Abstract
The nucleotide second messengers pppGpp and ppGpp [(p)ppGpp] are responsible for the global downregulation of transcription, translation, DNA replication, and growth rate that occurs during the stringent response. More recent studies suggest that (p)ppGpp is also an important effector in many nonstringent processes, including virulence, persister cell formation, and biofilm production. In Bacillus subtilis, (p)ppGpp production is primarily determined by the net activity of RelA, a bifunctional (p)ppGpp synthetase/hydrolase, and two monofunctional (p)ppGpp synthetases, YwaC and YjbM. We observe that in B. subtilis, a relA mutant grows exclusively as unchained, motile cells, phenotypes regulated by the alternative sigma factor SigD. Our data indicate that the relA mutant is trapped in a SigD “on” state during exponential growth, implicating RelA and (p)ppGpp levels in the regulation of cell chaining and motility in B. subtilis. Our results also suggest that minor variations in basal (p)ppGpp levels can significantly skew developmental decision-making outcomes.
INTRODUCTION
Genetic regulation makes it possible for cells possessing identical genomes to differentiate into subpopulations of phenotypically and physiologically distinct cell types. The Gram-positive bacterium Bacillus subtilis is an excellent model system in which to study the molecular mechanisms underlying differentiation because it produces several well-characterized cell types, including spores, competent cells, biofilm formers, nonmotile chained cells, and motile unchained cells. These different cell types are produced through both stochastic and responsive phenotypic-switching mechanisms (1–3). For example, the proportion of chained, nonmotile cells versus unchained, motile cells in a culture of exponentially growing wild-type B. subtilis bacteria is driven by stochastic switching between the “on” and “off” states of SigD (4, 5), an alternative sigma factor that drives expression of the genes responsible for daughter cell separation and motility. However, stochastic determinants are only one part of SigD regulation, as the proportion of cells in the SigD on state also increases as cells transition out of exponential growth (6–9). In this study, we focus on the role of a conserved, intracellular signaling molecule in triggering the switch to a SigD on state.
The bacterial “alarmone” (also called “magic spot”), consisting of pppGpp and ppGpp [here referred to as (p)ppGpp], acts as an important second-messenger molecule linking both intra- and extracellular environmental cues with global changes in transcription, translation, and DNA replication (10–12). In Escherichia coli and B. subtilis, (p)ppGpp levels are estimated to be less than 20 pmol per optical-density unit during exponential growth (13–16) but rapidly rise to millimolar levels in response to adverse growth conditions, such as amino acid starvation (15). High levels of (p)ppGpp lead to the arrest of DNA replication and cell division, reduced lipid synthesis, a global reduction in transcription and translation (10–12, 17), and increased tolerance for cell stress (11, 18, 19). Levels of (p)ppGpp are strongly correlated with the growth rate (13) and ribosomal pool size (20), and one study found that various substringent concentrations of (p)ppGpp regulate different sets of genes, suggesting that the (p)ppGpp concentration may be associated with graded responses in gene expression (21).
In B. subtilis, (p)ppGpp levels are predominately determined by the net activity of RelA, a dual-function (p)ppGpp synthetase and hydrolase, and two additional (p)ppGpp synthetase enzymes, YwaC and YjbM (22). (p)ppGpp is synthesized at low but detectable levels during exponential growth, primarily through the activity of RelA (22). RelA is also responsible for the high levels of (p)ppGpp that accumulate in B. subtilis during stringent response (22, 23). Less is known about the contribution of YwaC and YjbM activity to (p)ppGpp accumulation. ywaC is under the control of at least three stress response sigma factors, SigW, SigV, and SigM, and encodes the protein responsible for the increase in (p)ppGpp observed during alkaline shock (22, 24–26). Based on microarray data, yjbM is the second gene in an operon of otherwise experimentally uncharacterized genes in B. subtilis (27). Expression profiles suggest that the level of yjbM transcription peaks during rapid growth and transition states in rich media (22, 27). In addition, YjbM contributes, albeit less substantially than RelA, to the basal levels of (p)ppGpp present during exponential growth (22).
When B. subtilis has relA deleted or is depleted of a RelA variant that possesses hydrolase but not synthetase activity (RelAD264G), it grows slowly (22) and is prone to growth rate suppressors (22, 28, 29). RelA mutants have also been shown to exhibit pleiotropic phenotypes, including reduced competence (30) and delayed sporulation (30). Absolute levels of (p)ppGpp are likely elevated in the relA deletion and hydrolase mutants, since these mutants lack the major activity responsible for breaking down (p)ppGpp synthesized by the remaining synthetases, YwaC and YjbM (22, 28). Consistent with the idea that (p)ppGpp levels are elevated, the slow-growth phenotype of the relA mutant can be complemented by introduction of an allele of relA that encodes only the (p)ppGpp hydrolase and through loss-of-function mutations in ywaC and/or yjbM (22). It is likely that small differences in (p)ppGpp levels during exponential growth account for the observed phenotypes; however, any variances in (p)ppGpp levels present in the relA mutant and wild type during exponential growth (when stringent response is not induced) are not quantifiable using current methodologies.
In this study, we show that the relA mutant grows exclusively as swimming, unchained cells during the exponential growth phase; this unchained phenotype contrasts sharply with the wild-type domesticated strain PY79, which exhibits a primarily chained phenotype during exponential growth in rich laboratory media (31). We also find that the relA mutant exhibits an accelerated swim front in a swim plate assay, consistent with the increased population of swimming cells we observe microscopically. Analysis of SigD synthesis and activity indicate that the relA mutant is trapped in a SigD on state during exponential growth. Thus, our results show that cell chaining and motility in B. subtilis are regulated by RelA through its activity as a (p)ppGpp hydrolase.
MATERIALS AND METHODS
General methods.
A description of strains, plasmids, and oligonucleotides used in this study and a detailed description of plasmid constructions can be found in Tables S1, S2, and S3 in the supplemental material. The B. subtilis strains used in this study were grown at 37°C in CH medium (32) or Luria-Bertani (LB) broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl). All samples were grown in volumes of either 25 ml or 50 ml in 250-ml baffled flasks in a shaking water bath set at 280 rpm. LB plates were supplemented with 1.5% Bacto agar. E. coli strains DH5α and TG-1 were used for isolation of plasmids and were grown in LB medium. When needed, antibiotics were included at the following concentrations: 100 μg ml−1 spectinomycin, 7.5 μg ml−1 chloramphenicol, 10 μg ml−1 tetracycline, and 1 μg ml−1 erythromycin (Erm) plus 25 μg ml−1 lincomycin (MLS) for B. subtilis strains and 100 μg ml−1 ampicillin for E. coli strains.
Swim plate assay.
B. subtilis strains were grown at 37°C in baffled flasks containing CH medium to mid-log phase. The cells were pelleted and resuspended at an optical density at 600 nm (OD600) of 10 in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 0.5% India ink (Higgins). Ten microliters of the cell suspension was spotted on top of 150- by 15-mm LB plates fortified with 0.25% Bacto agar (100 ml/plate), dried for 30 min at room temperature on the benchtop to allow time for absorption, and incubated at 37°C in a loosely covered glass dish containing paper towels soaked with water to generate humid conditions. The swim radius was measured relative to the edge of the origin marked by the India ink.
Microscopy.
