Abstract
Systems biology is an approach to understanding living systems that focuses on modeling diverse types of high-dimensional interactions to develop a more comprehensive understanding of complex phenotypes manifested by the system. High throughput molecular, cellular, and physiologic profiling of populations is coupled with bioinformatic and computational techniques to identify new functional roles for genes, regulatory elements, and metabolites in the context of the molecular networks that define biological processes associated with system physiology. Given the complexity and heterogeneity of asthma and allergic diseases, a systems biology approach is attractive, as it has the potential to model the myriad connections and interdependencies between genetic predisposition, environmental perturbations, regulatory intermediaries, and molecular sequelae that ultimately lead to diverse disease phenotypes and treatment responses across individuals. The increasing availability of high-throughput technologies has enabled system-wide profiling of the genome, transcriptome, epigenome, microbiome, and metabolome, providing fodder for systems biology approaches to examine asthma and allergy at a more holistic level. In this article, we review the technologies and approaches for system-wide profiling as well as their more recent applications to asthma and allergy. We discuss approaches for integrating multiscale data through network analyses and provide perspective on how individually-captured health profiles will contribute to more accurate systems biology views of asthma and allergy.
Keywords: systems biology, network, asthma, allergy, atopic, genome, transcriptome, epigenome, microbiome, metabolome, individual health profile, big data
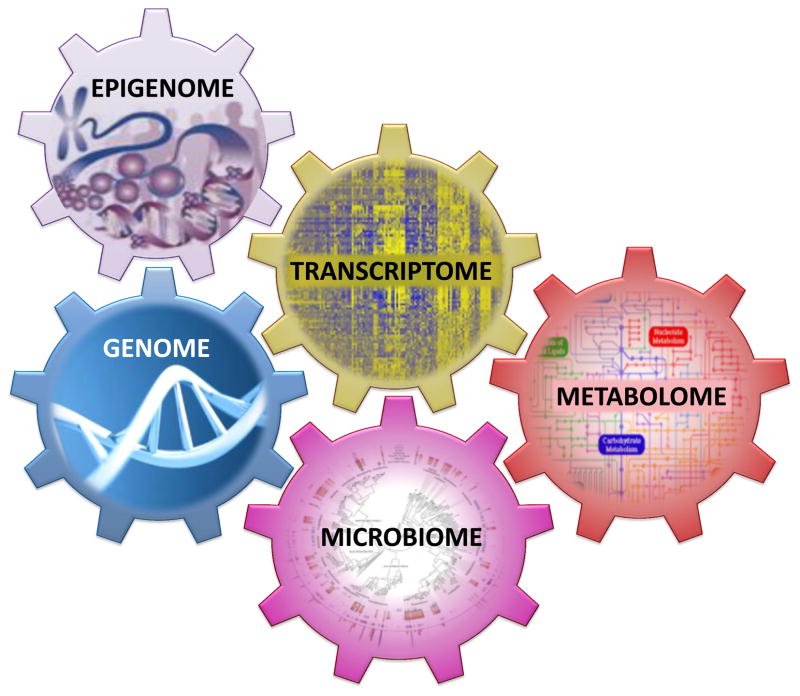
The complexity and heterogeneity of asthma and allergic diseases make both their clinical management and investigation challenging. Innumerable environmental and microbial exposures modulate variable degrees of genetic predisposition, yielding a spectrum of transcriptional and molecular sequelae, disease phenotypes, and treatment responses across individuals. The increasing availability of high-throughput technologies has enabled system-wide profiling of the genome, transcriptome, epigenome, microbiome, and metabolome, providing fodder for systems biology approaches to examine asthma and allergy at a more holistic level (Figure 1). In systems biology, large data sets collected by multiple modalities in populations, ideally with multiple dimensions of data for each individual, are used to generate networks that link phenotypic information to interdependent genetic, regulatory, metabolite, and environmental profiles. The resulting networks are used to predict behavior of the trait and generate novel, biologically relevant information. In this review, we provide an overview of the technologies and approaches for system-wide profiling and review their more recent applications to the study of asthma and allergy. We focus on findings from the past few years and provide perspective on their integration with increasingly available personal health profiles toward a systems view of asthma and allergy.
Figure 1. Relationship between system-wide profiles in asthma and allergy.
Data on the genome, transcriptome, epigenome, microbiome, and metabolome in asthma and allergy have been generated. These types of data are interdependent and interact. Systems biology approaches can be used to model the multidimensional interactions that characterize complex disease.
Genome
Since the completion of the Human Genome Project, oligonucleotide microarrays have enabled the simultaneous genotyping of millions of genetic variants (single nucleotide polymorphisms (SNPs)) scattered across the genome at low cost and with small amounts of starting DNA. The increasing technical and financial accessibility of this technology has led to direct-to-consumer genotyping services, expanding the pool of genotyped populations.1 Genome-wide association studies (GWAS), which examine for associations between genotype and phenotype, enable unbiased identification of genetic loci for asthma and allergy disease risk.2 The National Human Genome Research Institute provides a searchable catalog of published genome-wide association studies at www.genome.gov.3
More GWAS of asthma have been conducted compared to GWAS for other allergic diseases.3 The 17q21 locus4 has been associated with asthma with the greatest reproducibility. 4–7 Encompassing four genes (ORMDL3, GSDMB, ZPBP2, and IKZF3), this locus may affect endoplasmic reticulum mediated Ca2+ homeostasis and protein folding, resulting in the unfolded protein response as an endogenous inducer of inflammation.8 Other asthma susceptibility loci identified through GWAS include genes related to recruitment or activation of inflammatory cells (TSLP5, 9, 10, IL335, 9, IL1RL15, 6, 11), T-cell response and differentiation (HLA genes9, 10, IL2RB9, DENND1B12, IL6R7), and like the 17q21 locus, cell-signaling modulation (PDE4D13, SMAD39, CDHR314). GWAS have also identified genetic variants associated with drug treatment response in asthma.15, 16
Although GWAS have uncovered common variation in numerous genes for asthma, a large proportion of genetic risk for asthma remains unexplained. SNPs on genotyping arrays largely represent common variants that may tag rare variants incompletely. Sequencing can uncover rare variants, as it enables deeper coverage of genes and regions of interest, or of the entire genome in the case of whole genome sequencing. Sequencing is more cost prohibitive than genotyping arrays, however, and therefore practical at this time to implement for targeted gene regions (e.g. 9 candidate asthma genes in 965 subjects 17) or for discovery in limited numbers of individuals (e.g. whole genome sequencing in 16 subjects with well-characterized asthma18).
GWAS of atopic dermatitis (AD) support that mutations in the FLG gene encoding the epidermal structural protein filaggrin are risk factors for AD in Europeans and Asians.19–21 FLG mutations are not commonly found in those of African ancestry, as a study using whole exome sequencing demonstrated.22 In whole exome sequencing, the subset of DNA that encodes proteins (exons) is sequenced. The whole exome represents about 1% of the human genome and has been more frequently sequenced in family-based studies of rare diseases (such as primary immunodeficiencies23) with limited application to asthma and allergy.
