Abstract
Bacteria use a chemical communication process called quorum sensing to monitor cell density and to alter behavior in response to fluctuations in population numbers. Previous studies with Vibrio harveyi have shown that LuxR, the master quorum-sensing regulator, activates and represses >600 genes. These include six genes that encode homologs of the Escherichia coli Bet and ProU systems for synthesis and transport, respectively, of glycine betaine, an osmoprotectant used during osmotic stress. Here we show that LuxR activates expression of the glycine betaine operon betIBA-proXWV, which enhances growth recovery under osmotic stress conditions. BetI, an autorepressor of the V. harveyi betIBA-proXWV operon, activates the expression of genes encoding regulatory small RNAs that control quorum-sensing transitions. Connecting quorum-sensing and glycine betaine pathways presumably enables V. harveyi to tune its execution of collective behaviors to its tolerance to stress.
INTRODUCTION
The cell-cell communication process called quorum sensing provides bacteria a mechanism to monitor the density of the community. Quorum sensing involves the production, detection, and response to signal molecules called autoinducers (reviewed in reference 1). At low cell density (LCD) and, thus, at low autoinducer concentrations, bacteria express genes underpinning individual behaviors. As bacteria grow and divide to reach high cell density (HCD), autoinducers accumulate. In response to autoinducer buildup, bacteria synchronously transition into programs of gene expression driving group behaviors. In the marine bacterium Vibrio harveyi, three autoinducers are produced and detected by three cognate two-component membrane-bound receptors (1). At LCD, the receptors act as kinases, transferring phosphate to the response regulator LuxO (2, 3). Phosphorylated LuxO (LuxO∼P) activates transcription of genes encoding five homologous regulatory small RNAs (sRNAs) called the quorum regulatory RNAs (Qrr sRNAs) (Fig. 1) (2, 4, 5). The Qrr sRNAs repress translation of the master quorum-sensing transcription factor LuxR, and they activate translation of the master LCD quorum-sensing transcription factor AphA (5, 6). Thus, at LCD, high AphA and low LuxR levels produce a gene expression pattern that yields individual behaviors (7). At HCD, the quorum-sensing receptors bind accumulated autoinducers, and in the bound state, the receptors act as phosphatases. Phosphate is drained from LuxO (8, 9). Dephosphorylated LuxO cannot activate transcription of the qrr genes (5). Thus, at HCD, AphA is not made, while LuxR is maximally produced. LuxR, in turn, controls the gene expression pattern underpinning group behaviors (7). Several regulatory feedback loops exist to properly maintain the levels of the Qrr sRNAs and to facilitate transitions between LCD and HCD: the Qrr sRNAs repress translation of LuxO (10, 11), AphA represses luxR and represses qrr expression, and LuxR represses aphA and activates qrr expression (Fig. 1) (6, 7, 11, 12).
FIG 1.

Model for quorum-sensing regulation of the betIBA-proXWV operon in V. harveyi. Phosphorylated LuxO (LuxO∼P) activates transcription of the genes encoding the Qrr sRNAs at LCD. The Qrr sRNAs repress the translation of AphA and activate the translation of LuxR. LuxR activates the transcription of the betIBA-proXWV operon. BetI activates expression of the genes encoding the Qrr sRNAs.
Bacterial osmoregulation systems enable adaptation to extracellular osmolarity changes (reviewed in references 13 and 14). Under conditions of osmotic stress, for example, Escherichia coli accumulates osmoprotectant solutes that maintain cytoplasmic osmolarity homeostasis. The osmoprotectant glycine betaine is a preferred solute. In E. coli, glycine betaine is transported into the cell by two mechanisms: via the ProP transporter and via the ProU transport system. The latter is encoded by proV (ATP-binding subunit), proW (integral membrane protein), and proX (periplasmic glycine betaine binding protein) (15), and expression of these genes is regulated by osmotic stresses, such as increased salt (15, 16). If extracellular glycine betaine is unavailable, it can be synthesized from the precursor choline. In the choline-glycine betaine synthesis pathway, betIBA and betT are divergently transcribed from partially overlapping promoters (17). BetI is a TetR-type transcription factor that represses the bet genes (17–19) by binding to a 21-bp site spanning the −35 regions of the divergently oriented betT and betIBA promoters. BetI DNA binding affinity increases in the presence of choline (19). The betT gene encodes the choline transporter, betA encodes the choline dehydrogenase, and betB encodes the betaine aldehyde dehydrogenase (20). Increases in extracellular osmolarity cause induction of bet gene expression (17). Thus, both osmotic stress and the presence of choline contribute to regulation of the bet genes in E. coli.
Often, signaling pathways responsible for detection of environmental stimuli are linked to bacterial quorum-sensing systems. For example, several bacterial species respond to environmental stresses, such as changes in temperature, pH, and nutrient availability, through modulation of quorum-sensing components (21–23). Alterations in extracellular osmolarity could also be connected to bacterial quorum-sensing status. Vibrios experience fluctuations in salinity in the marine environment and also in eukaryotic hosts (24). In several Vibrio species, total bacterial abundance often correlates with salinity (24, 25). To adapt to changes in salinity, the genomes of vibrios encode osmoregulation systems, including homologs of the E. coli Bet and ProU systems (26).
