Abstract
Evidence over the last two decades from a number of disciplines has solidified some fundamental concepts in metastasis, a major contributor to cancer associated deaths. However, significant advances have been made in controlling this critical cellular process by focusing on targeted therapy. A key set of factors associated with this invasive phenotype is the nm23 family of over twenty metastasis-associated genes. Among the eight known isoforms, Nm23-H1 is the most studied potential anti-metastatic factor associated with human cancers. Importantly, a growing body of work has clearly suggested a critical role for Nm23-H1 in limiting tumor cell motility and progression induced by several tumor viruses, including Epstein-Barr virus (EBV), Kaposi’s sarcoma associated herpes virus (KSHV) and human papilloma virus (HPV). A more in depth understanding of the interactions between tumor viruses encoded antigens and Nm23-H1 will facilitate the elucidation of underlying mechanism(s) which contribute to virus-associated cancers. Here, we review recent studies to explore the molecular links between human oncogenic viruses and progression of metastasis, in particular the deregulation of Nm23-H1 mediated suppression.
Keywords: Nm23, Metastasis, Cancer, Tumor virus
INTRODUCTION
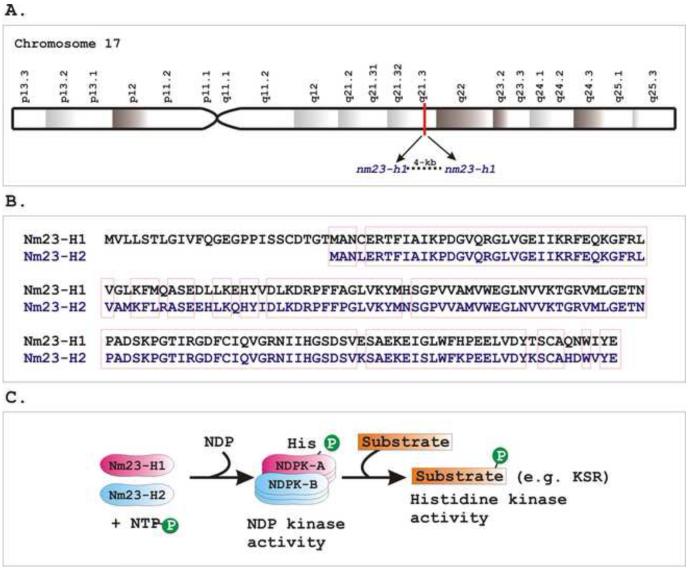
The metastatic suppressor nm23 gene family is highly conserved among a wide variety of eukaryotic species [1,2]. To date, eight genes have been identified in humans, namely, nm23-h1, nm23-h2, nm23-h3, nm23-h4, nm23-h5, nm23-h6, nm23-h7, and nm23-h8 [3-12]. However, it is yet unknown whether all nm23 family members can function as metastasis suppressors. The two most abundantly expressed and currently studied genes are nm23-h1 and nm23-h2 [13,14]. By utilizing somatic cell hybrid analysis and fluorescence in situ hybridization, nm23-h1 and nm23-h2 genes were found to be located at q21.3 position on chromosome 17 near the BRCA1 locus, which is known to be associated with early onset of familial breast and ovarian cancers (Figure 1A) [15]. The corresponding proteins, Nm23-H1 and -H2 possess 88% amino acids sequence homology (Figure 1B) [16,17], and together, they encode the A and B subunits of nucleoside diphosphate kinase (NDPK), a ubiquitously expressed enzyme that transfers the terminal phosphates from ATP to deoxy nucleoside diphosphates (dNTPs) (Figure 1C) [17]. The murine counterparts of these proteins both Nm23-M1 [18] and Nm23-M2 [19], respectively, share approximately 95% of amino acids sequence homology with human versions [14]. Although they are highly homologous and form a functional enzyme together, their cellular function and sub-cellular localization are significantly different [20]. Since their discovery, clinical studies regarding Nm23 expression, particularly Nm23-H1, at the protein and transcript levels in various types of cancer have been extensively reported [21,22]. An inverse correlation between its level of expression and the metastatic potential was observed for numerous cancers such as breast, melanoma, ovarian, hepatocellular, esophageal, oral squamous and colon cancer [16,23-27]. However, a positive association between tumor aggressiveness and Nm23 expression has also been reported for several other cancer types including cervical, testicular, prostate, thyroid, renal cell carcinoma and hematologic malignancies [16,28-30]. The metastasis-suppressive function of Nm23-H1 was established by ectopically expressing Nm23-H1 (or Nm23-M1) at physiological levels in highly aggressive cancer cell lines, which exhibited reduction in their metastatic potential [31]. However, there are numerous examples which suggest a dual role for Nm23-H1 expression in the course of cancer propagation. While the primary tumors are found to be associated with an elevated expression, dramatic down-regulation of Nm23-H1 expression was observed at later stages with high metastatic activities. Altogether, this information suggests that pathogenesis associated with Nm23-H1 regulation may be tumor specific, and thus elucidation of the regulatory mechanisms for Nm23-H1 expression would further enhance our understanding of Nm23-H1 associated type-specific cancer progression and so allow for the development of novel therapeutic strategies for metastatic cancer.
Figure 1. Nm23-H1 and -H2 represent the two best studied proteins of the human Nm23 family.
A) Both nm23-H1 and nm23-H2 mapped 4 kb apart at position q21.3 on chromosome 17. B) Nm23-H1 and Nm23-H2 share approximately 90% amino acids sequence homology. Sequence alignment was generated by a multiple sequence alignment program, ClustalW (http://www.ebi.ac.uk/FTP/). Red boxes depict similar amino acid residues. C) Nm23-H1 and -H2 encode A and B subunits, respectively, of nucleoside diphosphate kinase (NDPK) enzyme, which transfers the terminal phosphates from a nucleoside triphosphate (NTP) to nucleoside diphosphate (NDP). They form either homo- or hetero-hexamers of an active NDPK enzyme complex. Subsequently Nm23 transfers the phosphate moiety to another substrate protein. For example, the kinase suppressor of Ras oncoprotein (KSR) is a substrate for the histidine kinase activity of Nm23-H1.
