Abstract
Trehalose glycolipids are found in many bacteria in the suborder Corynebacterineae, but methyl-branched acyltrehaloses are exclusive to virulent species such as the human pathogen Mycobacterium tuberculosis. In M. tuberculosis, the acyltransferase PapA3 catalyzes the formation of diacyltrehalose (DAT), but the enzymes responsible for downstream reactions leading to the final product, polyacyltrehalose (PAT), have not been identified. The PAT biosynthetic gene locus is similar to that of another trehalose glycolipid, sulfolipid 1. Recently, Chp1 was characterized as the terminal acyltransferase in sulfolipid 1 biosynthesis. Here we provide evidence that the homologue Chp2 (Rv1184c) is essential for the final steps of PAT biosynthesis. Disruption of chp2 led to the loss of PAT and a novel tetraacyltrehalose species, TetraAT, as well as the accumulation of DAT, implicating Chp2 as an acyltransferase downstream of PapA3. Disruption of the putative lipid transporter MmpL10 resulted in a similar phenotype. Chp2 activity thus appears to be regulated by MmpL10 in a relationship similar to that between Chp1 and MmpL8 in sulfolipid 1 biosynthesis. Chp2 is localized to the cell envelope fraction, consistent with its role in DAT modification and possible regulatory interactions with MmpL10. Labeling of purified Chp2 by an activity-based probe was dependent on the presence of the predicted catalytic residue Ser141 and was inhibited by the lipase inhibitor tetrahydrolipstatin (THL). THL treatment of M. tuberculosis resulted in selective inhibition of Chp2 over PapA3, confirming Chp2 as a member of the serine hydrolase superfamily. Efforts to produce in vitro reconstitution of acyltransferase activity using straight-chain analogues were unsuccessful, suggesting that Chp2 has specificity for native methyl-branched substrates.
INTRODUCTION
In bacteria, glycolipids based on the disaccharide trehalose have been identified in Mycobacteria, Corynebacteria, Nocardia, and Rhodococcus (1). These members of the suborder Corynebacterineae have distinctive cell envelopes: exterior to the cytoplasmic membrane, they have not only a peptidoglycan layer but also an arabinogalactan polysaccharide layer and a second, outer bilayer membrane. Here, we use the term “cell envelope” to refer collectively to the assembly of membrane and polymer layers surrounding the cell, including the cytoplasmic membrane, peptidoglycan, arabinogalactan, and outer membrane. Trehalose lipids are found in the outer membranes of all of the aforementioned genera, but those with multiply methyl-branched acyl chains are specific to virulent strains of mycobacteria, including Mycobacterium tuberculosis, which causes tuberculosis (2). In the current model of the mycobacterial outer membrane, the inner leaflet is composed of extremely long-chain mycolic acids, which are covalently attached to the arabinogalactan layer, and the noncovalently associated outer leaflet is dominated by long branched-chain lipids, including di-, tri-, and pentaacyltrehaloses (3, 4). Consistent with their surface location, acyltrehaloses have proposed roles in the attachment of the carbohydrate capsule layer and have been identified as antigens, leading to their exploration as serodiagnostic markers with the potential to distinguish between tuberculous and nontuberculous mycobacteria (5–8).
The strain specificity of acyltrehaloses implies a role in virulence, and several studies support this idea. Diacyltrehaloses inhibit proliferation of naive T cells and downregulate cytokine production in activated monocytes, indicative of an immunosuppressive function (9–11). Also, impairment of acyltrehalose production in M. tuberculosis is associated with acidification of M. tuberculosis-containing phagosomes (12). Acyltrehaloses may thus contribute to phagosome-lysosome fusion arrest, a key factor in the survival of M. tuberculosis in host cells. Indeed, the gene cluster associated with acyltrehalose biosynthesis is upregulated during phagosome acidification (13). Overall, however, the functions of these lipids in M. tuberculosis physiology and pathogenesis are incompletely understood.
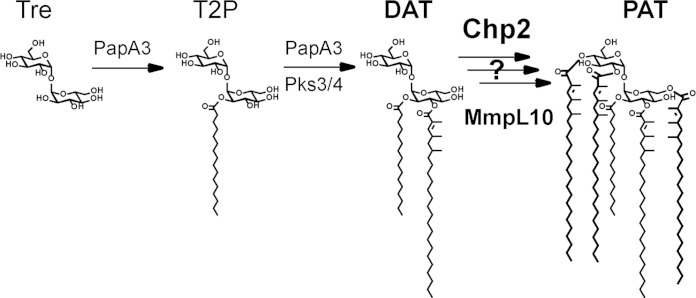
The dominant acyltrehalose in M. tuberculosis is polyacyltrehalose (PAT; also known as pentaacyl or polyphthienoyl trehalose), which comprises trehalose modified by one straight-chain fatty acid and four mycolipenic (phthienoic) acids (Fig. 1) (2). The mycolipenic acids are produced by Pks3/4 (also known as Msl3), and disruption of the pks3/4 gene results in the loss of PAT (4). Due to the cyclical processive mechanism of Pks3/4, the mycolipenic acids differ in the degree of methylation (1 to 3 methyl branches) as well as in the length of the saturated alkyl chain (C16 to C18) (14, 15). This variation gives rise to a diverse range of PAT structures, which are observed by mass spectrometry as a distinctive envelope of ions in the m/z 2,000 to 2,300 range (16). More recently, the papA3 gene was also shown to be essential for PAT biosynthesis and PapA3 was proposed as an acyltransferase that modifies trehalose with a saturated alkyl chain to form trehalose-2-palmitate (T2P) and then with a mycolipenic acid to form diacyltrehalose (DAT) (16). However, it remains unclear whether PapA3 is in fact a bifunctional enzyme in M. tuberculosis. One hypothesis is that PapA3 also catalyzes further esterifications to form the final PAT product, but, at least in vitro, this activity was not detected.
FIG 1.
The biosynthetic pathway for PAT is incomplete. The acyltransferase PapA3 converts trehalose into T2P and then to DAT using mycolipenic chains produced by Pks3/4. Chp2 and MmpL10 may be involved in the subsequent conversion of DAT into PAT.
As noted in the PapA3 study, the PAT biosynthetic gene cluster closely resembles that of sulfolipid 1 (SL-1; also known as Ac4SGL), a structurally similar trehalose glycolipid that is also unique to pathogenic mycobacteria. The SL-1 cluster contains two PapA3 homologues, PapA2 and PapA1, that esterify sulfotrehalose with straight-chain and methyl-branched phthioceranoic acids to form diacyl sulfotrehalose (also known as SL-1278) (16, 17). Recent work showed that subsequent modifications with phthioceranoic acid are mediated by a third enzyme, Chp1 (18). Chp1 is unrelated to the PapA acyltransferases but in its predicted structure and catalytic mechanism closely resembles the antigen 85 complex (Ag85A, Ag85B, and Ag85C), which catalyzes the transesterification of trehalose monomycolate to form trehalose dimycolate and free trehalose (19).