Here, we show that quorum sensing and osmotic stress regulation are linked in V. harveyi. In V. harveyi, the betIBA-proXWV operon encodes homologs of the E. coli Bet and ProU systems for the synthesis and transport of glycine betaine, respectively. LuxR activates expression of betIBA-proXWV at HCD by binding the betI promoter at two binding sites. In the absence of LuxR, V. harveyi grows poorly under low- and high-osmolarity conditions. Under conditions when the betIBA-proXWV operon is expressed, BetI influences quorum sensing by activating expression of the Qrr sRNA genes, which in turn repress luxR expression. Thus, quorum sensing activates osmoregulatory genes at HCD, and osmotic stress cues repress quorum sensing.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli strains S17-1λpir, BL21(DE3) (Life Technologies), DH10B (Life Technologies), and derivatives (see Table S1 in the supplemental material) were grown with aeration at 37°C in Luria-Bertani (LB) medium with 40 μg ml−1 kanamycin, 10 μg ml−1 tetracycline, and 10 μg ml−1 chloramphenicol, when appropriate. V. harveyi strain BB120 (BAA-1116) and derivatives (see Table S1 in the supplemental material) were grown with aeration at 30°C in Luria-marine (LM) medium with 100 μg ml−1 kanamycin, 5 μg ml−1 tetracycline (in liquid), 10 μg ml−1 tetracycline (on plates), and 10 μg ml−1 chloramphenicol, when required. Plasmids (see Table S2 in the supplemental material) were transferred from E. coli to V. harveyi by conjugation (6). Gene expression from plasmids containing the Ptac promoter was induced with isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) at the concentrations indicated below.
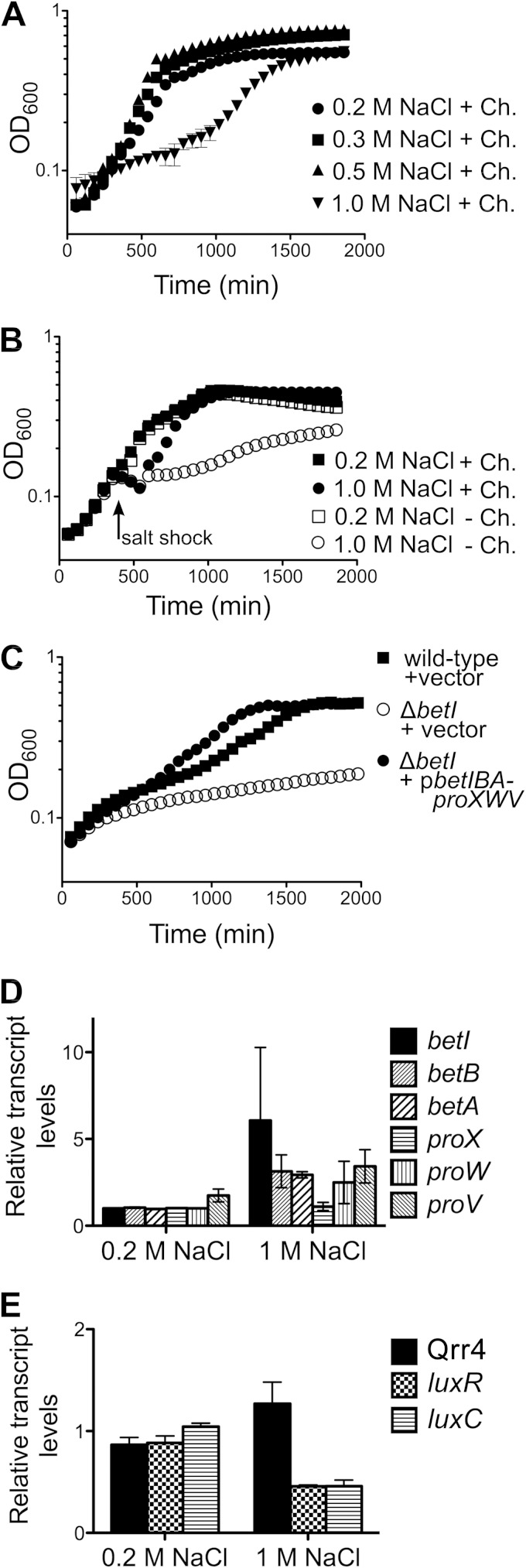
In experiments to study osmotic stress, V. harveyi strains were grown overnight in AB medium (27), and cells were subsequently diluted into low-osmolarity medium (LOM) (28) at pH 7 with 200 mM NaCl and 1 mM choline chloride, unless otherwise indicated. For growth analyses and salt shock experiments, overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 into 150 μl fresh LOM containing various concentrations of NaCl with 100 μl mineral oil overlay in 96-well plates. Cells were grown with shaking at 30°C in a BioTek Synergy H1 plate reader. At ∼6.5 h, NaCl was added at a final concentration of 1 M (30 μl of 5 M NaCl) or 0.2 M (in control cultures; 30 μl of 0.2 M NaCl), and cell growth (OD600) was monitored every 30 min (Fig. 5 and 6 show the results for the 60-min time points to enable discernment of symbols).
FIG 5.
Osmotic stress response in V. harveyi. (A) Wild-type V. harveyi (BB120) cultures were grown in LOM with 1 mM choline (Ch.) containing 0.2 M, 0.3 M, 0.5 M, or 1 M NaCl. (B) Wild-type V. harveyi (BB120) cultures were grown in LOM containing 0.2 M NaCl in the presence or absence of 1 mM choline. At 6.5 h, either 1 M NaCl or 0.2 M NaCl (final concentrations) was added to the cultures (designated with the arrow specifying salt shock). In panels A and B, data show the mean and standard error from three biological replicates and represent those from three independent experiments. (C) V. harveyi strains were grown in LOM with 1 mM choline and 1 M NaCl. The strains tested were wild-type V. harveyi carrying a control plasmid (BB120::pLAFR2), V. harveyi ΔbetI carrying a control vector (JC2212::pLAFR2), and V. harveyi ΔbetI carrying a plasmid encoding betIBA-proXWV (JC2212::pJV302). The data shown are the means and standard errors from three biological replicates and represent those from three independent experiments. (D, E) The transcript levels of betIBA-proXWV (D) and Qrr4, luxR, and luxC (E) were assayed by qRT-PCR 15 min following salt shock. Test cultures of BB120 received salt shock with 1 M NaCl, and control cultures received an equal volume of 0.2 M NaCl (final concentrations). The data shown are the means and standard errors from three biological replicates and represent those from three independent experiments.
