Abstract
Objective
To study the cytokine/chemokine profiles in response to HIV-1 viremia, and elucidate the pathways leading to HIV-1-induced inflammation.
Design/Methods
Plasma levels of 19 cytokines in individuals with early HIV-1 infection and individuals undergoing treatment interruptions were evaluated via multiplex assay. To investigate the cellular sources of relevant cytokines, sorted cells from HIV-1 infected individuals were assessed for mRNA expression. Relevant signaling pathways were assessed by comparing cytokine production patterns of PBMCs stimulated with intact HIV-1 or specific TLR stimulants with and without a TLR7/9 antagonist.
Results
IP-10 plasma concentration was most significantly associated with HIV-1 viral load and was the most significant contributor in a multivariate model. IP-10 mRNA was highly expressed in monocytes and mDCs and these cells were the dominant producers after in vitro stimulation with TLR7/8 ligands (CL097 and ssRNAGag1166), AT-2 HIV-1, and HIV-1NL43 virus. Partial least square discriminant analysis of culture supernatants revealed distinct cytokine/chemokine secretion profiles associated with intact viruses compared to TLR7/8 ligands alone, with IP-10 production linked to the former. A TLR7/9 antagonist blocked IP-10 production following whole virus stimulation, suggesting the involvement of TLR7/9 in the recognition of HIV-1 by these cells.
Conclusions
Monocytes and mDCs produce significant amounts of IP-10 in response to HIV-1 viremia and after in vitro stimulation with HIV-1. Stimulation with HIV-1-derived TLR7/8-ligands versus HIV-1 resulted in distinct cytokine/chemokine profiles, indicating additional pathways other than TLR7/8 that lead to the activation of innate immune cells by HIV-1.
Keywords: IP-10, immune activation, TLR, monocytes, mDCs
Introduction
Since the introduction of highly active antiretroviral therapy (HAART), people with HIV-1 have experienced dramatic improvements in life expectancy; however, they continue to have increased morbidity and mortality even with durable suppression of viral replication [1–9]. Several studies have demonstrated that HIV-1-induced immune activation is associated with worse clinical outcomes and may contribute to this excess in mortality (reviewed in [10]). In the pre-HAART era, higher levels of immune activation were observed in patients with faster progression to AIDS [11, 12]. Even with suppression of viral replication, the level of CD8+ and CD4+ T cell activation does not return to normal levels [13, 14], and higher levels of T cell activation have been associated with slower gains in CD4+ T cell count with the initiation of ART [15] and with failure to recover CD4+ T cell count despite a good response to ART [16]. In the Strategies for Management of Antiretroviral Therapy (SMART) trial [17], patients in the CD4+ T cell count-guided therapy arm where therapy was interrupted had higher risk of death from any cause than those in the continuous therapy arm. Elevated levels of IL-6 and D-dimer were significantly associated with increased risk of all cause mortality in both the continuous treatment and the treatment interruption arms [18]. Inflammatory markers such as soluble CD14 can also help predict mortality even after accounting for viral load [19, 20]. Taken together, the data indicate that immune activation contributes to both HIV disease progression in untreated individuals and to serious non-AIDS events and morbidity in patients with full virus suppression. The foundation of this HIV-1-associated immune dysregulation and immune activation is likely established early in HIV-1 infection when a host of plasma cytokines and chemokines rise sharply [21, 22].
In this study, we sought to gain insights into the immune dysregulation and activation that occur with HIV-1 by examining plasma cytokines and chemokines in early HIV-1 infection and during a structured treatment interruption (STI) trial of patients started on therapy during acute or early HIV-1 infection. We employed partial least squares regression analysis (PLSR)[23] to assess multi-variate differences in plasma cytokines and chemokines as related to viral load. We examined the cellular sources of the individual chemokine with the strongest association with viremia, IP-10, using both ex vivo analysis of mRNA in matched patient peripheral blood mononuclear cells (PBMCs) and in vitro stimulation of PBMCs from healthy individuals with HIV-1. To assess the pathways responsible for HIV-1 associated inflammatory cytokine and chemokine production, we compared the effects of HIV-1 stimulation to activation with Toll-like receptor (TLR) ligands and used a partial least squares discriminant analysis (PLSDA) model to identify patterns associated with either whole virus HIV-1 stimulation or stimulation with specific TLR ligands. Finally, we tested the effects of blocking TLR7/9 signaling in our in vitro assays to measure the contribution of these pathways to HIV-1 induced cytokine and chemokine production.
Methods
Study participants
Samples from the following three groups of participants were utilized in this study: HIV-1- negative individuals, men and women with acute or early HIV-1 infection (Table 1), and HIV-1- infected participants from a clinical trial evaluating structured treatment interruption in early infection (acute STI group, Table 1). All participants were enrolled in protocols that were approved by the Massachusetts General Hospital Institutional Review Board. Each participant gave written informed consent. Briefly, the fourteen participants in the acute STI group were identified during acute or early HIV-1 infection and were started on HAART before or shortly after seroconversion as previously described [24, 25]. Following durable viral suppression for a minimum of 2 months, all antiretroviral medications were stopped. HAART was restarted if the viral load was greater than 5000 RNA copies per ml on three consecutive visits or greater than 50,000 copies per ml on any occasion. Participants were eligible for additional treatment interruptions once viral suppression was again achieved.
Table 1.
Patient Characteristics
| Study Subjects with Early HIV-1 Infection | ||
|---|---|---|
| Participant | Viral Load (copies/ml) | Weeks after Enrollment |
| AC255 | 439 | 29 |
| AC253 | 1440 | 25 |
| AC259 | 3550 | 10 |
| AC258 | 5590 | 14 |
| AC260 | 15,800 | 15 |
| AC277 | 29,100 | 5 |
| AC270 | 32,300 | 2 |
| AC246 | 46,200 | 40 |
| AC264 | 88,100 | 29 |
| AC248 | 124,000 | 28 |
| AC278 | 640,000 | 0 |
| AC267 | 5,710,000 | 2 |
| Study Subjects in Structured Treatment Interruption Trial | |||
|---|---|---|---|
| Participant | No. of Interruptions | Viral Load at Initiation of ARVs During Acute Infection | CD4 Count at Initiation of ARVs |
| AC-10 | 1 | 40,700 | 919 |
| AC-15 | 1 | 27,000 | 413 |
| AC-45 | 1 | 2,090,000 | 377 |
| AC-02 | 2 | 4,850,000 | NA |
| AC-16 | 2 | 337,000 | NA |
| AC-13 | 2 | 5500 | 667 |
| AC-33 | 2 | 26,700 | 512 |
| AC-14 | 3 | 95,100 | 981 |
| AC-25 | 3 | 697,000 | 314 |
| AC-26 | 3 | 1,260,000 | 390 |
| AC-46 | 3 | 123,000 | NA |
| AC-04 | 4 | 9,620,000 | NA |
| AC-05 | 4 | 1,100,000 | 289 |
| AC-06 | 4 | 8,180,000 | 551 |
Measurement of cytokine and chemokine levels in cell culture supernatant or plasma
Plasma or cell culture supernatant cytokine and chemokine concentrations were measured using MILLIPLEX Human Cytokine/Chemokine kits (Millipore). The following cytokines and chemokines were measured using the high sensitivity cytokine kit (standards ranged from 0.13 to 2000 pg/ml) according to the manufacturer’s instructions: GM-CSF, IFN-γ, IL-1β, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (p70), IL-13, and TNFα. The following cytokines and chemokines were measured using the regular sensitivity kit (standards ranged from 3.2 to 10,000 pg/ml): IL-1α, IL-12 (p40), MCP-1α, IP-10, and Eotaxin. Each sample was run in duplicate on a Bio-Plex 200 system (Bio-Rad). Results were tabulated and analyzed with Bioplex software (Bio-Rad). Values for which the coefficient of variation (%CV) exceeded 30 were excluded. Low or high out of range values were set to half of the lowest standard or 1.5 times the highest standard, respectively. Values that were extrapolated out of the standard range were set to the highest or lowest standard. For the STI group, the baseline level used to calculate the fold change in plasma cytokine/chemokine level was the average of the specific cytokine/chemokine level at 3 time points prior to the interruption. The viral load was undetectable at these time points. All plasma cytokine and cell culture supernatant analyses were performed on frozen samples.