Structure-function relationship of Nm23-H1
According to the genomic architecture and phosphotransferase activity, human Nm23 genes have been classified into two different groups [32]. Group I is represented by the Nm23-H1 - H4 isoforms, whereas group II contains the Nm23-H5 - H8 isoforms [32,33]. Group I Nm23 proteins possess a highly conserved kinase active site. On the other hand there is no strict conservation of the kinase site motifs for group II proteins, and only Nm23-H6 has been reported to display kinase activity [33]. Nm23-H1 was first discovered as a metastatic suppressor gene by Steeg et al. [18], and different approaches have been employed to unravel the biochemical activities associated with Nm23-H1. Nm23-H1 is characterized by a wide variety of biological functions, including transcriptional regulation, differentiation, proliferation, and most importantly suppression of tumor metastasis [2]. To date, three major enzymatic functions have been attributed to Nm23-H1, namely, its nucleoside diphosphate kinase (NDPK), histidine kinase and 3’-5’exonuclease activities [31]. Besides these enzymatic activities, Nm23-H1 was shown to bind to many cellular as well as viral antigens [34]. Although, none of the Nm23-H1 binding partners have been subjected for clinical testing, several have prompted new translational approaches to metastatic disease [34,35]. The precise mechanism by which Nm23-H1 regulates metastasis is not fully understood. Thus, a better understanding of its structure-function relationship would provide clues to the mechanisms regulating Nm23-H1 associated cancer metastasis in a wide range of human cancers.
Histidine kinase activity of Nm23-H1
The histidine kinase activity has been shown to be a major factor in regulating its metastatic suppressive functions [36-38], as mutations in Nm23-H1 which ablate its histidine kinase activity, such as P96S and S120G, were shown to be ineffective in cell motility-suppressing activity [36,38]. Histidine protein kinases are well documented in prokaryotes and lower eukaryotes, where they form the ‘two component’ signal transduction system [34,35]. A number of molecular features clearly distinguished histidine kinases from the conventional serine, threonine or tyrosine protein kinases. As a histidine kinase, Nm23-H1 was first auto-phosphorylated (referred to as NDPK activity, see below) at histidine residue using ATP, and subsequently transfers the phosphate moiety to another protein on a histidine or other residue [34]. For example, the kinase suppressor of Ras oncoprotein (KSR) is a substrate for the histidine kinase activity of Nm23-H1 [38], suggesting a potential mechanism of cell motility suppression by regulating Ras-Raf-MEK-Erk signaling cascade [34]. However, a detailed characterization of downstream histidine kinase substrates as well as their associations to metastasis suppression is yet to be validated.
NDPK activity of Nm23-H1
The nucleoside diphosphate kinase (NDPK) activity of Nm23-H1 is not essential for metastasis suppression [39], although this is yet to be validated using an over-expression system as used in determining histidine kinase activity. The human nm23-h1 gene encodes the A subunit of NDPK that can be activated as homo or hetero-hexamers with B subunit encoded by nm23-h2 gene [17]. A nucleotide trans-phosphorylation activity covalently transfers the terminal phosphate from a nucleoside triphosphate to a nucleoside diphosphate and maintains cellular homeostasis [40]. For example, GTP is used as a source of energy for protein biosynthesis and is essential for activation of G-protein mediated signal transduction [41]. Nm23-H1 has explicitly been shown as a cellular GTP supplier in G-protein mediated signal transduction [40]. In addition, it was recently demonstrated that the secreted NDPK-A in breast cancer cells produced extracellular ATP which activates the PY2 receptor [42]. This receptor is associated with neo-vascularization [42]. In addition, Nm23-H1 can phosphorylate the aspartic acid residue of aldolase-C, although the precise function of the phosphorylated aldolase-C in tumor metastasis is stillunclear [35].
Nuclease activity of Nm23-H1
Exonuclease
In contrast to histidine kinase and NDPK activities, no reports have as yet implicated the Nm23-H1 3’-5’exonuclease activity as to a role in contributing to the metastatic potential. However, as 3'-5' exonucleases are typically important for the maintenance of genomic integrity; this activity represents a potential target for regulating the metastasis suppressor properties of Nm23-H1. A role for DNA repair activities in resisting cancer progression is supported indirectly by reports of accelerated mutation in highly metastatic clones versus weakly metastatic cells from the same tumor cell population. Moreover, development of metastasis depends on genetic instability of transformed cells that give rise to populations of cells capable of metastatic spread, apparently through a Darwinian-like selection process [31]. These observations suggest a novel anti-mutator activity that mediates metastasis suppressor activity of Nm23-H1, and that the down-regulation of Nm23-H1 may facilitate the spread of mutations, which are thought to be critical for metastasis progression [31].
Endonuclease
Furthermore, data supporting the association of Nm23-H1 as a constituent of the SET complex, a macromolecular complex, which is associated with the endoplasmic reticulum and is targeted by Granzyme-A during cytotoxic T lymphocyte induced apoptosis was presented by the Lieberman group [43]. Granzyme-A cuts the SET complex and subsequently activates the endonuclease activity of Nm23-H1. The importance of Nm23-H1 and SET in Granzyme-A -mediated cell death was confirmed by finding increased DNA damage and cell death in cells that over-express Nm23-H1 or have silenced SET and, conversely, by finding less cell death in targets with silenced Nm23-H1 or enhanced SET expression [43]. Furthermore, SET was thought to also translocate along with Nm23-H1, to the nucleus where the complex is then disrupted, SET is then degraded and Nm23-H1 now released from the complex can induce it DNA-nicking activities which leads to genomic instability and apoptotic activities. Following Granzyme-A treatment, the purified SET complex shows significant exonuclease activity on supercoiled plasmid DNA [44,45]. This activity could be mediated due to both Nm23-H1 and/or TREX1, another important 3’-5’ exonuclease [43]. Inclusion of TREX1 in the SET complex was shown to be specific, since the TREX1 homolog TREX2 was not found to be present in SET complex [43]. As similar to Nm23-H1, TREX1 is also not a Granzyme-A substrate [43] and functions synergistically with Nm23-H1 to destroy DNA during Granzyme-A-mediated cell death [43]. After Nm23-H1 nicks one strand, TREX1 removes bases from the free 3’-end to enhance the damage and prevents DNA end re-annealing and rapid repair [43]. Additional studies investigating complexes which include Nm23-H1 and SET also suggest that the observed DNA nicking activity is likely to be of an endonuclease which is a co-purifying component of the complex containing Nm23-H1 [46], and that the activity of Nm23-H1 in the complex has not been fully demonstrated.