FIG 6.
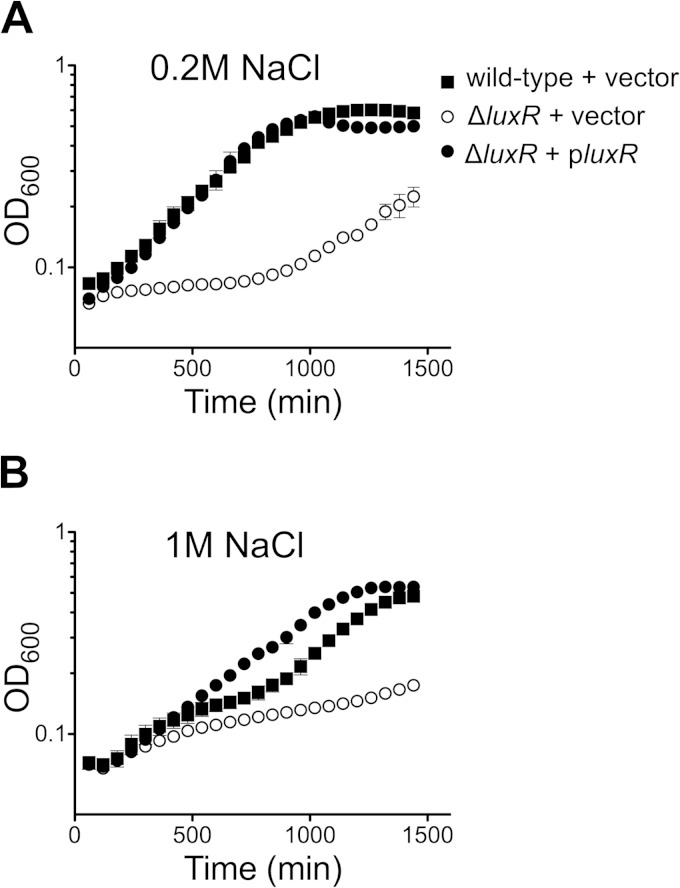
Quorum sensing regulates the osmotic stress response in V. harveyi. (A, B) V. harveyi strains were grown in LOM with 1 mM choline and 0.2 M (A) or 1 M NaCl (B). The strains are tested were wild-type V. harveyi carrying a control plasmid (BB120::pLAFR2), V. harveyi ΔluxR carrying a control vector (KM669::pLAFR2), and V. harveyi ΔluxR carrying a plasmid encoding luxR (KM669::pKM699). The data shown are the means and standard errors from three biological replicates and represent those from at least three independent experiments.
Molecular methods.
Plasmid cloning (see Table S2 in the supplemental material) was performed using E. coli strains S17-1λpir and DH10B, and cloning details and sequences are available upon request. iProof DNA polymerase (Bio-Rad) was used for PCRs. Oligonucleotides (see Table S3 in the supplemental material) were purchased from Integrated DNA Technologies. T4 polynucleotide kinase, T4 DNA ligase, restriction enzymes, and calf intestinal phosphatase (CIP) were purchased from New England BioLabs (NEB). Plasmid sequences were confirmed by Genewiz, Inc., or ACGT, Inc. RNA isolation and cDNA synthesis were performed as described previously (6). For quantitative real-time reverse transcription-PCR (qRT-PCR) analyses, the levels of expression from the samples were normalized to the level of expression of the internal standard, hfq, as described previously (11), and either the ΔΔCT threshold cycle (CT) method or the standard curve method was used for data analysis.
Gene deletion, complementation, and overexpression of luxR and betI.
The construction of the ΔbetI mutant strain was performed as described previously (6) using a cosmid containing a deletion of the betI open reading frame (ORF) (see Tables S1 and S2 in the supplemental material). The chloramphenicol resistance (Cmr) cassette used during strain construction was removed from all the final betI deletion strains using FLP-mediated recombination (29). Following removal of the Cmr cassette, each strain retained a scar of ∼100 bp at the FLP recombination site. For complementation of the ΔluxR strain, overnight cultures (BB120, KM669, KM669::pJV036, and KM669::pJV239) were diluted 1:100 and grown to an OD600 of 0.3, 10 μM IPTG was added, and cells were grown for an additional 2 h to an OD600 of 1.0. At that point, RNA extraction was performed. For complementation of the ΔbetI strain, overnight cultures (BB721::pJV298, JC2216::pJV298, and JC2216::pJV299) were diluted to an OD600 of 0.05 and grown to an OD600 of 0.2, and 10 μM IPTG was added. Cells were grown for an additional 2 h, after which they were collected for RNA extraction. For overexpression of betI, overnight cultures (BB721::pJV298 and BB721::pJV299) were diluted to an OD600 of 0.05 and grown to an OD600 of 0.2, 1 mM IPTG was added, and after 2 h of growth, cells were collected for RNA extraction.
Protein purification.