Quantification of T cell activation
Cryopreserved PBMCs from the subjects with early HIV-1 infection (Table 1) were chosen from time points matched (identical or within 1 week) to the plasma samples examined by multiplex cytokine analysis. After thawing, an aliquot of cells was stained for T cell subsets and activation markers (CD14 APC Cy7, HLADR PerCP, CD4 Pacific Blue, CD3 Alexa Fluor 700, CD69 FITC, CD8 PECy7, CD25 APC, CD38 PE all from BD Biosciences). Samples were acquired on a 4 Laser LSR II (BD Biosciences) and results were analyzed with FlowJo software version 9.6.2 (Tree Star). Level of activation was defined as percent of T cells (CD8+ T cell = CD14−CD3+CD4−CD8+; CD4+ T cell= CD14−CD3+CD4+CD8−) double positive for CD38 and HLA-DR.
mRNA expression
As above, matched cryopreserved PBMCs were used for analysis of mRNA levels. 500,000 bulk PBMCs were immediately stored in RNA lysis buffer (Qiagen) from each sample. In addition, cells were stained and sorted into T cells, B cells, mDCs, and monocytes on the FACS Aria (BD Biosciences) cell sorter (CD14 APCCy7, HLA-DR PerCP, CD56 PECy7, CD19 Alexa Fluor 700, all from BD Biosciences and CD3 Pacific Blue, CD11c APC from Biolegend). Sorted populations were stored in RNA lysis buffer. Total RNA was extracted using the Qiashredder for cell lysis followed by the Qiagen RNEasy Plus kit, including genomic DNA removal, as per the manufacturer’s instructions (all from Qiagen). Quantitative RT-PCR was performed using the QuantiFast SYBR Green RT-PCR kit with Quantitect primers for IP-10 and Beta-actin (all from Qiagen) and previously designed and validated primers for GAPDH and ISG15. cDNA amplification was detected on the Light Cycler 480 (Roche).
In vitro simulation of PBMCs with TLR ligands or HIV-1
Fresh PBMCs from healthy individuals were extracted from whole blood using Ficoll-Hypaque (Sigma). 1.5 million PBMCs per ml in RPMI plus 10% heat inactivated fetal calf serum were incubated for 20 hours at 37°C, 5% CO2, with the following: media alone, 1 μg/ml of TLR7/8 agonist CL097 (Invivogen), 15 μg/ml of HIV-1 single-stranded RNA (gag1166, 5′-UUGUUAAGUGUUUCAAUUGU-3′) as described by Meier and colleagues [26], 400 ng/ml by p24 of aldrithiol-2 inactivated virus (AT-2 HIV-1, lot P092), 40 ng/ml by p24 of HIV-1NL43 or HIV-1JRCSF. Cells were stimulated without a protein transport inhibitor for analysis of cell culture supernatant by multiplex cytokine assay as detailed below. A protein transport inhibitor was added to cell culture as described below for analysis with flow cytometry.
Inhibition of TLR signaling
A TLR7/9 oligonucleotide antagonist was kindly provided by Idera Pharmaceuticals. Prior to incubation with TLR agonists or HIV-1, PBMCs from healthy donors were pretreated for 1 hour with TLR7/9 antagonist (15 μg/ml for CL097, AT-2, and control and 30 μg/ml for HIV-1 strain JRCSF). IP-10, TNFα, and IFNα intracellular cytokine staining was then determined in monocytes, mDCs, and pDCs by flow cytometry as outlined below.
Analysis of cytokine production by multiparameter flow cytometry
Intracellular cytokine staining assays were performed on fresh PBMCs from HIV-1-negative individuals. One and a half million PBMCs were incubated with TLR ligands, virus or no stimulation for a negative control as above. Brefeldin A (5 μg/mL, Sigma) was added at the time of stimulation for CL097 and at 5 hours for AT-2 HIV-1 and HIV-1NL43. Following 20 hour incubation, the cells were washed and surface stained with one of 3 antibody panels. Panel 1 included CD3 FITC, CD4 PeCy5, CD8 V500, CD19 V450, CD56 Alexa Fluor 700, and CD16 APCCy7 (all from BD Biosciences). Panel 2 included CD3 Alexa Fluor 700, CD56 Alexa Fluor 700, CD19 Alexa Fluor 700, CD123 PeCy5, CD11c FITC, HLA-DR V500, CD14 APCCy7, and CD11b Pacific Blue (all from BD Biosciences). Panel 3 included CD3 Alexa Fluor 700, CD56 Alexa Fluor 700, CD19 Alexa Fluor 700, CD123 PeCy5, CD11c APC, HLA-DR V500, and CD14 APCCy7 (all from BD Biosciences). Panel 1 and 2 were used to identify the source of IP-10. Panel 3 was used in the TLR antagonist experiments. The cells were then intracellularly stained with IP-10-PE (BD Biosciences) and TNFα-PECy7 (BD Biosciences) for panels 1 and 2 and with IP-10-PE (BD Biosciences), TNFα-PECy7 (BD Biosciences), and IFNα-FITC (PBL InterferonSource) for panel 3. Results are reported as percentages of cells that produce cytokines following stimulation with the corresponding stimulant, following subtraction of the negative control values. Cell populations were defined as the following: monocytes (Lineage−HLA-DR+CD11c+CD123−CD14+), mDCs (Lineage−HLA-DR+CD11c+CD123−CD14−), pDCs (Lineage−HLA-DR+CD11c−CD123+CD14−), NK cells (CD3−CD56+CD16− or CD3−CD56+CD16+ or CD3−CD56−CD16+), CD4+ T cells (CD3+CD4+), CD8+ T cells (CD3+CD4+), and B cells (CD3−CD19+).
