, Eq. (7) can be used to calculate the Q ratio. It is indicated that within a certain range of Q2, the values of the Q ratio are only determined by the geometry of microfluidic channels, independent of Q or V.
The Q ratio is an important indicator of the trapping efficiency of the current design. Literature28 indicates that its suggested values should be higher than 1. With this goal in mind, the design variables of the channels were fine tuned for trapping efficiency optimization.
Device design and fabrication
The 2-D device design for photomask fabrication was drawn using L-edit (Tanner EDA Inc., USA). Subsequent prototyping and fabrication was conducted based on soft photolithography, including single-layer SU-8 mold fabrication and PDMS molding.
Briefly, the process of SU-8 molding first incorporates seed layer fabrication where SU-8 5 (MicroChem Corp., USA) was spin coated on clean glass slides (500 rpm 10 s, 2500 rpm 35 s, expected height is 5 μm), pre baked (65 °C 1 min, 95 °C 3 min), UV exposed without a photomask (15 mw/cm2 6 s), post-exposure baked (65 °C 1 min, 95 °C 1 min), and hard baked (175 °C 2 h).
Then SU-8 25 was spin coated on top of the seed layer to form a ∼25 μm thick cell trapping layer (500 rpm 10 s, 1800 rpm 35 s) or to form a ∼15 μm thick cell trapping layer (500 rpm 10 s, 2000 rpm 35 s), followed by pre baking (65 °C, 2 min; 95 °C, 5 min), UV exposure using a prepared photomask (15 mw/cm2 3.5 s), post-exposure bake (65 °C 1 min, 95 °C 3 min), development (SU-8 developer), and hard bake (175 °C 2 h). In the whole process of experiments, two photomasks were used to fabricate two configurations of the trapping array, one with H = W2 = 15 μm (array 1) and the other one with H = W2 = 25 μm (array 2). Other parameters of the two arrays were changed correspondingly.
For the PDMS molding, 10:1 elastomer base and curing agent (Sylgard 184, Dow Corning) were mixed to form PDMS which was then decanted into a petri dish whose bottom laid the SU-8 mold, followed by degassing (30 min) to remove any residual air bubbles. After curing the PDMS in an oven for 6 h at 80 °C, the mold was dissembled with tubing interconnects punched out manually. After being cleaned using absolute ethyl alcohol, the PDMS layer was irreversibly bonded to a glass slide using oxygen plasma based surface activation or reversibly pasted onto the glass slide by degassing for cell trap experiments.
Computational analysis and numerical simulation
In order to investigate the effects of geometric parameters on the hydrodynamic cell trapping efficiency, finite-volume based computational simulation was conducted based on ANSYS 14.0 (ANSYS Inc., USA). The Geometry package was used to create the 3-D geometry model, the Mesh package was then used to generate meshes and finally the Fluent package was used to solve the incompressible Navier-Stokes equation. In this model, boundary conditions included an inlet flow velocity between 0.0001 and 10 m/s and a zero pressure setup at the outlet. All the other surfaces were defined as the no-slip conditions. Living cells were assumed as solid spheres in contact with the traps.
As mentioned above, the Q ratio must be higher than 1 and the increase in the Q ratio can lead to a higher probability of cell trapping. Thus, in simulations, the effects of geometric parameters on the Q ratio were extensively investigated for parameter optimization.
The results of the Q ratio calculated by mathematical modeling were compared to the values obtained via numerical simulations, where 12 different combinations of geometric parameters were studied and categorized into three groups in line with three values of W1. Within each group, W2 = H = 25 μm remains constant where the default values are: L1 = 90 μm, L2 = 6 μm, L3 = 90 μm. For each simulation, only one variable was altered with respect to the default values, following the controlling variable method.
Cell culture and medium formulation
The HeLa (human cervical carcinoma) cell line and the HEK293T (human embryonic kidney cells) cell line were used in experiments. For HEK293T, cells were preliminarily transfected with a lentivirus stably expressing GFP (green florescent protein) for downstream fluorescent microscope observation. Both cell lines were maintained by passaging three times weekly with Dubelcco's Modified Eagle Medium (DMEM, Life Technologies, USA) supplemented with 10% fetal bovine serum (FBS, Life Technologies, USA), 100 U/mL penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich, USA).
For loading cells in single-cell trapping experiments, adherent cells were detached from 100 mm diameter culture dishes (Corning, USA) with 2 ml 0.05% trypsin EDTA (Gibco, USA). After the detachment process, an equal amount of DMEM + FBS was then added to deactivate remaining trypsin. Cells were then centrifuged at 1200 g for 5 min to a pellet and re-suspended in the DMEM medium. Single-cell trapping experiments were conducted immediately after re-suspension within 30 min to reduce non-specific adhesion to surfaces.