The PAT biosynthetic gene cluster includes rv1184c, a homologue of chp1 that we here call chp2. The coordinate upregulation of chp2 and other PAT biosynthetic genes upon phagosome acidification or expression of transcriptional regulators such as PhoP and WhiB3 supports the idea of a role for chp2 in PAT biosynthesis (20, 21). Therefore, we hypothesized that Chp2 is an acyltransferase that transforms DAT into PAT. To test this prediction, we characterized the M. tuberculosis Δchp2 knockout strain and showed that it does not produce PAT but accumulates DAT. We also showed that Chp2 is a serine-dependent enzyme that is associated with the cell envelope but that has cytosolic activity that requires the lipid transporter MmpL10. Finally, Chp2 activity was reduced in M. tuberculosis cells treated with the lipase inhibitor tetrahydrolipstatin (THL).
MATERIALS AND METHODS
Bacterial strains, growth media, vector construction, and oligonucleotide primers.
7H9 and 7H11 growth media and albumin-dextrose-catalase (ADC) and oleic acid-ADC (OADC) supplements were obtained from BD Biosciences. Mycobacterial strains used in these studies were M. tuberculosis Erdman (ATCC 35801) and M. smegmatis mc2155 (ATCC 700084). The growth medium was 7H9 (liquid) or 7H11 (solid) with 0.5% glycerol. Additional additives were 0.05% Tween 80 and 10% ADC supplement for M. smegmatis or 0.025% Tyloxapol and 10% OADC for M. tuberculosis. For selective media, antibiotic concentrations were 100 μg/ml ampicillin, 50 μg/ml kanamycin, or 100 μg/ml hygromycin for Escherichia coli and 25 μg/ml kanamycin or 50 μg/ml hygromycin for mycobacteria. See Tables S1 and S2 in the supplemental material for details on vector constructs and oligonucleotide primer sequences.
Sequence homology analysis and structure prediction.
The amino acid sequence for Chp2 (Rv1184c) was obtained from Tuberculist (http://genolist.pasteur.fr/TubercuList). Transmembrane helices were predicted by the TMHMM hidden Markov model (http://www.cbs.dtu.dk/services/TMHMM) (22) and signal peptides by SignalP (http://www.cbs.dtu.dk/services/SignalP/) (23). The Chp2 sequence was also submitted to Phyre (http://www.sbg.bio.ic.ac.uk/phyre) for protein fold and structure prediction (24).
Construction and complementation of gene deletion mutants.
The Δchp1 mutant strain was created by homologous recombination using specialized phage transduction (25). This deletion replaced 765 bp of chp2 (corresponding to amino acids [aa] 53 to 307) with a hygromycin resistance cassette. Flanking sequences for chp2 were amplified from H37Rv bacterial artificial chromosome (BAC) clone Rv7 (gift of Roland Brosch, Institut Pasteur). Recombinant clones were confirmed by PCR (see Table S2 in the supplemental material). ΔmmpL10 mutant M. tuberculosis Erdman strain jcm110 was a gift of Jeffrey Cox (University of California, San Francisco). The deletion was confirmed by PCR and sequencing and replaced 3,000 bp of mmpL10 (corresponding to aa 3 to 1002) with a hygromycin resistance cassette. Each deletion strain was complemented with a pMV306 integrating plasmid carrying the target gene with a native promoter corresponding to 1 kb upstream of the first gene in the corresponding putative operon (26). The S141A mutation was introduced into Chp2 by site-directed mutagenesis. For ΔmmpL10 complementation, additional constructs containing mmpL10 with other promoters (2 kb upstream of pks3/4; the hsp60 promoter [26]; or the glutamine synthase [gs] promoter [27]) or containing both chp2 and mmpL10 with a native promoter were also tested. The chp1 integrating plasmid for complementation and the ΔpapA3 mutant and complement strains were previously reported (16, 18).
Lipid extraction and mass spectrometry analysis.
M. tuberculosis strains were grown for 3 to 5 days to the late log phase. Cultures were diluted in 15-ml detergent-free medium to an optical density at 600 nm (OD600) of 0.3 to 0.4 and incubated at 37°C for 2 days. Cells were harvested and extracted in 1 ml hexane. The upper organic phase (“surface lipid” fraction) was removed and added to an equal volume of 1:1 chloroform/methanol. All extractions were repeated in at least three independent experiments. Extracts were dried by rotary evaporation and resuspended in 500 μl of 20 mM ammonium acetate in methanol. High-resolution mass spectra were obtained on an LTQ Orbitrap XL mass spectrometer with 100,000 full width at half maximum (FWHM) resolving power (28) and equipped with an electrospray ionization (ESI) source (ThermoFinnigan, San Jose, CA) operating in the negative-ion mode. Ions were introduced into the ion source via direct infusion at a rate of 30 μl/min. The ESI source tuning parameters were as follows: sheath gas at 20 arbitrary units, spray voltage at 6 kV, capillary temperature at 275°C, capillary voltage at −37 V, and tube lens voltage at −230 V. Spectra (m/z 500 to 3,000) were acquired using a resolution of 30,000 and a maximum injection time of 500 ms. Spectra represent the averages of the results of 100 scans acquired using Xcalibur (version 2.0.7 SP1; ThermoFinnigan). The spectra were externally calibrated using the manufacturer's standard calibration mixture and further calibrated internally with the lock mass tool using endogenous lipids phosphatidyl inositol (m/z 851.565473) and methoxymycolic acid (m/z 1,252.286917); such calibration provides mass accuracy within 2 ppm as previously reported (28).
14C metabolic labeling and lipid analysis by TLC.
M. tuberculosis strains were grown to the late log phase. For 14C labeling, 5 μCi [14C]propionic acid was added directly to 10 ml culture at an OD600 of ∼1. After overnight incubation, cell pellets were extracted in hexane as described above. Extracts were dried and resuspended in a 1/10 or 1/20 dilution of the original extraction volume. Volumes representing equivalent numbers of cells were spotted on silica plates (high-performance thin-layer chromatography [HPTLC] Silica Gel 60; EMD Chemicals) and developed in 92:8 petroleum ether/acetone or 90:10 chloroform/methanol to resolve PAT or DAT, respectively, followed by phosphorimaging (21, 29, 30). For THL treatment experiments, THL dissolved in dimethyl sulfoxide (DMSO) was added to 10-ml cultures at 0, 10, 20, and 40 μg/ml for 6 h followed by the addition of 5 μCi [14C]propionic acid with further incubation, processing, and analysis as described above.
Subcellular localization and immunoblot analysis of Chp2.