Statistical analyses
The relationship between the log viral load and fold change in plasma cytokine/chemokine level or plasma cytokine/chemokine concentration was determined using linear regression. Differences among stimulant groups for each cytokine/chemokine or fold change in cytokine/chemokine level from negative were tested using the two-sided non-parametric Kruskal-Wallis Test. For analyses that included tests of all 19 cytokines/chemokines, correction for multiple comparisons was performed using the Bonferroni correction, and a result was considered significant if the p value was less than 0.0026 (0.05/19). For the quantitative RT-PCR data, the relative expression of IP-10 mRNA was compared between cell types using the nonparametric Friedman’s test and Dunn’s test for multiple comparisons and relationship to viral load and serum IP-10 was assessed by linear regression. For the TLR antagonist experiments, results with and without TLR7/9 antagonist pretreatment were compared using a non-parametric Mann-Whitney test. Tests were considered significant if the p value was less than or equal to 0.05, and reported p-values for the TLR antagonist experiments were two-sided without adjustment for multiple comparisons.
Multivariate Analysis
Partial least squares regression (PLSR) was used to identify multivariate cytokine profiles associated with viral load in two separate models generated from data from individuals with early HIV-1 infection, and data obtained after structured treatment interruption (STI). In the case of the STI model, we included measurements from the first sample after each interruption. Samples where 3 or more cytokine values were missing were excluded from analysis. All data was mean-centered and variance scaled before analysis. Cross-validation was performed by iteratively omitting random data subsets (each comprised of ~20% of data) to generate the model, then testing model performance on remaining data. Variable Importance in Projection (VIP) scores were calculated to determine cytokines most associated with viral load. Cytokines with VIP scores greater that 1 were considered cytokines most associated with viral load. For the both early HIV and STI data, we also created 4 separate models, some with only one cytokine, one with all VIP cytokines, and one with all 19 measure cytokines, and calculated the RMSEC and RMSECV of each for comparison.
For cytokine measurements from in vitro studies, partial least square discriminant analysis (PLSDA) was used to determine multivariate cytokine/chemokine secretion profiles that best distinguish PBMC response to TLR agonists and HIV-1 strains. This analysis is similar to PLSR but is used with a dichotomous outcome, in this case stimulation with either TLR agonists or HIV-1. Data was normalized with mean centering and variance scaling, and cross-validation was performed by iteratively excluding random subsets (in groups of 5 samples) during model calibration, then using excluded data samples to test model predictions.
Results
Plasma cytokine/chemokine levels in patients with early HIV-1 infection and in patients with HIV-1 following structured treatment interruption
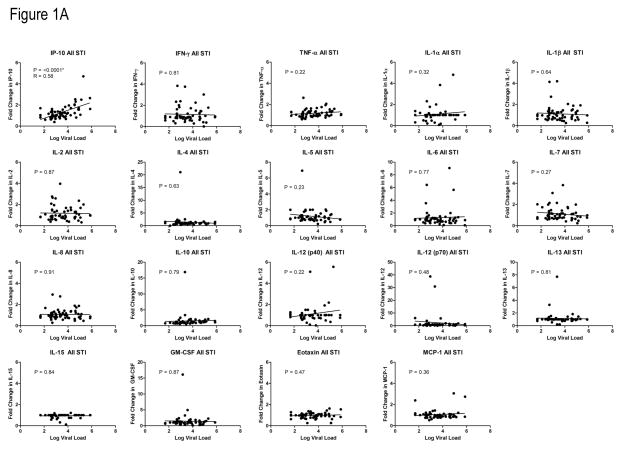
To assess the relationship between plasma viral load and plasma cytokine/chemokine concentration in early HIV-1 infection, twelve participants were studied (Table 1) within 1 year of enrollment in an acute and early infection cohort. Their plasma viral loads ranged from 439 to 5.7 million RNA copies per ml in the samples studied. None of the participants were co-infected with hepatitis C virus. Linear regression models with viral load and each individual cytokine or chemokine (Supplemental Figure 1A) were used. Log viral load and the concentrations of IP-10 (p = 0.006) and TNFα (p = 0.035) were the most related but were not significant after accounting for multiple comparisons.
We subsequently performed a similar analysis of plasma cytokine/chemokine levels during the onset of viremia following treatment interruptions in fourteen patients enrolled in clinical study of structured treatment interruption (Table 1)[24]. These patients were identified during acute HIV-1 infection and immediately started on combination ART either prior to or shortly after HIV-1 seroconversion. After their viral load became stably suppressed on therapy, they underwent a treatment interruption with the goal of enhancing the immune response with periodic antigen exposure. We studied the fold change in cytokine/chemokine level following treatment interruption as a function of viral load. The baseline cytokine level was defined as the average of 3 concentrations obtained at time points just prior to the cessation of treatment when the viral load was undetectable. As the viral load was increasing to its peak, the cytokine/chemokine levels were again determined. A median of 2 samples per subject was studied after an interruption. After the first treatment interruption, the only change in cytokine/chemokine level that was significantly associated with viral load was fold change in IP-10 (p = 0.0018, R = 0.58) (Supplemental Figure 1B). This relationship between IP-10 and viral load remained significant when data from all treatment interruptions were included (p <0.0001, R = 0.58) (Figure 1A). The larger sample size and comparison to baseline samples likely accounted for the increased statistical power of the STI analyses compared to the early infection group analysis. Taken together, these data show that plasma IP-10 concentration is closely associated with HIV-1 viral load levels in the absence of ART.
Figure 1. Relationship between plasma cytokine and chemokine levels and viral load.
A) Relationship between plasma cytokine and chemokine levels and viral load following all STIs. The relationship between fold change in cytokine or chemokine level and log viral load was determined by linear regression for each analyte. Data from all treatment interruptions was combined. Baseline cytokine or chemokine level was defined as the average of 3 levels just prior to stopping ART. P values were considered significant (*) if less than 0.0026 using a Bonferroni correction for multiple comparisons.
B) PLSR analysis of multivariate relationships between cytokine levels and plasma viral load. Analysis of all variables tested resulted in a model predictive of viral load, with relative contributions of various cytokines as indicated by a higher VIP score. In both early infection (left panel) and the STI groups, IP-10 contributed significantly to the model predicting viral load.
C) Plasma IP-10 levels are significantly related to CD8+ T cell activation. Matched PBMC samples (identical or within one week of plasma samples) were analyzed for levels of CD3+CD8+ T cell activation as indicated by expression of CD38 and HLADR. P value shown for linear regression analysis.