Microfluidic system operation
In each cell trapping experiment, the microfluidic device was first primed with the DMEM cell culture medium to remove air bubbles. Then cells were loaded by injecting cell suspensions into the microfluidic device positioned on the microscopic stage. For observation of the trapping process, photographs and videos were captured via a digital camera (Canon Corp., Japan) which was coupled on an inverted microscope (Nikon Eclipse Ti, Nikon Corp., Japan). GFP-labeled HEK293T cells were imaged via a green florescent channel filter.
Cell debris is a trouble-maker in cell handling, and thus we paid extra care to this issue in our experiment. Generally, in healthy conditions, cell types used in our experiment did not actually produce enough debris that would cause the experiment to totally fail. To minimize cell debris, we still took two measures. First, during cell suspension preparation, we washed cells in petri dish twice before trypsinizing for cell detachment. This procedure could eliminate most of cell debris which might potentially block the channels. If there were still considerable amounts of debris after detachment, we further centrifuged cell suspensions with 400 g for 3 min to remove the debris at the supernatant. Second, we actually fabricated funnel ratchets34 across the inlet channel to filter out cell debris while allowing cells to pass. These ratchets worked well in experiment to stop any residual debris in the medium.
RESULTS AND DISCUSSION
Numerical simulations
The design in Fig. 1 was simulated to obtain the Q ratio which indicates trapping efficiency for different geometric configurations of microfluidic channels. Treating the cell as a sphere model, the length W2 and width H of the cross section of the main channel were set equal in consistency with the diameter range of trapped cells. More specifically, the diameters of HeLa and HEK293T cells cultured in this study are about 10–20 μm, and thus W2 and H can be specified as 25 μm which is larger than the largest cells to avoid cell blocking. With constant values of W2 and H, L1, L2, L3, and W1 are four variables that can affect the trapping efficiency or the Q ratio, which were investigated in the simulation. A set of values for the four variables were chosen as default values based on a good guess from literatures: L1 = 90 μm, L2 = 6 μm, L3 = 90 μm, W1 = 9 μm.
With this set of parameters, simulations were first conducted to obtain the velocity contour and the pressure distribution in the channels as shown in Fig. 2. The visualization clearly shows that the constrictions undergo the greatest fluidic pressure drop. On the other hand, non-uniform distributions of fluidic velocities and pressure drops along the main channel are also revealed. Although in this figure the inlet flow rate 10 nl/min is rather low, we observed the same patterns of velocity contour and pressure distribution when the inlet flow rate was increased by 5-order of magnitude to 1 ml/min.
FIG. 2.
Computational simulation results for laminar flow (without particles or cells) in microfluidic channels. View was taken on the vertical middle plane (half way of the channel height) with perfusion at a flow rate of 10 nl/min in the inlet. (a) Velocity contour (m/s). (b) Dynamic pressure contour (Pa).
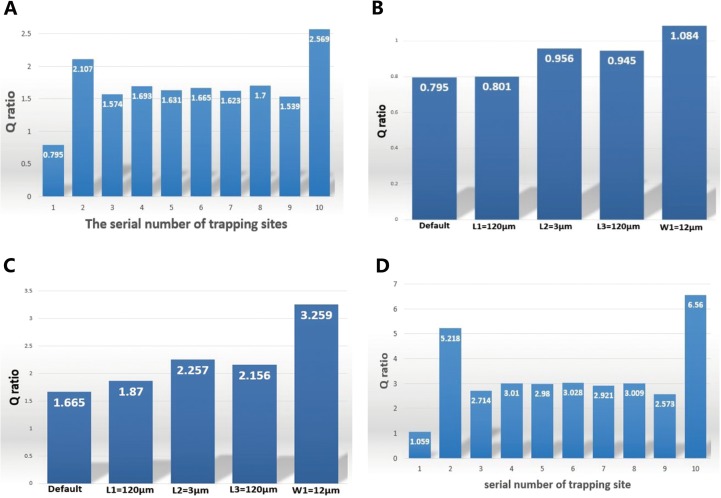
Using the simulation data for the default setup, the Q ratio values were calculated for ten trap sites and plotted in Fig. 3(a), which exhibits three representative patterns. (i) The first trap site is shown to have the lowest Q ratio value while the last one possesses the highest value, due to their boundary locations in the microfluidic channels. (ii) The second trap site is shown to have a relatively higher Q ratio value than the surrounding sites because of the influence of the first one. (iii) Intermediate sites (i.e., sites 3 to 9) have the similar level of the Q ratio, indicating that the microfluidic channel can be extended with an infinite number of trap sites without any significant loss in the trapping efficiency.
FIG. 3.
Trapping efficiency (Q ratio) for various configurations of the channel geometries by computational simulation. (a) Q ratio values of the ten trap sites with default parameters: L1 = 90 μm, L2 = 6 μm, L3 = 90 μm, W1 = 9 μm. W2 and H were set to be 25 μm and only one of L1, L2, L3, and W1 was modulated with respect to the default parameters each time. (b) Q ratio values of the first site when L1, L2, L3, and W1 were separately changed. (c) Q ratio values of the sixth site when L1, L2, L3, and W1 were separately changed. (d) Q ratio values of the optimized design with the following parameters: L1 = 120 μm, L2 = 6 μm, L3 = 120 μm, W1 = 10 μm.