Cellular binding partners of Nm23-H1
Recent insights into the ability of Nm23-H1 to interact with a wide range of cellular proteins and manipulating multiple cellular signaling have shed light on new avenues of research and possible drug targets in the field of metastasis. Nm23-H1 has been reported to interact with many proteins, including a number of important cellular proteins involved in critical biochemical pathways as well as tumor virus encoded essential oncoproteins. Table 1 represents a list of the Nm23-H1 interacting proteins thought to have themost significant alterations in their biological functions due to interactions with Nm23-H1.
Table 1.
Binding partners of Nm23-1 H1 and proposed biological consequences
| Binding partners | Biological consequences | References |
|---|---|---|
| A. Cellular | ||
| h-prune | Increases h-prune phosphodiesterase activity and cell motility. Directly correlates with high expression of h-prune and low expression of Nm23-H1 in aggressive tumor cells. |
[53,55,56] |
| STRAP | Negative regulation of TGF-β mediated signaling, Increases p53 activity. |
[51,52] |
| KSR | Direct substrate for histidine kinase activity of Nm23-H1, regulates cell motility by controlling Ras-Raf-MEK-Erk pathway. |
[38] |
| SET | Formation of an inhibitory SET complex, blocks Nm23-H1 mediated exonuclease and tumor metastatic activity. |
[43,47,62] |
| Rad | Maintains GDP-GTP cycling, acts as a GTPase-activating protein for Rad. |
[61,168] |
| Tiaml | Blocks Tiam1-Rac1 mediated signaling pathway, decrease cell motility and intercellular adhesion. |
[58] |
| Dbl-1 | Interaction results in loss of Dbl-1 function as a guanine nucleotide exchange factor for Cdc42, and blocks Nm23-H1 activity. |
[59] |
| ERα | Alters estrogen-responsive gene expression. | [60] |
| B. Viral | ||
| EBNA3C | Promotes Nm23-H1 nuclear localization, negatively regulates the metastatic potential of Nm23-H1; together EBNA3C and Nm23-H1 can block Necdin-mediated transcriptional repression and anti- angiogenic activities. In cooperation with Nm23-H1, EBNA3C can transcriptionally upregulate MMP-9, Cox-2 and α-V integrin expression levels. |
[13,63,106,107,125,136,148] |
| EBNA1 | Modulates Nm23-H1 associated metastatic activities | [13,105] |
| LANA | Induces Nm23-H1 nuclear translocation, re-expression of Nm23-H1 blocks KSHV induced cell invasiveness through inhibiting MAPK- signaling pathway. |
[167] |
| E7 | Blocks granzyme A-induced apoptosis and promotes cell invasiveness by inhibiting Nm23-H1 multidirectional activities. |
[157] |
STRAP
Serine-threonine kinase receptor-associated protein (STRAP) is a positive regulator of 3-phosphoinositide-dependent protein kinase-1 (PDK1), and negatively regulates TGF-β signaling by stabilizing the interaction between TGF-β receptor and Smad7 [47]. A recent study has shown that through interaction with STRAP, Nm23-H1 promotes STRAP induced inhibition of TGF-β signaling, suggesting a molecular link between TGF-β and Nm23-H1 regulated signaling pathways [48]. Later, the same group also demonstrated that Nm23-H1 and its interacting partner STRAP can directly interact with p53 and positively regulate its functions, including p53-induced apoptosis and cell-cycle arrest, probably through inactivation of Mdm2 functions [49].
H-prune
Another attractive Nm23-H1 binding protein that was identified in humans is prune (h-prune) [50]. Elevated levels of h-prune expression has been shown to be associated with a low expression level of Nm23-H1 in aggressive breast carcinomas, suggesting an inhibitory role for h-prune against the metastasis suppression function of Nm23-H1 [51]. Over-expression of h-prune in a breast cancer cell line stimulated cellular motility, however abrogated by the simultaneous expression of Nm23-H1 [52]. Other than breast cancer, h-prune expression has been correlated with tumor aggressiveness in gastric cancer and sarcoma [53,54]. Interestingly, a direct correlation between an increase in h-prune phosphodiesterase activity and cellular motility, as a result of a direct protein-protein interaction with Nm23-H1, was found in the breast cancer model [52]. Several specific chemical inhibitors of h-prune phosphodiesterase activity have been shown to block tumor cell motility, suggesting that these inhibitors could be used as ‘lead’ compounds in development of additional therapies against cancer metastasis [52].
KSR
Other reported Nm23-H1 binding proteins include the kinase suppressor of Ras (KSR), a scaffold protein for the mitogen-activated protein kinase (MAPK) cascade [38]. Nm23-H1 also interacts with Tiam1 (T-cell lymphoma invasion and metastasis 1), a specific guanine nucleotide exchange factor for Rac1, and down-modulates Tiam1-Rac1 mediated signaling [55]. The inhibitory effect of Nm23-H1 on Tiam1 activity can lead to reduce cell motility and intercellular adhesion [55]. Recently, we have shown that Nm23-H1 also interacts with another member of the guanine nucleotide exchange factor family, Dbl-1 [56]. This interaction results in the loss of Dbl-1 function as a guanine nucleotide exchange factor for Cdc42, thereby inhibiting the suppression of cell motility by Nm23-H1 [56]. Nm23-H1 was also shown to bind estrogen receptor α, which resulted in an altered pattern of estrogen induced gene expression [57]. Rad is the prototypic member of a new class of Ras-related GTPases [58]. Nm23-H1 was shown to interact with Rad and functions as a GTPase-activating protein for Rad [58].
SET
The DNA exonuclease activity of Nm23-H1 is likely to be central to its metastasis suppressor function (discussed above). Nm23-H1 forms an inhibitory complex with SET, a critical protein complex that can regulate a number of cellular pathways linked to tumorigenesis and metastasis [59]. SET may modulate the potential Nm23-H1 mediated exonuclease activity by sequestering Nm23-H1 into the cytosol, thereby promoting metastasis [59]. Further studies suggest that the DNA nicking activity may be due to the co-purification with another protein with nicking activity like UDG [46] and the role of Nm23-H1 in this complex still needs to be further explored.