In both the early HIV-1 infection and STI groups we employed partial least squares regression (PLSR) analysis relating cytokine/chemokine levels and viral load; covariance was used to identify linear combinations of the analytes that were grouped into latent variables used to predict viral load. Latent variables consistent of predictive groups of cytokines and chemokines indicate IP-10 was positively associated with viral load in both early HIV-1 infection and STI. In the early HIV-1 infection PLSR model (Figure 1B, left panel), in addition to IP-10, TNF-α, IL-5, MCP-1, and IL-12 also had VIP scores greater than 1, suggesting strongest association of these cytokines with viral load in acute HIV-1 infection. In the STI PLSR model, IP-10 again had the highest VIP score (Figure 1B, right panel) with IL-10, TNF-α, IL-5 and Eotaxin also reaching VIP scores greater than 1.. For the early HIV-1 infection PLSR model, a model with 2 latent variables was able account for 92% of the variance in viral load, suggesting these 19 cytokines may be useful measurements for predicting viral load. In the STI PLSR model, a model with 2 latent variables accounted for only 26% percent of variance in viral load, suggesting other factors or cytokines not tested here might additionally be useful predicting viral load (Supplemental Figure 2).
In order to compare the ability of our 19 measured cytokines to predict viral load to the ability of individual cytokines to predict viral load, we generated 3 different additional PLSR models each for the early and STI data sets. In each case we used a different set of cytokine features to predict viral load: 1) IFN-γ only, often used in ELISpot assays (identified as a non-VIP cytokine in both the original early and STI model), 2) IP-10 only (the cytokine with the highest VIP score for both the original early and STI models), 3) all cytokines with a VIP score greater than one in the original models (IP-10,TNF-α, IL-5, MCP-1, and IL-12 in the early model and IP-10, IL-10, TNF-α, IL-5 and Eotaxin). For each model we calculated the calibration error (a measure of model fit of this data) and cross-validation error (an indication of model performance on unknown data). In general, models with more cytokines were able to account for more variance in the viral load response variable and had better calibration error (Supplemental Figure 2) in both the early HIV and STI. Models with only VIP cytokines were better than models with all 19 cytokines because they additionally had low cross-validation error, likely due to elimination of cytokines data that were not useful for predicting viral load and added noise to the models (Supplemental Figure 2).
We next sought to determine whether IP-10 levels were in concert with other validated measures of immune activation including T cell activation. Using samples from the early infection group, we selected PBMCs from timepoints matched (identical date or within 1 week) to the plasma samples and measured T cell activation as defined by CD38 and HLADR expression. CD8+ T cell activation was significantly related to plasma IP-10 levels by linear regression and correlation analysis (Figure 1C).
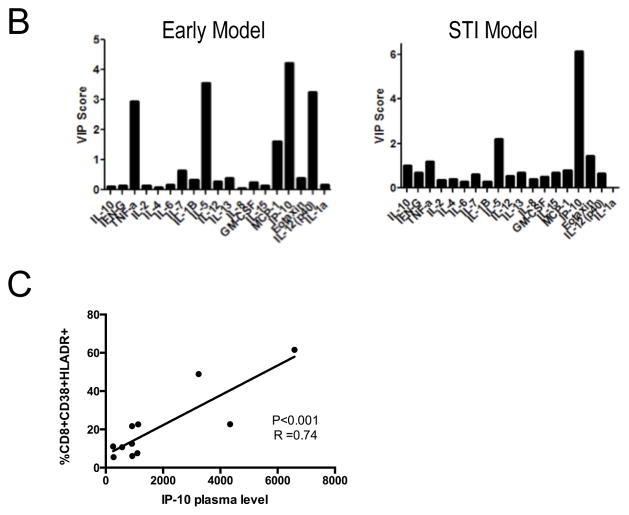
Determining the cellular source of IP-10
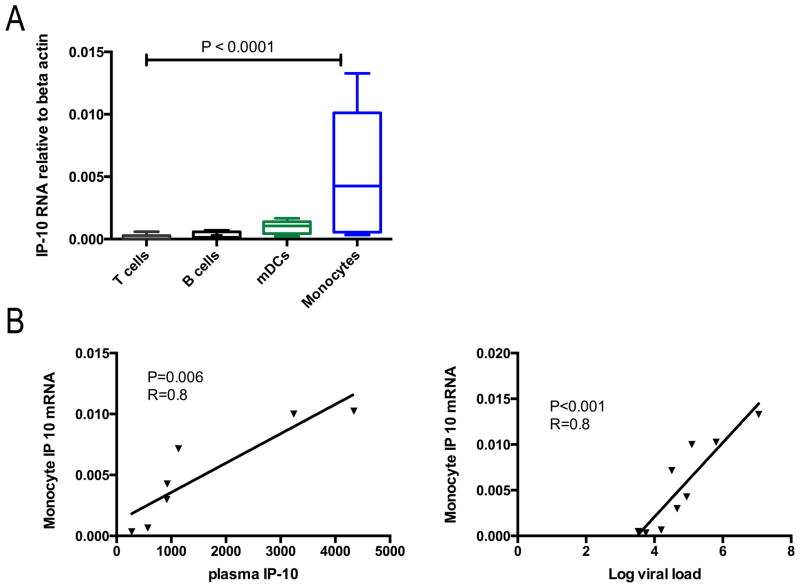
To identify the cell populations responsible for IP-10 production, we used the matched cryopreserved PBMC samples from the early infection group and sorted them into monocytes (CD14+HLADR+), mDCs (CD14−CD11c+HLADR+), T cells (CD11c−CD14− CD19−CD56− CD3+) and B cells (CD11c−CD14−CD3−CD56−CD19+). IP-10 mRNA was measured by quantitative RT-PCR for each cell subset and target gene copy numbers compared to beta actin and GAPDH. This demonstrated detectable IP-10 mRNA expression in mDCs and monocytes, with the latter demonstrating highest levels of expression (Figure 2A). Additionally, monocyte IP-10 mRNA levels were significantly related to plasma IP-10 levels and viral load (Figure 2B). We further tested the cell sources of IP-10 using an in vitro system. PBMCs from HIV-1-negative donors were stimulated with HIV-1NL43 or replication incompetent AT-2 HIV-1. We examined cytokine production by NK cells, B cells, CD4+ T cells, CD8+ T cells, monocytes, mDCs, and pDCs with intracellular cytokine staining. By flow cytometry analysis, monocytes and mDCs had the highest proportion of IP-10-producing cells following stimulation with HIV-1 (Figure 2C). The majority of monocytes and mDCs stained positive for IP-10 with 79% and 91% of monocytes and 62% and 72% of mDCs producing IP-10 following incubation with AT-2 HIV-1 or HIV-1NL43, respectively (Figure 2D, left panel). Geometric mean fluorescence intensity (MFI) values of the IP-10 producing cells indicate highest per cell expression of IP-10 in the monocytes and mDCs, consistent with the ex vivo mRNA analysis above (Figure 2D, right panel). Taken together, these results indicate that IP-10 is produced mainly by innate immune cells in response to HIV-1.
Figure 2. Cellular sources of IP-10.
A) IP-10 mRNA expression in sorted cell populations from individuals with early HIV-1 infection. IP-10 mRNA expression relative to beta actin was determined for each cell population. Box plots show 25th to 75th percentile with whiskers to the minimum and maximum and bisecting line at the median. Data was analyzed with nonparametric analysis of variance using the Friedman’s test with a p<0.0001 across the groups. When controlled for multiple comparisons with the Dunn’s test, the levels differed significantly between T cells and mDCs and monocytes and B cells and Monocytes.