To further study the effects on the Q ratio induced by the four variables, further simulations were conducted where the default set was changed. In each simulation, only one of the four variables with respect to the default values was changed and the Q ratio values for the first and the sixth trap site were calculated. The first site was chosen because it is the trigger for the trapping to initiate, featured with the smallest Q ratio in the channel. It is imperative in the design to guarantee a greater-than-one value of the Q ratio in order to retain the trapping efficiency. The sixth site was chosen to represent these intermediate sites. Figs. 3(b) and 3(c) show the Q ratio values for the first and sixth sites, respectively, when one of the variables was changed. It was found that the increase of L1, L3, and W1 can lead to the increase of the Q ratio while the decrease of L2 can have the same effect. This trend agrees with the qualitative indication of the mathematical model.
Guided by the above-mentioned findings, an optimal design was obtained based on the following parameters: L1 = 120 μm, L2 = 6 μm, L3 = 120 μm, W1 = 10 μm, for which the quantified Q ratio values of all trap sites were shown in Fig. 3(d). Clearly, Q ratio values of all trap sites are higher than 1 and except for the first site, all the others are higher than 2, producing high capturing performance.
To verify the single cell sequential trapping, the Q ratio distribution pattern was calculated when one, two, and three cells are trapped by the first site, the first two sites, and the first three sites, respectively. As shown in Table I, when one site is taken by one cell, the Q ratio of that particular site drops to nearly zero (highlighted in bold), losing the fluidic flow to attract any more cells and thus ensuring single cell capture. Meanwhile, the Q ratio of the next site is shown to remain higher than 1 with a moderate decrease (highlighted in italics). In other words, the next site can substitute in as the first site to enable single cell trapping.
TABLE I.
Q ratio values of the ten sites of the optimized model when zero, one, two and three cells were trapped, respectively. When one site was taken by a cell, the Q ratio of that site was noticed to drop to a very low level (highlighted in bold), losing the ability to capture a second cell, while the Q ratio of the next site remains higher than 1 (highlighted in italics), retaining the ability to trap the next coming cell.
| Trap site | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| No cell trapped | 1.056 | 1.059 | 2.714 | 3.010 | 2.980 | 3.028 | 2.921 | 3.009 | 2.573 | 6.560 |
| One | 0.468 | 1.919 | 3.334 | 2.839 | 3.003 | 3.005 | 2.917 | 2.997 | 2.557 | 6.550 |
| Two | 0.508 | 0.712 | 1.726 | 3.454 | 2.888 | 2.990 | 2.910 | 3.015 | 2.549 | 6.561 |
| Three | 0.510 | 0.821 | 0.676 | 1.738 | 3.523 | 2.891 | 2.952 | 3.019 | 2.586 | 6.522 |
To validate the assumption that in the mathematical model there exists a threshold of Q2 below which the Q ratio is independent of the flow rate, a group of simulations were conducted. Specifically, six values for the inlet velocity with increasing magnitudes were tested in simulation and the corresponding Q ratio values were obtained (Table II). It clearly shows that when the inlet velocity is below 0.01 m/s, the Q ratio remains constant regardless of the inlet velocity. When the inlet velocity exceeds 0.01 m/s, the increase in the inlet velocity leads to the decrease in the Q ratio, which is consistent with the mathematical model.
TABLE II.
Relationship between the Q ratio and the inlet velocity revealed by simulation.
| Inlet velocity (m/s) | 0.0001 | 0.001 | 0.01 | 0.1 | 1 | 10 |
|---|---|---|---|---|---|---|
| Q2 (nL/min) | 3.75 | 37.5 | 375 | 3750 | 3.75 × 104 | 3.75 × 105 |
| Q ratio | 1.665 | 1.665 | 1.665 | 1.657 | 1.506 | 1.127 |
In order to determine a proper value of K in the mathematical modeling, we obtained the pressure drop for each segment of both paths via the mathematical model and numerical simulation for the optimized structure. Through comparison, it was verified that the pressure drop differences between modeling and simulation for all the segments other than the narrow one are less than 5% and thus negligible. We further determined the value of K (K = 2.2) as the ratio of the pressure drop of the narrow segment of path 1 calculated by simulation to that by modeling.
With this K value, we compared the Q ratios obtained by modeling and simulation for different geometric parameters (mentioned in the section “Computational analysis and numerical simulation”), by keeping a typical inlet velocity (0.0001 m/s). The result is shown in Table III, from which we found that the error is not that significant with the maximum of ∼16% and the minimum of ∼2%.
TABLE III.