To date, multiple important cellular proteins were identified as Nm23-H1 binding partners as potential therapeutic targets (reviewed in [60]). However, due to the sticky nature of Nm23 proteins [60], excessive care is strongly suggested for investigators in designing their experiments to minimize potential artifacts. Subsequent studies are necessary to further establish the functional relevance to the binding activities, such as mutational analyses to show a loss of the activity as well as biological consequences due to protein-protein interaction or in vivo studies using mouse model systems.
Tumor viruses and Nm23-H1
A number of current studies have clearly suggested a critical role for Nm23-H1 in the suppression of tumor virus induced cell migration and cancer progression. Apparently this role ismediated by protein-protein interactions between Nm23-H1 and tumor virus encoded oncoproteins. Below, we discuss the current thoughts on the multiple tumor viruses encoded oncoproteins that associate and regulate Nm23-H1 associated functions to accelerate cancer progression due to their pro-metastatic activities (Figure 2).
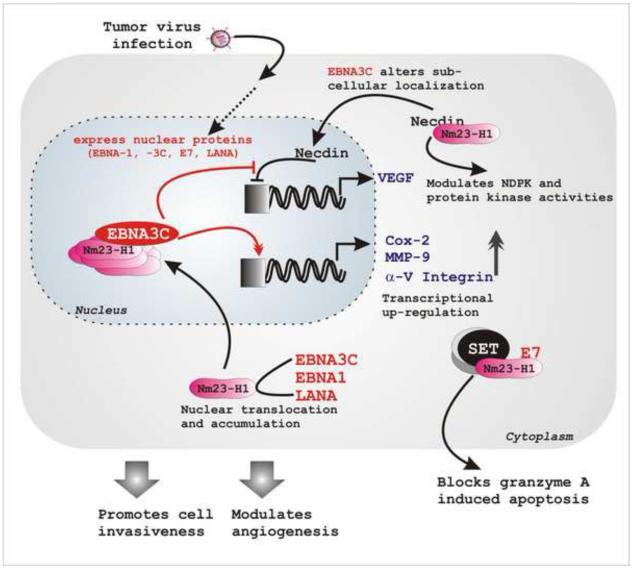
Figure 2. Tumor virus encoded oncoproteins can effectively modulate cancer metastasis by regulating Nm23-H1 associated activities.
Following infection, tumor viruses encoded oncoproteins (EBV EBNA3C and EBNA1; KSHV LANA and HPV E7) target multiple cellular pathways, including metastasis. The presence of different viral proteins (EBNA1, EBNA3C and LANA) results in change in sub-cellular localization of Nm23-H1 from cytoplasmic to predominantly nuclear. In nucleus they form a stable complex. Together with Nm23-H1, EBNA3C induces transcriptional up-regulation of several genes, inducing MMP-9, Cox-2 and α-V integrin. These activities further alter cellular invasiveness and angiogenesis. Coupled with Nm23-H1, EBNA3C also rescues Necdin-mediated downregulation of its downstream promoter (VEGF). Necdin can interact with Nm23-H1 that further modulates Nm23-H1 associated NDP kinase as well as protein kinase activities. HPV encoded E7 oncoprotein interacts with Nm23-H1 and blocks granzyme A induced apoptosis, thereby promoting cell invasiveness.
EBV and Nm23-H1
One of the best studied Nm23-H1 binding proteins is a latent antigen, EBNA3C, encoded by Epstein-Barr virus (EBV) [61,62]. EBV is a lymphotropic gamma-herpesvirus that asymptomatically persists in greater than 90% of the world population [63-65]. However, EBV occasionally causes a self-limiting disease, infectious mononucleosis in adolescents and has been found to be associated with the development of several B-cell lymphomas and epithelial cancers including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, AIDS-associated and transplant-associated immunoblastic lymphoma, and in some cases invasive breast carcinoma [13,63-65]. Recently, EBV has been shown to be related to gastric carcinoma (GC) with an occurrence rate varying from 1.3% to 20.1% in different countries [66]. In vitro, EBV can efficiently transform quiescent B-cells into continuously proliferating lymphoblastoid cell lines (LCLs) [63,65]. These latently infected LCLs carry the viral genome as extra-chromosomal episomes that express only a small subset of genes including six nuclear antigens (EBNA- 1, 2, 3A, 3B, 3C and LP), three integral membrane proteins (LMP- 1, 2A, and 2B), two non-translated RNAs (EBER-1 and -2) and several microRNAs [63,65,67]. Genetic studies using recombinant viruses established with a bacterial artificial chromosome (BAC) system, from a number of different groups have shown that EBNA-1, -2, -3A, -3C, -LP and LMP-1 are important for EBV mediated transformation of naïve B-cells in vitro, indicating that a complex cascade of molecular events is required to surpass normal growth controls [67-71].
EBNA1
EBNA1 is the only virus encoded antigen expressed in all EBV associated tumors [72]. Although, the oncogenic potential of EBNA1 in EBV-associated malignancies still remains elusive due to differences in the results from a number of groups [73-75], the importance of EBNA1 in EBV infection is unquestionable as it is absolutely essential for the episomal maintenance and replication in latently infected cells [69,76]. EBV episomal stability has been shown to be maintained through the interaction between EBNA1 and a cis-acting DNA element referred to as the origin of plasmid replication (OriP). Thisis composed of two separate regions, the family of repeats (FR) and a dyad symmetry (DS) element [77,78]. In addition to direct DNA binding ability through the C-terminal domain, EBNA1 tethers the EBV genome to metaphase chromosomes through two RGG-like motifs located at the N-terminal DNA binding domain. However, theprecise mechanism through which EBNA1 is attached to the metaphase chromosome is not completely understood [79-83]. EBNA1 is composed of unique amino-terminal (residues 1-89) and carboxy-terminal (residues 328-641) domains flanking a large Gly-Ala with no clear biochemical function [84]. However, this repeat region has a cis-acting capacity to suppress antigen presentation through MHC class I pathway and thus plays a central role in immune escape during EBV infection [85,86]. A number of studies identified a high frequency of sequence variation at a region (residues 466-527) within the DNA binding-dimerization domain of EBNA1 [66]. EBNA1 was also shown to induce genomic instability in malignant B-cells, as manifested by the occurrence of non-clonal chromosomal aberrations, oxidative DNA damage and activation of the DNA damage response [87,88]. In addition, EBNA1 was shown to regulate cellular gene expression and inhibits the canonical NF-κB pathway by inhibiting IKK phosphorylation, implicating a vital role for EBNA1 in the pathogenesis of nasopharyngeal carcinoma [89]. Moreover, EBNA1 expression in gastric carcinomas cells was shown to be associated with enhanced tumorigenicity [90]. A number of published studies are consistent with a role for EBNA1 in proliferation of EBV-positive cells [91-93]. For instance, interference with EBNA1 function in EBV-positive Burkitt's lymphoma cells, by ectopic expression of a dominant-negative EBNA1 mutant resulted in increased cell death [91]. Similarly, down-regulation of EBNA1 expression in a Burkitt's lymphoma cell line or EBV-positive epithelial cells by RNA interference exhibited reduction in cell proliferation [92,93].