All fluorescence microscopic analyses were performed with a Nikon Ti-E microscope equipped with a CFI Plan Apo lambda DM 100× objective and a Prior Scientific Lumen 200 Illumination system; C-FL UV-2E/C DAPI (4′,6-diamidino-2-phenylindole), C-FL GFP (green fluorescent protein) HC HISN Zero Shift, and C-FL Texas Red HC HISN Zero Shift filter cubes; and a CoolSnap HQ2 monochrome camera. Membranes were stained with either TMA-DPH [1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate; 0.02 mM] or FM4-64 (3 μg ml−1) (Life Technologies) and imaged with exposure times of 200 to 800 ms. All images were captured, analyzed, and processed using NIS Elements Advanced Research (version 4.10) and Adobe Photoshop (version 12.0).
Flagella were visualized by using the cysteine-reactive dye Alexa Fluor 488 C5 maleimide (Life Technologies) using a previously described method (33). Briefly, 1-ml samples of mid-logarithmic-phase cultures were collected at an optical density at 600 nm (OD600) of 0.5, and the cells were pelleted by centrifugation. The pellet was then washed with 1 ml of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4), resuspended in 50 μl of PBS containing 5 μg ml−1 Alexa Fluor 488 C5 maleimide, and incubated at room temperature for 5 min. The cells were then washed twice with 0.5 ml PBS. The pellet was resuspended in 50 μl of PBS containing 5 μg ml−1 FM4-64 and on a glass slide with a poly-l-lysine (Sigma)-treated coverslip.
Western blot analysis.
B. subtilis strains were grown to logarithmic growth phase (OD600, 0.4 to 0.7), and 1- or 2-ml samples were collected. The cells were pelleted by centrifugation, resuspended in 50 μl lysis buffer (20 mM Tris, pH 7.0, 10 mM EDTA, 1 mg ml−1 lysozyme, 10 μg ml−1 DNase I, 100 μg ml−1 RNase A, with 1 mM phenylmethylsulfonyl fluoride [PMSF]), and incubated for 15 min at 37°C. Fifty microliters of sample buffer (0.25 M Tris, pH 6.8, 4% SDS, 20% glycerol, 10 mM EDTA) containing 10% 2-mercaptoethanol was added to the lysates, and the samples were boiled for 5 min prior to loading. Proteins were separated by SDS-PAGE on precast 4 to 20% Tris-HCl gels (Bio-Rad), transferred onto a nitrocellulose membrane (Pall) at 100 V for 1 h in transfer buffer (20 mM Tris, 15 mM glycine, and 20% methanol [vol/vol]), and then blocked in 5% nonfat milk in PBS containing 0.5% Tween 20. The membranes were incubated overnight at 4°C with a 1:5,000 dilution of anti-SigD peptide antibody (CIRDDKNVPPEEKIM; Genscript), a 1:20,000 dilution of anti-SigA (provided by Masaya Fujita, University of Houston, Houston, TX), or a 1,40,000 dilution of anti-Hag antibody (provided by Daniel B. Kearns, Indiana University) and washed and incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad). After washing, the blots were incubated with SuperSignal West Femto Chemiluminescent substrate (Thermo) according to the manufacturer's instructions.
Flagellin labeling.
B. subtilis strains were grown to logarithmic growth phase (OD600, 0.5), and 1-ml samples were collected. The samples were labeled by Alexa Fluor 488 C5 maleimide labeling as described in “Microscopy” above, and cell lysates were prepared for SDS-PAGE analysis as described in “Western blot analysis” above. Proteins were separated by SDS-PAGE on precast 4 to 20% Tris-HCl gels (Bio-Rad) and visualized by scanning the gels with a Typhoon Trio (GE Healthcare) at 100-μm resolution and an excitation wavelength of 488 nm.
β-Galactosidase assays.
To assay expression from lacZ transcriptional fusions, B. subtilis strains were grown at 37°C in baffled flasks containing CH medium. One- or 2-ml samples were harvested at an OD600 of 0.5 by centrifugation in a room temperature tabletop centrifuge, and the cell pellets were frozen at −20°C or −80°C. To perform the assays, the pellets were thawed on ice, resuspended in 500 μl of Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl, 38 mM β-mercaptoethanol, and 0.2 mg ml−1 lysozyme), and incubated for 15 min at 30°C. The reaction was started by adding 100 μl of 4-mg ml−1 O-nitrophenyl β-d-galactopyranoside (in Z buffer) and stopped with 250 μl of 1 M Na2CO3 after a yellow color developed. The optical densities of the reaction mixtures were measured at 420 and 550 nm and used to calculate the β-galactosidase specific activity according to the following equation: 1,000 × {[OD420 − (1.75 × OD550)]/(time × OD600)}.
RESULTS
The relA mutant grows slowly but exponentially in liquid culture.
Previous studies showed that replacement of B. subtilis relA with an antibiotic resistance cassette leads to a slow-growth phenotype. relA is in an operon with dtd (27), encoding d-amino acid tRNA deacylase (34, 35), an enzyme that removes d-amino acids from mischarged tRNAs. In order to test if the slow-growth phenotype was attributable to loss of relA alone and not due to polar effects on dtd, we introduced ΔrelA into a strain harboring an intact copy of relA and its putative promoter region at an ectopic locus (amyE::PrelA-relA) and examined the growth rate of the resultant strain. The ΔrelA mutant grew more slowly than the wild type on LB plates, consistent with previous reports (22) (Fig. 1A). We determined that the ΔrelA mutant also grew more slowly than the wild type in liquid culture (Fig. 1B), with a generation time in liquid CH medium at 37°C of 50 min for the ΔrelA mutant compared to 30 min for the wild type (Fig. 1A). Introduction of amyE::PrelA-relA into the ΔrelA strain complemented the slow-growth phenotype both on plates and in liquid medium (Fig. 1A and B), indicating that the slow-growth phenotype was solely attributable to the loss of relA and not related to polar affects on dtd. Despite the longer generation time of the ΔrelA mutant, the linear phase of exponential growth mirrored that of the wild type, showing a decrease in the rate around an OD600 of 0.8 to 1.0. This result suggests that the cues triggering the transition out of exponential growth are intact in the ΔrelA mutant background.
FIG 1.