M. smegmatis containing pRibo Chp2-3xFLAG (31) was cultured overnight. The cells were pelleted and resuspended at an OD600 of 0.3 in 250 ml Sauton's medium with or without 2 mM theophylline and grown to an OD600 of 1. The cells were harvested by centrifugation at 4,000 × g for 30 min. The supernatant was sterilized by passage through a 0.22-μm-pore-size filter, concentrated ∼100-fold by centrifugation to ∼2.5 ml through a 10-molecular-weight-cutoff (MWCO) membrane, and stored at −20°C as the culture filtrate (CF) fraction. The cell pellet was washed twice with 50 ml phosphate-buffered saline (PBS). The final pellet was weighed and resuspended in PBS–1× protease inhibitor (Roche) at 2 ml/g. The suspension was lysed by sonication for 2 min 10 s on/off at power level 1 to 2 with a Model 550 Sonic Dismembrator (Fisher Scientific). Cell debris was removed by centrifugation at 10,000 × g for 10 min. After a sample was reserved for immunoblot analysis, the cleared total lysate was subjected to ultracentrifugation at 100,000 × g for 1 h. The supernatant was collected as the cytosol-enriched (cyt) fraction and the pellet as the cell envelope-enriched (CE) fraction, which included the cytoplasmic membrane, peptidoglycan, and arabinogalactan layers and the outer membrane. The protein concentration was determined by the bicinchoninic acid (BCA) assay (Pierce). For the anti-MspA blot, samples were prepared as previously described. Briefly, samples were adjusted to 1 μg/μl in 20 μl of 0.6% octyl-thioglucoside–PBS, heated at 98°C for 30 min, and centrifuged at 10,000 × g for 5 min to remove protein precipitate. For reducing SDS-PAGE, samples were loaded at 10 μg/well (anti-FLAG), 20 μg/well (anti-KatG), or 5 μg/well (anti-MspA). After transfer and blocking, membranes were probed with anti-FLAG (clone M2; Sigma) (1:1,000); anti-KatG (clone IT-57 [CDA4]; NR-13793 from BEI Resources, NIAID, NIH) (1:500); or anti-MspA (gift from Michael Niederweis, University of Alabama at Birmingham) (1:666). Secondary goat anti-mouse IgG-IR800 or goat anti-rabbit IgG–IR700 antibodies (Li-COR Biosciences) were prepared according to the manufacturer's recommendation, and the signal was detected with an Odyssey scanner (Li-COR Biosciences).
Enzyme activity of Chp2-βGal and Chp2-AP fusions.
M. smegmatis strains containing pRibo Chp2-alkaline phosphatase (AP) or Chp2-β-galactosidase (βGal) fusion constructs were grown in 7H9 medium containing 0 or 2 mM theophylline. Levels of βGal and AP activity in lysates and whole cells, respectively, were determined using the substrates 2-nitrophenyl-β-d-galactopyranoside and 4-nitrophenyl-phosphate essentially as previously reported (32, 33). Activity [reaction mixture OD420/(culture OD600 × Vs × min), where min represents the reaction time and Vs represents the volume of the original culture used in the reaction] is reported in Miller units. Control vectors pMB111 and pMB124 encoding a secretion signal and secreted AP, respectively, were a kind gift of Miriam Braunstein (University of North Carolina at Chapel Hill). To detect βGal activity on chromogenic medium, cells were streaked on 7H9 agar containing 50 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 2 mM theophylline, 10% ADC supplement, 0.5% glycerol, and 0.05% Tween 80.
Expression and purification of His-tagged Chp2 catalytic domain in E. coli.
A truncated construct of Chp2 lacking the first 25 amino acids (Chp2-cat) was cloned by InFusion (Clontech) into ligation-independent cloning vector 2BT (gift of Scott Gardia, University of California [UC], Berkeley) to yield Chp2-cat with an N-terminally fused 6×His purification tag. Following a 4-hour induction with 0.2 mM isopropyl-β-d-thiogalactopyranoside at 37°C, cells were lysed in 20 mM Tris (pH 7.4)–200 mM NaCl–1 mM EDTA–1 mM dithiothreitol (DTT). The clarified crude lysate was loaded onto a 5 ml HisTrap FF (GE Biosciences) column and washed with 100 ml 50 mM Tris (pH 7.4)–1 mM DTT–10% glycerol (buffer A). Bound protein was eluted over 15 column volumes in a gradient of 0% to 50% 1 M imidazole in buffer A. Fractions containing Chp2-cat were pooled and dialyzed overnight at 4°C into buffer A. Dialyzed Chp2-cat was loaded onto a 5 ml HiTrap Q column (GE Biosciences), washed with 25 ml buffer A, and eluted in over 15 column volumes in a gradient of 0% to 100% 1 M NaCl–buffer A. The fractions containing purified Chp2-cat were concentrated and stored at −80°C. The S141A mutation was introduced by site-directed mutagenesis, and the resulting Chp2-cat(SA) protein was purified as described above.
Enzymatic reactions.
PapA3 was expressed and purified as previously described (16). For reconstitution of acyltrehalose synthesis, reaction mixtures contained 2 μM PapA3, 20 μM [1-14C]palmitoyl coenzyme A ([1-14C]palmitoyl-CoA), 1 mM trehalose, and 4 μM Chp2-cat or Chp2-cat(SA) in 25 μl reaction buffer (100 mM sodium phosphate [pH 7.2], 1 mM DTT, 10% glycerol). Reaction mixtures were incubated at room temperature for 12 to 16 h and then quenched with an equal volume of ethanol. Reactions were analyzed by TLC (65:35 chloroform:methanol) and visualized by phosphorimaging. For biphasic reactions, twice the reaction volume of hexanes was slowly added as a separate phase on top of the aqueous reaction mixture immediately after all components had been combined. Biphasic reactions were not quenched; instead, the two phases were mixed immediately prior to spotting onto the TLC plate.
Reactivity of Chp2-cat with a serine hydrolase-specific activity-based probe.
TMR-FP (tetramethylrhodamine linked to fluorophosphonate via a polyethylene glycol [PEG] linker; gift of Benjamin Cravatt, Scripps Institute) (34) was incubated at 2 μM for 1 h at room temperature with 2 μM Chp2-cat or Chp2-cat(SA) in 50 mM Tris (pH 7.4)–1 mM DTT–10% glycerol either with or without preheating at 98°C for 5 min. For inhibition experiments, THL was added to each sample at 20 μM and incubated for 30 min at room temperature prior to labeling with TMR-FP as described above. Samples were separated by SDS-PAGE and scanned for TMR fluorescence with a Typhoon FLA 7000 scanner (GE Healthcare).