EBNA3C
It has been proposed that EBNA -3A, -3B, and -3C genes likely to be generated as a result of a series of gene duplication events [94]. They possess the same promoter, similar exons, codons, introns, and a homologous N-terminal domain, which interacts with the Notch-signaling mediator RBP-Jκ, a sequence-specific DNA binding transcription factor that mediates EBNA2 transcriptional up-regulation [63,94-97]. Despite these similarities, EBNA3A, EBNA3B, and EBNA3C have divergent roles in maintaining LCL growth, as EBNA3B is dispensable, whereas both EBNA3A and EBNA3C are absolutely required for LCL transformation [71,98]. Importantly, EBNA3C can only co-activate the viral LMP1 promoter in accordance with EBNA2 [99]. An EBNA3C SUMO interaction motif (SIM) and a domain that interacts with the PU.1 transcription factor have been implicated in LMP1 promoter co-activation with EBNA2 [99]. Conditional EBNA3C inactivation resulted in up-regulation of both p16INK4A and p14ARF tumor suppressors, suggesting a possible mechanism forEBNA3C mediated LCL growth regulation [98]. Beside this, EBNA3C has been shown to interact with multiple transcriptional regulators, including c-Myc, prothymosin α, histone deacetylase 1 (HDAC1), CtBP, DP103, SMN, SCFSkp2, p300, p53 and Nm23-H1as well as a number of important cell-cycle modulators such as Cyclin A, Cyclin D1, Cyclin E, pRb, Mdm2, and Chk2, implicating a complex regulatory role played by EBNA3C during EBV mediated cellular transformation [64,100-102]. The contribution of EBNA3C and Nm23-H1 to transcription regulation was further explored by investigating the role of these proteins on modulating transcription of Cox2, αv-integrin and the metalloproteinase (refs).
Role of EBV antigens in Nm23-H1 mediated metastasis
The EBV encoded essential latent oncoproteins EBNA1 and EBNA3C have been shown to specifically interact with Nm23-H1 [61,103]. The significance of these interactions was determined in the nude mice model using cancer cells expressing the EBV antigens and Nm23-H1 [13]. These in vivo studies showed that both the EBV antigens promoted the growth of transformed cells; however and interestingly their expression was less important at the later stage of tumor development [13]. As expected, the expression of Nm23-H1 critically affected the growth of cancer cells and suppressed their metastatic potential [13]. This effect was rescued by the expression of either EBV antigens. The pro-metastatic potential of EBNA3C was found to be higher when compared to EBNA1, which triggered a dramatic immune response, as indicated by increased spleen size and development of ascites in the mice [13]. This study was the first report linked to tumor virus mediated metastasis suppression during cancer development, and at least in part widens the range of potential drug targets. Furthermore, the molecular mechanisms by which these viral oncogenes functions as pro-metastatic factors are still been investigated and studies along these lines should also yield more interesting possibilities for targeted therapies against EBV associated cancers.
Anti-angiogenic activity of Necdin is blocked by EBNA3C coupled with Nm23-H1
The block to the anti-angiogenic activity of Necdin by EBNA3C and Nm23-H1 suggests another level of regulation through which these viral antigens can modulate Nm23-H1 activities. The domain of EBNA3C that specifically binds to Nm23-H1 has been previously identified and lies within the region comprising residues 637-675 of EBNA3C, flanked by the proline- and glutamine-rich domains [104]. A Blast analysis of this domain sequence shows a considerable homology to a cellular protein, called Necdin [105]. Necdin belongs to the MAGE family of proteins, which are shown to have multiple roles in different cellular processes, including cell-cycle regulation as well as apoptosis [106]. The most significant characteristic of the MAGE family of proteins the possession of a large central region which is referred to as the MAGE homology domain (MHD), whereas their amino- and carboxy-terminal domains are unique for the individual family members[106]. The sub-cellular localization of Necdin in differentiated neurons has been previously demonstrated to be predominantly cytoplasmic, with a salient change in location to the nucleus underspecific physiological conditions [107]. Necdin was also shown to act as a cellular transcription repressor by directly binding to multiple guanosine clusters within the promoter region of target genes [108]. It has also been suggested that Necdin can regulate transcription of other genes engaged in multiple cellular events either through direct binding with DNA or through its interaction with other transcription factors, such as p53 and E2F1 [106,109]. Importantly, in the context of cancer and tumor development, Necdin was shown to interact with and modulate the activity of Hif-1α, a major transcription regulator in hypoxia [110]. Thus, the regulation of Hif-1α expression by Necdin may have important consequences, including downstream effects on cell growth and proliferation [110]. Subsequently, we have shown that EBNA3C together with Nm23-H1 modulates the biological functions of Necdin in EBV infected cells [105]. In this study, we showed that Necdin levels were significantly lower in EBV-positive cells [105]. EBNA3C affects the sub-cellular localization of Necdin as well as rescues cells from anti-angiogenic and anti-proliferative effects mediated by Necdin [105]. Interestingly, both EBNA3C and Nm23-H1 were able to rescue not only Necdin-mediated transcriptional repression of the downstream vascular endothelial growth factor promoter but also its growth suppression and anti-angiogenic effects on cancer cells [105]. The majority of these responses were mediated through residues 191-222 of Necdin, known to be important for nuclear matrix targeting [105]. This above study further suggests a novel role for Necdin in regulation of downstream cellular targets in virus-associated human cancers.