Loci near C11orf3021, 24, 5q22.120, and 20q13.3320 have also been identified as AD risk variants by GWAS. A meta-analysis of 16 population-base cohorts identified risk loci for AD near OVOL1 and ACTL9, which are implicated in epidermal proliferation and differentiation; these loci were replicated in an independent study of Japanese subjects21, where additional variants were also identified. GWAS of AD overlapping with other conditions such as psoriasis and asthma have also been performed.25
The C11orf30 locus associated with AD was also found to be associated with allergic rhinitis through GWAS.26 A genome-wide association meta-analysis of Caucasian subjects27 examined the associations between genotype and self-reported allergies using data from a direct-to-consumer genotyping service 1 combined with a more traditionally recruited study cohort. Several loci overlapped with those for asthma, and there was one locus near HLA-DQA1 at 6p21.32 specifically associated with self-reported cat allergy.27 GWAS of ethnically diverse North American subjects identified distinct genome-wide significant loci among Latinos, with integrative genomic analyses showing enrichment of the identified GWAS loci for mitochondrial pathways.28
GWAS have also been performed for allergen sensitization. A European study focusing on grass sensitization specifically found that the AD-associated gene C11orf30 was also associated with grass sensitization, as were HLA-DRB4 and a locus near TMEM232 and SLC25A46.26 A subsequent larger-scale GWAS of food and environmental allergen sensitization combined data from 16 cohorts and identified 10 loci associated with sensitization to any allergen. These loci were scattered across the genome, with 6 previously associated with established roles in the immune system including STAT6, IL1RL1, BCL6, IL2, HLA-DQB1, and HLA-B-MICA.29
GWAS of eosinophilic esophagitis (EoE) showed that common variants at 5q22 encompassing TSLP and WDR36 were associated with the disease.30 Consistent with this, TSLP transcripts were higher in esophageal biopsy specimens from EoE subjects compared to unaffected controls, although this was not observed for WDR36.30 GWAS with an expanded cohort replicated the previously observed association at the 5q22 locus and identified an association at 2p23 spanning CAPN14, which was also specifically expressed in esophagus tissue.31
Transcriptome
The systematic, unbiased quantitative and qualitative characterization of RNA transcripts across the genome is called transcriptomics.32 A disease-relevant tissue is sampled, and then oligonucleotide microarrays or RNA sequencing (RNA-seq) technologies are used to systematically profile the tissue’s RNA transcripts. Transcriptomics offers a complementary and synergistic approach to GWAS for studying disease, as RNA reflects the more dynamic processes at play in a given tissue or tissues that underlie pathophysiology.
The majority of transcriptome profiling to date has been performed using oligonucleotide microarrays. With microarrays, a pool of fluorescently tagged cDNA generated from RNA samples is hybridized against a microarray surface studded with known oligonucleotide DNA sequences.32 The binding of each oligonucleotide probe with its complementary cDNA yields fluorescence that is quantitatively measured and corresponds to the relative abundance of the target RNA in the sample. A disadvantage of oligonucleotide microarrays is that gene expression measurement depends on efficient, unbiased hybridization of target sequence to a predefined, fixed probe set.32 Because selecting probes specific to a given gene across all genes in the transcriptome is not possible to achieve, most gene specific probe sets report on a complicated mixture of background noise, different isoforms of the targeted genes, and cross hybridization of different genes given shared sequence characteristics with the targeted gene.
Recent advances in sequencing technologies have made possible the comprehensive and in-depth characterization of transcriptomes via RNA sequencing (RNA-seq).33 Unlike microarrays, RNA-seq does not depend on predefined sequence-dependent hybridization. Sequence reads of ~30–500 bases in length for current next generation sequencing technologies, and into the thousands of bases for third generation single molecular sequencing technologies, are generated randomly from the target sample. Sequencing-based measures represent individual transcription reads and are free from the constraints of pre-specified probes. RNA-seq offers several advantages over oligonucleotide microarrays, including the ability to quantify (1) more RNA species, (2) novel non-coding variants (3) RNA at baseline, rather than only relative changes across conditions, and (4) a wider range of signal.34, 35 Downstream analysis of RNA-seq data is more involved and requires bioinformatics expertise to align raw sequence reads to a reference genome or to de novo reconstruct transcripts, annotate, and assign gene identity to the sequences. Experimental analysis may involve differential gene expression analysis, network modeling, clustering algorithms, and machine learning approaches.32 Methods for analyzing RNA-seq data are being actively developed.
The majority of gene expression studies to date have been conducted using oligonucleotide microarrays. Within asthma and allergy, these array-based studies have involved relatively small sample sizes, as others have reviewed.32 Here we focus on gene expression studies in asthma and allergy that have applied the more recent technology of RNA-seq.
RNA-seq was recently used to identify new gene transcripts associated with EoE.36 Using esophageal biopsies from 6 healthy controls and 10 patients with active EoE, investigators identified 1607 differentially expressed transcripts, 66% of which had not been previously identified by oligonucleotide microarray profiling.36
In asthma, RNA-seq profiles of lower as well as upper airway biospecimens distinguish subjects with the disease. A comparison of RNA-seq profiles of endobronchial biopsies from steroid-free atopic asthma patients and healthy nonatopic controls demonstrated 46 differentially expressed genes, including SLC26A4, POSTN, and BCL2.37 Gene expression differences were also seen when the investigators examined airway smooth muscle RNA-seq profiles, with a set of 8 differentially expressed genes specifically discriminating asthma from non-asthmatics, irrespective of atopic status.37 RNA-seq profiles of nasal brushings from 10 asthma and 10 control subjects showed that nasal airway gene expression profiles can identify subjects with asthma driven by IL33.38 IL33 was previously implicated in GWAS of asthma.5, 9
Transcriptome profiling in asthma has also been done on tissue obtained peripherally. An examination of the transcriptome of circulating CD19+ B lymphocytes from asthmatic patients with house dust mite allergy showed increased IL4R expression compared to controls, suggesting that B cell transcriptional deregulation is involved in allergic asthma.39
Potential mechanisms underlying glucocorticoid treatment in asthma have been examined using RNA-seq. In a double-blind intervention study of 12 steroid-free patients who underwent endobronchial biopsy before and after prednisolone or placebo treatment, oral prednisolone therapy was observed to alter gene expression in airway smooth muscle. 40 Fifteen genes were differentially expressed before and after treatment, with 2 of these genes (FAM129A and SYNPO2) also associated with methacholine challenge response. 40 Of note, SYNPO2 was associated with total serum IgE in asthmatics in an independent GWAS,41 suggesting roles for this gene in both asthma and allergy. The effect of another glucocorticoid, dexamethasone, on airway smooth muscle response was studied using RNA sequence profiles of airway smooth muscle cell lines treated with dexamethasone.34 316 differentially expressed genes were identified, including known (DUSP1, KLF15, PER1, TSC22D3) as well as less investigated (C7, CCDC69, CRISPLD2) glucocorticoid response genes.34
Single-molecule RNA-seq was recently used to study pediatric AD, where small biopsies yield limited mRNA. The technology was applied to interrogate the transcriptome of 26 children with AD and 10 nonatopic teenage controls.42 2430 differentially expressed genes were identified, and gene ontology-based analysis to these differentially expressed genes highlighted aberrations in extracellular space and lipid metabolism in atopic skin.42 Further effects were seen when subjects were stratified by mutations in FLG,42 the gene frequently implicated in AD in GWAS.
Epigenome
Heritable changes in gene expression can occur without direct alteration of DNA sequence. The study of such epigenetic changes complements genome-wide and transcriptomic approaches for studying disease, as it can characterize DNA sequence-independent modification contributing to transcriptomic variation and downstream phenotype.
