Abstract
Proteus mirabilis is a dimorphic motile bacterium well known for its flagellum-dependent swarming motility over surfaces. In liquid, P. mirabilis cells are 1.5- to 2.0-μm swimmer cells with 4 to 6 flagella. When P. mirabilis encounters a solid surface, where flagellar rotation is limited, swimmer cells differentiate into elongated (10- to 80-μm), highly flagellated swarmer cells. In order for P. mirabilis to swarm, it first needs to detect a surface. The ubiquitous but functionally enigmatic flagellar basal body protein FliL is involved in P. mirabilis surface sensing. Previous studies have suggested that FliL is essential for swarming through its involvement in viscosity-dependent monitoring of flagellar rotation. In this study, we constructed and characterized ΔfliL mutants of P. mirabilis and Escherichia coli. Unexpectedly and unlike other fliL mutants, both P. mirabilis and E. coli ΔfliL cells swarm (Swr+). Further analysis revealed that P. mirabilis ΔfliL cells also exhibit an alteration in their ability to sense a surface: e.g., ΔfliL P. mirabilis cells swarm precociously over surfaces with low viscosity that normally impede wild-type swarming. Precocious swarming is due to an increase in the number of elongated swarmer cells in the population. Loss of fliL also results in an inhibition of swarming at <30°C. E. coli ΔfliL cells also exhibit temperature-sensitive swarming. These results suggest an involvement of FliL in the energetics and function of the flagellar motor.
INTRODUCTION
Most bacteria are able to live a planktonic free-living lifestyle or in a surface-attached microbial community called a “biofilm.” The interchange between the motile and the sessile phases, referred to as the “swim-or-stick” switch, is not merely stochastic. Rather, the lifestyle change occurs in response to cues that a cell senses as it nears a surface (1). These surface signals are required and initiate biofilm formation (1). A fundamental question underlying the transition in lifestyle from motile to sessile phases is how does a bacterium sense a surface?
Studies of many different bacterial species support the idea that surface sensing often involves the bacterial flagellum (2), which also facilitates movement toward and attachment to a surface. However, it is generally agreed that motility and biofilm formation are mutually exclusive. Moreover, flagella are used not only for swimming in liquid but also for swarming over a solid surface. Many bacterial species swarm, and often, as in Proteus mirabilis, require a surface-induced differentiated cell type, called a swarmer cell, that is elongated and hyperflagellated (3).
P. mirabilis is a Gram-negative gammaproteobacterium belonging to the Enterobacteriaceae family. It is an opportunistic pathogen capable of causing urinary tract infections (UTI) (4–6). P. mirabilis is dimorphic and produces short vegetative swimmer cells (1.5 to 2.0 μm in length) with a single nucleoid and 4 to 10 peritrichous flagella when cultured in nutrient broth. Conversely, when cultured on nutrient agar or in viscous environments, P. mirabilis swimmer cells differentiate into nonseptated, elongated (10 to 80 μm in length) swarmer cells with multiple nucleoids and numerous flagella (4, 7). P. mirabilis cells monitor the rotation of their flagella to recognize and sense surface contact. When a swimmer cell encounters a solid surface or viscous environment, inhibition of flagellar rotation triggers differentiation into a swarmer cell (8). Furthermore, on a surface, swarmer cells align with one another to perform a multicellular coordinative movement, known as “swarming” (4). P. mirabilis swarmer cell differentiation is correlated with elevated expression of several virulence factors that aid in the invasion of uroepithelial cells in human urinary tracts (4, 9).
P. mirabilis swarming may be divided into four stages: (i) surface-induced swarmer cell differentiation, (ii) a lag period of ca. 3.25 h prior to swarming migration, (iii) active swarming migration, and (iv) a consolidation phase, during which swarmer cells stop moving and dedifferentiate into swimmer-like cells. The four stages are cyclic and give rise to the distinctive bull's-eye colony pattern when P. mirabilis is grown on nutrient agar (10, 11).
Both swimming motility and swarming motility require functional flagella. The bacterial flagellum is a complex, extracellular filamentous structure that consists of three parts: a rotary basal body associated with the membrane, a hook junction, and an extended helical filament (12). The hook-basal body (HBB) complex is a rotary motor powered by proton motive force (PMF), which is generated by proton translocation through the stator complex, encoded by motA and motB (12). Genes involved in flagellum biosynthesis are clustered in several operons that make up the flagellar regulon. In P. mirabilis and other enteric bacteria, expression of the flagellar regulon is governed by a three-tiered hierarchical control allowing the coordinate expression of flagellar genes and flagellum biosynthesis (13). The flhDC operon (expressed from a class 1 promoter) encodes the flagellar master transcriptional regulatory protein complex, FlhD4C2. FlhD4C2 activates expression of class 2 promoters that transcribe genes such as fliA, encoding the flagellum-specific σ28 factor, flgM, encoding FlgM, an anti-σ28 factor, and genes encoding proteins comprising the HBB complex and the type III flagellar secretion apparatus. FliA is required for transcription of class 3 promoters, whose genes include those required for the production of the flagellar filament, the stator complex, and those involved in chemotactic behavior (14–16).
FlhD4C2 plays a central role in P. mirabilis swarmer cell differentiation (17). When P. mirabilis swimmer cells differentiate into swarmer cells, flhDC transcription increases, and genes in the flagellar regulon are upregulated (16, 18, 19). Many regulatory and environmental factors control the expression and activity of flhDC and swarming. For example, studies in Escherichia coli have shown that transcription of the flhDC operon is negatively regulated by the RcsCDB phosphorelay system (20), composed of the sensor kinase, RcsC, which passes a phosphoryl group to its cognate response regulator, RcsB, via RcsD, a phosphotransferase. Phosphorylated RcsB negatively regulates flhDC (21). Mutations in the components of P. mirabilis Rcs system result in increased expression of flhDC and precocious swarming: e.g., the swarming lag phase is shortened and swarming migration initiates earlier than in wild-type cells (22–24). P. mirabilis contains the four genes umoA to -D, two of which are known to interact with the Rcs system to increase expression of flhDC. These umo (i.e., upregulator of the master operon) genes were first identified as suppressors of the swarming defect in an flgN mutant, which is defective in a flagellar chaperone (25). The Umo proteins are predicted to be located in the cell envelope (25). umoA, encoding a putative outer membrane protein, contains both σ70-dependent and σ28-dependent (flagellar class 3) promoters (25), hinting that umoA may be a novel member of the flagellar regulon. Morgenstein and Rather have suggested that UmoB and UmoD are in a pathway that activates the Rcs phosphorelay system (26). How the Umo proteins function to increase flhDC expression is unclear.
Most flagellar genes have well-understood functions in bacterial motility, but fliL is an exception, and the role of FliL in flagellar structure and energetics remains, at best, obscure. In many species, fliL is the first gene of a class 2 operon, fliLMNOPQR, which is controlled by both FlhD4C2 and FliA. This operon encodes C-ring proteins (FliMN), components of the rotor of the flagellar motor and situated at the cytoplasmic face of the inner membrane, and those (FliOPQR) involved in the export apparatus. FliL is a small transmembrane protein (ca. 18 kDa in most species) located adjacent to the basal body and in close proximity to MotAB (27–29). FliL has a single transmembrane (TM) domain (residues 12 to 38 of 160 residues in P. mirabilis) near its N terminus, such that the N terminus of FliL is in the cytoplasm, while the rest of the protein is in the periplasm.
Among bacterial species, the phenotype of fliL cells is diverse. In alphaproteobacteria, such as Rhodobacter sphaeroides, Silicibacter sp. strain TM1040, and Caulobacter crescentus, disruption of fliL results in nonmotile cells (30–32), which is also true for fliL defects in the spirochete Borrelia burgdorferi (29). In gammaproteobacteria, such as Escherichia coli, Salmonella enterica serovar Typhimurium (“S. enterica” herein), and P. mirabilis, fliL mutations result in a minor decrease in swimming but severely affect swarming (27, 33, 34).
Our laboratory is interested in understanding the mechanism used by P. mirabilis to detect a surface and subsequently produce a swarmer cell. Transposon mutagenesis has proven to be valuable in identifying genes important for swarming and swarmer cell differentiation of P. mirabilis (35). Due to the polar nature of the mutation, insertion of a transposon in a flagellar gene often results in nonswarming (Swr−) cells that do not differentiate on surfaces. However, a small class of Tn5 insertion mutants, notably those with mutation in fliL (e.g., strain BB2204), are different because these fliL defects produce elongated swarmer-like cells (Elo+ [pseudoswarmer cells]) in noninducing liquid environments (8). BB2204 pseudoswarmer cells phenocopy most of the hallmarks of swarmer cells, such as cell elongation and polyploidy, but they do not produce flagella due to polar effects on fliMNOPQR (8, 34). We use the pseudoswarmer Elo+ phenotype as a proxy indicating that the surface-sensing mechanism has detected and responded to a surface. As such, the production of a pseudoswarmer cell under normally noninducing conditions indicates that the surface-sensing mechanism has malfunctioned and inappropriately signaled that the cell is on a surface when it is not.
To alleviate the unwanted polarity associated with transposon insertions in fliL (and the resulting loss of flagellar synthesis), in a previous report (18), we selected for a spontaneous fliL Swr+ mutant (YL1001) that had a partial excision of Tn5 from its BB2204 parent. This mutation altered the last 14 amino acids of FliL and restored transcription of fliMNOPQR, flagellum synthesis, and swarming, while retaining the Elo+ phenotype. This result demonstrates that the C terminus of FliL is involved in surface sensing (8, 18, 34). More recently, we constructed a nonpolar fliL-null mutant (fliL::kan-nt 30 [YL1003]) that resulted in the production of a severely truncated FliL protein. This fliL mutant is Swr− Elo+, produces flagella (Fla+), and swims (Swm+), emphasizing that FliL is essential for swarming but not for swimming motility. Significantly, the loss of fliL also abolishes the response of the cell to viscosity (in this case, the concentration of agar in nutrient medium), suggesting the role of FliL in surface sensing (34). YL1003 does not swarm on a surface, irrespective of the concentration of agar in the medium (34). An increase in the number of flagella per cell surface area is a hallmark of P. mirabilis swarmer cell differentiation induced after the cell has sensed a surface. An unexpected result of the fliL mutation in YL1003 is a decrease in expression of flagellin (encoded by flaA) (34), which further emphasizes the loss of surface sensing in fliL mutations. Paradoxically, expression of flhD and fliA increases in YL1003 (34). Disruption of fliL has also been reported to affect flagellar synthesis in other organisms: e.g., Silicibacter sp. strain TM1040 and Pseudomonas putida DOT-T1E (32, 36). Since FliL is a structural component of the flagellum, this outcome suggests that the fliL mutation in YL1003, but not necessarily the loss of FliL, has an unknown negative effect on the regulation of flaA (34). Insertion of the kanamycin cassette in the fliL gene may perturb the structure of the fliL operon mRNA in YL1003, which in turn may repress flaA transcription through an unknown mechanism.