B) Monocyte IP-10 mRNA levels from patient PBMCs are related to both plasma IP-10 level and log viral load. Relative expression of IP-10 mRNA was compared by linear regression to plasma IP-10 levels (left panel) and log viral load (right panel) from matched samples.
C) Assessment of monocyte and mDC production of IP-10 in response to HIV-1. Proportion of monocytes (top row,) and mDCs (second row) that produce IP-10 quantified by intracellular cytokine staining following PBMC stimulation with media (negative, gray shaded histogram) or AT-2 HIV-1 or HIV-1NL43.(blue open histograms). Gating strategies as detailed in the methods section, these plots are representative experiments.
D) In)vitro IP-10 production from different cell types and geometric mean fluorescence intensity (MFI) of IP-10 producing cells. Proportion of monocytes, mDCs, pDCs, B cells, CD4+ T cells, CD8+ T cells, and NK cells that produce IP-10 quantified by intracellular cytokine staining following PBMC stimulation with AT-2 HIV-1 or HIV-1NL43. (left panel). MFI of the IP-10 producing cells from the 3 populations with the most significant production (right panel). Values are reported following background subtraction and represent the average of 5 experiments.
Cytokine and chemokine profile following PBMC stimulation with TLR ligands and HIV-1
IP-10 can be produced in response to type I and II interferons. [27, 28] Our analysis of bulk PBMC mRNA demonstrated a significant relationship between of IP-10 plasma levels and the interferon stimulated gene product ISG15, consistent with an in vivo relationship between type I interferon secretion and IP-10 production (p=0.04, R=0.8 by linear regression). IFNα is produced by pDCs in vitro in response to HIV-1-encoded TLR7/8 ligands, including ssRNAGag1166 [26, 29]. Therefore, we investigated whether IP-10 production is the result of signaling through the TLR 7/8 pathways. Because HIV-1 is known to cause a wide range of cytokine responses in addition to IP-10 in primary infection as shown in Figure 1, we examined a panel of 19 cytokines and chemokines following stimulation with HIV-1 (AT-2 HIV-1, HIV-1NL43) and TLR ligands (ssRNAGag1166, CL097).
When each cytokine was examined individually, multiple cytokines including TNFα, IL-1α, IL-1β, IL-2, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (both p40 and p70), IL-13, and GM-CSF, had significantly different concentrations in the cell culture supernatants depending on the type of stimulation (Supplemental Figure 3) even after accounting for multiple comparisons. When fold change from no stimulation instead of concentration was examined, the same cytokines differed among the stimulants with the exception of IL-1α which no longer significantly differed among the groups (Supplemental Figure 4). In these analyses, IP-10 concentration or fold change was greater following stimulation with HIV-1 as compared to TLR 7/8 ligands but this did not reach statistical significance.
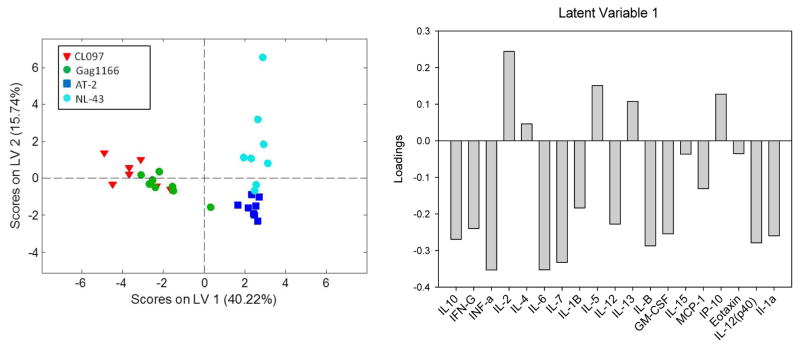
Partial least square discriminant analysis (PLSDA)[23] was employed to evaluate multi-variate differences in cytokine secretion in response to TLR and HIV-1 stimuli. Measures of co-variance were used to identify linear combinations of independent variables (here the 19 measured cytokine/chemokine secretion events), or latent variables, that best differentiate between dependent variables (in this case the stimuli classes CL097, ssRNAGag1166, AT-2 HIV-1, and HIV-1NL43). Every data point is assigned a score, which can be visualized in the latent variable space (Figure 3, left panel). Latent variable loadings (Figure 3, right panel), can then be used to identify cytokine secretion profiles associated with different stimuli. In our case, a model with 2 latent variables was sufficient to capture 55% of the variance in the X block, and provided excellent classification of TLR7/8 versus HIV-1 responses and good classification of AT-2 HIV-1 versus HIV-1NL43 responses (Figure 3, left panel). The overall model performed with average calibration and cross-validation accuracy of 90% and 87%, respectively, with most inaccuracy related to differentiating ssRNAGag1166, CL097 stimuli. The model was more accurate for differentiating the AT-2 response from all other responses with calibration and cross-validation accuracies of 96% and 93%, respectively, and the HIV-1NL43 response with calibration and cross-validation accuracies of 96% and 96% respectively.
Figure 3. Partial least square discriminant analysis of cytokine profiles following stimulation with TLR Ligands and HIV-1.
A PLSDA score plot (left) indicates 19 cytokine secretion measurements differentiate between TLR (CL097 and ssRNAGag1166) and HIV-1(AT-2 HIV-1 or HIV-1NL43) stimuli (separated by LV1) and also between AT-2 HIV-1 and HIV-1 strain NL-43 stimuli (separated by LV2). A loadings plot (right) illustrates that HIV stimuli (positive scores on LV1) are associated with a profile involving IP-10, IL-2, IL-4, IL-13, and IL-5 compared to TLR stimuli. LV2 (not shown) distinguishes between AT-2 and NL-43 HIV strains.
Examination of latent variable loadings revealed cytokine/chemokine secretion patterns that best differentiated responses to TLR and HIV-1 stimuli. Latent variable 1 (LV1) was critical for differentiating the TLR7/8 agonist responses from the HIV-1 responses and accounted for 40.2% of the variability among cytokine concentrations. Positive loadings on LV1 (IL-2, IL-4, IL-5, IL-13, and IP-10 pattern) indicate that IP-10, along with IL-2, IL-4, IL-5, and IL-13 comprise a cytokine/chemokine profile associated with HIV-1-induced cytokine expression, while negative loadings on LV1 indicate a profile associated with TLR agonist-induced expression (Figure 3 right panel). Latent variable 2 (LV2) differentiated HIV-1 replication competent (HIV-1NL43) and replication incompetent (AT-2) HIV-1 strains and accounted for 15.7 % of the variability among cytokine concentrations. Positive loadings and negative loadings on LV2 indicate cytokine secretion profiles associated with HIV-1NL43-and AT-2 -induced expression, respectively. Taken together these data indicate that distinct cytokine profiles are associated with HIV-1 versus TLR7/8 agonist stimulation of PBMCs and that IP-10 production is higher following viral stimulation compared to TLR7/8 stimulation. This model data further suggests that signaling through TLR7/8 alone likely accounts for some but not all of the HIV-1-induced IP-10 responses.