RESULTS
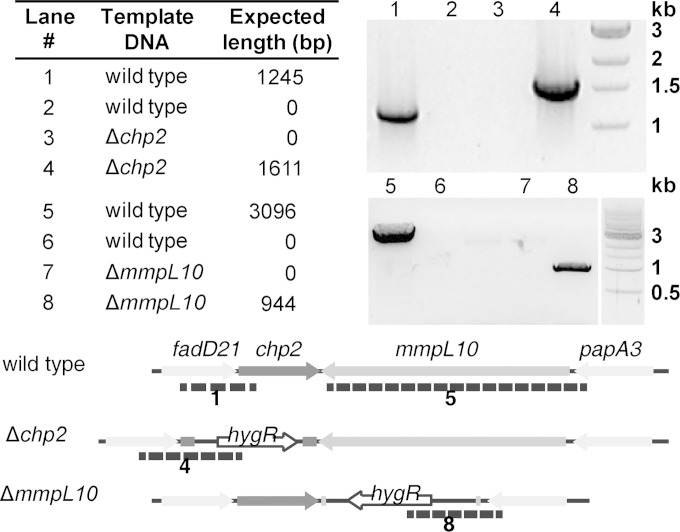
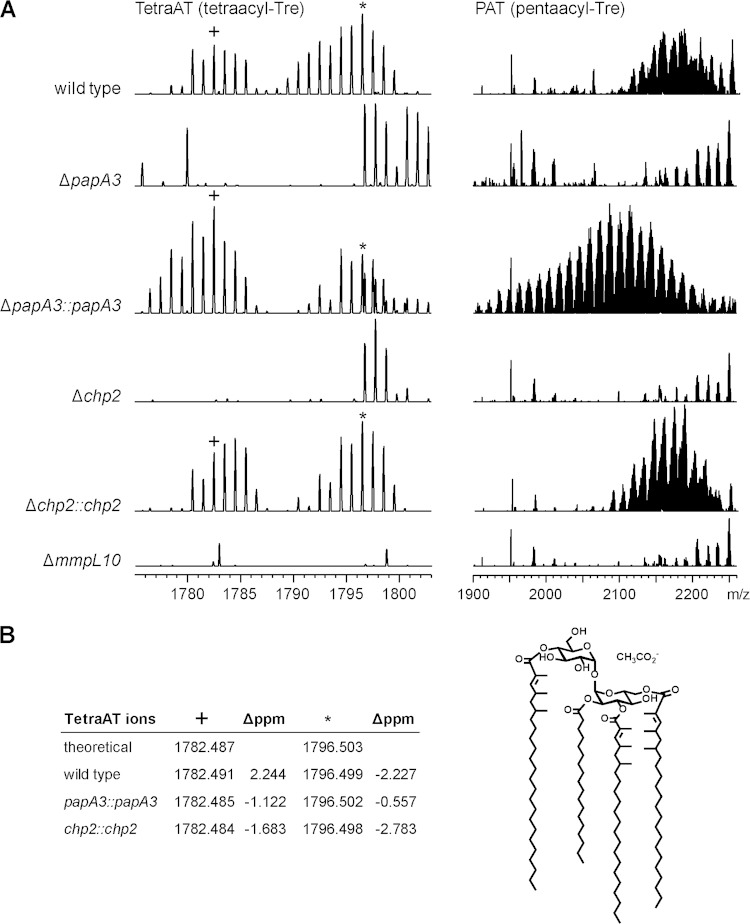
The chp2 gene in M. tuberculosis Erdman was replaced with a hygromycin resistance cassette to yield the knockout mutant Δchp2. Both this and the ΔmmpL10 strain were verified by PCR (Fig. 2). Extracts of surface lipids from the Δchp2 and ΔmmpL10 strains were analyzed by direct-infusion ESI-MS and compared with extracts from the wild type and a ΔpapA3 mutant strain (16). In the wild type, an envelope of ions was detected at m/z 1,900 to 2,200 that is characteristic of PAT (Fig. 3A) (16). Observed peaks were within 5 ppm of the predicted acetate pseudomolecular ion values (data not shown). In addition, a set of peaks centered at approximately m/z 1,790 was observed, which is consistent with tetraacyltrehaloses containing one fewer mycolipenic modification than PAT. Although multiple-stage mass spectrometry (MSn) analysis could not be completed on any of these ions due to low yield, the exact masses match the theoretical masses of predicted tetraacyltrehalose structures (Fig. 3B). To avoid confusion with the literature acronym TAT for triacyltrehalose, we refer to tetraacyltrehalose as TetraAT. The ions for PAT and TetraAT were not observed in the ΔpapA3, Δchp2, or ΔmmpL10 mutant strains.
FIG 2.
Confirmation of strains Δchp2 and ΔmmpL10 by PCR. For chp2, the 5′ primer annealed ∼900 bp upstream of chp2. For lanes 1 and 3, the 3′ primer annealed to a region of chp2 that is absent in the knockout strain. For mmpL10, the 5′ primer annealed ∼100 bp upstream of mmpL10. For lanes 5 and 7, the 3′ primer annealed to a region of mmpL10 that is absent in the knockout strain. For lanes 2, 4, 6, and 8, the 3′ primer annealed to the hygR marker that replaces chp2 and mmpL10 in the knockout strains. See Table S2 in the supplemental material for oligonucleotide sequences.
FIG 3.
Chp2 and MmpL10 are required for biosynthesis of PAT and the novel acyltrehalose TetraAT. M. tuberculosis wild-type, ΔpapA3, Δchp2, and ΔmmpL10 strains were analyzed by ESI-MS. (A) TetraAT and PAT were absent from all three mutants but were restored in the ΔpapA3 and Δchp2 complement strains. PAT appears as a characteristic envelope of peaks centered at approximately m/z 2,100 to 2,200. A representative segment of the TetraAT spectrum is shown in order to highlight two of the peaks used for assignment. (B) TetraAT was assigned by exact mass, and observed ions were within 3 ppm of the predicted m/z value. As an example, a possible structure for the m/z 1,782.487 [M+CH3CO2-] ion is shown.
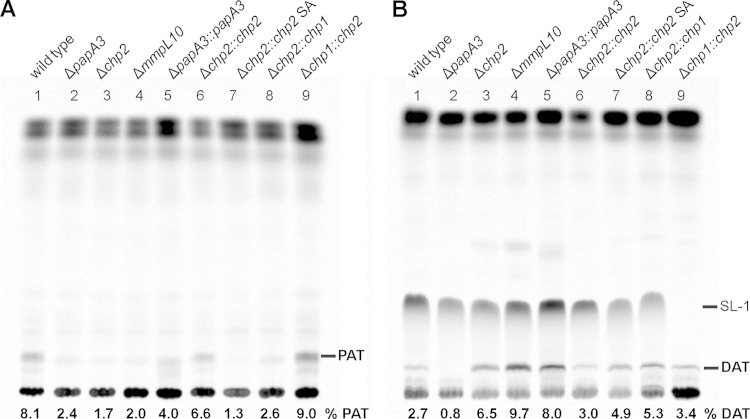
Complementary results were obtained by TLC and phosphorimage analysis of lipids from M. tuberculosis labeled metabolically with [14C]propionate, which is incorporated into methyl-branched lipids (Fig. 4). While the migration of TAT and TetraAT has not been assigned under the TLC conditions used, PAT and DAT were resolved. All the mutants lacked PAT, but while DAT was lost in the ΔpapA3 mutant, both strain Δchp2 and strain ΔmmpL10 still produced this biosynthetic intermediate. Ions corresponding to PAT are restored in the ΔpapA3::papA3 and Δchp2::chp2 complement strains. The PAT ion envelope is centered at a lower m/z in the ΔpapA3::papA3 complement than in either the wild-type strain or the Δchp2::chp2 strain and also migrates with a lower retention factor (Rf) on the corresponding TLC (Fig. 4). These data are indicative of PAT lipids that contain mycolipenic modifications with fewer methyl branch modifications on average. One explanation for this mass shift is that the mycolipenic acids have undergone fewer rounds of methyl branch modification by Pks3/4 before attachment to acyltrehaloses. The ΔpapA3 strain was complemented with a multicopy episomal plasmid expressing papA3 from the constitutive hsp60 GroEL promoter (16), in contrast to strain Δchp2, which was complemented by an integrating plasmid with a native promoter. The availability of lipid precursors is known to shift the mass of methylbranched lipids by altering the flux of substrates through the pathway. By similar reasoning, overexpression of PapA3 could lead to increased DAT levels that increase flux through subsequent steps, such that mycolipenic acids are released from Pks3/4 at a higher rate and therefore undergo fewer cycles of methyl branch modification. Finally, chp1 and chp2 did not cross-complement the respective Δchp2 and Δchp1 knockout strains in merodiploid strains containing a second copy of the corresponding gene. Multiple attempts and strategies designed to complement the ΔmmpL10 mutant, including the use of different promoters or chp2 coexpression, were not successful (data not shown).