The LuxR protein was purified as described previously (7). His-tagged BetI (His-BetI) was purified by overexpression in Rosetta(DE3)pLysS cells (Novagen) from plasmid pSQ004 expressing betI with an N-terminal 6× His tag. Cultures (2 liters) were grown with aeration at 30°C, induced with 1 mM IPTG at an OD600 of 0.4, and grown for an additional 4 h at 30°C. The cells were mechanically lysed using a cell cracker in Ni-nitrilotriacetic acid buffer (50 mM NaPO4, pH 7.4, 10 mM imidazole, 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride). His-BetI was purified on a HisTrap HP column (GE Healthcare), eluted with a linear 10 mM to 1 M imidazole gradient, and purified. His-BetI was dialyzed and stored at −80°C in storage buffer (50 mM NaPO4, pH 7.4, 300 mM NaCl, 10% glycerol).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) with purified LuxR protein were performed as described previously (7, 30) using the oligonucleotides listed in Table S3 in the supplemental material. EMSAs with purified His-BetI protein were performed as described previously (19), except that the BetI binding buffer was modified [10 mM Tris-HCl, pH 8.9, 50 mM KCl, 0.5 mM dithiothreitol, 0.1% Igepal CA-630, 8% Ficoll 400, 0.1 μg μl−1 bovine serum albumin, and 10 ng μl−1 poly(dI-dC)].
RESULTS
LuxR regulates expression of betIBA-proXWV.
Our previous microarray studies showed that LuxR controls ∼625 genes at HCD in V. harveyi (7). We examined those encoding LuxR-controlled transcriptional regulators to determine whether they drive a downstream tier of quorum-sensing gene expression. Among the putative transcription factors is VIBHAR_06181, a TetR-type transcriptional regulator, which shares 46% amino acid sequence identity with E. coli BetI. Microarray analyses indicated that LuxR activates expression of V. harveyi betI (VIBHAR_06181) (Fig. 2A) 3-fold at HCD (7). To verify these findings, we analyzed betI levels at HCD in the presence and absence of luxR by quantitative real-time reverse transcription-PCR (qRT-PCR), and indeed, an ∼3-fold decrease in betI expression compared to the level of expression in the wild type occurred in a ΔluxR strain (Fig. 2B). Production of LuxR from a plasmid restored betI expression to the level of the wild type (Fig. 2B), confirming that LuxR activates expression of betI. Consistent with a role for LuxR as an activator, betI transcript levels are high at HCD, and this requires the presence of LuxR (see Fig. S1A in the supplemental material) (7).
FIG 2.
LuxR activates expression of betIBA-proXWV in V. harveyi. (A) Chromosomal regions encoding the bet and pro genes in E. coli and V. harveyi. V. harveyi gene numbers are indicated below each gene. The positions of the LuxR binding sites (bp −338 and −41, denoted LuxR BS1 and BS2, respectively) and BetI binding site (bp −276, denoted BetI BS) are indicated relative to the translation start site of betI. (B) Transcript levels of V. harveyi genes were determined by qRT-PCR from wild-type V. harveyi (BB120), ΔluxR V. harveyi (KM669), the ΔluxR strain containing a control plasmid (pJV036), and the ΔluxR strain carrying the plasmid expressing luxR (pluxR; pJV239). Error bars represent standard errors for three biological samples, and the data represent those from three independent experiments. (C) EMSAs for reaction mixtures containing 0, 10, 100, or 1,000 nM LuxR incubated with a radiolabeled DNA substrate corresponding to the predicted LuxR binding sites (BS1 or BS2) upstream of betI.
BLAST analyses indicated that the organization of the osmoregulatory genes in V. harveyi differs from that in E. coli (Fig. 2A). Similar to their organization in E. coli, the V. harveyi genes encoding homologs of BetB (VIBHAR_06180) and BetA (VIBHAR_06179) lie directly downstream of betI. However, unlike in E. coli, no BetT homolog was colocated. There are six possible BetT candidates in the V. harveyi genome (VIBHAR_02455, VIBHAR_06429, VIBHAR_01883, VIBHAR_03056, VIBHAR_06536, and VIBHAR_05503) (26); however, each shares only modest homology with E. coli BetT (28 to 36% identity). We did not observe LuxR regulation of any of the possible betT-like genes (7). Again, unlike in E. coli, three additional genes are located downstream of betA in V. harveyi, and they encode proteins homologous to the E. coli ProU transport system: ProX (VIBHAR_06178), ProW (VIBHAR_06177), and ProV (VIBHAR_06176) (Fig. 2A). In E. coli, the proVWX genes are not located adjacent to bet genes. The V. harveyi genome encodes a second set of proXWV genes (26) (VIBHAR_06562, VIBHAR_06563, and VIBHAR_06564, respectively). None of these genes is regulated by LuxR (7). Preliminary transcriptome sequencing (RNA-seq) analysis of primary RNA transcripts indicated that the betIBA and proXWV genes are cotranscribed and thus form a single operon (betIBA-proXWV) (K. Papenfort, unpublished data). To determine if the V. harveyi betBA and proXWV genes are regulated by quorum sensing similarly to betI, we analyzed their expression levels in the presence and absence of luxR. LuxR activates expression of all of these genes 5- to 10-fold (Fig. 2B).
We previously determined the locations of the genomic LuxR binding sites by chromatin immunoprecipitation and deep sequencing (ChIP-seq) (30). Two LuxR binding sites (BS1 and BS2, located 338 bp and 41 bp upstream of the betI open reading frame [ORF], respectively) (Fig. 2A) were identified by our bioinformatic analyses of the LuxR ChIP-seq binding peaks (30). To confirm that LuxR binds to both sites in the betI promoter, we performed EMSAs with purified LuxR and found that LuxR bound both sites. LuxR showed a >50-fold greater affinity for BS1 than for BS2 (Fig. 2C). These data indicate that LuxR is a transcriptional activator of the betIBA-proXWV operon.
BetI represses expression of the betIBA-proXWV operon.