To eliminate possible effects caused by the alteration of fliL DNA sequence, we describe here the construction and phenotype of a mutant with complete deletion of fliL (ΔfliL). We discovered that ΔfliL cells swarm but have defects in their surface-sensing mechanism, such that they swarm precociously on soft agar where the parent strain does not move. Unlike the wild-type parent, swarming of ΔfliL cells is also affected by temperature and is inhibited at low temperature, hinting that the loss of FliL causes a temperature-dependent depowering of the flagellar motor. A ΔfliL mutant strain of E. coli also constructed as part of this study phenocopies the P. mirabilis ΔfliL strain in being Swr+ with an alteration in temperature-dependent motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. mirabilis BB2000 (37) is the wild-type stain and the parent of all mutants used in this study. P. mirabilis YL1006 is a ΔfliL strain with a deletion between nucleotide (nt) positions 4 and 459 in the fliL gene (construction described below). E. coli RP437 (38) is the parent of YL102 and YL103. E. coli DH5α λpir (39) and SM10 λpir (40) were used for construction of pKNG101-derivated plasmids and biparental mating, respectively. E. coli JW1928 (obtained from the CGSC at Yale University) (41) is the donor of the ΔfliL790::kan allele used to construct E. coli ΔfliL strains (methods described below). E. coli XL1-Blue (Stratagene) was used for plasmid manipulations. E. coli and P. mirabilis were maintained in Luria-Bertani (LB) broth as described previously (34). LSW− agar (34) was used to acquire single colonies of P. mirabilis. As required, media were supplemented with 100 μg ml−1 ampicillin, 40 μg ml−1 chloramphenicol, 50 μg ml−1 kanamycin, 50 μg ml−1 streptomycin, or 15 μg ml−1 tetracycline. Swimming motility was screened and analyzed in tryptone broth (T broth: Bacto tryptone, 10 g liter−1; NaCl, 5 g liter−1) or Mot agar (T broth containing 3 g liter−1 Bacto agar).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| Strains | ||
| P. mirabilis | ||
| BB2000 | Wild type; spontaneous Rfr from PRM1 | 37 |
| YL1006 | BB2000 fliLΔnt 4–459 (encoding residues 2–153) | This study |
| E. coli | ||
| DH5α λpir | supE44 ΔlacU169 (ϕlacZΔM15) recA1 endA1 hsdR17 thi-1 gyrA96 relA1 λpir phage lysogen | 39 |
| JW1928 | BW25113 ΔfliL790::kan (nt 4–444 replaced by kan) | CGSC (41) |
| RP437 | F− thr-1 araC14 leuB6(Am) fhuA31 lacY1 tsx-78 λ− eda-50 hisG4(Oc) rfbC1? rpsL136(strR) xylA5 mtl-1 metF159(Am) thiE1 | 38 |
| SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir phage lysogen; Kmr | 40 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| YL102 | RP437 fliL ΔfliL790::kan | This study |
| YL103 | RP437 fliLΔnt 4–444 (encoding residues 2–148) | This study |
| Plasmids | ||
| pACYC184 | Cloning vector; p15A ori; Cmr Tcr | 19 |
| pCP20 | Plasmid showing temperature-sensitive replication and containing yeast Flp recombinase gene, FLP; Apr Cmr | 45 |
| pKNG101 | Suicide vector, R6K ori; Smr | 42 |
| pYL30 | pACYC184 containing 699-bp DNA fragment containing PfliL (200 bp upstream from start codon), codon region, and 16-bp downstream fliL; Cmr | 34 |
| pYL68 | pACYC184 containing PflhD::lacZ transcriptional fusion; Cmr | This study |
| pYL70 | pACYC184 containing PflaA::lacZ transcriptional fusion; Cmr | 34 |
| pYL73 | pACYC184 containing PumoA::lacZ transcriptional fusion; Cmr | This study |
| pYL115 | pKNG101 containing 3,389-bp fragment from 1,480 bp upstream through 1,426 bp downstream of fliL gene; Smr | This study |
| pYL117 | pYL115 with deletion of nt 4–456 of fliL codon region; Smr | This study |
| pYL125 | pACYC184 containing PfliA::lacZ transcriptional fusion; Cmr | This study |
| pYL126 | pACYC184 containing PflaA-flaA′::′lacZ translational fusion; Cmr | This study |
| pYL128 | pACYC184 containing PmotA::lacZ transcriptional fusion; Cmr | This study |
Apr, Cmr, Kmr, Rfr, Smr, and Tcr indicate ampicillin, chloramphenicol, kanamycin, rifampin, streptomycin, and tetracycline resistance, respectively.
Growth curves.
A single colony was inoculated into 2 ml LB broth and incubated at 37°C with shaking (200 rpm) overnight. The overnight culture was mixed, and a 1:100 dilution was inoculated to fresh LB broth at a starting optical density at 600 nm (OD600) of 0.05 and incubated at 37°C with shaking. The OD600 of the culture was measured every hour using a Beckman DU640 spectrophotometer.
Swimming and swarming motility assay.
Swimming and swarming motilities were measured as previously described (34) with minor modifications. Mot agar was used for measurement of swimming motility. LB broth containing Bacto agar (0.3% to 2.5%) or Eiken agar (Eiken Chemical Co., Tochigin, Japan) (0.6%) plus 0.5% glucose was used to determine the swarming motility of P. mirabilis or E. coli, respectively. Swimming and swarming assays were conducted in an environmental chamber at 37, 30, or 25°C with 46% relative humidity.
Construction of the P. mirabilis ΔfliL mutant.
An in-frame deletion of P. mirabilis fliL was constructed using homologous recombination. A 3,389-bp fragment harboring a region from 1,480 bp 5′ of the start codon to 1,426 bp 3′ of the fliL stop codon was amplified from BB2000 genomic DNA (extracted using the DNeasy blood and tissue kit manufactured by Qiagen) and ligated into the BamHI and XbaI sites in the suicide vector pKNG101 (42, 43), resulting in pYL115. Reverse PCR was used to construct a deletion at nucleotides (nt) 4 to 456 and to introduce an NcoI site in fliL on pYL115. Subsequently, the PCR product was digested with NcoI and circularized by ligation to yield pYL117. pYL117 was transferred into YL1006 via biparental mating between E. coli SM10 λpir/pYL117 and BB2000, as previously described (37). The exconjugants were selected on LSW− agar containing streptomycin and tetracycline. Streptomycin-sensitive isolates resulting from a double crossover event were selected by sacB counterselection (43). The deletion of fliL was confirmed by PCR using combinations of primer pairs matching the flanking and the internal regions of the fliL gene. The primers used are listed in Table S1 in the supplemental material. The resulting strain, called YL1006, contained a deletion of nt 4 to 459 in the fliL coding region, which was confirmed by sequencing.
Construction of the E. coli ΔfliL mutant.
Deletion of fliL in E. coli RP437 was constructed by P1 transduction to replace the wild-type fliL allele with the ΔfliL::kan allele from JW1928, a single-gene-knockout strain in the Keio collection (41). The result was YL102 (ΔfliL::kan). The kanamycin-resistant cassette in YL102 was removed by FLP recombination target (FRT) recombination, which was achieved by the expression of the Flp recombinase (44) from pCP20 (45) and resulted inYL103 (ΔfliL). Each deletion was confirmed by PCR and sequencing.
Construction of transcriptional and translational fusion vectors.
Transcriptional fusions between the promoter regions of flhD, umoA, fliA, and motA (respectively) and lacZ were constructed in pACYC184 by previously described methods (34). The resulting plasmids are called pYL68 (PflhD::lacZ), pYL73 (PumoA::lacZ), pYL125 (PfliA::lacZ), and pYL128 (PmotA::lacZ). pYL68 harbors a transcriptional fusion between the flhD promoter (−499 to −1 with respect to the start codon of flhD) and lacZ. pYL73 is a transcriptional fusion between the umoA promoter (−497 to −1 with respect to the start codon of umoA) and lacZ. pYL125 carries a transcriptional fusion between the fliA promoter (−477 to +53 with respect to the start codon of fliA) and lacZ. A transcriptional fusion between the motA promoter (−498 to +24 with respect to the start codon of motA) and lacZ is carried on pYL128.
A translational fusion between flaA and lacZ (flaA′::′lacZ) on pACYC184 was constructed as follows. Two-step overlap extension PCR was used to fuse a region of flaA′ from −386 to +30 relative to the flaA start codon, and a ′lacZ fragment comprising +31 through the stop codon of lacZ). The resulting PCR product was ligated into the BamHI and HindIII sites of pACYC184, producing pYL126.
Measurement of transcription or translation by LacZ activity.
LacZ activity was measured to assess the expression of flhD, fliA, flaA, motA, or umoA in agar-grown cells. Plasmids carrying a lacZ fusion were transformed into BB2000 or YL1006 by electroporation (46). One hundred microliters of an overnight culture containing a reporter plasmid was spread on the surface of LB agar and incubated at 37°C, with cells harvested every hour thereafter for a period of 7 h. β-Galactosidase activity was measured using the Miller assay (47) as described previously (48).
Measurement of transcription by qRT-PCR.
The procedures for RNA extraction, cDNA synthesis, and quantitative reverse transcription-PCR (qRT-PCR) are described by Lee et al. (34). RNA samples were obtained from 2.75-h cultures grown in LB broth with shaking (200 rpm) and 4.5-h cultures grown on LB agar at 37°C. Changes in gene expression were calculated using the threshold cycle (2−ΔΔCT) formula (49), with rpoA serving as the reference transcript.
Protein sample preparation and Western blotting.
Protein samples were prepared using methods described previously (34), with minor changes. In brief, P. mirabilis cells were grown in LB broth or on the surface of LB agar at 37°C or 25°C. For samples prepared from broth culture, bacteria were harvested at 37°C for 2.75 h or at 25°C for 6 h, which were chosen as the times of maximum pseudoswarmer cell production (18). For samples prepared from agar cultures, bacterial cells were washed off every hour after incubation at 37°C or 25°C from the LB agar surface with 2 ml of 1× phosphate-buffered saline (PBS [pH 7.40]) (34). In both cases, the pellets were collected by centrifugation (6,000 rpm for 10 min at room temperature), washed with 1× PBS, and then resuspended in 1 ml of 1× PBS. Wide-bore tips were used to prevent shearing of the flagella. Whole-cell homogenates of the pellets were prepared using six cycles of sonication for 10 s each, followed by a pause on ice for 30 s. A bicinchoninic acid (BCA) protein assay (Pierce) was used to determine the concentration of protein in each sample, and 1 μg of total protein was applied to an Any kD Mini-protean TGX polyacrylamide gel (Bio-Rad). The resolved proteins were transferred to Hybond-P polyvinylidene difluoride (PVDF) membranes (GE Healthcare) and detected as previously described (34). Digital images of the blot were acquired using the scanner function of HP Officejet 4500. ImageJ software (National Health Institute) was used to determine the amount of protein in bands on the developed Western blots.