Epigenetic changes may include DNA methylation, DNA hydroxymethylation, histone modification, and microRNAs. DNA methylation refers to the covalent addition of a methyl group (-CH3) predominantly to a cytosine (C5) in a CpG dinucleotide. Occurring less commonly, DNA hydroxymethylation refers to the addition of 5-hydroxymethylcytosine (5-hmC) to C5 and has primarily been studied for its role in disordered cell function in neurological disorders, stem cell biology, and cancer. Hypermethylation at gene promoters is generally associated with downregulation of the gene, although DNA methylation may also play other roles. 42 More recently, epigenetic modifications in RNA have been identified and demonstrated to play regulatory roles, although protocols that enable routine detection of such modifications are still in the early stages of development.43 Other types of epigenetic modifications include the acetylation, methylation, phosphorylation, and/or ubiquitination of the tails of core histones. These histone modifications are associated with activation and repression of transcription. Another type of epigenetic regulation involves microRNAs, non-coding, single-stranded RNAs of about 22 nucleotides that function in post-transcriptional regulation of gene expression. They bind to complementary sequences in target mRNAs, leading to gene silencing by cleaving mRNA, shortening mRNA polyA tails, and reducing the efficiency of mRNA translation.44 The examination of epigenetic modifications in asthma and allergy has been of interest to investigators, as epigenetics may mediate links between environmental exposures and disease phenotype that have been epidemiologically evident. Most studies to date have used epigenetic approaches, targeting modifications to candidate genes and regions of interest in asthma and allergy. When DNA methylation, DNA hydroxymethylation, histone modification, or microRNAs are examined on a genome-wide basis, the term epigenomics is used.
Microarrays have been frequently used in epigenomics, with several platforms and protocols available for detecting 5-methylcytosine (5-mC), the most common type of DNA methylation.45 Array platforms have also been used to examine genome-wide histone modifications by chromatin immunoprecipitation (ChIP) followed by hybridization on microarrays (ChIP-chip). Analogous miRNAs platforms are also available (miRNA-chip).45 As in transcriptomics, next-generation sequencing has expanded epigenome assessment.46 These technologies have been applied to profiling histone marks (ChIP followed by high throughput sequencing (ChIP-seq)), microRNAs (miRNA-seq), open chromatin areas of the genome (FAIRE-seq), and spatial chromatin organization (3C-seq). Because bisulfite-converted DNA sequencing on the genome scale is expensive, most methylome profiling is still done on array platforms. 45 Emerging third generation sequencing technologies promise to provide a more comprehensive characterization of the epigenome, as they enable identification of all known chemical modifications that can occur in nucleic acid sequences at single molecule, single nucleotide resolution.47, 48
Recent studies on the epigenomics of asthma have examined several aspects of the disease and built upon epigenetic findings previously reviewed.45 Methylation array-based studies of bronchial brushings49 and mucosal biopsies50 have identified methylation marks associated with asthma and atopic status versus normal control. A recent study of genome-wide histone modification profiles in naïve, Th1, and Th2 cells from the peripheral blood of healthy and asthmatic individuals identified enhancers associated with Th cell subsets that differed in histone H3 Lys4 dimethyl (H3K4Me2) enrichment depending on asthma status.51 In a study of potential mechanisms for airway smooth muscle dysregulation in asthma, investigators observed that dysregulated trimethylation of histone H3 at lysine 9 (H3K9me3) at the vascular endothelial growth factor (VEGF) promoter may influence VEGF hypersecretion.52
To examine whether atopic risk due to maternal asthma and atopy is conveyed by epigenetic changes, research groups have applied genome-wide methylation profiling in murine and population-based studies. Splenic dendritic cells from mouse pups whose mothers had been ova-sensitized prenatally demonstrated more genome-wide DNA methylation changes at birth compared to controls.53 Subsequent allergen sensitization resulted in transcriptional changes at CpG loci, supporting that maternal transmission of atopic risk may occur through epigenetic means.53 Consistent with this, a separate human study showed that maternal asthma during pregnancy was associated with differential methylation at 70 CpG loci corresponding to 67 genes in infants’ peripheral blood DNA.54 The infants’ peripheral blood also showed that methylation at several loci was associated with intermediate phenotypes including maternal blood eosinophil level, maternal exhaled nitric oxide (eNO), maternal serum total IgE, and maternal inhaled corticosteroid use.54
Other studies have examined the epigenome as a potential mechanistic bridge between environmental exposures and the development of asthma. Prenatal smoke exposure was associated with methylation at 19 CpG loci in a study of 527 children ages 5–12 years with asthma,55 supporting that prenatal tobacco exposure may be associated with epigenetic changes that persist into childhood. 55 Human rhinovirus infection and gene expression response to infection were found to be associated with a methylation locus at SNORA12 in an in vitro study of nasal epithelial cells collected from asthmatics and healthy controls.56
Epigenetic changes were found to be specific to lesional epidermis in an epigenome-wide study of AD.57 AD-affected and healthy individuals were examined for methylation changes in DNA derived from epidermis (AD lesional, AD non-lesional, healthy non-lesional), whole blood, T cells, and B cells. Altered methylation and expression of genes important for keratinocyte differentiation, proliferation, and innate immune response were observed in lesional epidermis of AD patients but not in the other tissues examined.57 Null findings for peripheral blood-based epigenetic changes in AD patients were also reported in a genome-wide DNA methylation study of naïve CD4+ T cells from patients with psoriasis and AD.58 A methylation array-based study of cord blood DNA, however, found that prenatal smoke exposure was associated with methylation in TSLP,59 a gene previously associated with asthma5, 9, 10, AD60, allergen sensitization27, and EoE30 in GWAS and transcriptional studies. In keratinocytes from patients with AD, miRNA array profiles showed increased levels of miR-146a associated with inflammatory processes triggered by IFN-y and N-kB activation.61 Non-genome-wide studies of AD epigenetics have targeted loci such as the high-affinity IgE receptor gamma subunit (FCER1G) promoter 62, FLG63, TSLP64, and FOXP365.
In allergic rhinitis, genome-wide methylation profiling has been applied to phenotype patients. 66 Using DNA methylation data from peripheral blood CD4+ T cells of untreated allergic rhinitis patients and healthy controls, investigators used the aggregate methylation data to carry out principal component analysis resulting in the identification of principal components that could distinguish affected patients.66 Interestingly, they found that these methylation-based principal components more accurately predicted allergic rhinitis status than gene expression.
The association between genome-wide DNA methylation profiles from CD4+ T cells was also examined in food allergy. In a hypothesis-generating study of 12 children with food allergy and 12 controls, 179 and 136 probes were found to be differentially methylated at age 12 months and birth, respectively. 67 Pathway analysis of these genes suggested dysregulation of DNA methylation at MAPK signaling-associated genes.67
In EoE, miRNA profiling of esophageal biopsies identified several miRNA that were differentially expressed in EoE, with results supporting interaction between miR-21 and miR-223 in the polarization of adaptive immunity and regulation of eosinophilia.68 MiRNA profiling of esophageal tissue from EoE patients before and after treatment with steroids has also been performed.69
Microbiome
With an estimated composition of 100 trillion cells,70 our commensal microbiota outnumber us by at least 10-fold. Our immune system must contend with these symbionts through its innate and adaptive arms, and the same immune system also mediates asthma and allergic diseases. It is therefore compelling to consider that these complex communities of bacteria, viruses, fungi, and other commensal species play roles in asthma and allergy. Associations between culturable microbiota, asthma, and atopy have been studied71–73, but the recent availability of culture-independent tools has had a dramatic impact in the field. Specifically, systematic profiling of the collective microbial community is now possible by 16S rRNA and shotgun metagenomic sequencing of aggregate microbial genomes (aka microbiome), revealing far more microbiota and phylogenetic relationships than previously detectable. The bacterial 16S ribosomal RNA gene is a highly conserved locus of the bacterial genome, and DNA sequence differences within the hypervariable regions of the 16S rRNA gene allow for identification of bacterial species by reference to existing 16S rRNA gene sequence databases. 16S rRNA sequencing of the total genomic content of samples allows for precise identification of bacteria without being tied to the fastidious culture conditions of individual bacteria, and the fact that the majority of bacteria cannot be cultured. Metagenomic sequencing allows for the total DNA of the ecosystem to be sequenced, allowing for the inventorying of all microbiota in addition to bacteria. The introduction of sample barcoding, the decreasing cost of next-generation sequencing technologies, improvements in bioinformatics tools, and online databases have allowed researchers to more comprehensively examine microbes living in and on the human body.74 It is likely that the bacterial, viral, and fungal biomes interact with the human genome in complex ways to influence asthma and allergy. System biology approaches have been used to examine relationships between microbiota and host genomic profiles in other disease areas such as inflammatory bowel disease.75, 76 The role of the microbiome and its interaction with the host genome in asthma and allergic diseases can be examined via systems biology approaches.