FIG 4.
Chp2 and MmpL10 function downstream of DAT, and Chp2 is specific to the PAT biosynthesis. M. tuberculosis wild-type, ΔpapA3, Δchp2, and ΔmmpL10 strains were analyzed by metabolic labeling with [14C]propionate followed by TLC analysis of lipid extracts. In addition to loss of PAT in all mutants (lanes 2 to 4) (A), DAT was absent from the ΔpapA3 mutant (lane 2) (B). PAT and DAT were restored upon complementation (lanes 5 and 6), except in the Δchp2::chp2 SA mutant (lane 7). The chp1 and chp2 homologues did not cross-complement the Δchp2 and Δchp1 mutants (lanes 8 and 9). The mobile phase was 92:8 petroleum ether/acetone (A) and 90:10 chloroform/methanol (B) (21, 29, 30). The proportion of PAT or DAT is reported as the percent total integrated intensity in each lane.
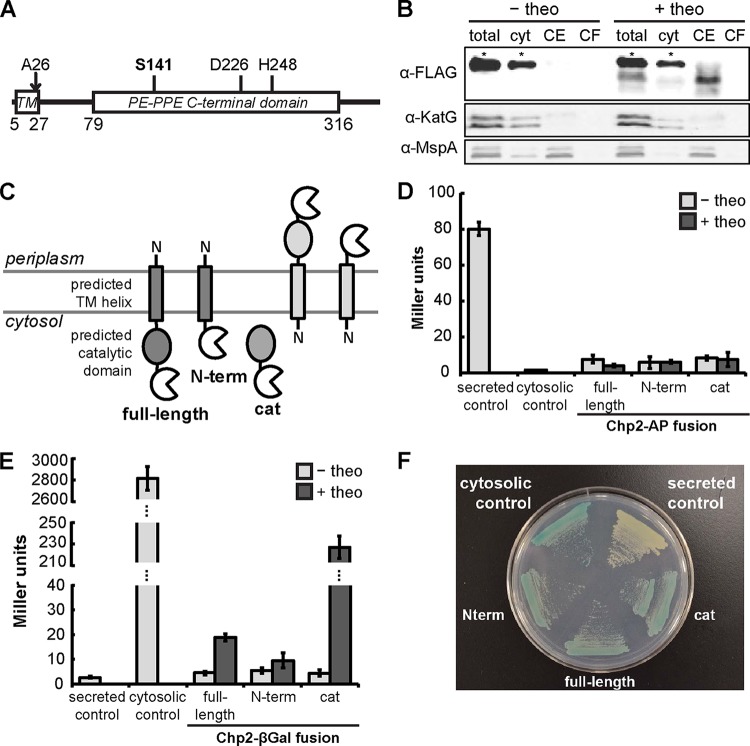
Bioinformatic analysis of the Chp2 amino acid sequence revealed several predicted features: (i) a signal sequence (aa 1 to 26); (ii) a transmembrane helix (aa 5 to 27); and (iii) a conserved PE-PPE C-terminal domain (aa 79 to 316) that is a predicted serine hydrolase (Fig. 5A). In addition, a structural homology model based on the Penicillium purpurogenum acetylxylan esterase structure (PDB identifier [ID] 1G66; 14% sequence identity with Chp2; 95% Phyre2-estimated precision with E value of 0.0093 [24]) identified the putative catalytic triad (S141-D226-H248; catalytic residue underlined), similar to a previously reported prediction based on the structure of a M. smegmatis lipase (35). To determine its subcellular location, Chp2 was expressed in M. smegmatis as a theophylline-inducible full-length C-terminal fusion to a 3× FLAG epitope tag. When cells were grown in theophylline-supplemented medium, FLAG-tagged Chp2 was enriched in the cell envelope fraction compared to the cytosolic and culture filtrate fractions (Fig. 5B). To determine whether the C-terminal catalytic domain is located in the cytosol or the periplasm, Chp2 was also expressed in M. smegmatis as a theophylline-inducible C-terminal fusion to either alkaline phosphatase (AP) or β-galactosidase (βGal), as has been performed previously to determine mycobacterial protein topology (Fig. 5C) (36–38). No AP activity was detected when Chp2 was expressed as the full-length protein, the N terminus (N-term), or the catalytic domain (cat) (Fig. 5D). On the other hand, βGal activity was confirmed in both cell lysates and whole cells (Fig. 5E and F). These data suggest that Chp2 has its C terminus located in the cytoplasm and is associated with the cytoplasmic membrane and possibly via an N-terminal transmembrane helix, similarly to Chp1 (18).
FIG 5.
Chp2 localizes to the cell envelope with the catalytic domain in the cytosol. (A) Primary structure of Chp2 with the predicted positions of the transmembrane helix (TM), signal peptide cleavage site (A27), conserved domain, and catalytic residues. (B) Immunoblot of subcellular fractions of M. smegmatis expressing theophylline (theo)-inducible Chp2-3xFLAG showed Chp2 enrichment in the cell envelope (CE) fraction. KatG and MspA are markers for the cytosol (cyt) and outer membrane, respectively. Each asterisk indicates a nonspecific α-FLAG-reactive band. (C) AP or βGal was expressed in M. smegmatis as a theophylline-inducible fusion(s) to the C terminus of full-length Chp2, the N-terminal domain, or the catalytic domain. (D to F) Enzymatic activity was not detected for the AP fusions (D) but was observed for the βGal fusions (E), which were further confirmed by growth on chromogenic medium containing X-Gal (F). Strains grown without theophylline or expressing AP or βGal with or without the Ag85B secretion signal served as negative and positive controls (48). Turnover of colorimetric substrates is expressed in Miller units.
To explore the proposed enzymatic activity of Chp2, the conserved C-terminal PE-PPE domain (aa 26 to 359) was expressed and purified as an N-terminally 6×His-tagged construct (Chp2-cat). Diverse strategies were pursued in an effort to reconstitute and detect trehalose glycolipid acyltransferase activity for Chp2-cat. The reaction of PapA3, trehalose, and palmitoyl-CoA yielded the expected mixture of trehalose monopalmitate and trehalose dipalmitate as analyzed by TLC and confirmed by MS (data not shown). Since DAT is the predicted substrate for Chp2, we hypothesized that Chp2 would modify the DAT analogue, trehalose dipalmitate, generated in situ by PapA3. Addition of Chp2-cat to the PapA3 reaction mixture did not, however, yield any Chp2 activity-dependent changes, as detected by either by TLC or MS analysis (data not shown). To control for possible inhibition of Chp2 by excess trehalose or palmitoyl-CoA from the PapA3 reaction, trehalose monopalmitate and dipalmitate were isolated by preparative TLC and incubated with Chp2-cat, but again, no additional products were observed. We further reasoned that the predicted products of Chp2 may aggregate or be inaccessible for further reaction in aqueous reaction buffer, since these lipids are neutral and highly hydrophobic. Although mixed organic:aqueous conditions have proven useful for promoting enzymatic wax ester synthesis (M. H. Touchette and J. C. Seeliger, unpublished results), applying a biphasic system to all of the reaction conditions described above did not yield Chp2-dependent products.