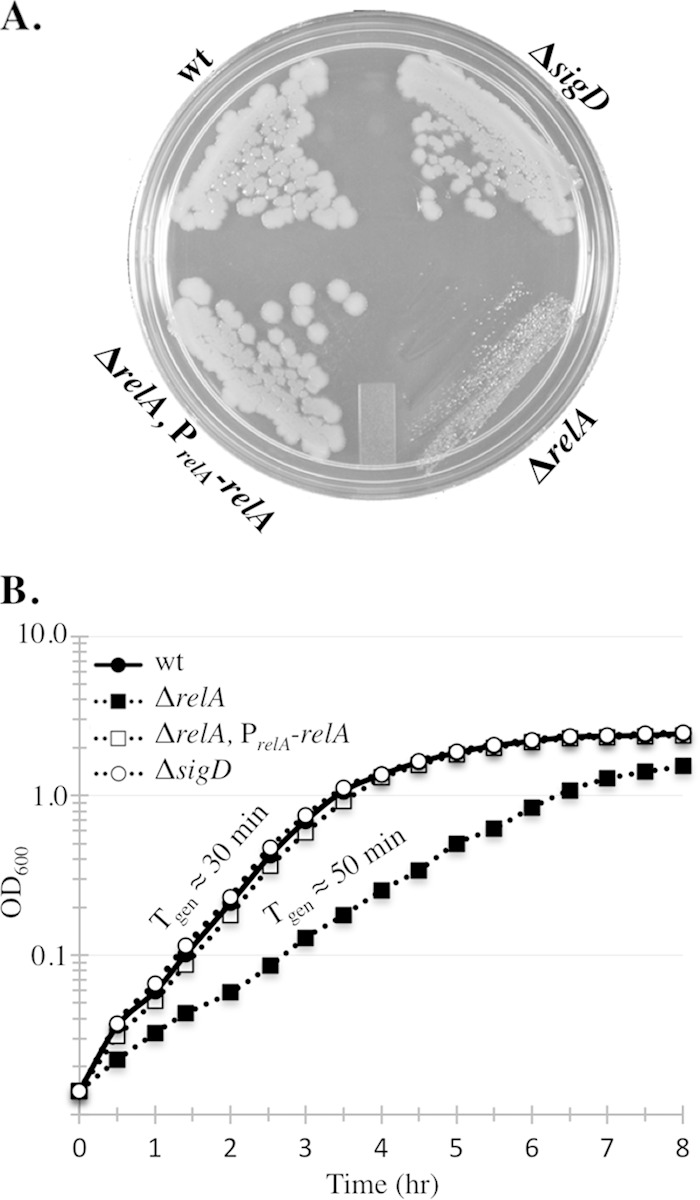
The relA mutant grows slowly and exponentially. (A) The indicated strains were streaked on an LB plate and incubated for 24 h at 37°C. (B) Growth curves of the same strains shown panel A grown in liquid CH medium. The average of three independent replicates is plotted for each strain. wt, wild type (BJH001); ΔrelA, BQA009; ΔrelA PrelA-relA, BQA068; ΔsigD, BQA022.
The slow-growing ΔrelA mutant was previously shown to be prone to the accumulation of growth rate suppressors in ywaC and yjbM, two other (p)ppGpp synthetase genes (22, 28, 29). When constructing and growing the ΔrelA strain, we also observed that spontaneous growth rate suppressors arose frequently on plates; sequencing of these suppressors revealed that the faster-growing strains always possessed mutations in ywaC and/or yjbM (data not shown), consistent with prior reports (22, 28, 29). Since the ΔrelA mutant frequently accumulates growth rate suppressors in ywaC and yjbM, the restoration of wild-type growth we observed in the presence of amyE::PrelA-relA could be attributable to the accumulation of one or more suppressor mutations in these genes as opposed to true complementation. To rule out this possibility, we sequenced the ywaC and yjbM regions of the complemented ΔrelA mutant and found that the regions contained the wild-type alleles (data not shown).
A ΔrelA mutant grows exclusively as unchained cells.
While examining the role of RelA in sporulation, we fortuitously observed that the ΔrelA mutant grew as a homogeneous population of unchained cells during exponential growth in rich medium (Fig. 2A and B). This unchained phenotype is in sharp contrast to PY79, the wild-type domesticated strain used in this study, which grows as a mixed population of 85% chained and 15% unchained cells under the same growth conditions (Fig. 2A and B). The chaining phenotype was restored in the relA complementation strain, and sequencing of genomic DNA from the same culture pictured in Fig. 2A confirmed that ywaC and yjbM had not accumulated suppressor mutations. These results indicate that the unchained phenotype is due to the absence of the relA gene product and not due to polar effects on dtd. Moreover, an inducible allele of relA that encodes the (p)ppGpp hydrolase activity but lacks synthetase activity (a D264G substitution in RelA) (22), restored not only the wild-type growth rate, as previously observed (22), but also the cell chaining in a ΔrelA background (Fig. 2A). In contrast, depletion of RelAD264G in the ΔrelA background resulted in growth as single cells (Fig. 2A). This result is consistent with the unchained phenotype being attributable to the loss of RelA's (p)ppGpp hydrolase function. The presence of chains in all the relA mutant cultures examined throughout this study correlated with growth rate suppressor mutations in the other (p)ppGpp synthetases (data not shown). Therefore, we found that the distinctive, nonchaining phenotype of the relA mutant served as a convenient marker to monitor the integrity of relA mutant strains used in our experiments. To determine if the unchained phenotype could be attributed to the slow growth of the ΔrelA mutant as opposed to a RelA-dependent activity, we examined wild-type cells grown at room temperature in identical media. Wild-type cells grown at room temperature had a generation time of ∼50 min, comparable to that of the ΔrelA mutant, but grew primarily as chained cells (data not shown), suggesting that a lower growth rate alone does not account for the unchained phenotype.
FIG 2.
The ΔrelA mutant grows exclusively as single cells. (A) Representative images of wt (BJH001), ΔrelA (BQA009), ΔrelA PrelA-relA (BQA068), ΔsigD (BQA022), and Pspac-relAD264G (EUB004) cells grown to mid-log phase in CH medium. For EUB004, cells grown in the presence (+RelAD264G) and absence (−RelAD264G) of 1 mM IPTG for 120 min are shown. The membranes were stained with TMA. (B) Frequencies of chained (three or more cells linked) and unchained (single and doublet) cells across a population. The cells in images from at least three independent cultures were counted, and no fewer than 1,500 cells were quantitated for each strain represented in the graph. (C) Representative images of ΔrelA ΔsigD (BQA059) and ΔrelA Phy-sigD ΔsigD (BQA083) cells grown to exponential phase in CH medium. The membranes were stained with TMA.
Deletion of ywaC or yjbM in a ΔrelA mutant restores cell chaining during exponential growth.
The ΔrelA mutant lacks the major (p)ppGpp hydrolase activity, so synthesis of (p)ppGpp from YwaC and/or YjbM should lead to a net accumulation of (p)ppGpp in a ΔrelA mutant background (22). We hypothesized that the unchained phenotype we observed in the ΔrelA mutant might be due to increased (p)ppGpp levels. Since the basal levels of (p)ppGpp present in the wild-type and ΔrelA mutant backgrounds fall outside the linear range of detection, we tested this idea genetically using (p)ppGpp synthetase mutants. We hypothesized that if higher levels of (p)ppGpp resulted in loss of chaining, then the ΔrelA ΔyjbM and ΔrelA ΔywaC double mutants should be more chained than the ΔrelA single mutant. We observed that in comparison to the wild type (85% chained) and the relA mutant strain (0% chained), the ΔrelA ΔyjbM and ΔrelA ΔywaC mutants were 78% and 34% chained, respectively (Fig. 2B). These results are consistent with the idea that accumulation of (p)ppGpp promotes the switch from chained to unchained cells and also suggest that YjbM is a more significant contributor to (p)ppGpp pools during exponential growth than YwaC.