Comparison of the Q ratio results between mathematical modeling and computational simulation when K = 2.2. 12 different dimension combinations were studied and categorized into three groups in line with the value of W1: A) W1 = 9 μm B) W1 = 12 μm C) W1 = 15 μm. Default parameters are: W2 = 25 μm, L1 = 90 μm, L2 = 6 μm, L3 = 90 μm. In each simulation, only one of L1, L2, L3 was altered with respect to the default parameters.
| A | Default | L1 = 120 μm | L2 = 3 μm | L3 = 120 μm |
|---|---|---|---|---|
| Simulation | 1.665 | 1.870 | 2.257 | 2.156 |
| Modeling | 1.428 | 1.632 | 2.629 | 1.836 |
| B | Default | L1 = 120 μm | L2 = 3 μm | L3 = 120 μm |
| Simulation | 3.259 | 3.812 | 4.122 | 3.947 |
| Modeling | 3.131 | 3.578 | 4.826 | 4.025 |
| C | Default | L1 = 120 μm | L2 = 3 μm | L3 = 120 μm |
| Simulation | 5.617 | 6.484 | 7.401 | 6.748 |
| Modeling | 5.455 | 6.295 | 8.241 | 7.134 |
Single cell trapping
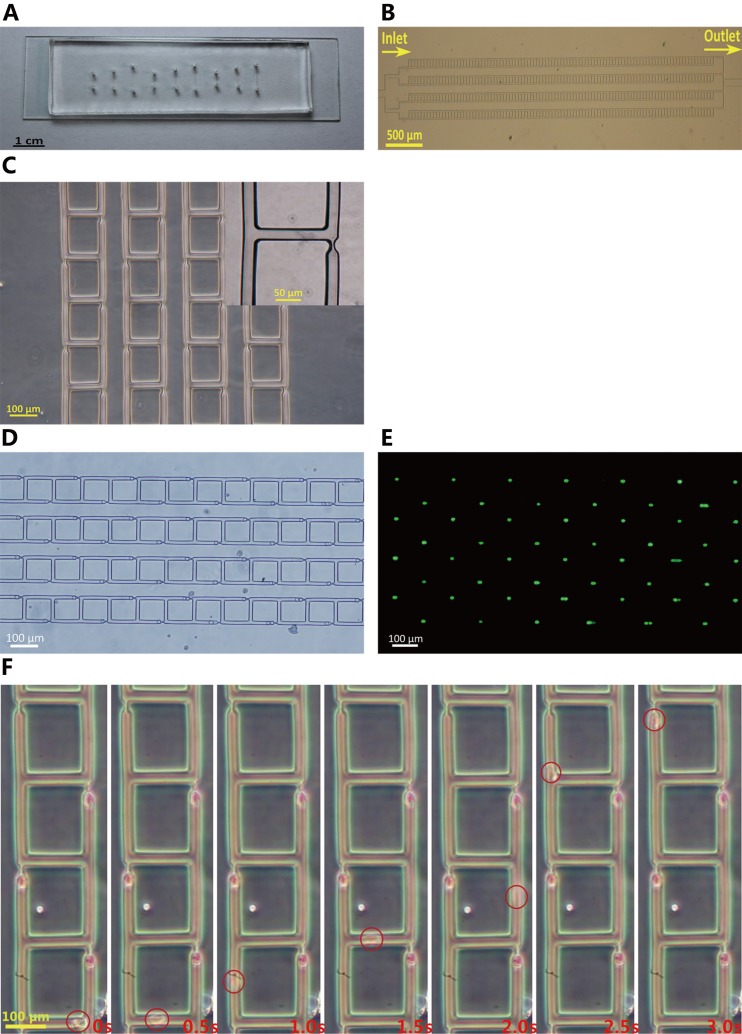
For the demonstration purpose, this work fabricated microfluidic devices with four parallel channels, as shown in Figs. 4(a)–4(c). Microfluidic devices based on the least flow resistance principle were proved to have high reliability and efficiency. Human cells including HeLa and HEK293T cells (HeLa 15.0 ± 4.6 μm, HEK293T 13.0 ± 3.9 μm) produced 100% cell trapping and ∼90% single cell immobilization. As one example, Figs. 4(d) and 4(e) show bright-field and fluorescent images of trapped HEK293T cells, confirming the feasibility of single-cell trapping. Fig. 4(f) illustrates the dynamic process of single-cell trapping, confirming that the trapped cells work as plugs to block the constrictions and prevent the coming of subsequent cells.
FIG. 4.
Single cell trapping experimental results. (a) A picture of the micro-fabricated PDMS micro-device (array 2). The PDMS layer was reversibly pasted onto the glass slide by degassing. (b) Microscopic picture of the single cell trapping array (array 2), featured with symmetrical microfluidic channel networks and microarrays of traps. The four channels are connected in parallel to a common inlet and outlet. (c) Zoom-in view of the microarray (array 2). (d) Bright-field microscopic image of cells trapped in the constrictions (array 1). Cells are on average of 13 μm in diameter. (e) Fluorescent microscopy image of the same cells (array 1) shown in (d). The green dots represent single cells trapped on site. (f) Time-elapsed view of trapping of a single cell (array 1).