Cox-2 mediated inflammatory response is augmented by the EBNA3C-Nm23-H1 complex
Similar to other tumor viruses, EBV is known to critically modulate the inflammatory response [63]. Prostaglandin (PG) G/H endoperoxidase synthases, also known as cyclooxygenases (COX), play a major role in inflammatory responses [111]. Among the two isoforms of COX (1 and 2), the inducible COX-2 molecule is highly stimulated in a variety of inflammatory diseases and in response to pro-inflammatory cytokines, growth factors and other tumor inducers [112-114]. Elevated expression of COX-2 has been reported in many premalignant lesions and epithelial cancers, such as colon cancer, lung cancer, breast cancer, gastric cancer, esophageal cancer, and head and neck cancers, suggesting that COX-2 plays a critical role in development of these cancers [115-119]. Up-regulation of COX-2 in cancer cells has also been linked to increased angiogenesis as well as cancer metastasis [120-122]. We and others have shown that COX-2 is frequently expressed in either EBV-positive nasopharyngeal tumors or detected at significantly higher levels in EBV-transformed LCLs, suggesting a role for COX-2 in EBV pathogenesis [123,124]. The EBV encoded oncoproteins, both LMP1 and EBNA3C, have been shown to up-regulate COX-2 expression utilizing different mechanistic pathways [123,124]. While, LMP1 induces COX-2 expression in an NF-κB-dependent manner [124], EBNA3C was shown to accelerate COX-2 expression and in cooperation with Nm23-H1 can significantly upregulate the expression [123]. Importantly, COX-2 was also shown to play a role in de novo infection of various DNA and RNA viruses, including herpesviruses, such as herpes simplex virus (HSV), human cytomegalovirus (HCMV), EBV, and murine gammaherpesvirus 68 (MHV-68) [63,125]. Nonetheless, a comprehensive study for COX-2 regulating EBV reactivation needs to be further addressed, particularly in cooperation with Nm23-H1. It would be interesting to know if Nm23-H1 in the absence of viral antigens can modulate Cox-2 expression with other cellular factors and whether or not the virus can target these factors to enhance the activities of these inflammatory molecules in EBV infected cells.
Regulation of metalloproteinases by the EBV essential latent antigen EBNA3C
An important step in invasion and metastasis is degradation of the basement membrane to enable tumor cells to escape from the primary growth site [126]. The matrix metalloproteinase (MMP) family of proteins is involved in the degradation of all components of the basement membrane, and members of every family of metalloproteinases have been implicated in malignancy and metastasis [126]. The activity of MMPs is tightly controlled, with regulation occurring mainly at the transcriptional level [126]. The proteins are further regulated through activation of the proenzyme and the tissue inhibitors of metalloproteinase [126]. A number of MMPs have been found to be associated with cell migration and invasion, including MMP-2 and MMP-9, both of which degrade type IV collagen [127,128]. The ability to degrade type IV collagen, a major component of the basement membrane, indicates that these proteins may play an important role in metastasis by degrading the basement membrane at sites of malignant tumor growth [127,128]. Specifically, MMP-9 has been implicated as an essential molecule required for tumor cell intravasation and extravasation during metastasis [129].
EBV-immortalized B-cells have been shown to synthesize MMP-9 and the addition of TIMP-1 to EBV-infected B cells has been shown to inhibit their migration in vitro [130,131]. Moreover, a number of studies have shown that EBV encoded LMP1 increases cell invasiveness through augmenting MMP-9 expression level [129,131-133]. LMP1-mediated MMP-9 expression occurs primarily through the NF-κB and Ap1 signaling pathways [133]. Subsequently, we have demonstrated that EBNA3C in cooperation with Nm23-H1 can induce MMP-9 expression at the transcript level as well as increase in MMP-9 gelatinolytic activity [134]. As seen in LMP1, specific mutations in the MMP-9 promoter showed that the Ap1 and NF-κB binding sites are important for EBNA3C mediated up-regulation [134]. This indicates that although these viral antigens are distinctly different in both structurally and functionally, they share similar mechanistic pathway for exerting their oncogenic properties.
The molecular association between Nm23-H1 and EBNA3C can regulate integrins expression and cell migration
Integrins are heterodimeric molecules composed of a non-covalently associated α- and β- subunit exclusively recognizing ligands via an RGD amino acid sequence [135]. The ligands for αvintegrins include vitronectin, bronectin, brinogen, von Willebrand factor and osteopontin [126,136-140]. Interestingly, αv integrins are expressed on migratory cells such as metastatic melanomas and breast cancer cells and integrin expression correlates with metastasis [141-143]. Additionally, αVβ3 integrin is directly associated with MMP2 on melanoma cells, which is thought to provide tumor cells with the coordinated matrix degradation and cellular motility, thus facilitating cellular invasion [144]. The role of Nm23-H1 in regulating α-V integrins has been poorly understood, with exceptions in some clinical reports where investigators have tried to correlate Nm23-H1 expression with αv integrin level. Importantly, elevated expression of αv integrins has been shown to be associated with EBV mediated immortalization of naïve B-cells [145]. Moreover, LMP-1, LMP-2 and EBNA-2 expression were shown to amplify αv integrins expression levels in LCLs [145]. In addition, we showed that through interacting with Nm23-H1, EBNA3C can lead to an increase expression of αv integrin [146]. The study showed that Nm23-H1 alone down-regulates αv integrin expression in a dose responsive manner, while in contrast, EBNA3C up-regulate αv integrin expression [146]. Furthermore, the study also showed that the association of the Sp1 and GATA transcription factors with Nm23-H1 is required for the transcriptional modulation of αv integrin promoter activity [146].
These findings provide at least in part, if not the complete scenario, potential molecular mechanisms which allows us to further understand how EBNA3C can reverse the anti-migratory effect of Nm23-H1 in virus infected cells. Thus, EBNA3C has pro-metastic properties through driving the upregulation of a number of these cellular targets known to be critical for cell migration and metastasis. These studies provide a framework which demonstrates the complexity of the potential interacting map which shows very clearly that EBV by using its essential antigens can strategically target specific cellular promoters to modulate their expression and so induce the transcription of the genes and so result in greater activity. The consequences of these activities are the enhanced metastatic potential of the viral infected cells.