To examine the regulatory role of BetI in expression of the betIBA-proXWV operon, we constructed a deletion of the betI ORF in several strain backgrounds and examined BetI function (see Table S1 in the supplemental material). To isolate the role of BetI, we used a ΔluxO strain of V. harveyi that constitutively mimics the HCD state (7). This strategy allowed us to exclude the possibility of regulatory effects from quorum-sensing components. Expression of the five genes (betB, betA, proX, proW, and proV) downstream of betI decreased ∼2-fold in the ΔbetI ΔluxO strain compared to that in the single ΔluxO strain (see Fig. S1B in the supplemental material), suggesting that there is a polar effect either from the betI deletion or as a consequence of the scar left following insertion and removal of the Cmr cassette during strain construction. Indeed, expression of betI from an inducible promoter complemented the expression of betI but not that of the downstream genes (see Fig. S1B in the supplemental material). Furthermore, modest overexpression of betI in the ΔbetI ΔluxO strain enhanced repression of betBA-proXWV expression (see Fig. S1B in the supplemental material). Consistent with these results, overexpression of betI in the ΔluxO strain containing an intact betIBA-proXWV operon repressed all five genes downstream of betI 2- to 3-fold (Fig. 3A). These data support a role for BetI as a transcriptional repressor of betIBA-proXWV.
FIG 3.
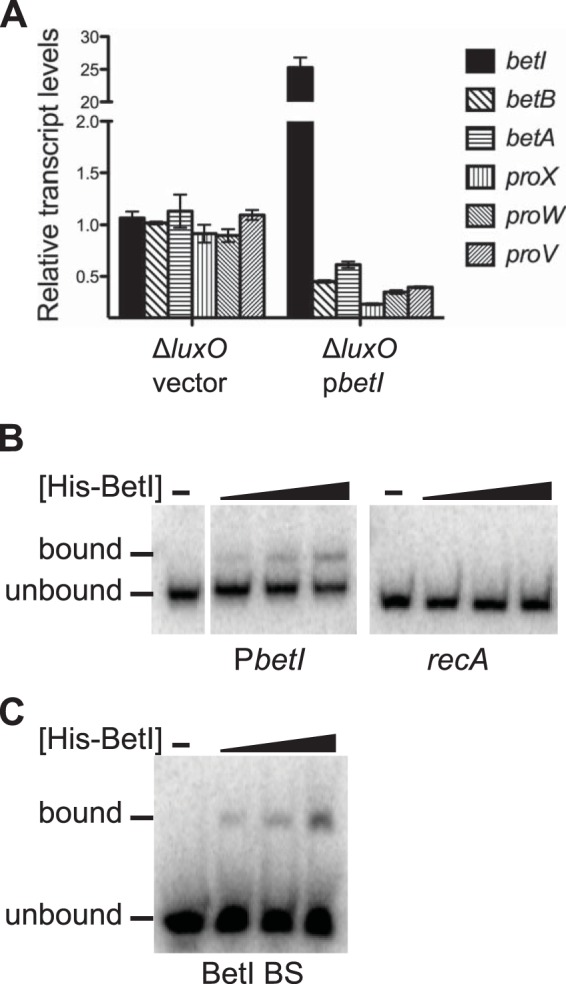
BetI represses the betIBA-proXWV operon. (A) The transcript levels of the betIBA-proXWV genes were assayed by qRT-PCR in the following V. harveyi strains induced with 1 mM IPTG: a ΔluxO strain carrying a control vector (BB721::pJV298) and a ΔluxO strain carrying a vector expressing betI from an IPTG-inducible promoter (BB721::pJV299). Error bars represent the standard errors of measurements from three biological replicates, and these data represent those from at least two independent experiments. (B, C) EMSAs for reaction mixtures containing 0, 250 nM, 500 nM, or 1,000 nM His-BetI incubated with a radiolabeled DNA substrate corresponding to the betI promoter region (PbetI; 359-bp promoter fragment, positioned immediately upstream of the start codon) or the recA ORF (300 bp) (B) or with the 45-bp fragment corresponding to the BetI binding site (BetI BS [see substrate C in Fig. S1C in the supplemental material], positioned 276 bp upstream of the start codon) (C).
To determine if BetI binds to the betI promoter, we purified His-tagged BetI (His-BetI) and performed EMSAs. His-BetI bound to a DNA fragment corresponding to the 359-bp region upstream of the betI start codon (Fig. 3B). As a control, we show that His-BetI did not bind to a DNA fragment corresponding to the recA ORF (Fig. 3B), indicating that binding to the betI promoter is specific. We could identify a possible BetI binding site using the E. coli BetI binding site as a reference (19). The putative site overlaps the −35 region and LuxR BS2 (see Fig. S1C in the supplemental material). This result is not surprising, given that BetI and LuxR are both TetR-type transcription factors (19, 30). However, what is surprising is that His-BetI did not bind this site (see Fig. S1C in the supplemental material; see substrate J). His-BetI also did not bind LuxR BS1 (see substrate A in Fig. S1C in the supplemental material). To identify the BetI binding site, we examined DNA fragments spanning the entire betI promoter region (see Fig. S1C in the supplemental material). His-BetI bound to a 45-bp fragment positioned 276 bp upstream of the betI start codon (see substrate C in Fig. S1C in the supplemental material). The affinity of BetI for this site was essentially equivalent to that for the full promoter region (compare Fig. 3B and C). These findings suggest that BetI represses transcription of the betIBA-proXWV operon by binding to the betI promoter but BetI binds to a sequence different from that to which E. coli BetI binds.
BetI regulates expression of quorum-sensing genes.