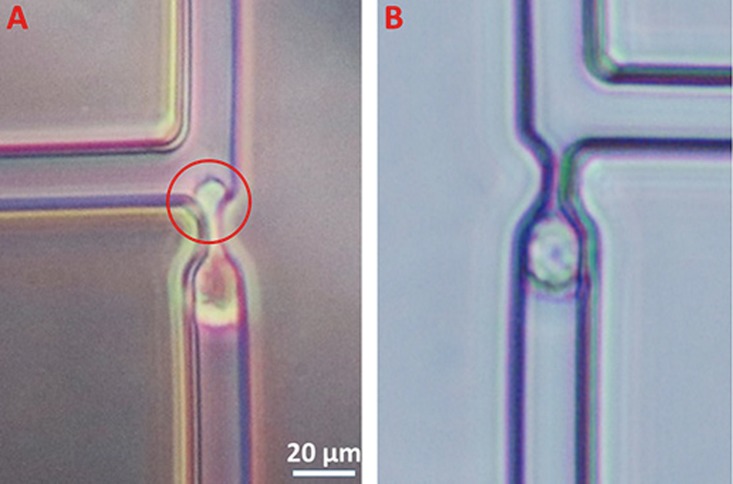
The flow rates were determined in consideration of two main factors: the throughput and the shear stress imposed on cells. If we consider the first factor alone, the flow rate can be set as high as possible so long as the Q ratio remains greater than 1. Table II indicates that we could at least set a flow rate of 375 μl/min, which makes sense for high-throughput. However, for trapping cells which are the focus of this work, we have to consider shear stress seriously. According to others' work,17 a mean flow velocity of 50 μm/s results in a shear stress maxima of 0.7 dyn/cm2 in similar trap site structure, while vascular endothelial cells could endure up to a shear stress of 10 dyn/cm2. As shear stress scales linearly with the velocity, the flow velocity in our experiment could be increased by ∼15 times to 750 μm/s. Initially, a syringe pump (Legato 200, KD Scientific Inc., USA) was used as the driving source to infuse the cell solution into the trapping array, which produced a relatively high flowing velocity with a minimum of ∼250 μm/s. However, such velocities can produce high shear stresses at the narrow constrictions, which squeeze cells through the constrictions with high-degree deformations as shown in Fig. 5(a). High shear stresses may also cause severe mechanical damages to the trapped cells and decrease cellular viabilities significantly.35
FIG. 5.
Comparison of trapped cell deformations for liquid driving forces based on (array 1) syringe pump (a) and gravity (b).
Fortunately, the trapping efficiency of the device proposed in this study is velocity independent under our experimental flow speed, suggested by both mathematical calculation and numerical simulations. Thus, the device can be operated at a low speed (≤100 μm/s) enabled by gravity based fluid flow, without the concern of cell deformation (Fig. 5(b)). Specifically, the syringe was removed from the pump and then adjusted with a relative height compared to the microfluidic device to provide a different driving force. In this way, a wide range of velocity from negative to zero to positive can be easily realized without the need of the pump. In our experiment, when the cell medium was fed into the channel at 100 μm/s, it took about 10 min to fill 400 trap sites with cells. Compared to previous work,17 which took about 30 min to fill 200 trap sites at a flow speed of 50 μm/s, this work reduced at least 3-fold operation time. Also very interestingly, our device was shown to be capable of releasing the trapped cells by reversely infusing culture solutions, which was achieved by placing the syringe lower than the microfluidic devices.
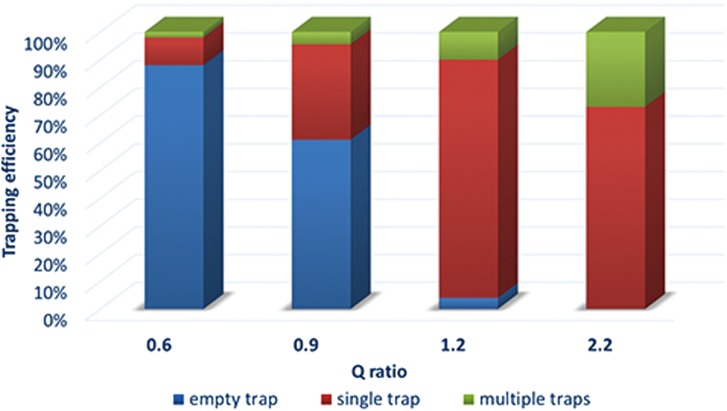
To verify the effect of the varying Q ratio values on cell trapping, the second device (array 2) with four arrays each having a different Q ratio was fabricated and experimentally tested to compare the trapping efficiency. As shown in Fig. 6, when the Q ratio was adjusted across the bar of 1.0 (say from 1.2 to 0.9), the empty trapping rate was shown to increase significantly, strongly supporting the effectiveness of the Q ratio as the determinant factor in cell trapping proposed by Tan and Takeuchi.28 On the contrary, when the Q ratio was increased to values much higher than 1.0 (say from 1.2 to 2.2), multiple-cells trapping within one trap site was observed. This happened because higher Q ratio values can reduce the plug effect of the trapped single cell, yielding a higher probability of trapping a second or more cells. In summary, it is not always true that higher Q ratio values can definitely lead to higher trapping performances. According to Fig. 6, Q ratio values ranging somewhere between 1.2 and 2.2 can achieve high efficiency for single cell trapping on site. To further optimize the Q ratio range, in future we plan to design and fabricate more devices with different Q ratio values ranging from 1.2 to 2.2 at a small interval (i.e., 0.1) and conduct more experiments with particles and cells.