The proportion of chained versus unchained cells in a population varies with strain and culture conditions and was previously shown to be regulated by the level of an activated form of the alternative sigma factor SigD (4). Active SigD stimulates transcription of several genes, including the cell wall amidase LytF, which promotes postseptational hydrolysis of the peptidoglycan connecting daughter cells (36). As previously shown, a sigD mutant grows exclusively as chains (Fig. 2A and B). In order to determine if the unchained phenotype observed in the relA mutant was dependent on SigD activity, we generated a ΔsigD ΔrelA double mutant by introducing ΔrelA into the ΔsigD background using transformation. We observed, using control DNA, that the transformation efficiency of the ΔsigD mutant was reduced more than 10-fold compared to the wild type. For unknown reasons, the efficiency of introducing the ΔrelA mutation into the ΔsigD mutant was even lower, so that we were able to obtain only one transformant after 15 attempts. In contrast, ΔrelA could be readily introduced into the ΔsigD background harboring a copy of PrelA-relA at an ectopic locus. These results hinted that the combination of ΔsigD and ΔrelA could be synthetically lethal. To further test this idea, we introduced ΔrelA into a SigD depletion strain in which the only copy of sigD is under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-regulated promoter. To test for synthetic lethality, the resulting strain was inoculated on media with and without inducer. Surprisingly, the strain grew similarly to the ΔrelA parent on plates whether sigD expression was induced with IPTG or not (data not shown). This result suggests that the reduced transformation efficiency we observed is due not to synthetic lethality but rather to a reduced ability to obtain transformants. Consistent with this, whole-genome sequencing of the viable ΔsigD ΔrelA mutant revealed no second-site suppressor mutations. Microscopic analysis of the ΔsigD ΔrelA mutant revealed an intermediate chaining phenotype different from that of either the ΔsigD or the ΔrelA single mutant (Fig. 2C). Moreover, the SigD depletion strain harboring the ΔrelA mutation appeared similar to the ΔrelA strain when SigD was expressed (Fig. 2C) and similar to the ΔsigD ΔrelA mutant when SigD was depleted (Fig. 2C). These results indicate that SigD is required for the homogeneous single-cell phenotype of the ΔrelA mutant.
The ΔrelA mutant displays flagella on the cell surface.
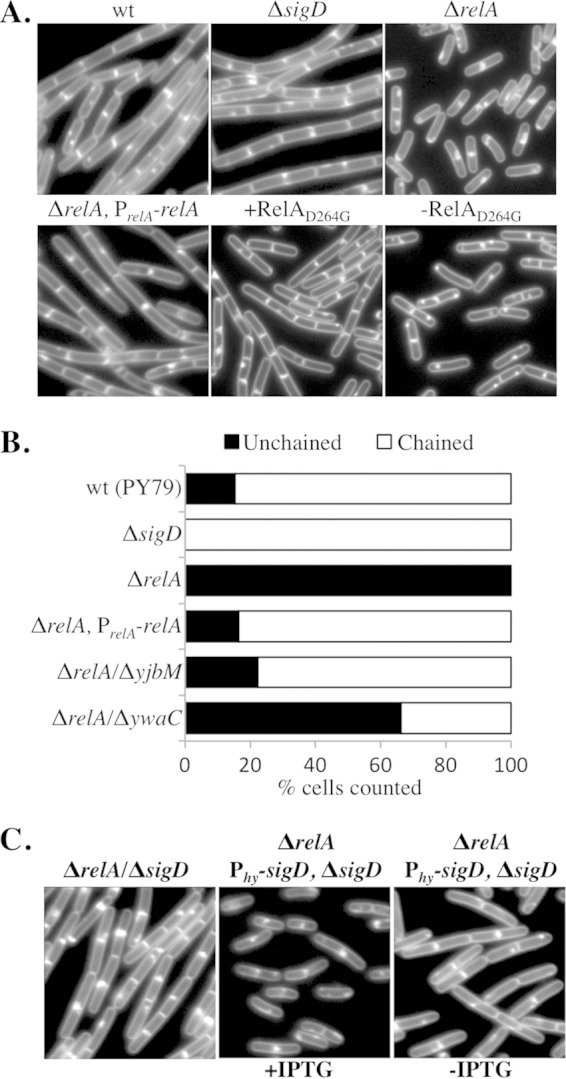
We observed that the unchained relA mutants observed by microscopy (Fig. 2A) appeared highly motile in wet mounts (data not shown), indicating that the unchained cells also possessed active flagella. As mentioned previously, accumulation of active SigD leads to the upregulation of genes responsible for the separation of chained cells into single cells (5, 36–38). Active SigD also coordinately upregulates the synthesis of late flagellar genes (39), including hag, the gene encoding flagellin (40). We hypothesized that since the relA mutants appeared exclusively unchained and motile by microscopy, the relA mutant would also be associated with more flagella than the wild type. In order to directly visualize flagella on intact cells, we engineered strains harboring a HagT209C change in the flagellin subunit, incubated intact cells with a cysteine-reactive dye that stains surface-exposed cysteine residues, and imaged the flagella by epifluorescence microscopy using methodology described previously (39). The relA mutant was unchained (as shown in Fig. 2A and B) and presented numerous peritrichous flagella (Fig. 3A). In contrast, our wild-type strain grew as both unchained cells that presented flagella and chained cells that generally lacked flagella (Fig. 3A). We were unable to determine the numbers of flagella present on the cell surfaces due to difficulty resolving individual filaments among the dense mass on the cell surface. In addition, we observed the presence of significant numbers of shed flagella in the samples that made it difficult to resolve whether the flagella were cell associated. We believe that the shed flagella likely break off due to the shear forces required to wash and centrifuge the samples during cysteine labeling (see below). We conclude that the presence of flagella on the surfaces of the unchained relA mutant cells suggests that SigD is more active in this strain background than in the wild type grown under identical conditions.
FIG 3.
The relA mutant is comprised of mostly flagellated cells (A) Representative images of mid-log-phase cultures of wt (BQA057) and ΔrelA (BQA062) strains. The membranes were stained with FM4-64 (red), and flagellin (Hag) was stained with Alexa Fluor 488 C5 maleimide (green). Both strains harbor a point mutation in the chromosomal copy of hag that creates HagT209C. (B) Comparison of flagellin (Hag) protein levels in wt (BQA057), ΔrelA (BQA062), ΔrelA PrelA-relA (BQA080), and Δhag (BQA076) strains. Samples were collected from mid-log-phase cultures grown in CH medium and stained with Alexa Fluor 488 C5 maleimide. Proteins from cell lysates were separated by SDS-PAGE, stained with Coomassie blue (left), and scanned with a laser scanner to visualize fluorescently labeled protein (right). The arrowhead indicates flagellin. (C) (Top) Western blot analysis showing flagellin levels associated with the culture supernatants (left; S) and cell pellets (right; P) of the indicated strains grown in CH medium to mid-log phase: wt (BJH001), ΔrelA (BQA009), ΔrelA PrelA-relA (BQA068), ΔsigD (BQA022), and Δhag (BQA076). It was necessary to dilute the cell pellet lysates to the indicated dilutions in order to assess the levels of protein associated with each sample without overexposure. (Bottom) SigA protein served as a loading control for the cell pellet samples.