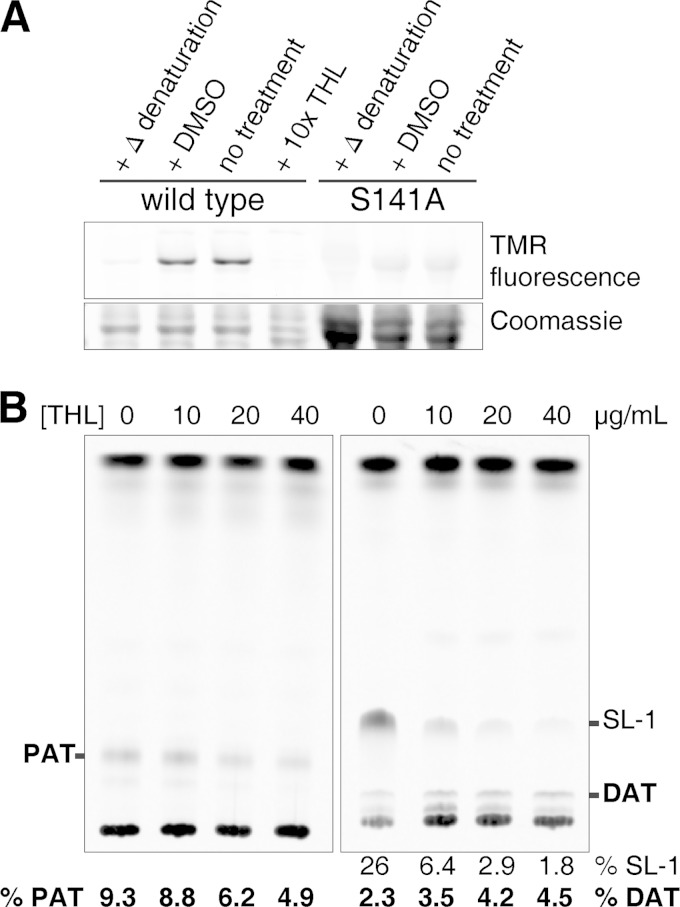
Chp2 enzymatic activity was instead confirmed using the activity-based probe tetramethylrhodamine-fluorophosphonate (TMR-FP), a fluorescent labeling reagent that specifically modifies the active-site residue of serine hydrolases (Fig. 6A) (34). No labeling was observed following heat denaturation or upon mutation of the Ser141 putative catalytic residue to Ala (Chp2-SA). These data are consistent with the inability of a chp2 allele containing this mutation (chp2 SA) to restore PAT biosynthesis in the M. tuberculosis Δchp2 knockout strain, as determined by analysis of 14C-labeled lipid extracts (Fig. 4). In addition, TMR-FP labeling of purified Chp2 catalytic domain was inhibited by preincubation with the lipophilic lipase inhibitor tetrahydrolipistatin (THL). Also, treatment of a wild-type M. tuberculosis mid-log-phase culture with THL prior to metabolic incorporation of [14C]propionate revealed a moderate dose-dependent inhibition of PAT biosynthesis and concomitant accumulation of DAT (Fig. 6B). In further attempts to reconstitute acyltransferase activity, additional constructs of Chp2 (aa 45 to 359, aa 52 to 359, and aa 79 to 359) were expressed as fusions to an N-terminal 6×His-maltose binding protein linked by a tobacco etch virus (TEV) protease site. In all cases, both the fusion and purified cleaved protein were active by TMR-FP labeling, but none yielded products in biochemical reactions with PapA3 (data not shown). Thus, the Chp2 constructs expressed in this study all reacted with the promiscuous label TMR-FP, which has been shown to modify a broad range of serine hydrolases (34), but apparently lacked the ability to turn over trehalose dipalmitate.
FIG 6.
Chp2 is a serine hydrolase-type enzyme that is inhibited by THL. (A) Labeling of purified Chp2-cat domain by TMR-FP was detected by fluorescence and inhibited by heat denaturation, treatment with THL, or mutation of the catalytic residue (S141A). (B) Lipid extracts from M. tuberculosis treated with different concentrations of THL for 6 h followed by [14C]propionate labeling revealed the dose-dependent, but incomplete, inhibition of PAT and accumulation of DAT (mobile phases were as described in the Fig. 4 legend). SL-1 inhibition by THL has been previously reported (18). The proportion of PAT or DAT is reported as percent total integrated intensity in each lane (SL-1 results are shown for comparison).
DISCUSSION
In this study, we confirmed that Chp2 has a C-terminal serine hydrolase domain that is inhibited by THL. Within the PAT biosynthesis pathway, THL inhibits Chp2 but not PapA3, leading to decreased levels of PAT and the accumulation of the DAT precursor in M. tuberculosis. Although we were unable to detect Chp2 activity on an acyltrehalose analogue, the biochemical data, along with the lipid profiling of the Δchp2 mutant, place Chp2 downstream of PapA3 as likely the sole acyltransferase that catalyzes the three esterifications necessary to convert DAT to PAT. We further identified a previously uncharacterized acyltrehalose, TetraAT, as a PAT precursor that is absent in the ΔpapA3, Δchp2, and ΔmmpL10 mutants.
The function of Chp2 is therefore directly analogous to the role of Chp1 in SL-1 sulfoglycolipid biosynthesis. Although Chp1 activity has been successfully reconstituted in vitro, Chp2 is not active under similar conditions, perhaps because it detects methyl branching and/or the α,ß-unsaturation on the mycolipenic chain and requires these features for activity (Fig. 1). The recognition of the conjugated planar geometry of the α,β-unsaturated ketone by Chp2 could contribute to its specificity for acyltrehaloses over sulfoglycolipids, which contain phthioceranoic chains that are also methyl branched but lack unsaturation and so have tetrahedral geometry at the α-carbon. Chp1 is more susceptible to THL inhibition than Chp2 upon treatment of M. tuberculosis (Fig. 4B) (18), but in the absence of structural data on these two enzymes, the origins of this specificity are not obvious.
In the SL-1 pathway, the membrane protein MmpL8 is a sulfoglycolipid transporter but also promotes Chp1 activity (39, 40). MmpL10 appears to act similarly within the PAT pathway, as the ΔmmpL10 mutant is defective in PAT biosynthesis but still produces DAT. We would further predict that DAT would be absent in the outer membrane of strain ΔmmpL10, but this could not be confirmed. While DAT was detectable by 14C radioisotope labeling, DAT ions were not observed by mass spectrometry and therefore could not be compared between the surface and cell pellet lipid fractions as done previously (18). Since MS analysis was performed without chromatographic separation, DAT was likely suppressed in our spectra by the presence of abundant ions in the same m/z range (e.g., theoretical m/z 1,057.777 for a DAT-acetate adduct with palmitate and C24 mycolipenic modifications). Since mmpL10 is the last gene in the predicted pks3/4-papA3-mmpL10 operon, its deletion is not expected to affect other loci, but it is nonetheless possible that mmpL10 disruption has a polar effect on the chp2 locus, which has opposite orientation but is separated from mmpL10 by only three nucleotides. However, complementation with both chp2 and mmpL10 or with mmpL10 under the control of promoters of various strengths did not restore PAT biosynthesis, and so the role of mmpL10 still requires additional confirmation.