FIG. 6.
Trapping efficiency vs different Q ratio values from 0.6 to 2.2 (array 2).
Several factors were found to influence the single-cell trapping efficiency of the device proposed in this study. The most important factor is the size variance of cells. Normally, the width and height of the microfluidic channel was designed big enough to house the largest cells. It is rare but still possible for largest cells to block the main channel and compromise the device. By contrast, once a cell with a significant small diameter is loaded into the microfluidic devices, it fails to fill the constriction adequately, which may lead to the trapping of more subsequent cells. From this perspective, it is always desirable to have rather uniform cells to obtain better trapping performance.
The second factor comes from the cell disaggregation in the trypsin treatment used for cell harvesting. In the cell harvesting process, EDTA was used for trypsin treatment and it was observed that sometimes several cells formed a cluster, leading to multiple-cells trapping. In order to deal with this issue, a more aggressive tissue dissociation solution may be adopted, e.g., accutase (Innovative Cell Technologies Corp., USA) which was reported36 to yield a better performance with a decreased occurrence of cell aggregates. Besides cell disaggregation, cell density in the cell suspension solution is also important. Though a higher density could lead to a faster trapping, it should be controlled carefully to ensure that multiple cells do not flow into the same trap site. In our experiment, a typical cell density of ∼5 × 105/ml was used.
The third factor is the height of the channel. When the ratio of height to cell diameter was changed from 1.0 to 1.8, the single cell trapping rate was shown to decrease from 87.5% to 53.0%, indicating that the channel height cannot be too higher than the diameter of cells. Like the effect of size variance, if the height to cell diameter ratio is too high, a small cell cannot fill a constriction completely and the corresponding medium leakage can bring another cell into the same site already taken by the small cell. This factor is compatible with the suggestion that the channel height should be set to 1–1.4 times of the cell diameter to achieve one-cell-on-one-trap raised by Tan and Takeuchi.28
CONCLUSION
A novel microfluidic single cell trapping device was introduced and demonstrated based on the differential flow resistance circuits. Both mathematical modeling and numerical simulations were conducted to guide geometry choice and optimization, leading to a high trapping efficiency. Applying the fine-tuned geometric parameters to the fabrication of two arrays, 100% cell trapping and ∼90% single cell trapping were achieved in a “deterministic” manner, which was relatively high among various microfluidic trapping devices. In summary, this microfluidic device bears great potential to advance various cell studies at the single-cell level such as metabolism, pharmacokinetics, drug toxicity, shear stress activation, chemical signaling pathway activation, and inhibition.
ACKNOWLEDGMENTS
This work was supported by the NSFC (Grant Nos. 61376120, 61201077, and 31170940), the One-Thousand Young Talent Program of China, and the National Instrumentation Project (No. 2013YQ19046701).
APPENDIX: DERIVATION PROCESS OF THE Q RATIO
Fluid can flow from junction A to B via path 1 or 2 and the distribution of volume flow rate in each path in the steady state is marked in Fig. 1(a). Equation (5) can be applied, respectively, to paths 1 and 2. For path 2, referring to the variables in Fig. 1(b), the pressure difference is represented as
| (A1) |
where , , , , , , .
For path 1, there are three fluidic pressure components corresponding to the channel's three geometric parts of wide, transitional and narrow segments. The pressure drops for wide and narrow segments resemble Eq. (A1), and the pressure drop for the transitional segment is represented by
| (A2) |
where , , , , , .
Thus, the total pressure applied on path 1 is represented as
| (A3) |
where , , , , , , .
The pressure drop from point A to B is the same for both paths, yielding
| (A4) |
Since it is rather difficult to get the analytical solution to the Q ratio from this equation, the mathematical software MATLAB (MathWorks Inc., USA) was used for obtaining its numerical solutions by varying the values of Q2 (or Q1) for default geometric parameters and the results are shown in Table IV.
TABLE IV.