Bacterial 16S rRNA sequenced from airway brushings support that the healthy bronchial tree is not sterile, in contrast to traditional medical teaching.77 The healthy bronchial tree contains a mean of 2,000 bacterial genomes per cm2, with Bacteroidetes predominating.78 Subjects with asthma have higher populations of Proteobacteria.78 The diversity of the bronchial epithelial microbiome appears to be greater in those with asthma and correlates with measures of bronchial hyperreactivity, as seen in a pilot study where greater representation of Proteobacteria was also observed among asthmatics.79 Greater bacterial diversity and high Proteobacteria representation were also observed when comparing 16S rRNA sequenced from induced sputum samples from asthmatic vs. non-asthmatic subjects.80 Microbiome examination of bronchial alveolar lavage samples from subjects with corticosteroid sensitive vs. corticosteroid-resistant did not show differences in diversity and community composition at the phylum level, although genus-level examination showed expansion of gram-negative bacteria in corticosteroid-resistant asthma.81 Bacteria from these genuses were experimentally observed to upregulate transforming growth factor-b-associated kinase and MAPK in cultured peripheral blood monocytes leading to reduced responses to corticosteroid.81
As the gut microbiome may influence immune responses that mediate allergy and asthma,82 researchers have also examined for associations between gut microbiota and atopic outcomes. In contrast to the associations between high diversity and asthma observed with airway microbiome studies, low total diversity of gut microbiota during infancy has been associated with asthma in children at 7 years of age83 and with AD during the first 18 months of life.84–86 In an open trial of potential treatment for reduced gut bacterial diversity in AD, mothers were supplemented with Bifidobacterium starting at 1 month before delivery followed by Bifidobacterium supplementation to the infant until 6 months of age. 87 Compared to unsupplemented controls, the supplemented infants were found to have reduced risk of developing AD during the first 18 months of life, and pyrosequencing analyses showed differences in gut microbiota in those who developed AD.87
16S-rRNA-sequencing-based analysis of the skin microbiome in AD has revealed its own findings. Temporal shifts in microbiota are seen in the lesional skin of AD patients at baseline, during disease flare, and following treatment.88 Specifically, Staphylococcus aureus and the skin commensal Staphylococcus epidermidis were increased with clinical disease activity, whereas Streptococcus, Propionibacterium, and Corynebacterium species were increased following therapy.88 Interestingly, skin at the popliteal fossae and elbow folds had lower diversity compared to the same areas in healthy controls.88
Initial studies of gut microbiota in food allergy support a potential disease-modifying role for the gut microbiome. Mice induced to have a food allergic phenotype through IL4raF709 mutation and oral sensitization to chicken egg ovalbumin exhibited a distinct gut microbiota compared to control animals.89 Interestingly, the food allergy phenotype could be transmitted by transferring the gut microbiota via fecal pellets from the affected mice to wildtype germ-free mice.89 In humans, a small study examining 16S rRNA gene V1-V3 hypervariable regions from the feces of food allergic infants (IgE mediated and non-IgE mediated) and controls showed reduced proportions of Bacteroidetes, Proteobacteria, and Actinobacteria in the food allergic children. 88 The study was limited by sample size and heterogeneity of the food allergy phenotype, as the case subjects had different food allergies and factors known to affect gut microbiome (feeding patterns and modes of delivery) were not factored into the analyses.
Metabolome
In metabolomics, the collection of low molecular weight compounds in biological samples is systematically assessed, identified, and quantified through pattern recognition methods. Metabolomic profiling can provide a snapshot of active physiology and has been used as a tool for biomarker discovery. Metabolite levels are typically measured by nuclear magnetic resonance (NMR) or liquid chromatography mass spectrometry (LC-MS). NMR can quantify limited numbers of molecules (hundreds), while LC-MS can analyze several hundreds to thousands.90 NMR analysis is not destructive, allowing for sample analysis in vivo more than once. NMR is not as sensitive as LC-MS, although LC-MS results tend to be less reproducible, more platform dependent, and subject to variability.91 The two methods are currently viewed as complementary.
Recent metabolomic studies of asthma have been conducted on exhaled breath condensate92–96, urine97–99, serum100, 101, and plasma.102 These studies have shown that metabolites from several pathways can be used to discriminate asthmatics from controls, asthma exacerbation from stable asthma, and severe asthma from nonsevere asthma. The implicated metabolites are numerous, including phosphatidylcholines, alkanes, aldehydes, ammonium ions, retinoic acid, adenosine, vitamin D, ercalcitriol, and urocanic acid, among others. Some have used metabolomics to identify possible sources of inflammation98, while others have sought to characterize metabolite profiles associated with subphenotypes of asthma, such as neutrophilic and eosinophilic asthma.92 Limitations of these studies include their small size and differences in baseline characteristics (such as medication use, race, and diet) between comparison groups.103
Although metabolomic strategies have been implemented in several studies of asthma, they have been less frequently applied to allergic diseases. A small, exploratory study on the metabolomics of AD measured urinary metabolite profiles by NMR in 32 children and found that these differed between children with and without AD.104
Building networks for asthma and allergy
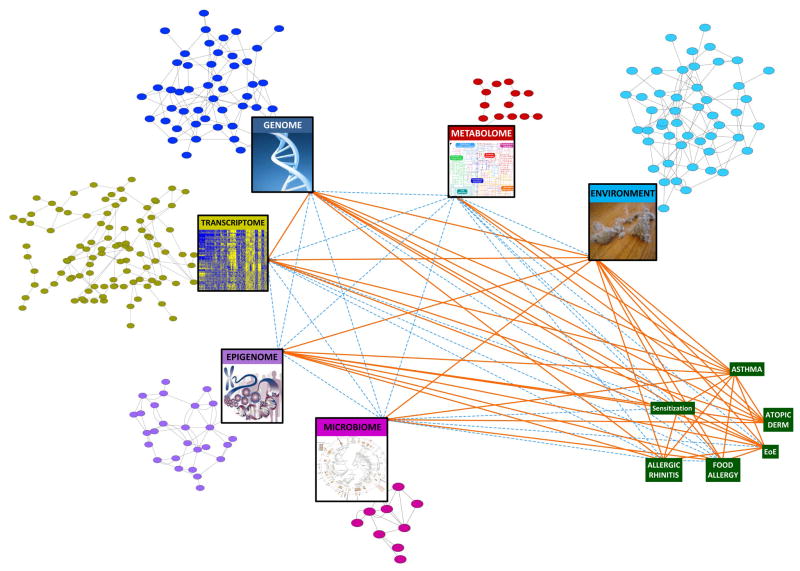
Data generated through genome-wide association, transcriptome, epigenome, microbiome, and metabolome studies have advanced our understanding of asthma and allergic diseases. As complex and heterogeneous diseases, however, it is unlikely that a single biomarker or even single high throughput profiling modality can capture the interdependent dynamics of the molecular networks involved in these diseases (Figure 2). Integrating these types of system-wide data is critical if we are to construct models that are predictive of complex biological interactions and systems, a necessary step to developing a more complete understanding of disease. Generating multiple dimensions of data on each individual would lend even more power to this approach. To make accurate disease predictions, we need a systems biology approach to create the networks capable of representing causal relationships among molecular features within a given cell or tissue type as well as between different tissues.105
Figure 2. Moving toward a systems biology view of asthma and allergic diseases.