TLR7/9 antagonist abrogates virus-induced IP-10 response
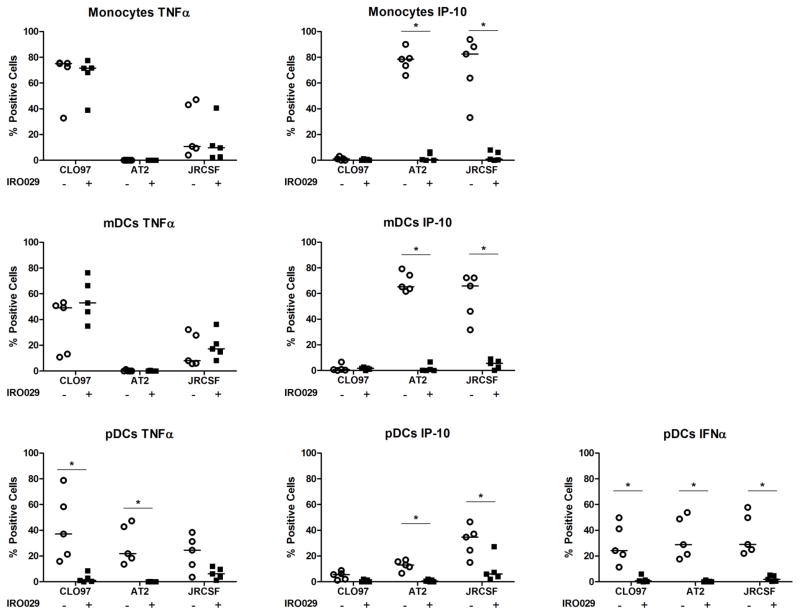
We postulated that IP-10 may be part of a second wave of cytokine responses following viral stimulation based on the kinetics of IP-10 production (data not shown). Because some IP-10 is produced following TLR7/8 stimulation though not to the same extent as following virus stimulation, we investigated whether HIV-1-induced IP-10 production may be downstream of TLR signaling by using a TLR7/9 antagonist and examining intracellular staining of IP-10, TNFα, and IFNα in monocytes, mDCs, and pDCs. Pretreatment of PBMCs with the TLR7/9 antagonist significantly reduced HIV-1-induced IFNα production by pDCs and abolished IP-10 production in monocytes and dendritic cells following incubation with HIV-1 (Figure 4). As expected, the TLR7/9 antagonist did not impact TNFα production by monocytes and mDCs as these responses are in part mediated through TLR8. Therefore, virus-induced IP-10 production may be at least partially a result of secondary activation following IFNα production in response to TLR7 stimulation.
Figure 4. Assessment of monocyte, mDC, and pDC production of TNFα or IP-10 with and without pre-treatment with a TLR7/9 antagonist.
Proportion of monocytes, mDCs, and pDCs that produce TNFα or IP-10 quantified by intracellular cytokine staining in response to CL097, AT-2 HIV-1 or JRCSF with (closed squares) or without (open circles) pre-treatment with a TLR7/9 antagonist. The horizontal lines are the medians. Data are reported following background subtraction. The stated p values were calculated using Mann-Whitney test for non-parametric data. P values were considered significant (*) if less than 0.05.
Discussion
HIV-1 is known to cause easly and persistent immune activation that contributes to disease progression and morbidity. In this study, we examined inflammatory plasma cytokine/chemokine levels in early HIV-1 infection and in the setting of a treatment interruption trial. We found that of the analytes tested, IP-10 was the most significantly related to viral load in both settings, and that mRNA for IP-10 was most highly expressed in monocytes and mDCs. Using an in vitro system, we determined that monocytes and mDCs had the highest proportion of cells producing IP-10 and highest MFI values following stimulation with HIV-1, using both replicating and non-replicating virus. To investigate the pathways involved in HIV-1-induced IP-10 production, we used multivariate analysis to examine cytokine/chemokine production following stimulation with TLR7/8 ligands and HIV-1, and found that TLR7/8 ligands and HIV-1 cause significantly different analyte profiles, with IP-10 secretion more associated with HIV-1-induced profiles, although all stimulants caused some production of IP-10. Finally, TLR7/9 blockade effectively blocked IP-10 production by monocytes and mDCs stimulated with HIV-1, suggesting the IP-10 production requires additional signals induced by IFNα or the JAK/STAT pathway. These data demonstrate that plasma IP-10 level mirrors viral load and that the increased level of IP-10 in HIV-1 viremia may in part be due to signaling through TLR7/9.
IP-10, also known as CXCL10, is an inflammatory chemokine and is a ligand for the receptor CXCR3 (reviewed by Groom and Luster [30]). IP-10 causes directional migration of different immune cells including CD4+ T cells [31] and CD8+ T cells [28, 32]. Our data is in keeping with other recent studies showing HIV-1 viremia induces elevated IP-10 plasma levels. IP-10 has been shown to be elevated in the plasma in primary HIV-1 infection in nearly all patients [21], following treatment interruption [33], in chronic untreated HIV-1 infection [34, 35] and in chronic treated HIV-1 infection [36, 37]. Also, higher pre-ART plasma levels of IP-10 have been associated with the development of immune reconstitution disease following initiation of ART [38–40]. In a follow up study to the SMART trial, IP-10 along with TNFα, IL-10, and IL-6 (by ELISA) were found to be elevated in the treatment interruption arm at month 2 compared to the continuous therapy arm, and increases in plasma TNFα, IL-10 and IP-10 and decreases in IL-17 were correlated with viral loads at month 2 in the unadjusted analysis [33]. We did not find these other cytokines to be significantly correlated with viral load following treatment interruption, possibly due to smaller sample sizes and differences in timepoints studied. Prior studies in acute [34, 41] and chronic [34] HIV-1 infection have also reported correlations between viral load and IP-10 concentrations, and although IP-10 plasma concentration in acute infection was not predictive of viral setpoint at 12 months, it has been reported as a significant predictor of disease progression [42].
In an effort to identify the immune cells producing IP-10, we measured mRNA for IP-10 in sorted cells from individuals in early HIV-1 infection, demonstrating highest expression in the monocyte and mDC populations. We further showed that when PBMCs from healthy donors were stimulated by HIV-1, monocytes and mDCs had the highest proportion of IP-10-producing cells. This is consistent with prior studies demonstrated that IP-10 mRNA increased following HIV-1 stimulation of PBMCs [43] and monocyte-derived DCs and monocyte-derived macrophages from healthy donors [31]. In addition, CD14+ monocytes in patients with HIV-1 have increased expression of the IP-10 gene [44] compared to monocytes from uninfected people; our data extend this finding, demonstrating a relationship between monocyte IP-10 mRNA levels and peripheral viral load. Foley and colleagues also found that IP-10 mRNA was expressed in the T cell region of lymph nodes prior to the initiation of ART and that this expression was eliminated following initiation of ART [31]. In an oral SIV infection model in rhesus macaques, elevated levels of IP-10 mRNA in lymph nodes and peripheral blood cells were furthermore associated with rapid disease progression [45]. Taken together, this suggests that in untreated HIV-1 infection, IP-10 produced by monocytes and mDCs may serve to recruit susceptible T cells to the lymph nodes thereby augmenting infection and contributing to immune activation and dysfunction.