Fluorescent staining and microscopic analysis.
Morphological changes and swarmer cell differentiation were assessed in LB broth and on soft LB agar (LB with 0.9% agar). Bacterial cells were harvested from LB broth and plates of soft LB agar every hour after incubation. Cells were fixed and fluorescently stained using previously described methods (34). Stained specimens were mounted in ProLong antifade (Invitrogen) and imaged using wide-field fluorescence microscopy with a Zeiss Axio Observer Z1 microscope using Zeiss filter sets, 10 shift free (F), 15 shift free (F), and 49 4′,6-diamidino-2-phenylindole (DAPI) shift free (E) and a Hamamatsu Orca-R2 charge-coupled device (CCD) camera. Acquired images were analyzed with Volocity 6.0.1 software (PerkinElmer). Channels for membrane and nucleoid staining were deconvolved using an iterative restoration function (with 25 iterations and a 98% confidence limitation). Color and contrast adjustment of processed images were done using Photoshop CS6 (Adobe).
Morphometric analyses of live bacterial cells were done using wet-mount suspensions viewed using phase-contrast light microscopy with an Olympus BX60 and a QImaging QCam CCD camera. Cell length was determined using a “skeletal length measurement” tool (Volocity), with 7 μm as the cutoff to define the minimum length of a swarmer cell (34). The mean and standard deviation of the cell length of each cell type and nonlinear fitting of the Gaussian distribution were calculated using Origin 9.0 software (OriginLab). A minimum of 500 cells were analyzed to determine the mean cell length for each population.
Measurements of swimming speed and behavior were obtained using an overnight culture grown in 2 ml T broth that was inoculated into fresh T broth at a 1:100 dilution and incubated with shaking (200 rpm) at 37°C for 5 h. The 5-h cultures were diluted in prewarmed T broth to an OD600 of 0.04 and quickly imaged. Videos of a ca. 5-s duration (∼25 frames per s) were captured. The motion of individual cells within a field was determined using the “track objects” function (with a trajectory variation tracking model [Volocity]). At least 150 tracks for each sample were analyzed. The minimum criterion defining a tumble was the change in direction of at least 90°.
Drop collapse assay.
A modification of the drop collapse assay of Be'er and Harshey (50) was used to determine the presence of surfactant in supernatants of bacterial cultures. One hundred microliters of an overnight culture was spread on the surface of soft LB agar and incubated at 37°C. At 5 h, the bacterial cells were scraped off the plate using a glass coverslip and resuspended in 200 μl deionized water, which was then transferred to a microcentrifuge tube. The cell pellet and the supernatant were separated by centrifugation at 8,000 rpm for 10 min at room temperature. Five microliters of the supernatant was carefully applied to the surface of a lid from a polystyrene petri dish (Fisher Scientific). After 1 min, a photograph of the side of the drop was taken with the camera lens parallel to the plastic surface. The contact angle, defined as the angle between the drop tangent contacting the plastic surface and the reflection line, was measured directly from the digital image using ImageJ software.
RESULTS
ΔfliL cells of P. mirabilis swarm.
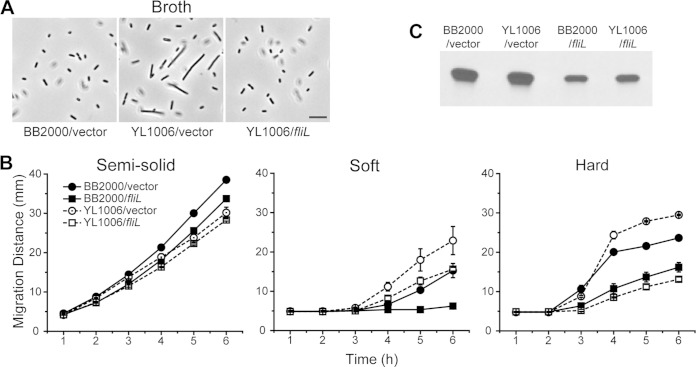
A mutant of P. mirabilis with 90% of fliL deleted (YL1006 [fliLΔnt 4–459]) was constructed (Materials and Methods). The growth of YL1006 is unaffected by the fliL mutation (see Fig. S1A in the supplemental material), and the deletion is nonpolar on the other genes in the fliL operon (see Fig. S1B). Based on previous experience with other FliL− mutants and the reports of fliL deletions in other bacteria (27, 34), we predicted that YL1006 would be Swr−; however, these ΔfliL cells swarmed nearly the same as parental cells (Fig. 1A, right panel), while possessing minor defects in swimming (Fig. 1A, left panel). Similar to other fliL mutants of P. mirabilis, YL1006 is Elo+, producing elongated cells in broth morphologically similar to pseudoswarmer cells seen in other fliL mutants (Fig. 1B). However, unlike YL1003, YL1006 pseudoswarmer cells are hyperflagellated (Fig. 1C; see Fig. S2 in the supplemental material) and for all purposes phenocopy wild-type swarmer cells obtained from agar. Thus, to distinguish them from YL1003 pseudoswarmer cells and wild-type agar-grown swarmer cells, we refer to the YL1006 cells as hyperflagellated pseudoswarmer cells (HPCs). Like other fliL pseudoswarmer cells, YL1006 HPC production is transient (with a maximum of 6.2% HPCs in the population at 2.75 h). As previously reported for other fliL pseudoswarmer cells, nucleoids in the ΔfliL HPCs are irregularly spaced, which implicates a function for FliL in cell septation and/or partitioning of the chromosome. These results show that FliL is not essential for P. mirabilis swarming.
FIG 1.
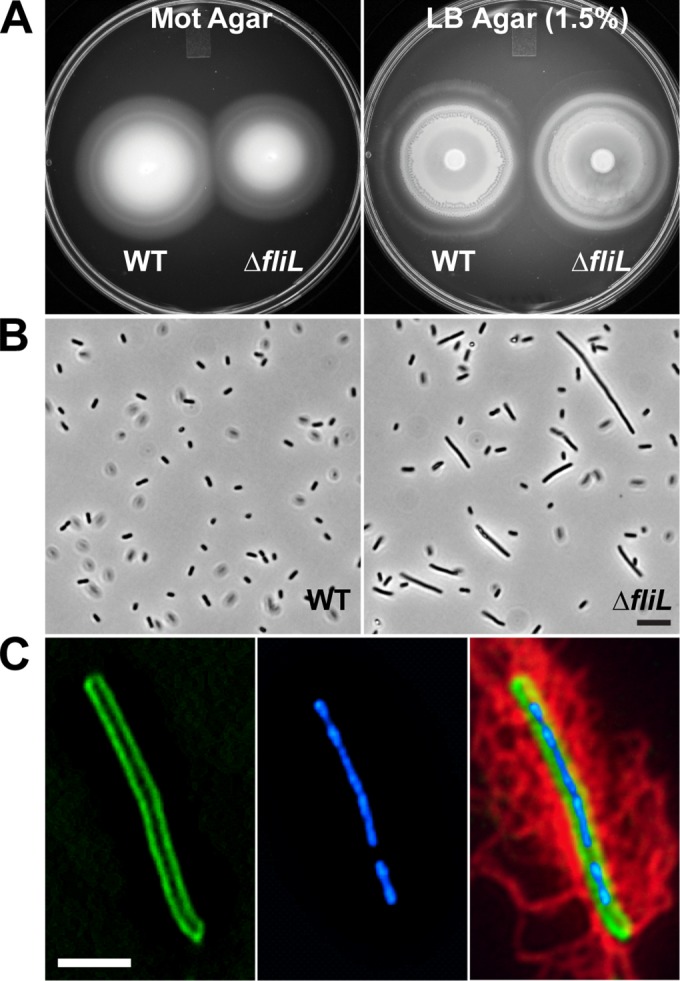
Phenotype of P. mirabilis ΔfliL strain YL1006. (A) Swimming and swarming motilities on Mot agar (left) and hard LB agar (right), respectively. (B) Phase-contrast micrographs of cells of broth-grown cells. Scale bar, 10 μm. WT, wild type. (C) A representative ΔfliL pseudoswarmer cell showing (from left to right) membrane (FM1-43 [green]), nucleoids (DAPI [blue]), and flagella (anti-FlaA immunostaining [red]). Scale bar, 5 μm.
ΔfliL cells precociously swarm on low-viscosity surfaces.
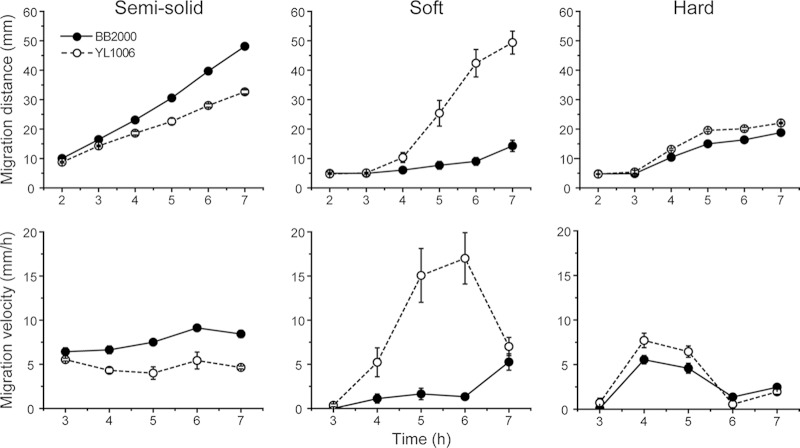
Our previous results have implicated FliL in viscosity-dependent surface sensing (34), such that defects in fliL alter the cell's response to a surface. Swimming and swarming motility were examined in wild-type and YL1006 cells using LB broth containing different concentrations of agar, with the concentration of agar acting as a proxy for viscosity. We define “semisolid agar” as broth containing 0.3% agar: “soft” agar is broth with 0.9% agar, and “hard” agar contains 1.5% agar. The results of these analyses are shown in Fig. 2 (see Fig. S3 in the supplemental material). As shown in Fig. 2, YL1006 swarms proficiently on LB with agar at 0.9% or above. As expected, swarming declines as viscosity increases. As is true for wild-type cells, YL1006 does not swarm when the viscosity of the surface is <0.9% agar, presumably because at this viscosity flagella rotate normally and rotation is unimpaired, and thus the cells do not sense that this is a surface. These results show that swarming of YL1006 is viscosity dependent.
FIG 2.
Comparison of wild-type and ΔfliL mutant cell motilities as affected by the concentration of agar in the medium. Shown is migration of the wild-type (solid circles) and ΔfliL (open circles) cells on LB with 0.3% (semisolid), 0.9% (soft), and 1.5% (hard) agar. (Top row) Migration distance; (bottom row) migration velocity. Means ± standard deviations from three independent experiments are shown.