The colored circular nodes represent genetic, regulatory, metabolite, and environmental entities associated with asthma and allergic disorders. Their identification and potential connectivity can be assessed by the profiling represented in the large rectangular nodes. Green rectangular nodes represent diseases of interest. Orange lines (edges) denote evidence for associations between the implicated nodes in asthma and allergy, many of which are reviewed in this article. Dashed blue edges denote relationships that are currently less well-studied. Examination of the network’s collective nodes and edges, or a substantial subset thereof, would move us toward a systems biology understanding of asthma and allergy.
We have made steps toward drawing the connections to map biological interactions relevant to asthma and allergy (Figure 2). While GWAS identify genetic susceptibility loci, the biologic relevance of these variants is sometimes not clear. Integrating genotype data with transcriptional profiles can prioritize causal variation,28–30 as genetic variants that are associated with RNA transcript change in disease-relevant tissue are more likely to be disease relevant than those that do not affect transcript levels. Expression quantitative trait loci (eQTL) and expression single nucleotide polymorphism (eSNP) analyses are statistical techniques to match variation in mRNA expression levels with DNA variation at specific genomic loci. Assessing which eQTLs and eSNPs are likely to be regulatory can be done by integrating epigenetic profiles (e.g. FAIRE-PCR/Seq, ChIP PCR/Seq), as has been done in asthma for novel susceptibility loci.106
We can build upon the level of data integration thus far toward systems biology network models that can capture the connectivity and interactions between the genome, transcriptome, epigenome, microbiome, and metabolome in asthma and allergy. Systems biology is an approach to understanding living systems that focuses on modeling diverse types of high-dimensional complex interactions to develop a more comprehensive understanding of biology at multiple scales: molecular, cellular, tissue, organ, organism, and community. Comprehensive, multi-scale profiles of populations such as those gained from genomic technologies are coupled with bioinformatic and computational techniques to build network models. Networks provide a framework for exploring the context in which genes, gene products, metabolites, and other variables operate. They are mathematical models comprised of nodes and edges that model the connectivity and complex interactions between variables that associate with one another. Nodes in a network typically represent genes, gene products, metabolites, other molecular entities, or higher order physiological features such as clinical features and disease state. Edges (or links) between any two nodes indicate a relationship between the two entities. By building networks, we create maps of potential connections among components that can suggest new functional roles for specific genes, regulatory elements, and metabolites.107 Networks can place genes or molecules of interest in the context of biologic pathways and molecular interactions, and the best nodes from a network can be used to assess the disease, assess disease risk or progression, and serve as targets for therapeutic intervention.
Systems biology approaches such as probabilistic causal network analysis can move us beyond association to inferring causal directionality. In such networks, Bayesian methods are used to infer causal relationships between molecular interactions by considering thousands of molecular or clinical variables and using statistical techniques to select a consensus model that best fits the data and identifies directionality of relationships between the variables.108 Mapping the connectivity structure of networks in disease helps us understand how biological processes are defined at the molecular level, how they are disrupted in disease, and how we can assess disease risk and potentially intervene.108 Causal network approaches have been applied to several disease areas outside of asthma and allergy, resulting in new genetic associations and mechanistic understandings for metabolic disease109, obesity110, and Alzheimer’s disease111. The implementation of such systems biology approaches in asthma and allergy is our next challenge.
Large-scale data management and analysis
The success of systems biology approaches in asthma and allergy research will depend on our ability to properly manage and interpret large-scale, multi-dimensional data sets. Individual laboratories are currently able to generate terabyte and petabyte scales of data at reasonable cost.112 However, multiple steps are involved in processing and integrating such high-dimensional datasets to yield meaningful results, including data transfer, access control and management, standardization of data formats, and accurate modeling of biological systems through data integration.112 Computational and bioinformatics infrastructure are necessary for each of these steps. Fields that regularly handle big data (such as climatology) and IT companies such as Amazon, Google, Facebook, and Microsoft point to computational solutions for big data management and analysis, including high performance computing systems, cloud-based computing, and high-speed, low-cost heterogeneous computation environments.112 For example, Amazon has led the way with cloud computing (EC2), high performance data storage (S3), and archival storage (Glacier). Large-scale efforts include the cloud-based clinical operating system Nanthealth, the data science and software centers Craig Venter is building to process a million human genomes, and BGI’s cloud-based bioinformatics platforms. Although institutions are increasingly building and bolstering their computational and bioinformatics infrastructures to reap the benefits of big data, ensuring individual investigators’ access to such resources is one of the challenges facing systems biology approaches in allergy and immunology (Table 1). To this end, companies such as Genalice are making next generation sequence data storage and analysis accessible to investigators without supercomputers by creating software to process and store big data on general purpose hardware. Continuing toward solutions to the computational and bioinformatics challenges facing systems biology will propel research on asthma and allergy forward, as systems biology can provide the global, quantitative understanding of high-dimensional, complex interactions in asthma and allergy that traditional reductionist approaches cannot capture.
Table 1.
Challenges and solutions for systems biology approaches to asthma and allergy.
| Challenges | Solutions |
|---|---|
| Access to system-wide profiling technologies | Core facilities |
| Cost of system-wide profiling | Costs decreasing with technological advances |
| Data transfer and management | High performance computing systems Cloud-based computing High-speed, low-cost heterogeneous computation environments Software innovations |
| Data quality and control | Standardization Data-sharing |
| Accurate modeling of biological systems through data integration | Development of tools and software platforms. Iterative integration by investigators. |
| Perceived as “fishing” and riskier than traditional reductionist approaches. | Education. Novel causal relationships elucidated by systems biology. |
The next wave in big data
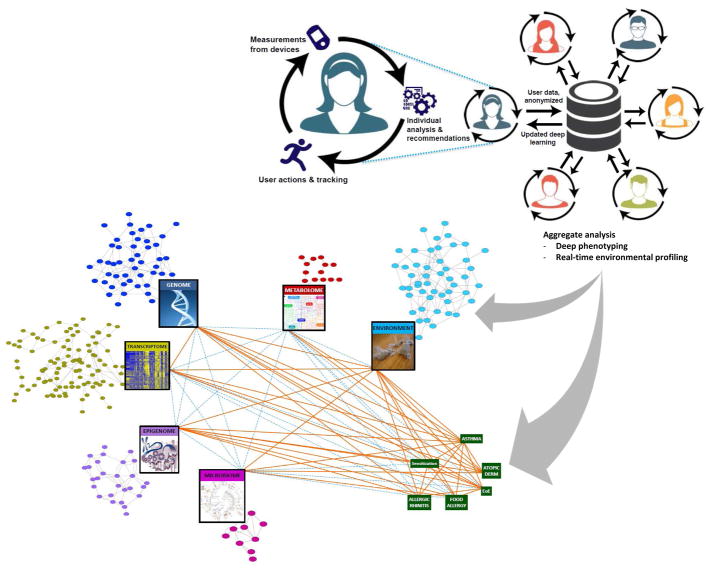
Personal health profiles captured by individuals themselves are the next wave of data that will contribute to a systems biology view of asthma and allergy (Figure 3). Although system-wide profiling technologies have yielded more data than ever on the genome, transcriptome, epigenome, microbiome, and metabolome in asthma and allergy, these data represent snapshots in time from individuals at patient or research subject visits. Yet the intrinsic and extrinsic variables shaping asthma and allergy pathobiology affect individuals continuously in real-time. In order for the molecular networks to inform more maximally on disease conditions, richer sets of phenotypes that enable a more direct linkage between these networks and the pathophysiological features of disease are needed. To gain the deeper phenotype and environmental data that will complement the system-wide profiles we are building in asthma and allergy, we should seek to reap the rich data that individuals are now enabled to regularly capture from wearable devices and mobile health apps.