Chp2 has a high-probability signal sequence (based on empirical algorithms for Gram-negative bacteria [23]) that indicates extracytoplasmic export, although this sequence also has the properties of a transmembrane helix, as is typical for signal peptides. Chp2 was found in the cell envelope fraction, consistent with export into the cell wall or membrane localization. The results of the complementary βGal/AP-fusion activity assay indicate that Chp2 is oriented with its C-terminal catalytic domain in the cytosol and therefore is most likely anchored in the cytoplasmic membrane (Fig. 5C).
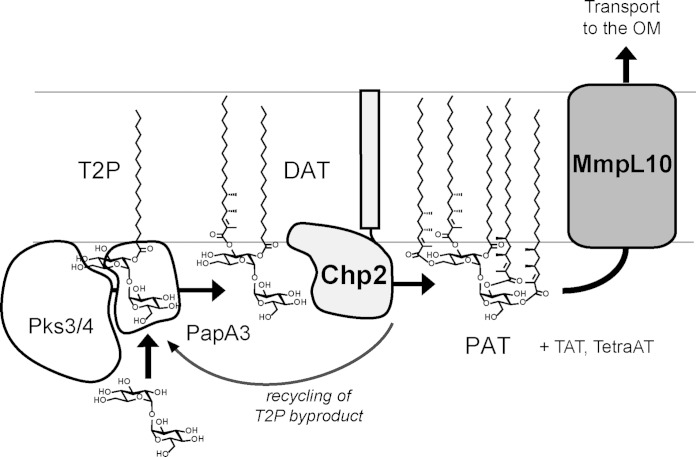
Based on these data, we conclude that PAT biosynthesis in M. tuberculosis proceeds similarly to sulfoglycolipid biosynthesis, in line with the organizational similarity of the two biosynthetic gene clusters. PapA3 modifies trehalose to yield DAT, which is then further esterified by Chp2 to form TAT, TetraAT, and PAT (Fig. 1). Given their sequence and predicted structural similarity, Chp2 likely functions by a mechanism similar to that of Chp1 and Ag85; that is, Chp2 catalyzes transesterification using DAT as the acyl donor and generating T2P as a side product. We therefore propose a model in which Chp2 at the cytoplasmic leaflet synthesizes PAT and in the process produces T2P that can immediately reenter PAT biosynthesis via modification by PapA3 (Fig. 7).
FIG 7.
Proposed model for acyltrehalose biosynthesis and transmembrane transport. The data are most consistent with a model in which Chp2 completes PAT biosynthesis in the cytosolic leaflet and MmpL10 transports acyltrehaloses across the membrane. The AcTre side products generated by Chp2 can be recycled by PapA3. (Note that the mycolipenic groups are truncated in this figure for simplicity.) OM, outer membrane.
As with Chp1 and MmpL8, the activity of Chp2 may be facilitated by the MmpL10 transporter. Because DAT, TAT, TetraAT, and PAT are all found at the cell surface, this model implies that MmpL10 transports all of these lipids across the cytoplasmic membrane. This substrate promiscuity would distinguish MmpL10 from previously characterized MmpLs such as MmpL7 and MmpL3, which exclusively transport phthiocerol mycocerosate (PDIM) and trehalose monomycolate (TMM), respectively (41–43). On the other hand, in sulfolipid 1 biosynthesis, when Chp1 is absent, the diacylated sulfotrehalose intermediate SL1278 (also known as Ac2SGL) is detected at the cell surface, suggesting that MmpL8 is capable of transporting this intermediate in addition to SL-1 and shares the ability to transport multiple substrates with MmpL10, which is a closely related homologue (58% sequence identity). Interestingly, both mmpL10 and chp2 have been characterized as essential M. tuberculosis genes in vivo (44), but an M. tuberculosis strain disrupted in pks3/4 (and therefore lacking all PAT-related acyltrehaloses) was not attenuated for growth in a mouse infection (45). These data suggest that the accumulation of DAT compromises M. tuberculosis survival in vivo.
In summary, we have assigned Chp2 as a DAT acyltransferase and thereby completed the biosynthetic pathway for PAT. While the overall synthetic scheme is now defined for PAT, as well as for other virulence-associated outer membrane lipids such as SL-1 and PDIM (46), many questions still remain regarding how MmpLs influence the activity of biosynthetic enzymes and how lipids are transported and assembled in the outer membrane. Elucidation of these mechanisms will have implications for our understanding of how the structure and composition of the outer membrane are regulated, what structural and functional roles these lipids play in M. tuberculosis survival and pathogenesis, and the consequences of targeting cell wall-associated processes for tuberculosis drug therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI051622 (to C.R.B.) and 1S10RR023558-01 (to J.A.L.) and a Stony Wold-Herbert Foundation grant-in-aid (to J.C.S.).
We thank Teresa Garrett, Sahar Bilal, Miriam Braunstein, and Bela Ruzsicska for technical assistance, reagents, and helpful discussions.
ADDENDUM
While this work was under review, Belardinelli et al. published their characterization of Chp2 and MmpL10, which agrees with our conclusions that both are involved in PAT biosynthesis downstream of PapA3 (47). They further confirm that, consistent with our model, acyltrehaloses were not transported to the outer membrane in a ΔmmpL10 strain and that this phenotype, as well as PAT biosynthesis, could be complemented. Chp2 was also shown to modify purified DAT. This supports the idea that Chp2 activity requires native (or at least native-like) substrates. Belardinelli et al. also conclude that Chp2 has the opposite topology from that proposed here. While their results are consistent with the detection of DAT, TAT, and PAT in the outer membrane, the resulting model of PAT biosynthesis requires the monoacyl byproduct T2P to undergo retrograde transport to reenter the biosynthetic pathway.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02015-14.