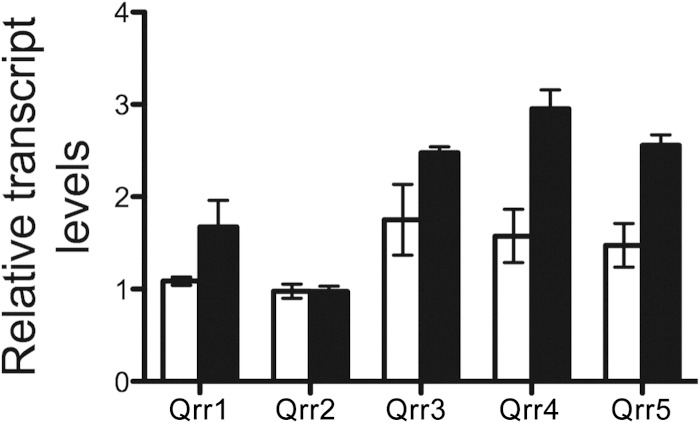
Bacteria frequently employ feedback loops to comodulate pathways (10, 29, 31–34). Given that quorum sensing controls the Bet system, we wondered if the Bet system likewise controls quorum sensing. To test this notion, we assayed BetI regulation of the core quorum-sensing regulatory components in V. harveyi following overexpression of betI on a plasmid. We used strains that constitutively mimic the HCD state (the ΔluxO strain) or the LCD state (the luxO D47E strain) (7). First, we measured transcript levels of aphA (at LCD) and luxR (at HCD) and found that they were unchanged following BetI overproduction (see Fig. S2A and B in the supplemental material). We next measured Qrr1 to Qrr5 levels at LCD. BetI activated the expression of qrr1, qrr3, qrr4, and qrr5 but not that of qrr2 (Fig. 4). To investigate if BetI regulation of the qrr genes is direct, we assayed His-BetI binding to the promoters of qrr3 and qrr4 using EMSAs. No specific binding occurred (see Fig. S2C in the supplemental material). These results suggest that BetI control of the qrr genes is indirect. We wondered if their regulation could occur through BetI control of LuxO. We measured luxO levels at LCD with and without BetI overproduction and found no change (see Fig. S2D in the supplemental material). Taken together, these data indicate that BetI is linked to the quorum-sensing system via indirect activation of expression of qrr genes.
FIG 4.
BetI regulates quorum-sensing genes. The transcript levels of Qrr1 to Qrr5 in V. harveyi strains induced with 1 mM IPTG were assayed by qRT-PCR. The strains tested were a luxO D47E strain carrying a control vector (white bars; JAF548::pJV298) and a luxO D47E strain carrying a vector expressing betI from an IPTG-inducible promoter (black bars; JAF548::pJV299). Error bars represent the standard errors of measurement from three biological replicates, and these data represent those from two independent experiments.
BetI controls the osmotic stress response in V. harveyi.
We examined osmotic stress tolerance in V. harveyi to define the role of BetI. First, we measured the growth of wild-type V. harveyi in low-osmolarity medium (LOM) containing choline and various concentrations of NaCl. V. harveyi grew at similar rates in 0.2 M, 0.3 M, and 0.5 M NaCl but exhibited slowed growth in 1 M NaCl (Fig. 5A). We next tested the effect of salt shock by growing V. harveyi to mid-exponential phase in medium containing 0.2 M NaCl, and at that point, we added NaCl to a final concentration of 1 M to rapidly change the osmolarity of the medium. Upon salt shock, the OD600 of the cultures decreased modestly prior to recovery, and growth resumed after ∼2.5 h (Fig. 5B, solid circles). Control cultures were unaffected by addition of the same volume of 0.2 M NaCl (Fig. 5B, solid squares). The ∼22% decline in the OD600 following addition of 1 M NaCl is likely due to the cell shrinkage that occurs upon hyperosmotic shock (35, 36). We also performed the salt shock experiment in medium lacking the osmolyte choline to test for a requirement for exogenous choline. The absence of choline considerably slowed growth following salt shock, and shocked cultures did not achieve the same optical density as cultures containing choline (Fig. 5B, open circles). The ΔbetI V. harveyi strain grew similarly to the wild type in 0.2 M and 0.5 M NaCl (see Fig. S3A and B in the supplemental material) but displayed an even more pronounced growth deficiency in 1 M NaCl than the wild type (Fig. 5C). Because we cannot restore expression of the betIBA-proXWV operon in the ΔbetI strain with betI on a plasmid (see Fig. S1B in the supplemental material), we introduced a fragment of the genome containing the entire betIBA-proXWV operon and its promoter, and we show that growth was restored to wild-type levels in medium containing 1 M NaCl (Fig. 5C).
We assessed the effect of osmotic shock on the transcription of osmoregulatory and quorum-sensing components. The transcript levels of the genes in the betIBA-proXWV operon increased roughly 3- to 6-fold within 15 min following the 1 M NaCl shock (Fig. 5D). Transcription of qrr also increased after salt shock, and in agreement with our understanding of how the quorum-sensing circuit functions (Fig. 1), luxR and luxC (luciferase) expression decreased (Fig. 5E). These findings support the conclusion that BetI activation of qrr4 occurs upon osmotic stress.
Quorum sensing controls the osmotic stress response.