Figure 3. Personal health profiles captured by individuals themselves will add the next big data dimension to understanding asthma and allergic diseases.
Data that individuals gather passively through wearable devices and mobile apps for personal health will enable deeper phenotyping and real-time profiling of environmental exposures. Combined with continued advances in system-wide profiling, network models of asthma and allergic disease will become more accurate for disease prediction and therapeutics.
Health and environmental data that directly impact an individual’s asthma and allergy control can be captured essentially passively with wearable devices and health apps. Already, consumer sensor technologies allow for measurement of activity, blood-pressure, glucose, heart rate, EKG, EEG, and EMG tracings, ultrasound images, and medication adherence through smart phones and wearable devices.113 Such individual-driven monitoring can be expanded to include ongoing measures of lung function, symptom scores, diet, and relevant environmental exposures such as ambient pollutant and allergen concentrations. In fact, today there are Bluetooth enabled peak flow meters and spirometry devices providing a mobile health app, and Bluetooth enabled inhalers that let asthma patients better monitor their condition. The environmental context for an individual’s health could be additionally complemented by freely available data relevant to the person’s GPS-identified location, such as ambient ozone concentration, weather conditions, and traffic patterns. The path for including such real-time data in healthcare is informed by existing models, from the Oracle Team for the America’s Cup to the Honda Indy Race Team to several NBA and NFL sport teams, where data from hundreds of sensors are already integrated to advise on all aspects of performance, even enabling fine tuning in real time to maximize performance. Although this level of integration is not yet commonplace in asthma and allergy, or even in medicine overall, the increasing role of data in our lives makes inevitable the incorporation of individually-generated data into systems biology approaches. Individuals are already leading the data-gathering effort through consumer health wearables and mobile platforms, and we should be ready to capitalize on the availability of this additional dimension of big data.
The deeper phenotype and environmental profiles captured by individuals themselves, coupled with ongoing advances and wider implementation in system-wide profiling (such as single-cell sequencing), will increase our power to build networks that accurately model the complex connectivity and interactions that lead to asthma and allergic disease in real-world settings. Such systems biology approaches will elucidate connections that will uncover relevant biological pathways and relationships among physiologic, biologic, and environmental processes in asthma and allergy that would otherwise remain hidden. Focusing on these networks and connections will enable improved disease assessment, risk monitoring, and therapeutic intervention.
Abbreviations
- 3C
chromosome conformation capture
- 5-hmC
5-hydroxymethylcytosine
- 5-mC
5-methylcytosine
- AD
atopic dermatitis
- ChIP
chromatin immunoprecipitation
- CpG
cytosine-phosphate-guanine
- EoE
eosinophilic esophagitis
- eQTL
expression quantitative trait loci
- eNO
exhaled nitric oxide
- eSNP
expression single nucleotide polymorphism
- FAIRE
formaldehyde assisted isolation of regulatory elements
- GWAS
genome-wide association study
- LC-MS
liquid chromatography mass spectrometry
- miRNA
microRNA
- NMR
nuclear magnetic resonance
- RNA-seq
RNA sequencing
- rRNA
ribosomal RNA
- seq
sequencing
- SNP
single nucleotide polymorphism
GLOSSARY
- SYSTEMS BIOLOGY
An approach to understanding living systems that focuses on modeling diverse types of high-dimensional interactions to develop a more comprehensive understanding of complex phenotypes manifested by the system. High throughput molecular, cellular, and physiologic profiling of populations is coupled with bioinformatic and computational techniques to identify new functional roles for genes, regulatory elements, and metabolites in the context of the molecular networks that define biological processes associated with system physiology
- GENOME
The complete set of genetic information for an organism, including genes and non-coding sequences. The genome contains the information needed to build and maintain the organism. The human genome is over 3 billion DNA base pairs
- GENOME-WIDE ASSOCIATION STUDY (GWAS)
A study in which DNA variants across the genome are simultaneously analyzed for association with a trait of interest
- MICROARRAY
A technology used to study genotype, gene expression, methylation, miRNAs, and chromatin marks of thousands of genes at once. Known specific nucleic acid sequences (probes) are studded on a glass slide. A sample containing cDNA or cRNA is then placed. Complementary base pairing between the sample and the sequences on the chip yield fluorescence that is measured and correlates with the amount of nucleic acid present
- WHOLE GENOME SEQUENCING
The process of determining the complete sequence of an organism’s chromosomal and mitochondrial DNA at a single time
- WHOLE EXOME SEQUENCING
The process of determining the sequence of an organism’s protein-encoding DNA (exons). The whole exome represents about 1% of the human genome
- TRANSCRIPTOME
The set of RNA molecules produced in one or a population of cells. The transcriptome can be profiled by oligonucleotide microarrays and RNA sequencing (RNA-seq)
- EPIGENETICS
Modifications to DNA that affect gene expression and occur without direct alteration of DNA sequence. Epigenetic changes may include DNA methylation, DNA hydroxymethylation, histone modification, and microRNAs. These changes can be heritable but are also reversible because the genetic code remains unchanged
- HUMAN MICROBIOME
The collection of commensal, symbiotic, and pathogenic microorganisms and their genomes that are found in the human body
- METABOLOMICS
High-throughput characterization of metabolites found in an organism. Nuclear magnetic resonance (NMR) or liquid chromatography mass spectrometry (LC-MS) are typically used. Metabolites can be specific to certain body fluids, such as urine, serum, plasma, and exhaled breath condensate
- NETWORK
A framework for exploring the context in which genes, gene products, metabolites, and other variables operate. Networks are mathematical models comprised of nodes and edges that model the connectivity and complex interactions between variables in a system that associate with one another. Nodes in a network typically represent genes, gene products, metabolites, or other important molecular entities. Edges (or links) between any two nodes indicate a relationship between the two entities
- WEARABLE DEVICE
Clothing and accessories incorporating sensors and electronic technologies to convey data. Health-related wearable devices currently on the market include bracelets and watches that monitor activity, nutrition, and sleep, tattoos that monitor pH and lactate content in sweat, diapers with urinalysis monitors, ingestible sensors that track medication adherence and response, and clothing that measures EKG, EEG, and EMG signals
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathias RA. Introduction to genetics and genomics in asthma: genetics of asthma. Adv Exp Med Biol. 2014;795:125–55. doi: 10.1007/978-1-4614-8603-9_9. [DOI] [PubMed] [Google Scholar]
- 3.Hindorff LAMJEBI, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA European Bioinformatics Institute. [Accessed July 8, 2014];A Catalog of Published Genome-Wide Association Studies. 2014 Available at: www.genome.gov/gwastudies.