As can be observed in Fig. 2 (see Fig. S3 in the supplemental material), YL1006 initiates swarming earlier (precocious swarming) and swarms significantly faster (3.2×) than the wild type on soft agar, while the difference in swarming is much less when on hard agar, i.e., YL1006 swarms slightly faster (1.4×) than the wild type on 1.5% agar. On soft agar, the maximum migration rate for YL1006 is 17.0 ± 2.9 mm/h compared with 5.3 ± 0.9 mm/h for the wild type, and YL1006 starts swarming at 3 h, compared to 6 h for wild-type cells. We conclude that ΔfliL cells have enhanced swarming over low-viscosity surfaces that otherwise prevent swarming of the wild type.
Similar to the wild type, YL1006 is able to swim through an agar matrix when the concentration is reduced to 0.3%, but ΔfliL cells swim at a slower speed than do wild-type cells (Fig. 2, top left panel). Why is YL1006 less effective in swimming through semisolid agar than BB2000? One possibility is that the fliL defect decreases swimming speed or alters the swimming behavior of the cell. Such changes in the swimming speed and behavior of individual cells could affect overall migration through semisolid agar. This hypothesis was tested by measuring the speed and direction of live cells using microscopy and cell tracking analyses. As shown in Table 2, the average swimming speed of YL1006 is ca. 10% less than the speed of the wild type (14.2 compared with 15.6 μm/s), while the tumbling frequency of the mutant is slightly higher (12.9%) than that of the wild type (11.9%). These differences are not statistically significant but may subtly reduce the overall migration rate in semisolid agar. However, as shown in Fig. 3, YL1006 cultures contain a higher percentage of poorly motile cells (61.0%) than do cultures of BB2000 (46.1%): i.e., the number of ΔfliL cells that swim fast decreases, while the percentage of cells that swim slowly increases (Fig. 3). These results suggest that the fliL defect in YL1006 reduces swimming speed, resulting in a population skewed toward more slowly swimming cells and helps explain why ΔfliL hampers swimming through semisolid agar.
TABLE 2.
Analysis of swimming motilities of BB2000 and YL1006 cells at 37°C
| Strain | Swimming speed (μm/s) | Tumbling rate (%) | % of motile cells |
|---|---|---|---|
| BB2000 (wild type) | 15.5 ± 2.5 | 11.9 | 46.1 |
| YL1006 (ΔfliL mutant) | 14.2 ± 3.0 | 12.9 | 61.0 |
FIG 3.
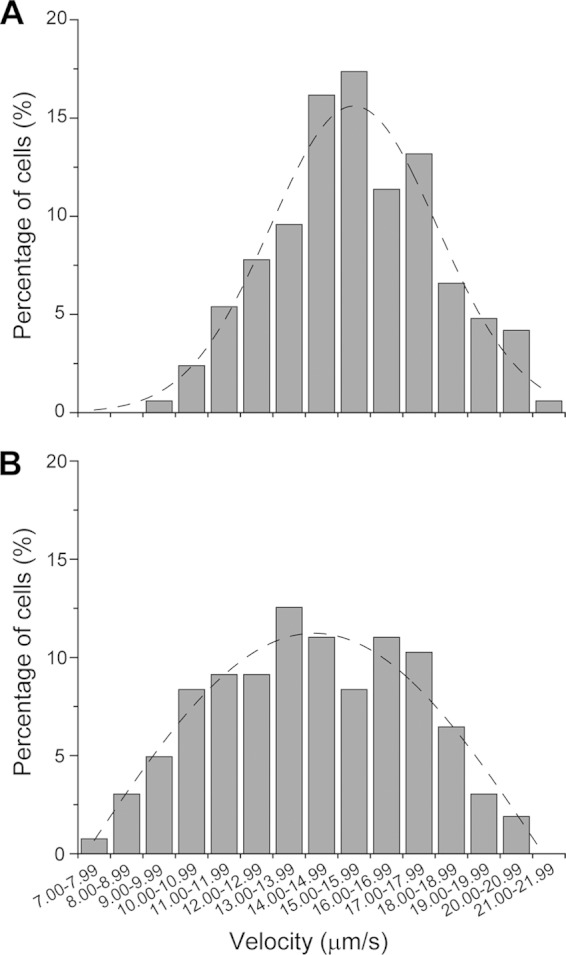
Comparison of swimming speeds between wild-type and ΔfliL mutant populations. Shown is a histogram of the percentage of swimming cells of the wild-type (A) versus the ΔfliL mutant (B). The dashed line in each panel is a nonlinear fitting of the Gaussian distribution of the data in each graph.
ΔfliL results in a minor increase of flaA expression on soft agar.
Why does YL1006 swarm proficiently over soft agar when BB2000 does not? Answers to this question are particularly important to our understanding of FliL's function, as paradoxically the same mutation hampers motility in lower-viscosity environments (i.e., semisolid agar). In approaching this problem, we note that bacteria employ common strategies to overcome the lack of wetness and friction associated with a surface that include producing wetting agents (surfactants), increasing the number of flagella, and increasing the length of the cell (51). The fliL mutation may affect any one of these factors to promote YL1006 swarming over soft agar.
We assessed surfactant production using a drop collapse assay, in which a reduction in contact angle of the drop is correlated with more surfactant. As shown in Fig. S4 in the supplemental material, the contact angles between drops of supernatant from wild-type cells (contact angle of 71°) and ΔfliL cells (contact angle of 72°) are the same within the detection limits of this assay. Therefore, the fliL mutation has no demonstrable effect on surfactant production, and both YL1006 and wild-type cells produce the same amount of wetting agent, ruling out surfactant production as a cause of YL1006 swarming over soft agar.
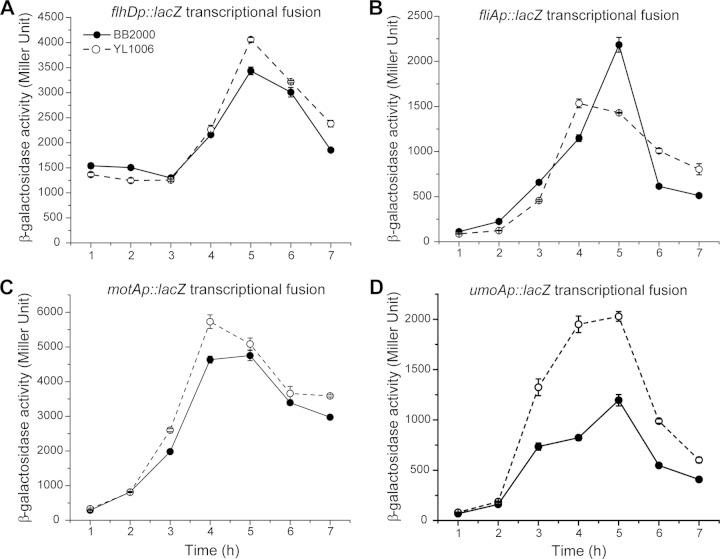
To test if an increase in flagella allowed ΔfliL cells to swarm over soft agar, we measured flagellin (flaA) expression in cells grown on soft agar and found that wild-type and ΔfliL cells have nearly identical levels of expression of flaA during the time period between 1 h and 4 h after inoculation (Fig. 4A). As the incubation progressed beyond 4 h, a difference in flaA transcription became noticeable: e.g., wild-type flaA expression was higher (1.3×) than flaA expression in YL1006, and flaA transcription remained higher in wild-type cells for the remainder of the time course.
FIG 4.
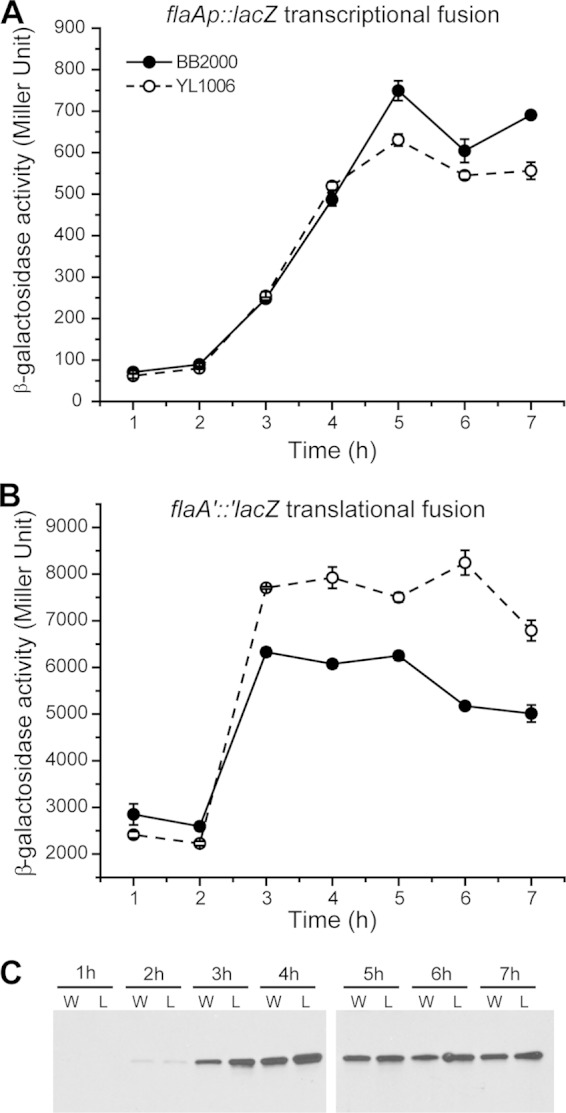
Expression of flagellin (FlaA) in wild-type and ΔfliL mutant cells. Shown are transcription (A) and translation (B) of flaA, measured as β-galactosidase activity from a PflaA::lacZ transcriptional fusion and flaA′::′lacZ translational fusion, respectively, in wild-type (solid circles) and ΔfliL (open circles) cells. Means ± standard deviations are shown (n = 3). (C) Immunoblot to flagellin in wild-type (W) and ΔfliL (L) cells.
A more dramatic difference was seen when comparing translation of flaA in wild-type and ΔfliL cells (Fig. 4B). Mirroring the transcription data, from 1 to 3 h after inoculation, translation of flaA increases dramatically in both YL1006 and BB2000, with flaA translation in ΔfliL cells increasing faster than the rate in wild-type cells (3.9× versus 2.5× from the 2-h to 3-h time points). After this, flaA translation leveled out; it ultimately declined at the end of the time course (7 h) and was ca. 1.4× greater in ΔfliL cells than in wild-type cells (Fig. 4B).
Flagellin production parallels flaA translation and is very low (almost undetectable) prior to 3 h, after which flagellin significantly increases (Fig. 4C). Comparable to flaA translation, after 3 h, FlaA protein in ΔfliL cells was 1.2 to 1.4× higher than that in wild-type cells (Fig. 4C). While the shapes of the transcription and translation curves (respectively) and overall patterns of flagellin protein levels are similar in YL1006 and wild-type cells, increased translation of flaA and FlaA levels in YL1006 are evident. We conclude that YL1006 produces more flagella than wild-type cells on soft agar, and this increase promotes precocious swarming.