We examined whether quorum sensing influences the V. harveyi response to osmotic stress by assaying the salt shock response in V. harveyi strains containing deletions of luxR and aphA as well as strains constitutively mimicking the LCD and HCD quorum-sensing states (luxO D47E and ΔluxO strains, respectively [7]). All strains grew more rapidly in medium containing 0.5 M NaCl than in medium containing 0.2 M or 1 M NaCl (see Fig. S3 in the supplemental material). The two strains with the lowest luxR levels (the luxO D47E and ΔluxR strains) showed impaired growth in medium containing low salt (see Fig. S3A in the supplemental material). Conversely, the HCD-locked strain (ΔluxO), which constitutively produces LuxR, was the strain that grew the most rapidly in all concentrations of NaCl (see Fig. S3 in the supplemental material). To verify that deletion of luxR caused the reduced growth rate in 0.2 M and 1 M NaCl, we compared the growth profiles of the ΔluxR strain to those of the ΔluxR strain complemented with luxR expressed under the control of its native promoter on a low-copy-number plasmid (Fig. 6). Complementation restored growth to wild-type levels in 0.2 M NaCl (Fig. 6A). Indeed, the complemented strain grew better than the wild type in 1 M NaCl (Fig. 6B), suggesting that slight overexpression of luxR improves growth in high salt. Collectively, these results show that LuxR plays a crucial role in osmotolerance in V. harveyi.
DISCUSSION
Marine salinity can vary widely; however, the average ocean salinity is 3.5% and is constituted primarily of Na+ and Cl− ions (∼0.6 M NaCl) (37, 38). Our observation that V. harveyi grows optimally under similar conditions, specifically, at 0.5 M NaCl, is therefore consistent with this notion. Nonetheless, as a free-living marine bacterium, V. harveyi presumably must have the capacity to adapt to fluctuations in osmolarity. We find that, following rapid changes in salt concentration, V. harveyi cells activate expression of the betIBA-proXWV operon, likely for osmoprotection through synthesis and/or transport of the osmoprotectant glycine betaine. V. harveyi strains with decreased betIBA-proXWV expression display decreased growth rates in high salt. We suspect that V. harveyi is capable of using choline to synthesize glycine betaine because cultures grown in the absence of choline had a marked growth defect under high-salt conditions compared to the growth of cultures with choline in the medium. This result also suggests that V. harveyi possesses an enzyme that functions analogously to E. coli BetT. We were unable to identify a V. harveyi BetT homolog regulated by either LuxR or BetI using microarray analyses (7) (data not shown). An alternative possibility is that, similar to E. coli, V. harveyi imports choline via the ProXWV transport system when choline is available at high concentrations (e.g., 1.5 mM) (39, 40).
The V. harveyi BetIBA and ProXWV systems appear to function similarly to their counterparts in E. coli. However, a striking difference between E. coli and V. harveyi is the consolidation of the betIBA-proXWV genes into a single locus in V. harveyi. Preliminary RNA-seq data suggest that there are two transcription start sites in this locus: one located 46 bp upstream and one located 9 bp downstream of the betI start codon (Papenfort, unpublished). We suspect that the upstream transcription start site (at position −46) (Fig. 2) controls transcription of betIBA-proXWV, while the downstream transcription start site (at position +9) (Fig. 2) controls transcription of the betBA and proXWV genes. In support of this idea, deletion of the betI ORF and, thus, the downstream transcription start site may have had a polar effect and decreased expression of betBA-proXWV. Also, the strength of LuxR regulation differs between betI and the five downstream genes, suggesting distinct regulatory mechanisms. We posit that the presence of two transcription start sites enables increases in betBA and proXWV expression to occur independently from increases in expression of the betI repressor. Future experiments will examine the role of the downstream transcriptional start site on expression of betBA-proXWV.
V. harveyi BetI represses betIBA-proXWV. Mechanistically, however, V. harveyi BetI functions differently than the E. coli BetI. Addition of choline to DNA binding reactions did not stimulate increases in His-BetI-DNA complex formation, as observed with E. coli BetI (data not shown) (19). Furthermore, V. harveyi His-BetI did not bind to the predicted binding sequence in the betI promoter in our in vitro assays. Rather, BetI bound to a region 276 bp upstream of the translation start site.
Our results show that quorum sensing is linked to the V. harveyi osmotic stress response. First, the master quorum-sensing regulator, LuxR, activates expression of the betIBA-proXWV operon 3- to 10-fold at HCD. LuxR binds to two sites in the betI promoter, as is common for genes activated by LuxR (30). Our finding that LuxR binds to BS2 with a higher affinity than to BS1 is consistent with the finding of a large LuxR binding peak centered around BS2, described in the ChIP-seq data obtained in a previous study (30). Conversely, no significant binding peak near BS1 was detected by the ChIP-seq analysis. Only bioinformatic scanning of the betI promoter revealed a site similar to the LuxR consensus binding sequence (BS1) (30). We do not yet know if both binding sites are required for betI activation. Because LuxR BS2 overlaps the −35 region of the promoter for the downstream transcription start site, it is possible that LuxR represses expression from this transcription start site while activating expression from the upstream start site via binding at BS1. We found that LuxR is critical for V. harveyi growth under both low- and high-NaCl conditions, suggesting a requirement for LuxR activation of the betIBA-proXWV genes for proper adaptation to osmotic stress. We hypothesized that the growth defect in the ΔluxR strain could be complemented by overexpression of betIBA-proXWV. We performed this experiment, but no change in growth occurred (data not shown). Two possibilities are that (i) the levels of betIBA-proXWV from the plasmid (4 to 7 copies per cell [41–43]) remain below the levels achieved by LuxR activation in the wild type (5- to 10-fold) (Fig. 2B) and/or (ii) LuxR activates expression of another factor (e.g., a BetT homolog) that is necessary for growth under conditions of osmotic stress.