HPV and Nm23-H1
The human papillomaviruses (HPVs) are the causative agents of the most common sexually transmitted diseases that infect both females and males [147,148]. HPVs commonly infect mucosal genital epithelia, with an estimated 75% of humans being affected during life [149]. HPV infection is very common among men and women across all geographical, racial and socio-economic subgroup worldwide [148]. So far, more than a hundred different types of HPV have been identified and approximately forty types can infect the anogenital region [148]. Anogenital HPV types have been further classified into ‘low-risk’, which are associated with anogenital warts and mild dysplasia, and ‘high-risk’ types, which are associated with high-grade dysplasia and anogenital cancers, such as cervical and anal carcinoma [148,150]. The HPV genome is divided into long control regions (LCR), which plays a role in regulating gene expression and DNA replication; open reading frames (ORF), which are involved in expression of the early gene proteins E1-E8; and the late genes encoding structural proteins L1-L2 [63]. HPV encoded antigens are engaged in viral replication, cellular transformation, checking viral as well as cellular gene transcription, and also generation of viral progeny [63]. Through encoding two major antigens, E6 and E7, HPV establishes cancer particularly through the ubiquitin-proteasome mediated degradation of two tumor suppressor proteins, p53 and pRb [63,151]. These two oncoproteins, E6 and E7 also play a central role in virus reproduction as well in inducing immortalization and transformation by re-programming cell-cycle control in differentiating host epithelial cells [151]. Through interacting with several important cellular proteins, both E6 and E7 have been shown to induce genomic instability and mitotic defects when introduced into cells [152]. The E7 oncoprotein is considered essential for ‘high-risk’ HPVs transforming properties, while a cooperating role is played by its oncogenic partner E6 [153,154]. Since E6 and E7 oncoproteins are critical but not sufficient for complete cellular transformation [151], additional genetic alteration is also required. A greater understanding of the role of HPV antigens in cancer propagation will eventually aid in the development of antiviral treatment, as well as unveil a more general mechanism of HPV mediated oncogenesis.
Similar to EBNA3C, the HPV encoded E7 oncoprotein has also been shown to interact with Nm23-H1 [155]. The results showed that E7 expressing cells are resistance to Granzyme A-induced apoptosis and promotes cell invasiveness [155]. The authors further suggested that disrupting the multifunctional activity of Nm23-H1 by E7 promotes cell transformation as well as tumor progression [155]. It would be interesting to know whether or not the activity of E7 and Nm23H1 can hold up in vivo using a mouse model system similar to that shown for the EBV studies. One would suspect that the same would hold true for most of the antigens encoded by well-known tumor viruses.
KSHV and Nm23-H1
The Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi's sarcoma (KS) [156]. Despite the reduced incidence of HIV-associated KS in the era of highly active anti-retroviral therapy, KS is still considered to be one of the most common HIV-associated tumors in the modern era and an important cause of morbidity and mortality in patients receiving solid organ transplants [157-159]. In addition to KS tumors, KSHV has also been causally associated with two types of lymphoproliferative diseases, primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD) [156,160]. Attainment of an invasive phenotype represents one of the major hallmarks of KSHV-infected cells and is thought to be a key pathogenic mechanism for the development of the KS phenotype [161]. Similar to EBV, KSHV infection is also largely latent and during latency, only a small subset of viral genes is expressed [63,156]. KSHV encoded latency-associated nuclear antigen (LANA) is one of the crucial genes expressed during latency and is necessary and sufficient for episome persistence in the absence of other virus encoded genes [63]. In addition to episomal maintenance, LANA functions as a transcriptional modulator of numerous cellular and viral promoters, including its own promoter [63,156]. LANA mediated transcriptional activation was extensively reported to be accompanied by many cellular proteins including ATF, AP-1, CAAT, and Sp1 [156]. LANA has also been shown to have transcriptional repressive effects through interacting with many cellular co-repressors including mSin3, SAP30, CIR, MeCP2 and SUV39H1 [156].
A large body of evidence supports a role for LANA in regulation of many metastasis suppressor genes during KSHV-mediated pathogenesis [162-164]. Recently, a more direct function for LANA in connection withmetastasis regulation has been established [165]. The study showed that LANA augments expression as well as nuclear translocation of Nm23-H1 [165] similar to that seen for the EBV antigens EBNA1 and EBNA3C. The study also suggested that activation of the Ras-BRaf-MAPK signal transduction pathway, secretion of pro-migratory factors associated with this pathway, and KSHV-induced cell invasiveness are critically dependent on deregulation of Nm23-H1 activity [165]. Interestingly, induction of cytoplasmic expression of Nm23-H1 using a pharmacologic inhibitor of DNA methylation has shown marked reduction in activation of KSHV-associated Ras-BRaf-MAPK pathway and cell invasiveness, suggesting a potential therapeutic approach against KSHV associated human cancers [165].
The link between viral antigens and Nm23-H1 has clearly piqued interest as a potential mechanism by which these tumor viruses can also regulate the aggressiveness of the tumors. Viral associated tumors are known to be highly invasive and when associated with high risk types of tumor viruses including HPV 16 and 18 in cervical carcinomas or EBV associated nasopharyngeal carcinomas these tumors tend towards been more aggressive and invasive (refs). It would be interesting to know whether or not these tumors also have changes in the levels of Nm23-H1 and that they can usurp the anti-metastatic functions of Nm23-H1 as pro-metastatic agents.
CONCLUSION
Studies exploring the mechanisms important for the metastatic process have advanced tremendously in the last two decades, largely due to the collaboration of a diverse cohort of investigators who have focused their expertise within several disciplines. The ultimate goal of these studies is to develop novel anti-metastatic therapies which are based upon the mechanistic controlsunderlying the metastatic process. Hence, a complete understanding of the biological phenomena of metastasis suppressor genes and the corresponding proteins is fundamental to achieving this goal. Tumor viruses encoded antigens not only can initiate the development of certain types of cancers but are also critically engaged in propagation of the primary tumors from the initial sites by targeting several metastasis suppressor genes [61,63,155,165]. Thus, these viral antigens could also serve as potential candidates in development of prophylactic vaccination strategies and along with the metastasis suppressor genes could also be critical for targeted therapeutic development against tumor virus associated human cancers. Promising steps towards this goal have recently been taken. For example, investigators have been able to restore the normal expression of metastasis suppressor genes, specifically Nm23-H1, in cancer cells [34,35]. Nonetheless, whether or not drug induced activation of metastasis suppressor genes can also be attained in clinical trials remains one of the major open questions and is yet to be elucidated.