ΔfliL enhances cell elongation and umoA transcription on soft agar.
Expression of flaA is dependent upon FlhD4C2 and FliA and increases along with the expression of other class 3 transcripts, such as motA and umoA. Does the fliL mutation in YL1006 affect the expression of other class 3 transcripts when cells are swarming over soft agar? We compared the levels of expression of four genes (flhD, fliA, motA, and umoA) in YL1006 and BB2000 cells grown on soft agar. Overall, the kinetics of expression of flhD (class 1 promoter), fliA (both class 2 and 3 promoters), motA and umoA are similar to those of flaA (compare Fig. 4 and 5), with the exception that transcription of these four genes decreases between 5 and 6 h, whereas flaA expression does not. Expression of umoA is markedly higher (2 to 5×) in YL1006 compared with the wild type (Fig. 5D). An equivalent increase in umoA transcription was also detected in broth-grown ΔfliL cells (Table 3) (18). qRT-PCR analysis showed that transcription of flhDC in broth-grown cells at 2.75 h (when YL1006 produces the maximum number of HPCs) is not different from that of the wild type (data not shown). Since the function of umo genes is thought to take part in flhDC expression, we measured the expression of umoB and umoD, which are also involved in swarming motility (26). As observed with umoA, umoD expression increased in ΔfliL cells compared with wild-type cells, but by a smaller amount (1.1 to 1.5×), while umoB expression remained unchanged (Table 3). These data suggest an indirect role of FliL in the control of umoA and umoD expression. We conclude that the expression of umoA and umoD is upregulated when ΔfliL cells swarm on soft agar and helps to explain precocious swarming.
FIG 5.
Comparison of the levels of expression of flhD, fliA, and motA in wild-type (solid circles) and ΔfliL (open circles) cells. Transcription of flhD (A), fliA (B), motA (C), and umoA (D) in cells grown on soft LB agar was measured as β-galactosidase activity from their respective lacZ transcriptional fusions. Means ± standard deviations (n = 3) are shown.
TABLE 3.
Fold change in expression of umoA and umoD in YL1006 compared with BB2000 cells in broth- or soft surface-grown cultures
| Gene | Broth |
Agar |
||
|---|---|---|---|---|
| Fold change in expressiona | P value | Fold change in expressiona | P value | |
| umoA | 1.8 ± 0.4 | 0.10 | 1.8 ± 0.4 | 0.05 |
| umoB | 0.9 ± 0.1 | 0.14 | 0.9 ± 0.1 | 0.15 |
| umoC | NDb | NAc | ND | NA |
| umoD | 1.1 ± 0.1 | 0.21 | 1.5 ± 0.3 | 0.04 |
The fold change in expression (YL1006 versus BB2000) was calculated to compare the expression of each gene (relative to the expression of rpoA, the reference gene) in YL1006 (the ΔfliL mutant) with the expression of the same gene (relative to rpoA expression) in BB2000 (the wild type). A value of >1 indicates that expression of the gene is greater in YL1006 than in BB2000, while a value of <1 means that expression decreases in YL1006. Means ± standard deviations from three independent measurements from three biological samples are presented.
ND, not done.
NA, not applicable.
Our third hypothesis to explain why YL1006 swarms on soft agar is that the fliL mutation causes an increase in the length of the cell or in the number of swarmer cells in the population. Using phase-contrast microscopy of living cells obtained from soft agar, we found that swarmer cells (i.e., cells >7 μm in length) comprised 10.8% of the YL1006 population, while only 3.4% of wild-type cells were swarmer cells (Fig. 6A). The population of YL1006 cells grown on soft agar also produces ca. 3× more preswarmer cells (defined as those cells with lengths between 4 and 7 μm) during the first 3 h of growth (Fig. 6B). Thus, a ΔfliL population swarming on soft agar has 3× more swarmer cells and 3× more preswarmer cells than its wild-type counterpart. While there are more swarmer cells in YL1006 populations grown on soft agar, there is no statistical difference in the mean cell lengths of ΔfliL and wild-type swarmer cells: the YL1006 swarmer cell length is 12.18 ± 3.13 μm, while wild-type swarmer cells average 10.42 ± 2.86 μm. These results suggest that ΔfliL cells swarm on soft LB agar by increasing the number of swarmer cells aided by precocious swarmer cell development.
FIG 6.
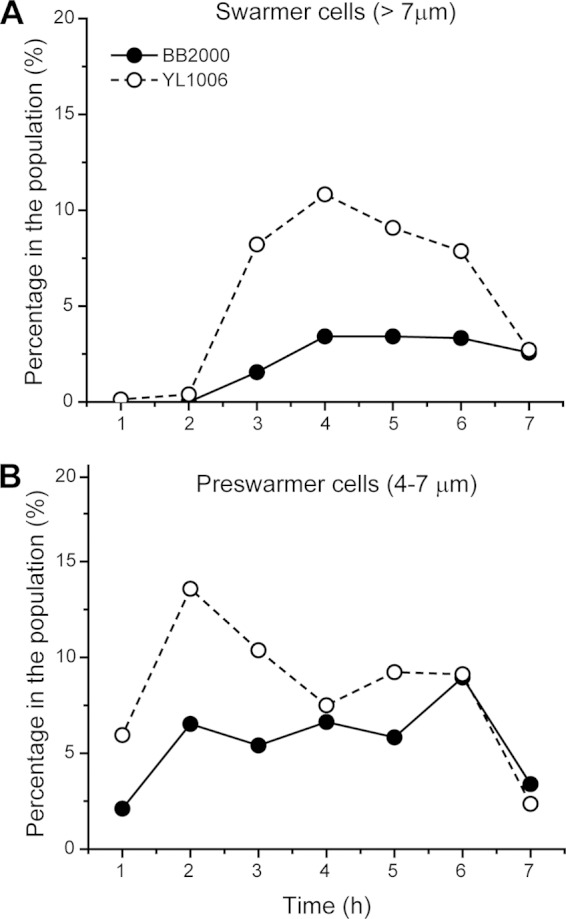
Comparison of rates of swarmer cell synthesis in wild-type and ΔfliL populations grown on soft LB agar. The percentages of swarmer cells (>7 μm) (A) and preswarmer cells (4 to 7 μm) (B) in the entire population (n > 500) were examined.
ΔfliL results in a low-temperature inhibition of swarming.
While conducting experiments with YL1006, we observed that, unlike in the wild type, ΔfliL cells failed to swarm on hard agar when left on the lab bench at room temperature. This observation was further explored, and the results are shown in Fig. 7. Although ΔfliL cells swarm on hard agar at 37°C, unlike BB2000, their swarming is inhibited at temperatures below 37°C. YL1006 has a significant reduction in swarming at 30°C (Fig. 7, middle panel) and completely loses the ability to swarm over hard agar at 25°C (Fig. 7, lower panel). One explanation for the failure of YL1006 to swarm at 25°C is that low temperature may prevent swarmer cell differentiation. This is not the case, as ΔfliL cells grown at 25°C produce HPCs in broth and differentiate into normal swarmer cells on hard agar (Fig. 8A). The percentage of YL1006 HPCs and swarmer cells produced at 25°C is roughly equal to those produced at 37°C. When in broth cultures, 5.6% of the YL1006 population are HPCs, while on hard agar, 36% of the cells are differentiated swarmer cells. At 25°C, the number of flagella per μm of cell surface on either HPCs or swarmer cells is the same as when grown at 37°C, and ΔfliL cells have wild-type production of flagellin at the lower temperature (Fig. 8B). These results lead us to conclude that inhibition of YL1006 swarming at low temperature is not due to defects in swarmer cell differentiation or flagellar synthesis. Another possible explanation for the lack of swarming is that the flagella of ΔfliL cells are more fragile at 25°C; however, this is not the case, as we did not detect a significant amount of detached flagellin by Western blotting in the supernatant fraction harvested from the agar surface (data not shown).
FIG 7.
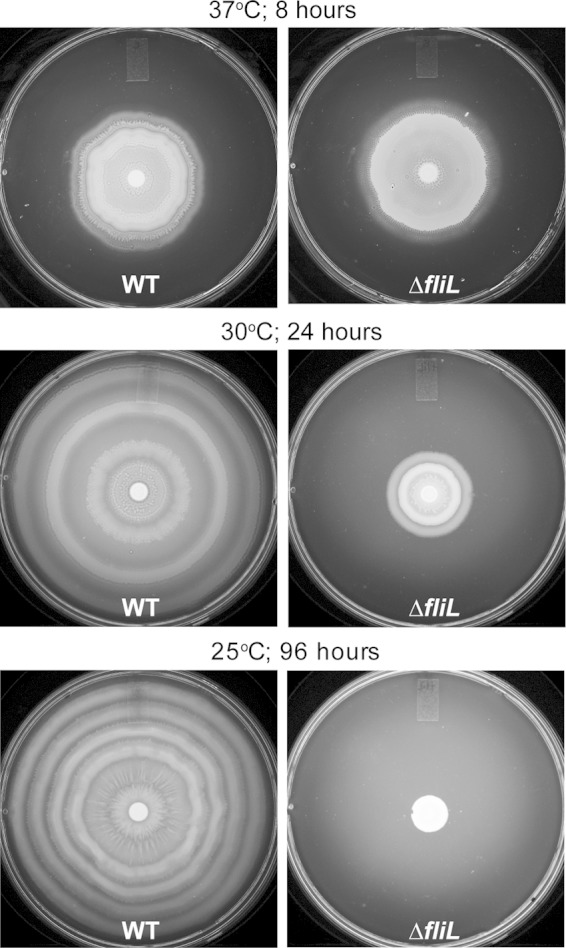
Effect of incubation temperature on the swarming of wild-type and ΔfliL mutant strains on hard LB agar.
FIG 8.