REFERENCES
- 1.Khan AA, Stocker BL, Timmer MS. 2012. Trehalose glycolipids–synthesis and biological activities. Carbohydr Res 356:25–36. doi: 10.1016/j.carres.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Daffé M, Lacave C, Lanéelle MA, Gillois M, Lanéelle G. 1988. Polyphthienoyl trehalose, glycolipids specific for virulent strains of the tubercle bacillus. Eur J Biochem 172:579–584. doi: 10.1111/j.1432-1033.1988.tb13928.x. [DOI] [PubMed] [Google Scholar]
- 3.Ortalo-Magné A, Lemassu A, Lanéelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol 178:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubey VS, Sirakova TD, Kolattukudy PE. 2002. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol 45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- 5.Rousseau C, Neyrolles O, Bordat Y, Giroux S, Sirakova TD, Prevost M-C, Kolattukudy PE, Gicquel B, Jackson M. 2003. Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell Microbiol 5:405–415. doi: 10.1046/j.1462-5822.2003.00289.x. [DOI] [PubMed] [Google Scholar]
- 6.Tórtola MT, Lanéelle MA, Martín-Casabona N. 1996. Comparison of two 2,3-diacyl trehalose antigens from Mycobacterium tuberculosis and mycobacterium fortuitum for serology in tuberculosis patients. Clin Diagn Lab Immunol 3:563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muñoz M, Lanéelle M-A, Luquin M, Torrelles J, Julián E, Ausina V, Daffé M. 1997. Occurrence of an antigenic triacyl trehalose in clinical isolates and reference strains of Mycobacterium tuberculosis. FEMS Microbiol Lett 157:251–259. doi: 10.1016/S0378-1097(97)00483-7. [DOI] [PubMed] [Google Scholar]
- 8.Julián E, Matas L, Alcaide J, Luquin M. 2004. Comparison of antibody responses to a potential combination of specific glycolipids and proteins for test sensitivity improvement in tuberculosis serodiagnosis. Clin Diagn Lab Immunol 11:70–76. doi: 10.1128/CDLI.11.1.70-76.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saavedra R, Segura E, Leyva R, Esparza LA, López-Marín LM. 2001. Mycobacterial di-o-acyl-trehalose inhibits mitogen- and antigen-induced proliferation of murine T cells in vitro. Clin Diagn Lab Immunol 8:1081–1088. doi: 10.1128/CDLI.8.6.1-91-1088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saavedra R, Segura E, Tenorio EP, Lopez-Marin LM. 2006. Mycobacterial trehalose-containing glycolipid with immunomodulatory activity on human CD4+ and CD8+ T-cells. Microbes Infect 8:533–540. doi: 10.1016/j.micinf.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Lee K-S, Dubey VS, Kolattukudy PE, Song C-H, Shin AR, Jung S-B, Yang C-S, Kim S-Y, Jo E-K, Park J-K, Kim H-J. 2007. Diacyltrehalose of Mycobacterium tuberculosis inhibits lipopolysaccharide- and mycobacteria-induced proinflammatory cytokine production in human monocytic cells. FEMS Microbiol Lett 267:121–128. doi: 10.1111/j.1574-6968.2006.00553.x. [DOI] [PubMed] [Google Scholar]
- 12.Neyrolles O, Guilhot C. 2011. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91:187–195. doi: 10.1016/j.tube.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Minnikin DE, Dobson G, Sesardic D, Ridell M. 1985. Mycolipenates and mycolipanolates of trehalose from Mycobacterium tuberculosis. J Gen Microbiol 131:1369–1374. [DOI] [PubMed] [Google Scholar]
- 15.Besra GS, Bolton RC, McNeil MR, Ridell M, Simpson KE, Glushka J, van Halbeek H, Brennan PJ, Minnikin DE. 1992. Structural elucidation of a novel family of acyltrehaloses from Mycobacterium tuberculosis. Biochemistry 31:9832–9837. doi: 10.1021/bi00155a040. [DOI] [PubMed] [Google Scholar]
- 16.Hatzios SK, Schelle MW, Holsclaw CM, Behrens CR, Botyanszki Z, Lin FL, Carlson BL, Kumar P, Leary JA, Bertozzi CR. 2009. PapA3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. J Biol Chem 284:12745–12751. doi: 10.1074/jbc.M809088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P, Schelle MW, Jain M, Lin FL, Petzold CJ, Leavell MD, Leary JA, Cox JS, Bertozzi CR. 2007. PapA1 and PapA2 are acyltransferases essential for the biosynthesis of Mycobacterium tuberculosis virulence factor sulfolipid-1. Proc Natl Acad Sci U S A 104:11221–11226. doi: 10.1073/pnas.0611649104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeliger JC, Holsclaw CM, Schelle MW, Botyanszki Z, Gilmore SA, Tully SE, Niederweis M, Cravatt BF, Leary JA, Bertozzi CR. 2012. Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. J Biol Chem 287:7990–8000. doi: 10.1074/jbc.M111.315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 20.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJC. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 22.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 25.Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. [DOI] [PubMed] [Google Scholar]
- 26.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 27.Harth G, Horwitz MA. 1997. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J Biol Chem 272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 28.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. 2006. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffé M, Gicquel B, Martín C, Jackson M. 2006. The virulence-associated two-component phop-phor system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem 281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 30.Slayden RA, Barry CE III. 2001. Analysis of the lipids of Mycobacterium tuberculosis. Methods Mol Med 54:229–245. doi: 10.1385/1-59259-147-7:229. [DOI] [PubMed] [Google Scholar]
- 31.Seeliger JC, Topp S, Sogi KM, Previti ML, Gallivan JP, Bertozzi CR. 2012. A riboswitch-based inducible gene expression system for mycobacteria. PLoS One 7:e29266. doi: 10.1371/journal.pone.0029266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillmann D, Eschenbacher I, Thiel A, Niederweis M. 2007. Expression of the major porin gene mspA is regulated in Mycobacterium smegmatis. J Bacteriol 189:958–967. doi: 10.1128/JB.01474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin TJ IV, Parsons L, Leschziner AE, DeVost J, Derbyshire KM, Grindley ND. 1999. In vitro transposition of tn552: a tool for DNA sequencing and mutagenesis. Nucleic Acids Res 27:3859–3865. doi: 10.1093/nar/27.19.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. 2001. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1:1067–1071. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Sultana R, Tanneeru K, Guruprasad L. 2011. The PE-PPE domain in mycobacterium reveals a serine α/β hydrolase fold and function: an in silico analysis. PLoS One 6:e16745. doi: 10.1371/journal.pone.0016745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn MY, Raman S, Anaya M, Husson RN. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigI regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol 187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryndak MB, Wang S, Smith I, Rodriguez GM. 2010. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J Bacteriol 192:861–869. doi: 10.1128/JB.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider JS, Sklar JG, Glickman MS. 2014. The Rip1 protease of Mycobacterium tuberculosis controls the SigD regulon. J Bacteriol 196:2638–2645. doi: 10.1128/JB.01537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE III. 2004. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem 279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 40.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. 2003. Mmpl8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci U S A 100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox JS, Chen B, McNeil M, Jacobs WR. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 42.Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE 3rd, Boshoff HI. 2012. SA109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passemar C, Arbues A, Malaga W, Mercier I, Moreau F, Lepourry L, Neyrolles O, Guilhot C, Astarie-Dequeker C. 2014. Multiple deletions in the polyketide synthase gene repertoire of Mycobacterium tuberculosis reveal functional overlap of cell envelope lipids in host-pathogen interactions. Cell Microbiol 16:195–213. doi: 10.1111/cmi.12214. [DOI] [PubMed] [Google Scholar]
- 46.Quadri LE. 2014. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit Rev Biochem Mol Biol 49:179–211. doi: 10.3109/10409238.2014.896859. [DOI] [PubMed] [Google Scholar]
- 47.Belardinelli JM, Larrouy-Maumus G, Jones V, Sorio de Carvalho LP, McNeil MR, Jackson M. 2014. Biosynthesis and translocation of unsulfated acyltrehaloses in Mycobacterium tuberculosis. J Biol Chem 289:27952–27965. doi: 10.1074/jbc.M114.581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonough JA, Hacker KE, Flores AR, Pavelka MS Jr, Braunstein M. 2005. The twin-arginine translocation pathway of mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J Bacteriol 187:7667–7679. doi: 10.1128/JB.187.22.7667-7679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.