Q ratio values calculated by mathematical modeling for Q2 of different orders of magnitude with default geometric parameters.
| Q2(nL/min) | 6 | 60 | 600 | 6000 | 6 × 104 | 6 × 105 |
|---|---|---|---|---|---|---|
| Q ratio | 1.43 | 1.43 | 1.43 | 1.38 | 1.27 | 1.01 |
Table IV indicates that when Q2 < 600 nl/min, the Q ratio does not change as Q2. Indeed, when Q2 < 600 nl/min, the difference between C and C(α) is less than 0.6%, leading to the following approximations:
Such approximations can be used for simplifying Eq. (A4).
References
- 1.Toriello N. M., Douglas E. S., Thaitrong N., Hsiao S. C., Francis M. B., Bertozzi C. R., and Mathies R. A., “ Integrated microfluidic bioprocessor for single-cell gene expression analysis,” Proc. Natl. Acad. Sci. U.S.A. 105, 20173–20178 (2008). 10.1073/pnas.0806355106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poste G., Tzeng J., Doll J., Greig R., Rieman D., and Zeidman I., “ Evolution of tumor cell heterogeneity during progressive growth of individual lung metastases,” Proc. Natl. Acad. Sci. U.S.A. 79, 6574–6578 (1982). 10.1073/pnas.79.21.6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irish J. M., Kotecha N., and Nolan G. P., “ Mapping normal and cancer cell signalling networks: Towards single-cell proteomics,” Nat. Rev. Cancer 6, 146–155 (2006). 10.1038/nrc1804 [DOI] [PubMed] [Google Scholar]
- 4.Graf T. and Stadtfeld M., “ Heterogeneity of embryonic and adult stem cells,” Cell Stem Cell 3, 480–483 (2008). 10.1016/j.stem.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 5.Walling M. A. and Shepard J. R. E., “ Cellular heterogeneity and live cell arrays,” Chem. Soc. Rev. 40, 4049–4076 (2011). 10.1039/c0cs00212g [DOI] [PubMed] [Google Scholar]
- 6.Lawrenz A., Nason F., and Cooper-White J. J., “ Geometrical effects in microfluidic-based microarrays for rapid, efficient single-cell capture of mammalian stem cells and plant cells,” Biomicrofluidics 6, 024112 (2012). 10.1063/1.4704521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Chen Z. Z., and Li Q. L., “ Microfluidic techniques for dynamic single-cell analysis,” Microchim. Acta 168, 177–195 (2010). 10.1007/s00604-010-0296-2 [DOI] [Google Scholar]
- 8.Yin H. and Marshall D., “ Microfluidics for single cell analysis,” Curr. Opin. Biotechnol. 23, 110–119 (2012). 10.1016/j.copbio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 9.Lecault V., White A. K., Singhal A., and Hansen C. L., “ Microfluidic single cell analysis: From promise to practice,” Curr. Opin. Chem. Biol. 16, 381–390 (2012). 10.1016/j.cbpa.2012.03.022 [DOI] [PubMed] [Google Scholar]
- 10.Pethig R., “ Review article-dielectrophoresis: Status of the theory, technology, and applications,” Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benhal P., Chase J. G., Gaynor P., Oback B., and Wang W. H., “ AC electric field induced dipole-based on-chip 3d cell rotation,” Lab Chip 14, 2717–2727 (2014). 10.1039/c4lc00312h [DOI] [PubMed] [Google Scholar]
- 12.Lu X., Huang W. H., Wang Z. L., and Cheng J. K., “ Recent developments in single-cell analysis,” Anal. Chim. Acta 510, 127–138 (2004). 10.1016/j.aca.2004.01.014 [DOI] [Google Scholar]
- 13.Carlo D. D., Wu L. Y., and Lee L. P., “ Dynamic single cell culture array,” Lab Chip 6, 1445–1449 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- 14.Gao X. H., Zhang X., Xu H., Zhou B. P., Wen W. J., and Qin J. H., “ Regulation of cell migration and osteogenic differentiation in mesenchymal stem cells under extremely low fluidic shear stress,” Biomicrofluidics 8, 052008 (2014). 10.1063/1.4896557 [DOI] [Google Scholar]
- 15.Karimi A., Yazdi S., and Ardekani A. M., “ Hydrodynamic mechanisms of cell and particle trapping in microfluidics,” Biomicrofluidics 7, 21501 (2013). 10.1063/1.4799787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L., Chu Y. S., Thiery J. P., Lim C. T., and Rodriguez I., “ Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns,” Lab Chip 13, 714–721 (2013). 10.1039/c2lc41070b [DOI] [PubMed] [Google Scholar]
- 17.Frimat J. P., Becker M., Chiang Y. Y., Marggraf U., Janasek D., Hengstler J. G., Franzke J., and West J., “ A microfluidic array with cellular valving for single cell co-culture,” Lab Chip 11, 231–237 (2011). 10.1039/c0lc00172d [DOI] [PubMed] [Google Scholar]