Although the use of cysteine-reactive dye to observe B. subtilis flagella was previously published (39) and we found that no fluorescent signal was associated with wild-type cells lacking HagT209C (data not shown), we performed an additional control to visualize the complement of protein species labeled by the cysteine-reactive dye. Intact cells were treated with dye as they would be prior to microscopy and then subjected to SDS-PAGE and laser scanning. A predominant band emitting in the green channel and running at the approximate molecular mass of the flagellin monomer (32 kDa) was present in strains harboring HagT209C for the wild type, the relA mutant, and the relA-complemented strain, but the band was absent from the flagellin mutant (Δhag) and the wild-type PY79 strain, which lacks the HagT209C variant (Fig. 3B and data not shown). The amounts of flagellin varied greatly even between identical strains in independent trials, despite the fact that samples were normalized by optical density; total-protein staining confirmed that approximately equal amounts of protein were loaded for each sample (Fig. 3B). We believe the variability seen in flagellin levels is due to shearing of the flagella during the sample handling required for the labeling reaction. We conclude that the fluorescence observed by microscopy is due primarily to labeled flagella and that, while analysis of labeled proteins using SDS-PAGE followed by laser scanning is useful for determining the apparent molecular weight of stained proteins, it is not useful as a quantitative measure of total flagellin levels in B. subtilis.
The ΔrelA mutant produces more flagellin than the wild type.
In order to quantitate the amount of flagellin associated with the relA mutant relative to the wild type, we collected cells and supernatants from exponentially growing cultures and examined flagellin levels using Western blot analysis (Fig. 3C). The supernatants were collected so that we could also assess flagellin that might accumulate in the supernatants due to shedding, shearing, or cell lysis. A predominant band running at the approximate molecular mass of the flagellin monomer (32 kDa) was present in both supernatants and cell lysates of the wild type, the relA mutant, and the relA complemented strain but was absent from the flagellin mutant (Δhag) and the sigD mutant (ΔsigD). The amount of flagellin associated with the supernatants was negligible compared to the amount associated with the cell bodies, so it was necessary to dilute the cell body lysates 50-fold to visualize all of the samples on the same Western blot without overexposure (Fig. 3C). Since the amount of flagellin present in the supernatant was minor compared to that in the cell bodies, we did not analyze these data further. The amounts of flagellin present in the wild-type control and the complemented relA strain were similar in each of four independent trials and showed no statistically significant difference in flagellin levels. The relA mutant, in contrast, showed a 5-fold increase in flagellin levels compared to the wild type, and it was necessary to dilute the sample 5-fold to avoid overexposure (Fig. 3C). The SigA antibody served as a loading control, confirming that the diluted sample was loaded in the relA mutant lane. These results are consistent with what we observed microscopically (Fig. 3A) and suggest that the unchained relA mutant population consists primarily of flagellated cells in a SigD on state.
The ΔrelA mutant strain exhibits increased mobility on swim plates.
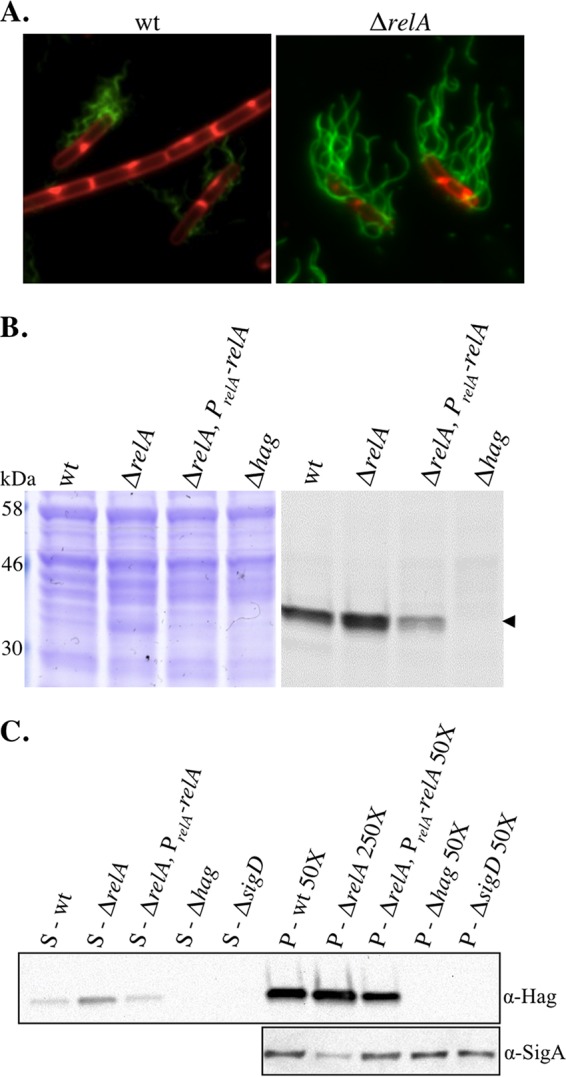
Since the relA mutant population is composed exclusively of unchained, flagellated cells, we expected that it would also produce a more motile population than the wild type. The laboratory strain used in this study (PY79) does not swarm (33), a type of surfaced-based group motility usually facilitated by a surfactant produced by bacteria (33, 41). Therefore, in order to obtain a quantitative assessment of the motility behavior of the population, we performed a swim plate assay that utilizes a lower concentration of agar and does not require surfactant production (42). All cultures were grown to mid-log phase and spotted in equal volumes and densities on swim plates. The visible edge of the culture, which we refer to as the “swim front,” was measured relative to the edge of the original spot over a time course (Fig. 4A). An example of a swim plate at the 3.5-h time point is shown in Fig. 4B. The absolute distance between the edge of the original spot and the swim front (the “swim radius”) varied slightly from assay to assay (standard deviations are shown in 4A); however, the maximal rate of migration, calculated from linear regions of the data in Fig. 4A, remained essentially constant from experiment to experiment.
FIG 4.
Loss of relA leads to increased mobility on swim plates. (A) Quantitation of swim expansion from plate assays performed with wt (BJH001), ΔrelA (BQA009), and ΔrelA PrelA-relA (BQA068) strains on LB fortified with 0.25% agar. Each symbol indicates the average of measurements from five independent experiments with standard deviations. Measurements were recorded every 30 min for 8 h. (B) wt (BJH001), ΔrelA (BQA009), ΔrelA PrelA-relA (BQA068), ΔsigD (BQA022), Δhag (BQA076), ΔrelA ΔyjbM (BQA081), and ΔrelA ΔywaC (BQA082) strains were inoculated on LB fortified with 0.25% agar. After 3.5 h of incubation at 37°C, the plates were scanned against a black background. The images were inverted to better show contrast.
The relA mutant exhibited two notable phenotypes in the swim plate assay. First, unlike the wild type, which exhibited an ∼2-h lag before reaching its maximal swim front migration rate of 2.8 mm/h, the relA mutant had a constant migration rate throughout the course of the experiment (30 min to 8 h) of 4.9 mm/h. Wild-type cells from exponential-phase cultures, early transition phase (OD600, 1.2), and mid-transition phase (OD600, 1.9) all exhibited similar lags (data not shown). Lag periods for swarming had also been observed previously and may represent a physiological adjustment to growth on a more solid surface (41). The second observable phenotype was a maximal migration rate of the relA mutant of 4.9 mm/h, significantly higher than that of the wild type at 2.8 mm/h. The complemented relA strain behaved similarly to the wild type (Fig. 4A). These results suggest that both the absence of the lag period and the overall increase in the maximal swim front migration rate can be attributed to relA function. As expected, the swim fronts of strains deficient in either SigD (ΔsigD) or flagellin (Δhag) production did not migrate a measurable distance from the culture origin (Fig. 4B). The SigD and flagellin nonswimming controls exhibited wild-type growth rates in liquid culture (Fig. 1A and data not shown), suggesting that the growth rate has little effect on the distance traveled by the swim front in the 8-h time course. We conclude that, consistent with the unchained and flagellated phenotype we observed microscopically, the relA mutant population is poised to swim during the exponential growth phase.