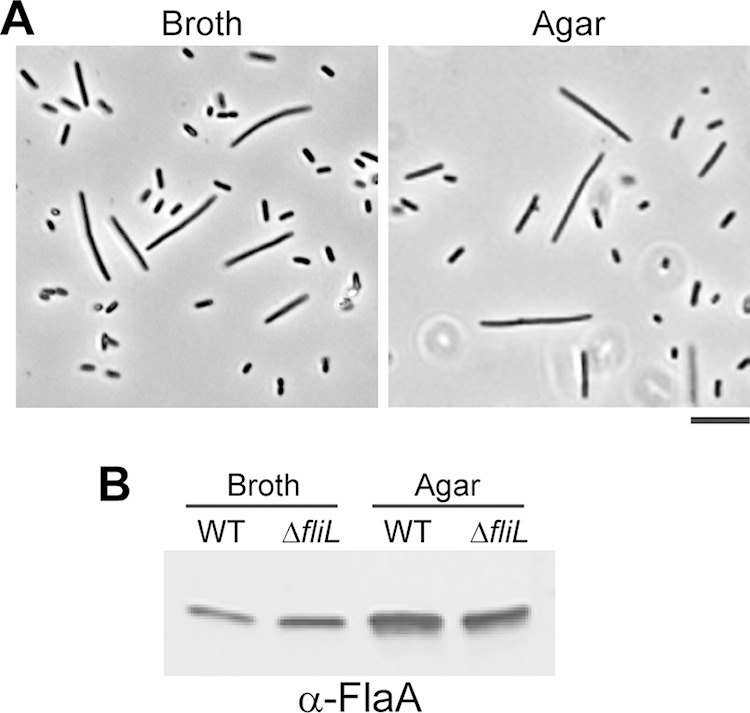
Cell morphology and flagellin expression of wild-type and ΔfliL mutant cells at 25°C. (A) Phase-contrast micrographs (400× magnification) of ΔfliL cells obtained from LB broth after 6 h or hard LB agar at 8 h of growth at 25°C. Scale bar, 10 μm. (B) Immunoblot to flagellin in the wild-type (WT) and ΔfliL cells grown in LB broth for 6 h or LB agar for 8 h.
We questioned whether low temperature also inhibits motility of ΔfliL cells on semisolid agar, which would suggest that loss of FliL in YL1006 may somehow deenergize the flagellar motors. The results are shown in Table 4 and in Table S2 in the supplemental material. In general, ΔfliL cells are poorer swimmers than wild-type cells, irrespective of temperature, and swim at ca. 70% the speed of wild-type cells at 37°C (Table 4). At 25°C, ΔfliL cell swimming is further reduced to ca. 30% of the wild-type rate (Table 4; see Table S2). As can be seen in Table 4, while the mean migration velocity of wild-type cells in semisolid agar at 25°C is 30% of the swimming speed of the same strain at 37°C (2.2 ± 0.2 versus 7.4 ± 1.6 mm/h), incubation at the lower temperature has a much greater effect on ΔfliL cells than it does on wild-type cells (0.7 ± 0.3 versus 5.0 ± 0.7 mm/h), reducing the swimming speed of ΔfliL cells at 25°C to ca. 14% of the velocity at 37°C. We conclude that the flagellar motors of ΔfliL cells are more negatively affected at 25°C than are the motors of wild-type cells and suggest that these results support a role for FliL in flagellar motor performance or energetics.
TABLE 4.
Migration velocity of BB2000 and YL1006 on semisolid LB agar
| Strain | 37°C |
25°C |
||||
|---|---|---|---|---|---|---|
| Migration velocity (mm/h) | ΔfliL mutant/wild-type ratio | P value | Migration velocity (mm/h) | ΔfliL mutant/wild-type ratio | P value | |
| BB2000 (wild type) | 7.4 ± 1.6 | 2.2 ± 0.2 | ||||
| YL1006 (ΔfliL mutant) | 5.0 ± 0.7 | 0.7 | 7.15E−04 | 0.7 ± 0.3 | 0.3 | 3.23E−07 |
fliL+ in trans partially complements the ΔfliL phenotype.
Three phenotypic characteristics are unique to ΔfliL cells: (i) the production of HPCs, (ii) precocious swarming on soft agar, and (iii) low-temperature inhibition of swarming. We asked whether fliL+ in trans could complement the YL1006 fliL mutation and return the ΔfliL phenotype to wild type. Plasmid pYL30, containing fliL under the control of its native promoter, was introduced into YL1006, and the resulting phenotype was determined (Fig. 9).
FIG 9.
Cell morphology, migration, and flagellin production of wild-type and ΔfliL mutant cells affected by fliL+. (A) Phase-contrast micrographs of wild-type and ΔfliL cells containing the empty cloning vector and fliL expressed in trans. Scale bar, 10 μm. (B) Migration of wild-type and ΔfliL cells as affected by the agar concentration in the presence and absence of fliL. Shown are the migration distances of the wild-type (solid symbols) and ΔfliL (open symbols) cells containing the empty cloning vector (circles) and fliL expression plasmid (squares) on semisolid, soft, and hard LB agar. Means ± standard deviations from three independent measurements are shown. (C) Comparison of flagellin production in wild-type and ΔfliL cells in the presence and absence of fliL. Shown is an immunoblot to flagellin in wild-type (BB2000) and ΔfliL (YL1006) cells containing empty cloning vector grown on the surfaces of soft LB agar at 37°C for 5 h.
First, YL1006/pYL30 cells lost their ability to form HPCs in broth and resembled wild-type swimmer cells (Fig. 9A), demonstrating that a functional FliL complemented the ΔfliL mutation and prevented HPC formation by ΔfliL cells. Second, on soft agar fliL+ complemented the fliL mutation, and YL1006/pYL30 cells swarmed the same as wild-type cells, losing their precocious swarming nature (Fig. 9B, middle panel). In conducting these experiments, we consistently observed that expression of fliL+ in trans resulted in a marked decrease in both swimming and swarming over all surfaces tested (Fig. 9B). Inhibition of motility is most pronounced when the cells are swarming, with a 40 to 80% reduction in swarming compared with a 10% reduction in swimming migration. One reason for the decrease in motility is that expression of fliL+ in trans results in a decrease in flagella (Fig. 9C) and occurs when fliL is driven from either its native promoter or a heterogenous promoter, such as Ptrc (data not shown).
The third phenotype of ΔfliL cells, inhibition of swarming at low temperature, was not altered by expression of fliL+ in trans. We suspect that this is due to repression of flagellar synthesis when fliL+ is expressed from a plasmid (as shown in Fig. 9C). We conclude that while expression of fliL+ in trans does not fully recapitulate the wild-type phenotype, two of the three unique phenotypes of YL1006 are complemented. Furthermore, these results point to an unexpected repression of motility and flagellar production by expression of fliL+ in trans.
E. coli ΔfliL partially phenocopies P. mirabilis ΔfliL.
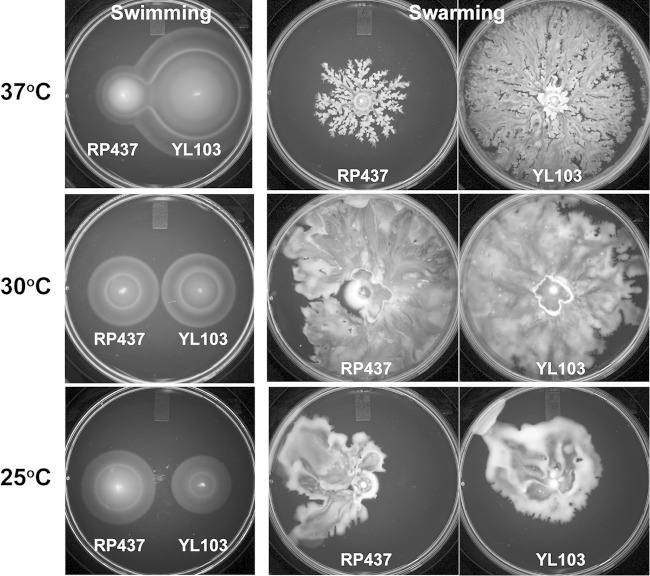
Many reports, including our previous papers, have underscored the importance of fliL in swarming, not just in P. mirabilis, but in other enteric bacteria as well (8, 27, 34). We wondered if the Swr+ phenotype that we observed in YL1006 is unique to P. mirabilis or if instead it is more universal and apparent in other enteric bacteria. This was tested by constructing an E. coli ΔfliL strain with a nearly identical in-frame deletion to that in P. mirabilis YL1006. The result is E. coli YL103 (fliLΔnt 4–444), a strain that swarms well (Fig. 10). Furthermore, like P. mirabilis YL1006, loss of fliL in YL103 results in altered temperature effects on motility. For both P. mirabilis and E. coli, loss of fliL enhances swarming at 37°C (Fig. 10). For YL103, loss of fliL results in a >220% increase in swarming at 37°C (compared to the wild-type control). This result suggests that fliL+ may repress motility of E. coli, similar to that observed in P. mirabilis. Indeed, as shown in Fig. 11, expression of fliL+ in trans also inhibits swarming of E. coli ΔfliL cells. On the other hand and unlike what we observed with P. mirabilis ΔfliL cells, growth of YL103 at lower temperatures (i.e., 30°C and 25°C) results in more moderate to no effects. We conclude that E. coli ΔfliL partially phenocopies P. mirabilis ΔfliL. This result emphasizes the universal nature of the ΔfliL phenotype in enteric bacteria.
FIG 10.
Swimming and swarming characterization of wild-type (RP437) and ΔfliL (YL103) E. coli cells. Swimming was measured on semisolid Mot agar, and swarming was measured on LB agar containing 0.6% Eiken agar and 0.5% glucose. Swimming plates were incubated for 8 h at 37°C and 30°C and for 24 h at 25°C. Swarming plates were incubated for 24 h at all temperatures.
FIG 11.
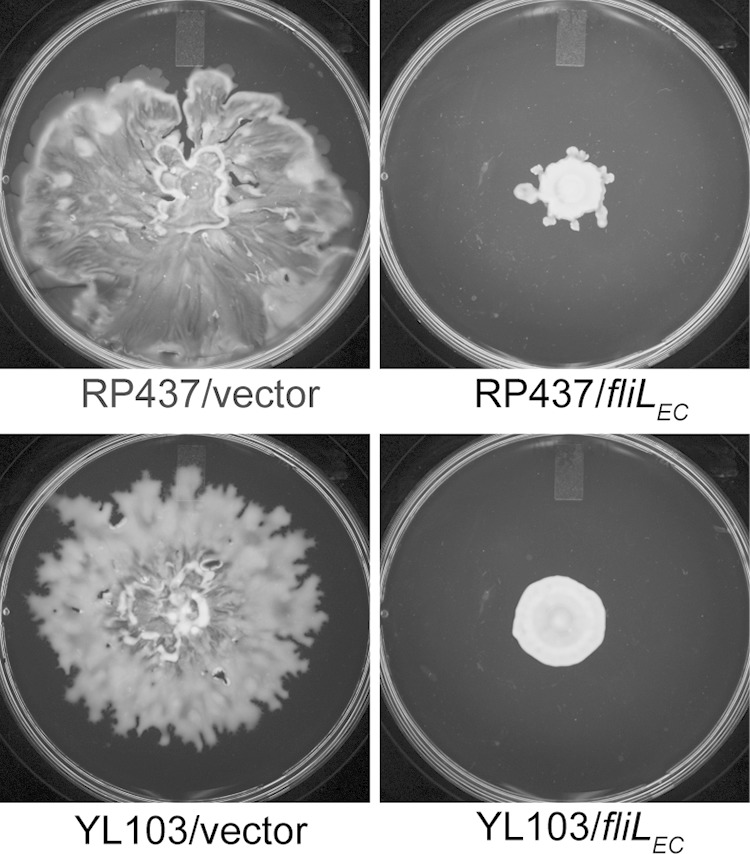
Swarming of wild-type and ΔfliL mutant E. coli cells in the presence and absence of E. coli fliL. Swarming was measured on LB agar containing 0.6% Eiken agar and 0.5% glucose incubated at 30°C for 24 h.