- 18.Chung K., Rivet C. A., Kemp M. L., and Lu H., “ Imaging single-cell signaling dynamics with a deterministic high-density single-cell trap array,” Anal. Chem. 83, 7044–7052 (2011). 10.1021/ac2011153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin R. S., Root P. D., and Spence D. M., “ Microfluidic technologies as platforms for performing quantitative cellular analyses in an in vitro environment,” Analyst 131, 1197–1206 (2006). 10.1039/b611041j [DOI] [PubMed] [Google Scholar]
- 20.Tanyeri M., Johnson-Chavarria E. M., and Schroeder C. M., “ Hydrodynamic trap for single particles and cells,” Appl. Phys. Lett. 96, 224101 (2010). 10.1063/1.3431664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Start P. R., Hudson S. D., Hobbie E. K., and Migler K. B., “ Breakup of carbon nanotube flocs in microfluidic traps,” J. Colloid Interface Sci. 297, 631–636 (2006). 10.1016/j.jcis.2005.11.044 [DOI] [PubMed] [Google Scholar]
- 22.Lin C. M., Lai Y. S., Liu H. P., Chen C. Y., and Wo A. M., “ Trapping of bioparticles via microvortices in a microfluidic device for bioassay applications,” Anal. Chem. 80, 8937–8945 (2008). 10.1021/ac800972t [DOI] [PubMed] [Google Scholar]
- 23.Hur S. C., Mach A. J., and Di Carlo D., “ High-throughput size-based rare cell enrichment using microscale vortices,” Biomicrofluidics 5, 22206 (2011). 10.1063/1.3576780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Li C. W., and Yang J., “ Cell docking and on-chip monitoring of cellular reactions with a controlled concentration gradient on a microfluidic device,” Anal. Chem. 74, 3991–4001 (2002). 10.1021/ac025536c [DOI] [PubMed] [Google Scholar]
- 25.Carlo D. D., Aghdam N., and Lee L. P., “ Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays,” Anal. Chem. 78, 4925–4930 (2006). 10.1021/ac060541s [DOI] [PubMed] [Google Scholar]
- 26.Gossett D. R., Weaver W. M., Ahmed N. S., and Carlo D. D., “ Sequential array cytometry: multi-parameter imaging with a single fluorescent channel,” Ann. Biomed. Eng. 39, 1328–1334 (2011). 10.1007/s10439-010-0199-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Chouc C. K., Xia X. F., Hung M. C., and Qin L. D., “ Block-cell-printing for live single-cell printing,” Proc. Natl. Acad. Sci. U.S.A. 111, 2948–2953 (2014). 10.1073/pnas.1313661111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan W. H. and Takeuchi S., “ A trap-and-release integrated microfluidic system for dynamic microarray applications,” Proc. Natl. Acad. Sci. U.S.A. 104, 1146–1151 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akagi J., Khoshmanesh K., Evans B., Hall C. J., Crosier K. E., Cooper J. M., Crosier P. S., and Wlodkowic D., “ Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos,” PLoS One 7, e36630 (2012). 10.1371/journal.pone.0036630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobel S., Valero A., Latt J., Renaud P., and Lutolf M., “ Optimization of microfluidic single cell trapping for long-term on-chip culture,” Lab Chip 10, 857–863 (2010). 10.1039/b918055a [DOI] [PubMed] [Google Scholar]
- 31.Judy J., Maynes D., and Webb B. W., “ Characterization of frictional pressure drop for liquid flows through microchannels,” Int. J. Heat Mass Transfer 45, 3477–3489 (2002). 10.1016/S0017-9310(02)00076-5 [DOI] [Google Scholar]
- 32.Liu D. and Garimella S. V., “ Investigation of liquid flow in microchannels,” J. Thermophys. Heat Transfer 18, 65–72 (2004). 10.2514/1.9124 [DOI] [Google Scholar]
- 33.Kohl M. J., Abdel-Khalik S. I., Jeter S. M., and Sadowski D. L., “ An experimental investigation of microchannel flow with internal pressure measurements,” Int. J. Heat Mass Transfer 48, 1518–1533 (2005). 10.1016/j.ijheatmasstransfer.2004.10.030 [DOI] [Google Scholar]
- 34.McFaul S. M., Lin B. K., and Ma H., “ Cell separation based on size and deformability using microfluidic funnel ratchets,” Lab Chip 12, 2369–2376 (2012). 10.1039/c2lc21045b [DOI] [PubMed] [Google Scholar]
- 35.Chisti Y., “ Hydrodynamic damage to animal cells,” Crit. Rev. Biotechnol. 21, 67–110 (2001). 10.1080/20013891081692 [DOI] [PubMed] [Google Scholar]
- 36.Merten O. W., “ Cell detachment,” Encyclopedia of Industrial Biotechnology (John Wiley & Sons, Inc., 2010), pp. 1–22. [Google Scholar]