The increased chaining we observed in the ΔrelA ΔyjbM and ΔrelA ΔywaC double mutants (Fig. 2C) is consistent with a shift toward a SigD off state relative to the relA single mutant. If there is reduced SigD activity in the double mutants, then we would also expect a decrease in their ability to swim compared to the relA single mutant. To test this hypothesis, we assayed the swim front migration of the ΔrelA ΔyjbM and ΔrelA ΔywaC strains. We observed that both the ΔrelA ΔywaC and ΔrelA ΔyjbM strains possessed smaller swim radii than the ΔrelA parent alone but larger radii than the wild type (Fig. 4B; see Fig. S1A in the supplemental material). The ΔrelA ΔywaC strain, which possesses an intact copy of exponentially expressed yjbM, consistently exhibited a larger swim radius than the ΔrelA ΔyjbM strain (Fig. 4B; see Fig. S1A in the supplemental material), indicating that it more closely resembles the ΔrelA strain than the ΔrelA ΔyjbM strain. These results are consistent with the idea that the switch from the nonmotile (SigD off) to the motile (SigD on) state is promoted by increasing (p)ppGpp levels; at the same time, we do not rule out the possibility that the switch is stimulated by another mechanism unrelated to (p)ppGpp accumulation.
Loss of relA leads to an increase in SigD levels and activity.
The relA mutant grows exclusively as unchained, motile cells during exponential growth, consistent with the hypothesis that the relA mutant possesses higher levels of active SigD than the wild type during exponential growth. In order to test this hypothesis, we first examined the levels of SigD protein by performing Western blot analysis on strains grown to mid-log phase. Relative to the wild type, the relA mutant consistently showed a nearly 10-fold increase in SigD levels (Fig. 5A). Approximately wild-type SigD levels were restored in the relA complementation strain (Fig. 5A), indicating that the increase in SigD levels in the relA mutant are due to the absence of a functional RelA protein. We conclude that a RelA-dependent function is required to prevent the accumulation of SigD protein during exponential growth.
FIG 5.
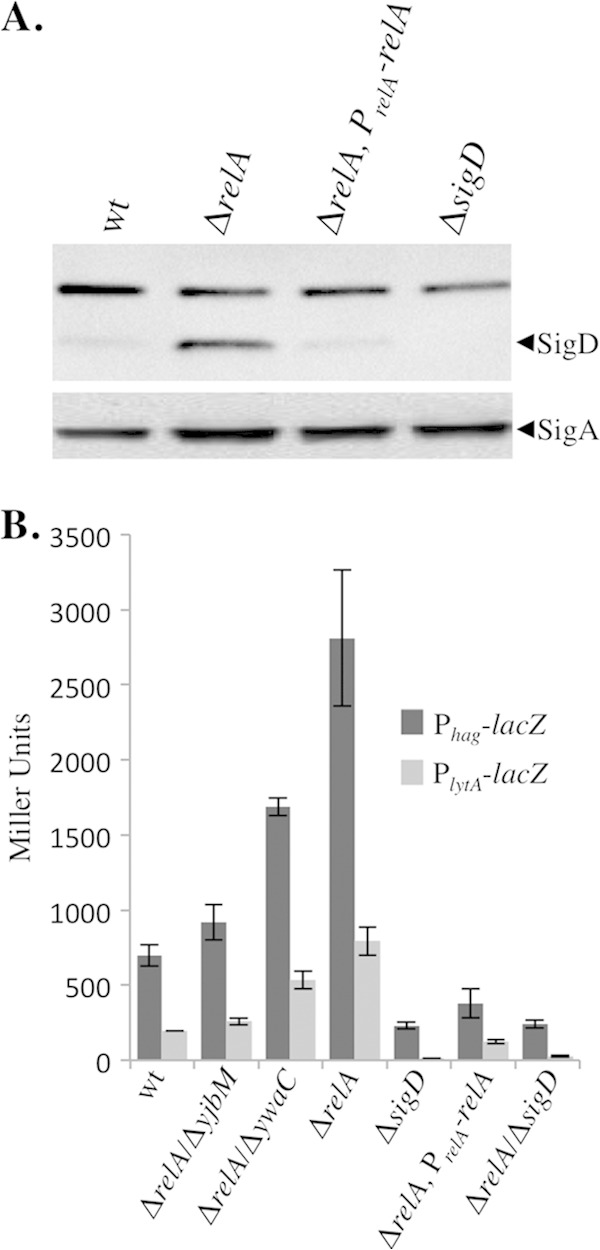
Loss of relA leads to an increase in SigD levels and activity. (A) Comparison of SigD protein levels in wt (BJH001), ΔrelA (BQA009), ΔrelA PrelA-relA (BQA068), and ΔsigD (BQA022) strains. Samples were collected from mid-log-phase cultures grown in CH medium. Proteins from cell lysates were separated by SDS-PAGE and analyzed by Western blot analysis by probing with either anti-SigD or anti-SigA antibody, as indicated. (B) β-Galactosidase assays of Phag-lacZ and PlytA-lacZ transcriptional activities conducted on wt (BJH046 and BJH047), ΔrelA (BQA050 and BQA051), ΔrelA ΔyibM (BQA086 and BQA087), ΔrelA ΔywaC (BQA088 and BQA089), ΔsigD (BQA071 and BQA072), ΔrelA PrelA-relA (BQA073 and BQA074), and ΔrelA ΔsigD (BQA084 and BQA085) strains. Samples were collected from mid-log-phase cultures grown in CH medium. The data shown are the means of three independent replicates with standard deviations.
Although SigD protein levels are correlated with SigD activity, the relationship is not linear, and several levels of posttranslational regulation ultimately determine actual SigD activity (41, 43). In order to determine if the increase in SigD levels we observed also corresponded to an increase in SigD activity, we analyzed expression from two SigD-dependent promoters using transcriptional fusions to a lacZ reporter gene. Based on the unchained, flagellated phenotype, the increase in mobility observed on swimming plates, and the increase in SigD abundance, we expected that the relA mutant would exhibit significantly higher levels of SigD activity than the wild type. The results of β-galactosidase assays performed on samples collected from mid-log-phase growth are shown in Fig. 5B. As expected, both Phag, the flagellin promoter and PlytA, which drives expression of proteins important for flagellar activation through peptidoglycan hydrolysis (37), showed significantly higher levels of transcription in the relA mutant than in the wild-type control. The increased expression in the relA mutant was dependent on the presence of sigD (Fig. 5B, ΔrelA ΔsigD). In contrast, expression was reduced to approximately wild-type levels in the relA-complemented strain. The ΔrelA ΔyjbM and ΔrelA ΔywaC mutants exhibit intermediate levels of SigD activity (Fig. 5B), consistent with the intermediate chaining (Fig. 2B) and swimming motility (Fig. 4B) phenotypes we observed. We conclude that a RelA-dependent activity regulates the amount of active SigD that accumulates during the exponential growth phase and that the ΔrelA mutant is shifted to a SigD on state during exponential growth.