DISCUSSION
In this study, we investigated the function of fliL in P. mirabilis. We discovered that (i) cells without fliL are able to swarm (Swr+), (ii) loss of fliL alters P. mirabilis sensing of surfaces, (i.e., a ΔfliL cell is more responsive to low-viscosity agar), (iii) loss of fliL enhances P. mirabilis swarmer cell differentiation (i.e., more swarmer cells are produced through precocious differentiation, and (iv) the motility of cells lacking fliL is temperature dependent.
It came as a surprise to find that ΔfliL cells of P. mirabilis swarm on hard agar at 37°C (Fig. 1A), as the phenotype of our previous fliL mutants—for example, YL1003 (fliL::kan-nt 30), an FliL null mutant—is Swr− (34). Why would the two fliL mutations result in opposite swarming phenotypes? These two fliL strains were constructed using different genetic manipulations that affect the fliL coding sequence in different ways. YL1003 has a nonpolar group II intron insertion (TargeTron) between nucleotides 30 and 31 of fliL and retains all of the fliL DNA (34), while YL1006 has a deletion of most of the fliL gene. It is apparent that these two mutations have different effects on flagellar synthesis, such that YL1006 produces HPCs reminiscent of wild-type swarmer cells, while flagellin synthesis is reminiscent of swimmer cell levels in YL1003 (Fig. 1C; see Fig. S2 in the supplemental material) (34). Why then is flagellar synthesis affected in YL1003 but not in YL1006? Expression of flaA and flagellin is significantly reduced in YL1003 (34), yet they are expressed at wild-type levels in YL1006 (Fig. 4), and both fliL mutants are incapable of synthesizing a functioning FliL protein. These facts lead us to conclude that the defects in swarming and flagellin expression observed in YL1003 are not due to the loss of FliL protein itself but rather are due to the presence of fliL DNA sequence. We speculate that swarming is wild type in YL1006 due to the loss of fliL DNA sequence that otherwise results in repressed flagellar synthesis. This idea is supported by our current results in both P. mirabilis and E. coli showing that in trans expression of fliL+ inhibits motility and flagellin synthesis (Fig. 9 and 11). Thus, we hypothesize that fliL has two functions. First, the gene encodes the FliL protein, whose function will be discussed subsequently. Second, fliL DNA (or possibly fliL mRNA) has a regulatory function that, when present, directly or indirectly represses flaA expression. This finding suggests that some fliL mutants constructed in other laboratories may give results similar to YL1003 and mislead us about the function of FliL. We conclude that the fliL gene but not the FliL protein is essential for P. mirabilis swarming over hard agar.
FliL is also not necessary for E. coli swarming, a result that is counter to the reports of others (27). The Swr+ phenotype of E. coli ΔfliL strain YL103 is reproducible, and the mutation with its associated Swr+ phenotype has been reconstructed several times in independent experiments in our laboratory (data not shown), emphasizing that this characteristic is not due to second site mutations or other spontaneous events that could have led to a Swr+ phenotype. What is the difference between YL103 and other E. coli ΔfliL strains, such as UA332 (27), that are Swr−? The size and the location of the deleted region within fliL are the major differences that distinguish these strains. YL103 deletion removes nt 4 to 444 (of 465 nucleotides) in fliL, while UA332 has a smaller deletion (Δnt 61–402) (27). We hypothesize that the additional DNA sequence of the fliL allele in UA332 may be causing its Swr− phenotype by causing defects in flagellin expression as we observed in P. mirabilis YL1003.
It has been proposed in our previous studies that P. mirabilis FliL plays a role in sensing surfaces via the effect of medium viscosity on impaired flagellar motor function (34). Our present results further support this function. We have demonstrated in this work that ΔfliL cells have an altered response to viscosity. Importantly, ΔfliL cells do not lose their ability to sense viscosity or respond to surfaces, but rather loss of fliL changes the low-end threshold of the response to a “surface”: e.g., the “window” of surface sensing is expanded to now include low-viscosity environments that normally are not sensed by wild-type P. mirabilis, resulting in a ΔfliL cell that responds to a greater range of viscosities. At the same time, the high end of the surface-sensing response of ΔfliL cells remains the same as that in wild-type cells.
What is the connection between the ΔfliL mutation and enhanced cellular differentiation of HPCs? Our results suggest that FlhD4C2 plays a central role in this differentiation process (16). The phenotype of P. mirabilis flhDC-overexpressing strains has been reported by the Hughes (University of Cambridge) and Rather (Emory University) groups (16, 24, 52). In these studies, overexpression of flhDC resulted in a reduction in the swarming lag period (what we call “precocious” swarming) and enhanced swarming motility, which was due to an increased expression of flagellin and elongated swarmer cells (16, 24, 52). Additionally, heightened amounts of FlhD4C2 give rise to swarmer-like cells (what we call “HPCs”) in broth. The phenotype of a cell that overexpresses flhDC is identical to that of ΔfliL cells and suggests that control of FlhD4C2 is the endpoint of the FliL surface-sensing pathway. In support of this, previous results (34) and the present report clearly show that flhD expression increases in fliL mutants (Fig. 5). Similar to flhD expression, expression of umoA and umoD is also upregulated in ΔfliL cells (Table 3). Dufour et al. have shown that overexpression of umo genes induces differentiation of swarmer cells, suggesting that umoA (and perhaps umoD) plays a role in the swarmer cell elongation (25). Moreover, other P. mirabilis fliL mutants (e.g., BB2204, YL1001, and YL1003) have increased umoA expression (18), suggesting the role of FliL in the umoA signaling pathway. Since FliL is not a DNA-binding regulatory protein, changes in gene expression caused by the ΔfliL mutation are most likely due to indirect, downstream effects leading to control of flhDC. Our data implicate UmoA and UmoD as being two proteins that function in the surface-sensing pathway. In support of our hypothesis, Morgenstein and Rather have suggested UmoB and UmoD are involved in activation of the Rcs system that acts to control swarming (26). Furthermore, we have shown that mutations in the Rcs system, specifically those in rcsC or rcsD (formerly rsbA), result in an Elo+ cells and precocious swarming (22) that partially phenocopy ΔfliL cells. The similarity between these mutant phenotypes suggests a possible involvement of the Umo proteins and the Rcs system in the FliL-mediated surface-sensing pathway.
As can be seen in Fig. 2, the loss of FliL has effects on swimming motility, such that the motility of ΔfliL cells in semisolid agar is slower than that of wild-type cells (see Fig. S3 in the supplemental material). The results from the present study suggest that the impairment of swimming results from a decreased number of fast-swimming motile ΔfliL cells (Fig. 3) that tumble more than wild-type cells (Table 2). However, other possibilities exist to explain the decrease in ΔfliL swimming, and they include the following: (i) the loss of FliL may cause slight structural defects in the HBB, and (ii) multiple flagellar filaments on ΔfliL HPCs may impair swimming motility due to tangling and ineffective coordination of multiple filaments. These factors undoubtedly contribute to the decrease in YL1006 overall migration.
In understanding the role FliL plays in surface sensing and flagellar motor function, it is important to emphasize two important findings from this study. First, there is the marked difference in response to viscosity displayed by ΔfliL cells compared to wild-type cells. This is best seen by comparing the swimming of ΔfliL cells to that of BB2000 in media with different viscosities. In very-low-viscosity nutrient broths (LB or T broth, for example), ΔfliL cells are only 10% slower than the wild type, but when the viscosity of the liquid is increased, as in semisolid agar, swimming of the ΔfliL mutant was markedly reduced (30% slower) compared with its parent. This suggests that the loss of FliL adversely affects the motor when under high torque.
Second, swarming of ΔfliL cells is sensitive to temperature and is inhibited at low temperatures. Similar temperature-dependent effects on P. mirabilis swarming have been noted by Armbruster et al. (53). The temperature-sensitive motility of ΔfliL cells also hints at a role for FliL in modulating motor energetics and suggests that loss of FliL underpowers the motor at low temperatures. At low loads, torque on the motor is exponentially dependent on temperature, while under high-load and high-torque conditions, motor speed is independent of temperature (54, 55). However, under high-load and low-torque conditions, motor speed is temperature dependent (55). When a bacterial cell migrates in a low-viscosity environment, its flagellar motor rotates under low-load conditions. On the other hand, highly viscous environments mimic high-load conditions. As such, the temperature-dependent motility of ΔfliL cells hints at a deenergizing of the flagellar motors that presumably produce less power, resulting in a motile force that is insufficient to overcome the physical barriers imposed by the surface.
A substantial body of evidence links the performance of the motor with temperature. For example, point mutations in S. enterica fliG, fliM, and fliN, encoding three proteins of the C-ring flagellar rotor, exhibit a temperature-sensitive (TS) motile phenotype (56). TS mutations have also been found in Na+-driven motors, and certain mutants with mutations in PomA of Vibrio alginolyticus, a MotA stator homologue, also exhibit a TS phenotype (57). Thus, the temperature-dependent phenotype of ΔfliL cells offers a clue that suggests FliL interacts with a component of the motor (either stator or rotor) that is involved in torque generation. Moreover, this interaction appears to be torque dependent, prompting us to speculate that FliL may be acting as a governor to control motor function and/or energetics under high-torque conditions. Although we favor this hypothesis, at present we cannot rule out the possibility that ΔfliL cells result in flagellar structural or assembly defects and that it is these defects that result in the temperature-dependent phenotype. Defining the mechanism underlying this FliL temperature-dependent phenotype is an important goal for future research.
With what motor proteins might FliL interact? Several studies have shown structural and functional relationships between FliL, the HBB complex, and the stator. In P. mirabilis, mutations not only in fliL but also in fliG (C-ring) and fliF (MS ring) result in the Elo+ phenotype (8), phenotypically linking FliL, FliG, and FliF and suggesting the three proteins may function in the same “pathway.” Fluorescence resonance energy transfer (FRET) data obtained from E. coli demonstrate a weak interaction between FliL and FliG (58), cryo-electron tomography images of the flagellar basal body of B. burgdoferi have localized FliL adjacent to the stator (29), and genetic data obtained from R. sphaeroides (30) and S. enterica (59) suggest that FliL interacts with the stator (30, 59, 60).
While evidence of direct interactions between FliL and the stator is lacking, we speculate that FliL enhances the performance of the motor by acting as a molecular governor to limit or control proton flow through the stator when the motor is operating under high-torque conditions, (i.e., in high-viscosity environments), as was recently suggested (61). This hypothesis presumes that under such conditions where flagellar motors are working at peak force, the cell risks acidification of its cytoplasm, which would lead to a potential hyperpolarization of the membrane and death, if left uncontrolled. In this model, FliL interacts with the stator to control the amount of protons flowing through the proton channel of the motor. We hypothesize that in the absence of FliL, unregulated proton flow across the membrane occurs, resulting in deenergizing flagellar motors. One prediction of this hypothesis is that the performance of the ΔfliL motor in viscous environments can be improved by increasing energy flow or PMF of the cell, perhaps through efflux of excess H+ to the external milieu. Investigations testing this hypothesis are under way in our laboratory.