Our experiments also demonstrate that BetI indirectly activates expression of the Qrr sRNA genes, with the exception of qrr2. This result was unexpected, given that the Qrr sRNAs play nearly identical regulatory roles. Nonetheless, although the Qrr gene sequences are highly conserved, their promoters are distinct and a different transcription factor possibly controls qrr2 (5). Regulation of the Qrr genes does not occur through BetI control of LuxO, suggesting that some unknown factor that links BetI to the Qrr genes must exist. The Qrr sRNAs, in turn, repress LuxR. BetI feedback on LuxR via Qrr regulation may serve to keep LuxR levels in check and, in so doing, prevent LuxR from overactivating the betIBA-proXWV operon. One possible advantage for LuxR activation of the osmotic stress pathway is that environments that support the growth of V. harveyi to high population densities also routinely contain high salt and therefore require the upregulation of osmotic stress genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Bochman for insightful comments and discussions. We thank Kai Papenfort for analyses of the transcripts of the betIBA-proXWV operon by RNA-seq.
This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) grant 5R01GM065859, and National Science Foundation (NSF) grant MCB-0343821 to B.L.B. and by funds from Indiana University to J.C.V.K.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02246-14.
REFERENCES
- 1.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 3.Freeman JA, Bassler BL. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol 181:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilley BN, Bassler BL. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 5.Tu KC, Bassler BL. 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell 18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Swem LR, Swem DL, Wingreen NS, Bassler BL. 2008. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng SW, Schaffer JN, Tu KC, Mehta P, Lu W, Ong NP, Bassler BL, Wingreen NS. 2011. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol Syst Biol 7:491. doi: 10.1038/msb.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. 2010. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu KC, Waters CM, Svenningsen SL, Bassler BL. 2008. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol 70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 14.Wood JM. 2006. Osmosensing by bacteria. Sci STKE 2006:pe43. doi: 10.1126/stke.3572006pe43. [DOI] [PubMed] [Google Scholar]
- 15.Stirling DA, Hulton CS, Waddell L, Park SF, Stewart GS, Booth IR, Higgins CF. 1989. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol 3:1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 16.Cairney J, Booth IR, Higgins CF. 1985. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J Bacteriol 164:1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamark T, Rokenes TP, McDougall J, Strom AR. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J Bacteriol 178:1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol 5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 19.Rkenes TP, Lamark T, Strom AR. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J Bacteriol 178:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andresen PA, Kaasen I, Styrvold OB, Boulnois G, Strom AR. 1988. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J Gen Microbiol 134:1737–1746. [DOI] [PubMed] [Google Scholar]
- 21.Joelsson A, Kan B, Zhu J. 2007. Quorum sensing enhances the stress response in Vibrio cholerae. Appl Environ Microbiol 73:3742–3746. doi: 10.1128/AEM.02804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, Wen Y, Son JS, Lee KH, Kim KS. 2013. The Fur-iron complex modulates the expression of the quorum-sensing master regulator SmcR to control the expression of virulence factors in Vibrio vulnificus. Infect Immun 81:2888–2898. doi: 10.1128/IAI.00375-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh JL, Fries JS, Noble RT. 2008. Dynamics and predictive modelling of Vibrio spp. in the Neuse River Estuary, North Carolina, USA. Environ Microbiol 10:57–64. doi: 10.1111/j.1462-2920.2007.01429.x. [DOI] [PubMed] [Google Scholar]
- 26.Naughton LM, Blumerman SL, Carlberg M, Boyd EF. 2009. Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl Environ Microbiol 75:2802–2810. doi: 10.1128/AEM.01698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg EP, Hastings JW, Ulizur S. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol 120:87–91. doi: 10.1007/BF00409093. [DOI] [Google Scholar]
- 28.Cairney J, Booth IR, Higgins CF. 1985. Salmonella typhimurium proP gene encodes a transport system for the osmoprotectant betaine. J Bacteriol 164:1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. 2009. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol 7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. 2013. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4:e00378–13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svenningsen SL, Waters CM, Bassler BL. 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev 22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin D, Lee EJ, Huang H, Groisman EA. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 33.Strauch MA, Wu JJ, Jonas RH, Hoch JA. 1993. A positive feedback loop controls transcription of the spoOF gene, a component of the sporulation phosphorelay in Bacillus subtilis. Mol Microbiol 7:967–974. doi: 10.1111/j.1365-2958.1993.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 34.Sen S, Garcia-Ojalvo J, Elowitz MB. 2011. Dynamical consequences of bandpass feedback loops in a bacterial phosphorelay. PLoS One 6:e25102. doi: 10.1371/journal.pone.0025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch AL. 1984. Shrinkage of growing Escherichia coli cells by osmotic challenge. J Bacteriol 159:919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldwin WW, Sheu MJ, Bankston PW, Woldringh CL. 1988. Changes in buoyant density and cell size of Escherichia coli in response to osmotic shocks. J Bacteriol 170:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickson AG, Goyet C. 1994. Handbook of methods for the analysis of the various parameters of the carbon dioxide system in sea water, version 2. US Department of Energy Carbon Dioxide Survey Science Team, Oak Ridge, TN. [Google Scholar]
- 38.Sharqawy MH, Lienhard V JH, Zubair SM. 2010. Thermophysical properties of seawater: a review of existing correlations and data. Desal Water Treat 16:354–380. doi: 10.5004/dwt.2010.1079. [DOI] [Google Scholar]
- 39.Lamark T, Styrvold OB, Strom AR. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol Lett 75:149–154. [DOI] [PubMed] [Google Scholar]
- 40.Styrvold OB, Falkenberg P, Landfald B, Eshoo MW, Bjornsen T, Strom AR. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J Bacteriol 165:856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman AM, Long SR, Brown SE, Buikema WJ, Ausubel FM. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 43.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A 77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.