DISCUSSION
Nucleotide second messengers, like cyclic di-GMP, cyclic AMP (cAMP), and the bacterial alarmone (p)ppGpp, are well-characterized examples of intracellular signaling molecules that play important roles in modulating gene expression in response to environmental and physiological fluxes in bacteria (11, 44, 45). (p)ppGpp is responsible for the global downregulation of transcription, translation, DNA replication, and growth rate that occurs during the stringent response to amino acid limitation (10–12). In more recent years, (p)ppGpp has been implicated in a number of nonstringent processes, including virulence (46–48), persister cell formation (49–51), biofilm production (52), maintaining a balanced growth rate (53), and regulating GTP pools in response to nutrient downshift (54).
A role for (p)ppGpp in modulating cell motility has also been previously reported for several Gram-negative bacteria, including Vibrio cholerae (55), Legionella pneumophila (56), and E. coli (57–59). For example, (p)ppGpp and the regulatory protein DksA have been shown to act directly on flagellar biosynthesis promoters to negatively regulate flagellum production during E. coli stationary-phase growth (59). In this study, we observed that a B. subtilis relA mutant deficient for (p)ppGpp hydrolysis (22) grows as a homogeneous population of unchained, motile cells during exponential growth in rich medium, a time when our wild-type laboratory strain is predominantly chained and nonmotile. These results suggest that increased (p)ppGpp levels or decreased GTP levels promote differentiation of B. subtilis (Fig. 6).
FIG 6.
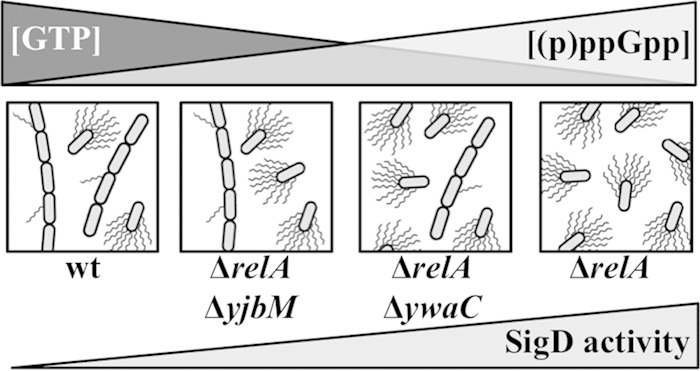
Model depicting the observed relationships between cell motility and chaining, SigD activity, GTP concentration (22), and putative (p)ppGpp levels in various (p)ppGpp synthetase/hydrolase backgrounds.
Since the levels of (p)ppGpp present in the strain backgrounds and under the growth conditions presented in this study fall outside the current quantitative-detection range, we tested our model genetically using (p)ppGpp synthetase mutants. We found that an increase in cell motility (Fig. 4B) and loss of cell chaining (Fig. 2B) directly correlate with the (p)ppGpp levels predicted to be produced in ΔrelA (highest), ΔrelA ΔywaC (intermediate), and ΔrelA ΔyjbM (lowest) mutants during exponential growth (Fig. 2 and 6). In contrast, the increase in cell motility and loss of chaining inversely correlate with the GTP levels measured in the ΔrelA (lowest), ΔrelA ΔywaC (intermediate), and ΔrelA ΔyjbM (highest) backgrounds (22). Our data also demonstrate that a ΔrelA mutant has elevated levels of SigD activity compared to the wild type (Fig. 5B), consistent with the homogeneous population of unchained and flagellated cells we observed microscopically. These results support a model in which (p)ppGpp levels act as a modulator of SigD activity and, subsequently, cellular differentiation (Fig. 6). More specifically, our data suggest that even slight increases in (p)ppGpp may promote a SigD on activation state during exponential growth phase (Fig. 6). The absence of RelA activity clearly has a profound effect on bacterial decision making, dramatically shifting the bias of exponentially growing B. subtilis cells from a nonmotile, chained wild-type population (85%) to a homogeneous population of unchained, motile cells in the ΔrelA mutant (Fig. 2A and B).
How might changes in (p)ppGpp levels and/or a decrease in GTP levels promote the activation of SigD and thus promote the increased frequency of unchained, swimming cells we observed in relA mutant backgrounds? SigD activity is regulated at the level of protein synthesis (43) and by being held in an inactive state through the action of the anti-sigma factor FlgM (60, 61). We showed that, compared to the wild type, SigD is both more abundant and more active in the relA mutant (Fig. 5A and B). Since the global regulator DegU has been shown to repress SigD production (9) and promote FlgM synthesis (62), determining the role of DegU in the single-cell, swimming phenotype we observed is a potential avenue for future investigations. Interestingly, we observed that a ΔrelA ΔsigD mutant does not grow as long chains like the ΔsigD parent (Fig. 2C). This result suggests that the absence of relA promotes activation of some SigD-independent autolysins (63). One attractive idea that could explain both the ΔrelA and the ΔrelA ΔsigD phenotypes is that elevated (p)ppGpp levels in B. subtilis may promote sigma factor switching such as has been reported in E. coli (64–66). In E. coli, (p)ppGpp has been shown to bind to RNA polymerase and, in concert with DksA, to directly regulate gene transcription (15). B. subtilis lacks a known DksA homolog, and direct regulation of RNA polymerase by (p)ppGpp has not been observed (66). Moreover, B. subtilis lacks conservation in the RNA polymerase (RNAP) residues that make contact with ppGpp in E. coli (67), suggesting that if (p)ppGpp directly regulates RNAP in B. subtilis, then the mechanisms will diverge significantly.
The B. subtilis ΔrelA mutant harbors reduced levels of GTP (22, 28), which may also contribute to the motile, unchained phenotype we observed. (p)ppGpp synthesis affects GTP levels through at least two independent mechanisms. First, GDP, GTP, and ATP are the substrates for (p)ppGpp-synthetic enzymes, and (p)ppGpp synthesis is correlated with a subsequent drop in GTP levels (8, 15, 30, 68). Second, a recent study in B. subtilis showed that (p)ppGpp can also negatively regulate cellular GTP levels by binding directly to two key enzymes in the GTP synthesis pathway, HptR and Gmk, thereby negatively regulating their ability to synthesize GMP and GDP precursors, respectively (54).
It is possible that lower GTP levels present in the relA mutant relieve transcriptional repression of some promoters in the SigD regulon through CodY derepression. CodY is a GTP-sensing protein that binds DNA and positively or negatively regulates the expression of over 100 B. subtilis gene products (69). Although CodY has been shown to repress expression from hag (7), a recent global analysis found no evidence for binding of CodY to the hag promoter in vivo (69). Previous studies also showed that transcription can be affected by shifts in the GTP and ATP pools of cells. In B. subtilis undergoing stringent response through amino acid starvation, (p)ppGpp and ATP levels rise while GTP levels fall (54, 70). Promoters beginning with a +1 G are apparently particularly sensitive to GTP pools, as GTP is rate limiting for initiation (70, 71). Therefore, it is possible that the lower GTP levels of the relA mutant lead to decreased expression from +1 G promoters. Both of the fla-che promoters PD-3 and PA, which regulate the bulk of sigD expression, begin with a +1 A (72), while the flgM promoter begins with a +1 G (70).