HIV-1 is known to signal through TLR7/8 [26, 46]. However, cytokine/chemokine profiles differed significantly in our study between stimulating with TLR ligands alone and HIV-1 itself, whether replication competent or incompetent HIV-1 was used. This could be due to several factors such as sensing of HIV-1 through other innate immune receptors, differences in secondary signaling cascades, or additional interactions with viral components. IP-10 was one of the cytokines associated with the viral stimulant cytokine profile versus the TLR7/8 ligands profile, though the levels were elevated in the cell culture supernatants after all of the stimulants. Blockade of TLR7/9 effectively eliminated IP-10 expression by monocytes and mDCs suggesting that type I interferons are important elements of this signaling. Rempel and colleagues also found that IFNα-induced gene expression in monocytes in vitro is correlated with IP-10 gene expression in CD14+ monocytes from HIV-1-infected persons with high viral loads[44]. Padovan and colleagues showed that monocyte-derived DCs stimulated with IFNα in vitro produced IP-10 [28]. However, one study has shown that IP-10 induction in myeloid derived macrophages infected with HIV-1 was independent of IFNα [31]. Our work supports the conclusion that HIV-1 derived TLR stimulants are one pathway leading to inflammatory mediators including IFNα that amplify production of IP-10 from monocytes and mDCs.
Importantly, while IP-10 was the analyte most associated with viral load in early HIV-1 infection and after STI, in both cases multivariate relationships were better for prediction of viral load than individual cytokine/chemokine levels. Our multivariate analysis offers new insight in to the network of inflammation associated with HIV-1 viral load and highlights differences between acute infection and STI; cytokines measured in acute HIV infection were more predictive of viral load than cytokines measured after STI. Of note, these models are not adequate for independent prediction of viral load (i.e. given measurements of the 19 cytokines we would not be able to precisely predict viral load). Applying the a similar modeling analysis to the in vitro stimulation assays offered important insight into differences between pure TLR agonists and whole virus stimulation, highlighting the distinct but overlapping inflammatory patterns.
In conclusion, we showed that HIV-1 viral load in early infection and following treatment interruption was associated with increasing plasma levels of IP-10. This chemokine was produced by monocytes and mDCs following stimulation with HIV-1 in vitro, consistent with detectable IP-10 mRNA in these populations ex vivo in HIV-1 infected individuals. High levels of IP-10 are linked to measures of T cell activation and its production may in part be due to Type I IFNs produced as a result of signaling though TLR7/8 by HIV-1.
Supplementary Material
Acknowledgments
We would like to thank Sheila Keating, John Heitman, and Phillip Norris at the Blood Systems Research Institute for their generous assistance with the techniques for cytokine and chemokine measurement. We would like to thank Idera Pharmaceuticals for providing the TLR antagonist. This publication was supported by the NIH/NIAID (R01 AI078784) and resulted (in part) from research supported by the Harvard University Center for AIDS Research (CFAR), an NIH funded program (P30 AI060354), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH NIA, NCCAM, FIC, and OAR.
References
- 1.Bhaskaran K, Mussini C, Antinori A, Walker AS, Dorrucci M, Sabin C, et al. Changes in the incidence and predictors of human immunodeficiency virus-associated dementia in the era of highly active antiretroviral therapy. Ann Neurol. 2008;63:213–221. doi: 10.1002/ana.21225. [DOI] [PubMed] [Google Scholar]
- 2.Crum NF, Riffenburgh RH, Wegner S, Agan BK, Tasker SA, Spooner KM, et al. Comparisons of causes of death and mortality rates among HIV-infected persons: analysis of the pre-, early, and late HAART (highly active antiretroviral therapy) eras. J Acquir Immune Defic Syndr. 2006;41:194–200. doi: 10.1097/01.qai.0000179459.31562.16. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Phillips AN. HIV infection, antiretroviral treatment, ageing, and non-AIDS related morbidity. BMJ. 2009;338:a3172. doi: 10.1136/bmj.a3172. [DOI] [PubMed] [Google Scholar]
- 4.Harrison KM, Song R, Zhang X. Life expectancy after HIV diagnosis based on national HIV surveillance data from 25 states, United States. J Acquir Immune Defic Syndr. 2010;53:124–130. doi: 10.1097/QAI.0b013e3181b563e7. [DOI] [PubMed] [Google Scholar]
- 5.Lewden C, May T, Rosenthal E, Burty C, Bonnet F, Costagliola D, et al. Changes in causes of death among adults infected by HIV between 2000 and 2005: The “Mortalite 2000 and 2005” surveys (ANRS EN19 and Mortavic) J Acquir Immune Defic Syndr. 2008;48:590–598. doi: 10.1097/QAI.0b013e31817efb54. [DOI] [PubMed] [Google Scholar]
- 6.Lohse N, Hansen AB, Pedersen G, Kronborg G, Gerstoft J, Sorensen HT, et al. Survival of persons with and without HIV infection in Denmark, 1995–2005. Ann Intern Med. 2007;146:87–95. doi: 10.7326/0003-4819-146-2-200701160-00003. [DOI] [PubMed] [Google Scholar]
- 7.May M, Gompels M, Delpech V, Porter K, Post F, Johnson M, et al. Impact of late diagnosis and treatment on life expectancy in people with HIV-1: UK Collaborative HIV Cohort (UK CHIC) Study. BMJ. 2011;343:d6016. doi: 10.1136/bmj.d6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palella FJ, Jr, Baker RK, Moorman AC, Chmiel JS, Wood KC, Brooks JT, et al. Mortality in the highly active antiretroviral therapy era: changing causes of death and disease in the HIV outpatient study. J Acquir Immune Defic Syndr. 2006;43:27–34. doi: 10.1097/01.qai.0000233310.90484.16. [DOI] [PubMed] [Google Scholar]
- 9.Phillips AN, Neaton J, Lundgren JD. The role of HIV in serious diseases other than AIDS. AIDS. 2008;22:2409–2418. doi: 10.1097/QAD.0b013e3283174636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt PW. HIV and inflammation: mechanisms and consequences. Curr HIV/AIDS Rep. 2012;9:139–147. doi: 10.1007/s11904-012-0118-8. [DOI] [PubMed] [Google Scholar]
- 11.Giorgi JV, Liu Z, Hultin LE, Cumberland WG, Hennessey K, Detels R. Elevated levels of CD38+ CD8+ T cells in HIV infection add to the prognostic value of low CD4+ T cell levels: results of 6 years of follow-up. The Los Angeles Center, Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr. 1993;6:904–912. [PubMed] [Google Scholar]