Recent reports have demonstrated that E. coli motor stators function as mechanosensors as well as torque-generating units, and the recruitment and assembly of the stator occur in a load-dependent manner (62, 63). This underscores the putative connection between flagellar mechanosensing, FliL, and components of the flagellar motor. Lele, Hosu, and Berg further demonstrated that FliL is not involved in stator recruitment (63). These results do not contradict our findings and emphasize that the function of FliL involves events that occur after the initial stage of motor localization.
While our understanding of the role FliL plays is slowly coming into focus, the function of fliL and the protein it encodes remain strangely enigmatic, especially in light of how much is known about the function of bacterial flagellar proteins in general. Two factors appear to complicate our efforts: (i) the function of FliL appears to be at least partially species dependent, as described in the introduction, and (ii) fliL defects result in conflicting phenotypes that depend on the how the mutation was constructed. The best examples of this are YL1003 (a group II intron insertion in fliL) and YL1006 (ΔfliL). The former strain is Swr−, while YL1006 is Swr+, the only apparent difference between them being the presence of fliL DNA in YL1003. The simplest explanation for this difference is that fliL (DNA or mRNA) has a second, regulatory function that controls transcription of flaA and other flagellar genes, although how this occurs is currently unknown. Nevertheless, what all P. mirabilis fliL mutants have in common is the elongated-cell phenotype (Elo+) in broth cultures, as well as a loss of the C-terminal domain of the FliL protein, which appears to play an important role in surface sensing (18).
Many studies emphasize a role of the flagellum in mechanosensing of surfaces (2, 8, 64, 65), and the present results demonstrate the importance of FliL in the surface-sensing sensory transduction pathway. To summarize our present results, the data reveal that loss of FliL alters the sensitivity of P. mirabilis cells to viscosity by lowering the low-viscosity threshold of the surface-sensing response without affecting the cell's high-viscosity response. Using these results, we have created a working model describing the function of FliL in surface sensing. We hypothesize that FliL functions as a structural component of the flagellum that acts as a governor to control proton flow when the motor is under high-torque conditions. This control is exerted through an interaction between the periplasmic C-terminal domain of FliL and a protein component of the motor, perhaps the plug region of MotB (30). This action is predicted to prevent acidification of the cytoplasm and maintain PMF and cellular energetics under these high-torque conditions. We speculate that a bacterial cell may sense a change in PMF, membrane potential, or a pH gradient when the rotation of its flagella becomes inhibited during contact with a surface. This “surface” signal then initiates swarmer cell differentiation through a pathway that minimally includes UmoA, UmoD, and Rcs proteins and results in increased expression of the flagellar master regulator, FlhD4C2. It is noteworthy that swarmer cells can be induced in broths by lowering the pH of the medium (66), adding credibility to this prediction.
In conclusion, our study provides a better understanding of bacterial biofilm formation, surface sensing, and the role FliL plays in these process. It also raises several important questions. For example, what protein-protein interactions are important for FliL function, and are these interactions viscosity dependent? How does FliL affect motor function in a temperature-dependent manner? Is PMF, membrane potential, or cytoplasmic pH altered by the ΔfliL mutation? What is the molecular signal that a cell responds to upon flagellar mechanosensing? The answers to these questions not only will help us to understand FliL, bacterial swarming, and flagellar motor function, but are likely to provide better insight into the mechanism(s) underlying surface sensing and biofilm formation in other bacterial species. The knowledge gained from answering these questions may be useful in the development of strategies to prevent biofilms and novel therapeutic agents to prevent diseases resulting from bacterial biofilms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sandy Parkinson and Rasika Harshey for the gift of E. coli strains. We also thank Rayford Payne and Peter Norris for editorial comments and two anonymous reviewers for helpful suggestions.
This work was supported by award MCB-0919820 from the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02235-14.
REFERENCES
- 1.Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol 17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Belas R. 2013. When the swimming gets tough, the tough form a biofilm. Mol Microbiol 90:1–5. doi: 10.1111/mmi.12354. [DOI] [PubMed] [Google Scholar]
- 3.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mobley HL, Belas R. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol 3:280–284. doi: 10.1016/S0966-842X(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 5.Coker C, Poore CA, Li X, Mobley HL. 2000. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect 2:1497–1505. doi: 10.1016/S1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. 2008. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev 21:26–59. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alavi M, Belas R. 2001. Surface sensing, swarmer cell differentiation, and biofilm development. Methods Enzymol 336:29–40. doi: 10.1016/S0076-6879(01)36575-8. [DOI] [PubMed] [Google Scholar]
- 8.Belas R, Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol 187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison C, Lai HC, Hughes C. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol Microbiol 6:1583–1591. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 10.Williams FD, Schwarzhoff RH. 1978. Nature of the swarming phenomenon in Proteus. Annu Rev Microbiol 32:101–122. doi: 10.1146/annurev.mi.32.100178.000533. [DOI] [PubMed] [Google Scholar]
- 11.Rauprich O, Matsushita M, Weijer CJ, Siegert F, Esipov SE, Shapiro JA. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J Bacteriol 178:6525–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 13.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176:7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Matsumura P. 1996. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol Microbiol 21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 16.Furness RB, Fraser GM, Hay NA, Hughes C. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J Bacteriol 179:5585–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgenstein RM, Szostek B, Rather PN. 2010. Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis. FEMS Microbiol Rev 34:753–763. doi: 10.1111/j.1574-6976.2010.00229.x. [DOI] [PubMed] [Google Scholar]
- 18.Cusick K, Lee YY, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol 194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson MM, Rasko DA, Smith SN, Mobley HL. 2010. Transcriptome of swarming Proteus mirabilis. Infect Immun 78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 21.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2004. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 22.Belas R, Schneider R, Melch M. 1998. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol 180:6126–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaw SJ, Lai HC, Ho SW, Luh KT, Wang WB. 2001. Characterisation of p-nitrophenylglycerol-resistant Proteus mirabilis super-swarming mutants. J Med Microbiol 50:1039–1048. [DOI] [PubMed] [Google Scholar]
- 24.Clemmer KM, Rather PN. 2007. Regulation of flhDC expression in Proteus mirabilis. Res Microbiol 158:295–302. doi: 10.1016/j.resmic.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Dufour A, Furness RB, Hughes C. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol 29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstein RM, Rather PN. 2012. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol 194:669–676. doi: 10.1128/JB.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attmannspacher U, Scharf BE, Harshey RM. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol 68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- 28.Schoenhals GJ, Macnab RM. 1999. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology 145:1769–1775. doi: 10.1099/13500872-145-7-1769. [DOI] [PubMed] [Google Scholar]
- 29.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suaste-Olmos F, Domenzain C, Mireles-Rodriguez JC, Poggio S, Osorio A, Dreyfus G, Camarena L. 2010. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J Bacteriol 192:6230–6239. doi: 10.1128/JB.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenal U, White J, Shapiro L. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol. 243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 32.Belas R, Horikawa E, Aizawa S, Suvanasuthi R. 2009. Genetic determinants of Silicibacter sp. TM1040 motility J Bacteriol 191:4502–4512. doi: 10.1128/JB.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raha M, Sockett H, Macnab RM. 1994. Characterization of the fliL gene in the flagellar regulon of Escherichia coli and Salmonella typhimurium. J Bacteriol 176:2308–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YY, Patellis J, Belas R. 2013. Activity of Proteus mirabilis FliL is viscosity dependent and requires extragenic DNA. J Bacteriol 195:823–832. doi: 10.1128/JB.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belas R, Erskine D, Flaherty D. 1991. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J Bacteriol 173:6279–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura A, Duque E, Hurtado A, Ramos JL. 2001. Mutations in genes involved in the flagellar export apparatus of the solvent-tolerant Pseudomonas putida DOT-T1E strain impair motility and lead to hypersensitivity to toluene shocks. J Bacteriol 183:4127–4133. doi: 10.1128/JB.183.14.4127-4133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belas R, Erskine D, Flaherty D. 1991. Transposon mutagenesis in Proteus mirabilis. J Bacteriol 173:6289–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol 151:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolter R, Inuzuka M, Helinski DR. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 41.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 44.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 46.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. 1982. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J 1:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Lee YY, Barker CS, Matsumura P, Belas R. 2011. Refining the binding of the Escherichia coli flagellar master regulator, FlhD4C2, on a base-specific level. J Bacteriol 193:4057–4068. doi: 10.1128/JB.00442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−DDCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Be'er A, Harshey RM. 2011. Collective motion of surfactant-producing bacteria imparts superdiffusivity to their upper surface. Biophys J 101:1017–1024. doi: 10.1016/j.bpj.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Partridge JD, Harshey RM. 2013. Swarming: flexible roaming plans. J Bacteriol 195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hay NA, Tipper DJ, Gygi D, Hughes C. 1997. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol 179:4741–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armbruster CE, Hodges SA, Mobley HL. 2013. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess l-glutamine. J Bacteriol 195:1305–1319. doi: 10.1128/JB.02136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan J, Berg HC. 2010. Thermal and solvent-isotope effects on the flagellar rotary motor near zero load. Biophys J 98:2121–2126. doi: 10.1016/j.bpj.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Berg HC. 2000. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J 78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mashimo T, Hashimoto M, Yamaguchi S, Aizawa S. 2007. Temperature-hypersensitive sites of the flagellar switch component FliG in Salmonella enterica serovar Typhimurium. J Bacteriol 189:5153–5160. doi: 10.1128/JB.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukuoka H, Yakushi T, Homma M. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J Bacteriol 186:6749–6758. doi: 10.1128/JB.186.20.6749-6758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol 80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 59.Partridge JD, Harshey RM. 2013. More than motility: Salmonella flagella contribute to overriding friction and facilitating colony hydration during swarming. J Bacteriol 195:919–929. doi: 10.1128/JB.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabela S, Domenzain C, De la Mora J, Osorio A, Ramirez-Cabrera V, Poggio S, Dreyfus G, Camarena L. 2013. A distant homologue of the FlgT protein interacts with MotB and FliL and is essential for flagellar rotation in Rhodobacter sphaeroides. J Bacteriol 195:5285–5296. doi: 10.1128/JB.00760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4(4):e00551–13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol 90:6–21. doi: 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol 20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 66.Fujihara M, Obara H, Watanabe Y, Ono HK, Sasaki J, Goryo M, Harasawa R. 2011. Acidic environments induce differentiation of Proteus mirabilis into swarmer morphotypes. Microbiol Immunol 55:489–493. doi: 10.1111/j.1348-0421.2011.00345.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.