In summary, our results show that during exponential growth, RelA activity leads to the modulation of gene expression in B. subtilis and support the idea that even minor fluxes in (p)ppGpp levels may greatly influence cellular decision making. More specifically, we show that the B. subtilis ΔrelA mutant is trapped in a SigD on state, biasing cells toward a motile, unchained phenotype during exponential growth. Future experiments will be aimed at elucidating the molecular mechanisms underlying this developmental outcome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Masaya Fujita for providing α-SigA antibody, Daniel B. Kearns for providing anti-Hag antibody and technical advice regarding the microscopy-based cysteine-labeling experiment, and members of the Herman laboratory and the Center for Phage Technology for providing helpful feedback.
We acknowledge the Center for Phage Technology and the Department of Biochemistry and Biophysics at Texas A&M University for providing funding.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02063-14.
REFERENCES
- 1.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 2.Shank EA, Kolter R. 2011. Extracellular signaling and multicellularity in Bacillus subtilis. Curr Opin Microbiol 14:741–747. doi: 10.1016/j.mib.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirouze N, Desai Y, Raj A, Dubnau D. 2012. Spo0A∼P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet 8:e1002586. doi: 10.1371/journal.pgen.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cozy LM, Phillips AM, Calvo RA, Bate AR, Hsueh YH, Bonneau R, Eichenberger P, Kearns DB. 2012. SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of sigma(D) in Bacillus subtilis. Mol Microbiol 83:1210–1228. doi: 10.1111/j.1365-2958.2012.08003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirel DB, Estacio WF, Mathieu M, Olmsted E, Ramirez J, Marquez-Magana LM. 2000. Environmental regulation of Bacillus subtilis sigma(D)-dependent gene expression. J Bacteriol 182:3055–3062. doi: 10.1128/JB.182.11.3055-3062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergara F, Ibarra C, Iwamasa J, Patarroyo JC, Aguilera R, Marquez-Magana LM. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J Bacteriol 185:3118–3126. doi: 10.1128/JB.185.10.3118-3126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amati G, Bisicchia P, Galizzi A. 2004. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol 186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnusson LU, Farewell A, Nystrom T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol 13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 12.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 13.Sarubbi E, Rudd KE, Cashel M. 1988. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet 213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- 14.Ryals J, Little R, Bremer H. 1982. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol 151:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cashel M, Gentry DM, Hernandez VJ, Vinella D. 1996. The stringent response, p 1458–1496. In Neidhardt FC. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 16.Nishino T, Gallant J, Shalit P, Palmer L, Wehr T. 1979. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J Bacteriol 140:671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Haldenwang WG. 2003. RelA is a component of the nutritional stress activation pathway of the Bacillus subtilis transcription factor sigma B. J Bacteriol 185:5714–5721. doi: 10.1128/JB.185.19.5714-5721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura M, Gourse R, Baughman G. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem 53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 21.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol 67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith I, Paress P, Cabane K, Dubnau E. 1980. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet 178:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 24.Czarny TL, Perri AL, French S, Brown ED. 2014. Discovery of novel cell wall-active compounds using PywaC, a sensitive reporter of cell wall stress in the model Gram-positive Bacillus subtilis. Antimicrob Agents Chemother 58:3261–3269. doi: 10.1128/AAC.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eiamphungporn W, Helmann JD. 2008. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol 67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guariglia-Oropeza V, Helmann JD. 2011. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J Bacteriol 193:6223–6232. doi: 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 28.Natori Y, Tagami K, Murakami K, Yoshida S, Tanigawa O, Moh Y, Masuda K, Wada T, Suzuki S, Nanamiya H, Tozawa Y, Kawamura F. 2009. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J Bacteriol 191:4555–4561. doi: 10.1128/JB.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inaoka T, Ochi K. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J Bacteriol 184:3923–3930. doi: 10.1128/JB.184.14.3923-3930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Guttenplan SB, Blair KM, Kearns DB. 2009. Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol 191:5775–5784. doi: 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, New York, NY. [Google Scholar]
- 33.Patrick JE, Kearns DB. 2009. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol 191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutourina O, Soutourina J, Blanquet S, Plateau P. 2004. Formation of D-tyrosyl-tRNATyr accounts for the toxicity of D-tyrosine toward Escherichia coli. J Biol Chem 279:42560–42565. doi: 10.1074/jbc.M402931200. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Zheng G, Peng X, Qiang B, Yuan J. 2003. d-Amino acids and d-Tyr-tRNA(Tyr) deacylase: stereospecificity of the translation machine revisited. FEBS Lett 552:95–98. doi: 10.1016/S0014-5793(03)00858-5. [DOI] [PubMed] [Google Scholar]
- 36.Margot P, Pagni M, Karamata D. 1999. Bacillus subtilis 168 gene lytF encodes a gamma-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, sigmaD. Microbiology 145:57–65. doi: 10.1099/13500872-145-1-57. [DOI] [PubMed] [Google Scholar]
- 37.Lazarevic V, Margot P, Soldo B, Karamata D. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol 138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 38.Blackman SA, Smith TJ, Foster SJ. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144:73–82. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 39.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 40.Mirel DB, Chamberlin MJ. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J Bacteriol 171:3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patrick JE, Kearns DB. 2012. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol Microbiol 83:14–23. doi: 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cozy LM, Kearns DB. 2010. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol 76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 45.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 50.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Jayaraman R. 2008. Bacterial persistence: some new insights into an old phenomenon. J Biosci 33:795–805. doi: 10.1007/s12038-008-0099-3. [DOI] [PubMed] [Google Scholar]
- 52.He H, Cooper JN, Mishra A, Raskin DM. 2012. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potrykus K, Murphy H, Philippe N, Cashel M. 2011. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol 13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK. 2012. Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J Bacteriol 194:5638–5648. doi: 10.1128/JB.00518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol 191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol 189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caramori T, Barilla D, Nessi C, Sacchi L, Galizzi A. 1996. Role of FlgM in sigma D-dependent gene expression in Bacillus subtilis. J Bacteriol 178:3113–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bertero MG, Gonzales B, Tarricone C, Ceciliani F, Galizzi A. 1999. Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM. J Biol Chem 274:12103–12107. doi: 10.1074/jbc.274.17.12103. [DOI] [PubMed] [Google Scholar]
- 62.Hsueh YH, Cozy LM, Sham LT, Calvo RA, Gutu AD, Winkler ME, Kearns DB. 2011. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol Microbiol 81:1092–1108. doi: 10.1111/j.1365-2958.2011.07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262. [DOI] [PubMed] [Google Scholar]
- 64.Osterberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu Rev Microbiol 65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 65.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol 67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 66.Sharma UK, Chatterji D. 2010. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev 34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 67.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krasny L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belitsky BR, Sonenshein AL. 2013. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci U S A 110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krasny L, Tiserova H, Jonak J, Rejman D, Sanderova H. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol 69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 71.Tojo S, Hirooka K, Fujita Y. 2013. Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control. J Bacteriol 195:1656–1665. doi: 10.1128/JB.02131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.West JT, Estacio W, Marquez-Magana L. 2000. Relative roles of the fla/che P(A), P(D-3), and P(sigD) promoters in regulating motility and sigD expression in Bacillus subtilis. J Bacteriol 182:4841–4848. doi: 10.1128/JB.182.17.4841-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