- 4.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 5.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–8. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13. 5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 9.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 12.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 13.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–5. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 15.Dahlin A, Tantisira KG. Integrative systems biology approaches in asthma pharmacogenomics. Pharmacogenomics. 2012;13:1387–404. doi: 10.2217/pgs.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake KA, Torgerson DG, Gignoux CR, Galanter JM, Roth LA, Huntsman S, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133:370–8. doi: 10.1016/j.jaci.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torgerson DG, Capurso D, Mathias RA, Graves PE, Hernandez RD, Beaty TH, et al. Resequencing candidate genes implicates rare variants in asthma susceptibility. Am J Hum Genet. 2012;90:273–81. doi: 10.1016/j.ajhg.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell CD, Mohajeri K, Malig M, Hormozdiari F, Nelson B, Du G, et al. Whole-genome sequencing of individuals from a founder population identifies candidate genes for asthma. PLoS One. 2014;9:e104396. doi: 10.1371/journal.pone.0104396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2012;44:187–92. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 21.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 22.Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in african-american children with atopic dermatitis. J Invest Dermatol. 2014;134:2272–4. doi: 10.1038/jid.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt C, Geha RS, Chou J. Gene hunting in the genomic era: Approaches to diagnostic dilemmas in patients with primary immunodeficiencies. J Allergy Clin Immunol. 2014;134:262–8. doi: 10.1016/j.jaci.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 25.Weidinger S, Willis-Owen SA, Kamatani Y, Baurecht H, Morar N, Liang L, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum Mol Genet. 2013;22:4841–56. doi: 10.1093/hmg/ddt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 27.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–11. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunyavanich S, Schadt EE, Himes BE, Lasky-Su J, Qiu W, Lazarus R, et al. Integrated Genome-wide Association, Coexpression Network, and Expression Single Nucleotide Polymorphism Analysis Identifies Novel Pathway in Allergic Rhinitis. BMC Med Genomics. 2014;7:48. doi: 10.1186/1755-8794-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–6. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sordillo J, Raby BA. Gene expression profiling in asthma. Adv Exp Med Biol. 2014;795:157–81. doi: 10.1007/978-1-4614-8603-9_10. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, et al. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9:e99625. doi: 10.1371/journal.pone.0099625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014 doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yick CY, Zwinderman AH, Kunst PW, Grunberg K, Mauad T, Dijkhuis A, et al. Transcriptome sequencing (RNA-Seq) of human endobronchial biopsies: asthma versus controls. Eur Respir J. 2013;42:662–70. doi: 10.1183/09031936.00115412. [DOI] [PubMed] [Google Scholar]
- 38.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. Journal of Allergy and Clinical Immunology. 2014;133:670–8. e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascual M, Roa S, Garcia-Sanchez A, Sanz C, Hernandez-Hernandez L, Greally JM, et al. Genome698 wide expression profiling of B lymphocytes reveals IL4R increase in allergic asthma. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Yick CY, Zwinderman AH, Kunst PW, Grunberg K, Mauad T, Fluiter K, et al. Glucocorticoid-induced changes in gene expression of airway smooth muscle in patients with asthma. Am J Respir Crit Care Med. 2013;187:1076–84. doi: 10.1164/rccm.201210-1886OC. [DOI] [PubMed] [Google Scholar]
- 41.Kim JH, Cheong HS, Park JS, Jang AS, Uh ST, Kim YH, et al. A genome-wide association study of total serum and mite-specific IgEs in asthma patients. PLoS One. 2013;8:e71958. doi: 10.1371/journal.pone.0071958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole C, Kroboth K, Schurch NJ, Sandilands A, Sherstnev A, O’Regan GM, et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salam MT. Asthma epigenetics. Adv Exp Med Biol. 2014;795:183–99. doi: 10.1007/978-1-4614-8603-9_11. [DOI] [PubMed] [Google Scholar]
- 45.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. J Allergy Clin Immunol. 2012;130:1243–55. doi: 10.1016/j.jaci.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–30. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 47.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012;30:1232–9. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schadt EE, Banerjee O, Fang G, Feng Z, Wong WH, Zhang X, et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 2013;23:129–41. doi: 10.1101/gr.136739.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanowicz D, Hackett TL, Garmaroudi FS, Gunther OP, Neumann S, Sutanto EN, et al. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PLoS One. 2012;7:e44213. doi: 10.1371/journal.pone.0044213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim YJ, Park SW, Kim TH, Park JS, Cheong HS, Shin HD, et al. Genome-wide methylation profiling of the bronchial mucosa of asthmatics: relationship to atopy. BMC Med Genet. 2013;14:39. doi: 10.1186/1471-2350-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol. 2014;15:777–88. doi: 10.1038/ni.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol. 2012;189:819–31. doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikhaylova L, Zhang Y, Kobzik L, Fedulov AV. Link between epigenomic alterations and genome-wide aberrant transcriptional response to allergen in dendritic cells conveying maternal asthma risk. PLoS One. 2013;8:e70387. doi: 10.1371/journal.pone.0070387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunawardhana LP, Baines KJ, Mattes J, Murphy VE, Simpson JL, Gibson PG. Differential DNA methylation profiles of infants exposed to maternal asthma during pregnancy. Pediatr Pulmonol. 2013 doi: 10.1002/ppul.22930. [DOI] [PubMed] [Google Scholar]
- 55.Breton CV, Siegmund KD, Joubert BR, Wang X, Qui W, Carey V, et al. Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One. 2014;9:e99716. doi: 10.1371/journal.pone.0099716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McErlean P, Favoreto S, Jr, Costa FF, Shen J, Quraishi J, Biyasheva A, et al. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Med Genomics. 2014;7:37. doi: 10.1186/1755-8794-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez E, Baurecht H, Wahn AF, Kretschmer A, Hotze M, Zeilinger S, et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol. 2014;134:1873–83. doi: 10.1038/jid.2014.87. [DOI] [PubMed] [Google Scholar]
- 58.Han J, Park SG, Bae JB, Choi J, Lyu JM, Park SH, et al. The characteristics of genome-wide DNA methylation in naive CD4+ T cells of patients with psoriasis or atopic dermatitis. Biochem Biophys Res Commun. 2012;422:157–63. doi: 10.1016/j.bbrc.2012.04.128. [DOI] [PubMed] [Google Scholar]
- 59.Wang IJ, Chen SL, Lu TP, Chuang EY, Chen PC. Prenatal smoke exposure, DNA methylation, and childhood atopic dermatitis. Clin Exp Allergy. 2013;43:535–43. doi: 10.1111/cea.12108. [DOI] [PubMed] [Google Scholar]
- 60.Liu YJ. Thymic stromal lymphopoietin: master switch for allergic inflammation. J Exp Med. 2006;203:269–73. doi: 10.1084/jem.20051745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rebane A, Runnel T, Aab A, Maslovskaja J, Ruckert B, Zimmermann M, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Liang Y, Wang P, Zhao M, Liang G, Yin H, Zhang G, et al. Demethylation of the FCER1G promoter leads to FcepsilonRI overexpression on monocytes of patients with atopic dermatitis. Allergy. 2012;67:424–30. doi: 10.1111/j.1398-9995.2011.02760.x. [DOI] [PubMed] [Google Scholar]