- 12.Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 13.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T Cell Activation Is Associated with Lower CD4+ T Cell Gains in Human Immunodeficiency Virus-Infected Patients with Sustained Viral Suppression during Antiretroviral Therapy. Journal of Infectious Diseases. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 14.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt PW, Deeks SG, Rodriguez B, Valdez H, Shade SB, Abrams DI, et al. Continued CD4 cell count increases in HIV-infected adults experiencing 4 years of viral suppression on antiretroviral therapy. AIDS. 2003;17:1907–1915. doi: 10.1097/00002030-200309050-00009. [DOI] [PubMed] [Google Scholar]
- 16.Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis. 2011;204:1217–1226. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Sadr WM, Lundgren JD, Neaton JD, Gordin F, Abrams D, Arduino RC, et al. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. 2006;355:2283–2296. doi: 10.1056/NEJMoa062360. [DOI] [PubMed] [Google Scholar]
- 18.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5:e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulware DR, Hullsiek KH, Puronen CE, Rupert A, Baker JV, French MA, et al. Higher levels of CRP, D-dimer, IL-6, and hyaluronic acid before initiation of antiretroviral therapy (ART) are associated with increased risk of AIDS or death. J Infect Dis. 2011;203:1637–1646. doi: 10.1093/infdis/jir134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann DE, Lichterfeld M, Altfeld M, Addo MM, Johnston MN, Lee PK, et al. Limited durability of viral control following treated acute HIV infection. PLoS medicine. 2004:1. doi: 10.1371/journal.pmed.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, et al. Immune control of HIV-1 after early treatment of acute infection. Nature. 2000;407:523–526. doi: 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- 26.Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larner AC, Petricoin EF, Nakagawa Y, Finbloom DS. IL-4 attenuates the transcriptional activation of both IFN-alpha and IFN-gamma-induced cellular gene expression in monocytes and monocytic cell lines. J Immunol. 1993;150:1944–1950. [PubMed] [Google Scholar]
- 28.Padovan E, Spagnoli GC, Ferrantini M, Heberer M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J Leukoc Biol. 2002;71:669–676. [PubMed] [Google Scholar]
- 29.Chang JJ, Altfeld M. Innate immune activation in primary HIV-1 infection. J Infect Dis. 2010;202 (Suppl 2):S297–301. doi: 10.1086/655657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley JF, Yu CR, Solow R, Yacobucci M, Peden KW, Farber JM. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J Immunol. 2005;174:4892–4900. doi: 10.4049/jimmunol.174.8.4892. [DOI] [PubMed] [Google Scholar]
- 32.Agostini C, Facco M, Siviero M, Carollo D, Galvan S, Cattelan AM, et al. CXC chemokines IP-10 and mig expression and direct migration of pulmonary CD8+/CXCR3+ T cells in the lungs of patients with HIV infection and T-cell alveolitis. Am J Respir Crit Care Med. 2000;162:1466–1473. doi: 10.1164/ajrccm.162.4.2003130. [DOI] [PubMed] [Google Scholar]
- 33.Cozzi-Lepri A, French MA, Baxter J, Okhuysen P, Plana M, Neuhaus J, et al. Resumption of HIV replication is associated with monocyte/macrophage derived cytokine and chemokine changes: results from a large international clinical trial. AIDS. 2011;25:1207–1217. doi: 10.1097/QAD.0b013e3283471f10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keating SM, Golub ET, Nowicki M, Young M, Anastos K, Crystal H, et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of women. AIDS. 2011;25:1823–1832. doi: 10.1097/QAD.0b013e3283489d1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauce D, Larsen M, Fastenackels S, Pauchard M, Ait-Mohand H, Schneider L, et al. HIV disease progression despite suppression of viral replication is associated with exhaustion of lymphopoiesis. Blood. 2011;117:5142–5151. doi: 10.1182/blood-2011-01-331306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamat A, Misra V, Cassol E, Ancuta P, Yan Z, Li C, et al. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PLoS One. 2012;7:e30881. doi: 10.1371/journal.pone.0030881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stylianou E, Aukrust P, Bendtzen K, Muller F, Froland SS. Interferons and interferon (IFN)-inducible protein 10 during highly active anti-retroviral therapy (HAART)-possible immunosuppressive role of IFN-alpha in HIV infection. Clin Exp Immunol. 2000;119:479–485. doi: 10.1046/j.1365-2249.2000.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver BG, Elliott JH, Price P, Phillips M, Saphonn V, Vun MC, et al. Mediators of innate and adaptive immune responses differentially affect immune restoration disease associated with Mycobacterium tuberculosis in HIV patients beginning antiretroviral therapy. J Infect Dis. 2010;202:1728–1737. doi: 10.1086/657082. [DOI] [PubMed] [Google Scholar]
- 39.Crane M, Oliver B, Matthews G, Avihingsanon A, Ubolyam S, Markovska V, et al. Immunopathogenesis of hepatic flare in HIV/hepatitis B virus (HBV)-coinfected individuals after the initiation of HBV-active antiretroviral therapy. J Infect Dis. 2009;199:974–981. doi: 10.1086/597276. [DOI] [PubMed] [Google Scholar]
- 40.Oliver BG, Price P, Wand H, French MA. Increased plasma CXCL10 may be a marker of increased risk of immune restoration disease associated with nonviral pathogens. J Acquir Immune Defic Syndr. 2012;59:e47–49. doi: 10.1097/QAI.0b013e3182427796. [DOI] [PubMed] [Google Scholar]
- 41.Roberts L, Passmore JA, Williamson C, Little F, Bebell LM, Mlisana K, et al. Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS. 2010;24:819–831. doi: 10.1097/QAD.0b013e3283367836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liovat AS, Rey-Cuille MA, Lecuroux C, Jacquelin B, Girault I, Petitjean G, et al. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One. 2012;7:e46143. doi: 10.1371/journal.pone.0046143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wetzel MA, Steele AD, Henderson EE, Rogers TJ. The effect of X4 and R5 HIV-1 on C, C-C, and C-X-C chemokines during the early stages of infection in human PBMCs. Virology. 2002;292:6–15. doi: 10.1006/viro.2001.1249. [DOI] [PubMed] [Google Scholar]
- 44.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS. 2010;24:1415–1423. doi: 10.1097/QAD.0b013e32833ac623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durudas A, Chen HL, Gasper MA, Sundaravaradan V, Milush JM, Silvestri G, et al. Differential innate immune responses to low or high dose oral SIV challenge in Rhesus macaques. Curr HIV Res. 2011;9:276–288. doi: 10.2174/157016211797635928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meier A, Alter G, Frahm N, Sidhu H, Li B, Bagchi A, et al. MyD88-Dependent Immune Activation Mediated by Human Immunodeficiency Virus Type 1-Encoded Toll-Like Receptor Ligands. J Virol. 2007;81:8180–8191. doi: 10.1128/JVI.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.