- 63.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013;27:e420–3. doi: 10.1111/jdv.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo Y, Zhou B, Zhao M, Tang J, Lu Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin Exp Dermatol. 2014;39:48–53. doi: 10.1111/ced.12206. [DOI] [PubMed] [Google Scholar]
- 65.Weisse K, Winkler S, Hirche F, Herberth G, Hinz D, Bauer M, et al. Maternal and newborn vitamin D status and its impact on food allergy development in the German LINA cohort study. Allergy. 2013;68:220–8. doi: 10.1111/all.12081. [DOI] [PubMed] [Google Scholar]
- 66.Nestor CE, Barrenas F, Wang H, Lentini A, Zhang H, Bruhn S, et al. DNA methylation changes separate allergic patients from healthy controls and may reflect altered CD4+ T-cell population structure. PLoS Genet. 2014;10:e1004059. doi: 10.1371/journal.pgen.1004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics. 2014:9. doi: 10.4161/epi.28945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–75. e9. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, et al. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS One. 2012;7:e40676. doi: 10.1371/journal.pone.0040676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 71.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–52. e1–5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 72.Hormannsperger G, Clavel T, Haller D. Gut matters: microbe-host interactions in allergic diseases. J Allergy Clin Immunol. 2012;129:1452–9. doi: 10.1016/j.jaci.2011.12.993. [DOI] [PubMed] [Google Scholar]
- 73.Gilstrap DL, Kraft M. Asthma and the host-microbe interaction. Journal of Allergy and Clinical Immunology. 2013;131:1449–50. e3. doi: 10.1016/j.jaci.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol. 2012;129:1204–8. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–63. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang YJ, Nelson CE, Brodie EL, DeSantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. Journal of Allergy and Clinical Immunology. 2011;127:372–81. e3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. Journal of Allergy and Clinical Immunology. 2013;131:346–52. e3. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLoughlin RM, Mills KHG. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. Journal of Allergy and Clinical Immunology. 2011;127:1097–107. doi: 10.1016/j.jaci.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 83.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 84.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. Journal of Allergy and Clinical Immunology. 2012;129:434–40. e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 85.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–34. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 86.Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, et al. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy. 2008;6:11. doi: 10.1186/1476-7961-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Enomoto T, Sowa M, Nishimori K, Shimazu S, Yoshida A, Yamada K, et al. Effects of Bifidobacterial Supplementation to Pregnant Women and Infants in the Prevention of Allergy Development in Infants and on Fecal Microbiota. Allergol Int. 2014 doi: 10.2332/allergolint.13-OA-0683. [DOI] [PubMed] [Google Scholar]
- 88.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noval Rivas M, Burton OT, Wise P, Zhang Y-q, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. Journal of Allergy and Clinical Immunology. 2013;131:201–12. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reisdorph N, Wechsler ME. Utilizing metabolomics to distinguish asthma phenotypes: strategies and clinical implications. Allergy. 2013;68:959–62. doi: 10.1111/all.12238. [DOI] [PubMed] [Google Scholar]
- 91.Scrivo R, Casadei L, Valerio M, Priori R, Valesini G, Manetti C. Metabolomics approach in allergic and rheumatic diseases. Curr Allergy Asthma Rep. 2014;14:445. doi: 10.1007/s11882-014-0445-5. [DOI] [PubMed] [Google Scholar]
- 92.Ibrahim B, Marsden P, Smith JA, Custovic A, Nilsson M, Fowler SJ. Breath metabolomic profiling by nuclear magnetic resonance spectroscopy in asthma. Allergy. 2013;68:1050–6. doi: 10.1111/all.12211. [DOI] [PubMed] [Google Scholar]
- 93.Sinha A, Krishnan V, Sethi T, Roy S, Ghosh B, Lodha R, et al. Metabolomic signatures in nuclear magnetic resonance spectra of exhaled breath condensate identify asthma. Eur Respir J. 2012;39:500–2. doi: 10.1183/09031936.00047711. [DOI] [PubMed] [Google Scholar]
- 94.Gahleitner F, Guallar-Hoyas C, Beardsmore CS, Pandya HC, Thomas CP. Metabolomics pilot study to identify volatile organic compound markers of childhood asthma in exhaled breath. Bioanalysis. 2013;5:2239–47. doi: 10.4155/bio.13.184. [DOI] [PubMed] [Google Scholar]
- 95.Carraro S, Giordano G, Reniero F, Carpi D, Stocchero M, Sterk PJ, et al. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy. 2013;68:110–7. doi: 10.1111/all.12063. [DOI] [PubMed] [Google Scholar]
- 96.Caldeira M, Perestrelo R, Barros AS, Bilelo MJ, Morete A, Camara JS, et al. Allergic asthma exhaled breath metabolome: a challenge for comprehensive two-dimensional gas chromatography. J Chromatogr A. 2012;1254:87–97. doi: 10.1016/j.chroma.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 97.Loureiro CC, Duarte IF, Gomes J, Carrola J, Barros AS, Gil AM, et al. Urinary metabolomic changes as a predictive biomarker of asthma exacerbation. Journal of Allergy and Clinical Immunology. 2014;133:261–3. e5. doi: 10.1016/j.jaci.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 98.Mattarucchi E, Baraldi E, Guillou C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed Chromatogr. 2012;26:89–94. doi: 10.1002/bmc.1631. [DOI] [PubMed] [Google Scholar]
- 99.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, et al. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. J Allergy Clin Immunol. 2011;127:757–64. e1–6. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 100.Ried JS, Baurecht H, Stuckler F, Krumsiek J, Gieger C, Heinrich J, et al. Integrative genetic and metabolite profiling analysis suggests altered phosphatidylcholine metabolism in asthma. Allergy. 2013;68:629–36. doi: 10.1111/all.12110. [DOI] [PubMed] [Google Scholar]
- 101.Jung J, Kim SH, Lee HS, Choi GS, Jung YS, Ryu DH, et al. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin Exp Allergy. 2013;43:425–33. doi: 10.1111/cea.12089. [DOI] [PubMed] [Google Scholar]
- 102.Fitzpatrick AM, Park Y, Brown LAS, Jones DP. Children with severe asthma have unique oxidative stress–associated metabolomic profiles. Journal of Allergy and Clinical Immunology. 2014;133:258–61. e8. doi: 10.1016/j.jaci.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amaral AFS. Metabolomics of asthma. Journal of Allergy and Clinical Immunology. 2014;133:1497–9. e1. doi: 10.1016/j.jaci.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 104.Assfalg M, Bortoletti E, D’Onofrio M, Pigozzi R, Molinari H, Boner AL, et al. An exploratory (1) H-nuclear magnetic resonance metabolomics study reveals altered urine spectral profiles in infants with atopic dermatitis. Br J Dermatol. 2012;166:1123–5. doi: 10.1111/j.1365-2133.2011.10711.x. [DOI] [PubMed] [Google Scholar]
- 105.Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma S, Zhou X, Thibault DM, Himes BE, Liu A, Szefler SJ, et al. A genome-wide survey of CD4 lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat Immunol. 2014;15:118–27. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schadt EE, Bjorkegren JL. NEW: network-enabled wisdom in biology, medicine, and health care. Sci Transl Med. 2012;4:115rv1. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 109.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–7. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Computational solutions to large-scale data management and analysis. Nat Rev Genet. 2010;11:647–57. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Topol E, Gardner E. Inventing the future. Health Data Manag. 2014;22:48. [PubMed] [Google Scholar]