ACKNOWLEDGMENTS
This work was supported by grants from the Leukemia and Lymphoma Society of America and Public Health Service grants from the NCI (CA108461, CA137894, and CA091792). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Rinker-Schaeffer CW, O'Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin Cancer Res. 2006;12:3882–9. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:247–58. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- [3].van Golen KL, Risin S, Staroselsky A, Berger D, Tainsky MA, Pathak S, Price JE. Predominance of the metastatic phenotype in hybrids formed by fusion of mouse and human melanoma clones. Clin Exp Metastasis. 1996;14:95–106. doi: 10.1007/BF00121206. [DOI] [PubMed] [Google Scholar]
- [4].Rosengard AM, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–80. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- [5].Stahl JA, Leone A, Rosengard AM, Porter L, King CR, Steeg PS. Identification of a second human nm23 gene, nm23-H2. Cancer Res. 1991;51:445–9. [PubMed] [Google Scholar]
- [6].Venturelli D, et al. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proc Natl Acad Sci U S A. 1995;92:7435–9. doi: 10.1073/pnas.92.16.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Milon L, Rousseau-Merck MF, Munier A, Erent M, Lascu I, Capeau J, Lacombe ML. nm23-H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localised on chromosome 16p13. Hum Genet. 1997;99:550–7. doi: 10.1007/s004390050405. [DOI] [PubMed] [Google Scholar]
- [8].Munier A, Feral C, Milon L, Pinon VP, Gyapay G, Capeau J, Guellaen G, Lacombe ML. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 1998;434:289–94. doi: 10.1016/s0014-5793(98)00996-x. [DOI] [PubMed] [Google Scholar]
- [9].Mehus JG, Deloukas P, Lambeth DO. NME6: a new member of the nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Hum Genet. 1999;104:454–9. doi: 10.1007/s004390050987. [DOI] [PubMed] [Google Scholar]
- [10].Tsuiki H, et al. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J Cell Biochem. 1999;76:254–69. doi: 10.1002/(sici)1097-4644(20000201)76:2<254::aid-jcb9>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- [11].Padma P, Hozumi A, Ogawa K, Inaba K. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene. 2001;275:177–83. doi: 10.1016/s0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- [12].Nallamothu G, Woolworth JA, Dammai V, Hsu T. Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol Cell Biol. 2008;28:1964–73. doi: 10.1128/MCB.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81:10352–61. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, Daniel JY, Lacombe ML. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–45. doi: 10.1093/jnci/dji143. [DOI] [PubMed] [Google Scholar]
- [15].Backer JM, et al. Chromosomal localization and nucleoside diphosphate kinase activity of human metastasis-suppressor genes NM23-1 and NM23-2. Oncogene. 1993;8:497–502. [PubMed] [Google Scholar]
- [16].Tee YT, Chen GD, Lin LY, Ko JL, Wang PH. Nm23-H1: a metastasis-associated gene. Taiwan J Obstet Gynecol. 2006;45:107–13. doi: 10.1016/S1028-4559(09)60206-0. [DOI] [PubMed] [Google Scholar]
- [17].Bosnar MH, De Gunzburg J, Bago R, Brecevic L, Weber I, Pavelic J. Subcellular localization of A and B Nm23/NDPK subunits. Exp Cell Res. 2004;298:275–84. doi: 10.1016/j.yexcr.2004.04.018. [DOI] [PubMed] [Google Scholar]
- [18].Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–4. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- [19].Urano T, Takamiya K, Furukawa K, Shiku H. Molecular cloning and functional expression of the second mouse nm23/NDP kinase gene, nm23-M2. FEBS Lett. 1992;309:358–62. doi: 10.1016/0014-5793(92)80807-s. [DOI] [PubMed] [Google Scholar]
- [20].Lee E, Jeong J, Kim SE, Song EJ, Kang SW, Lee KJ. Multiple functions of Nm23-H1 are regulated by oxido-reduction system. PLoS One. 2009;4:e7949. doi: 10.1371/journal.pone.0007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Okabe-Kado J, Kasukabe T, Honma Y, Hanada R, Nakagawara A, Kaneko Y. Clinical significance of serum NM23-H1 protein in neuroblastoma. Cancer Sci. 2005;96:653–60. doi: 10.1111/j.1349-7006.2005.00091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sgouros J, Galani E, Gonos E, Moutsatsou P, Belechri M, Skarlos D, Dionyssiou-Asteriou A. Correlation of nm23-H1 gene expression with clinical outcome in patients with advanced breast cancer. In Vivo. 2007;21:519–22. [PubMed] [Google Scholar]
- [23].Ilijas M, Pavelic K, Sarcevic B, Kapitanovic S, Kurjak A, Stambrook P, Gluckman J, Pavelic Z. Expression of nm23-h1 gene in squamous-cell carcinoma of the cervix correlates with 5-year survival. Int J Oncol. 1994;5:1455–7. doi: 10.3892/ijo.5.6.1455. [DOI] [PubMed] [Google Scholar]
- [24].Ouatas T, Salerno M, Palmieri D, Steeg PS. Basic and translational advances in cancer metastasis: Nm23. J Bioenerg Biomembr. 2003;35:73–9. doi: 10.1023/a:1023497924277. [DOI] [PubMed] [Google Scholar]
- [25].Bevilacqua G. NM23 gene expression and human breast cancer metastases. Pathol Biol (Paris) 1990;38:774–5. [PubMed] [Google Scholar]
- [26].Hennessy C, Henry JA, May FE, Westley BR, Angus B, Lennard TW. Expression of anti-metastatic gene nm23. Br J Cancer. 1991;63:1024. doi: 10.1038/bjc.1991.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Florenes VA, Aamdal S, Myklebost O, Maelandsmo GM, Bruland OS, Fodstad O. Levels of nm23 messenger RNA in metastatic malignant melanomas: inverse correlation to disease progression. Cancer Res. 1992;52:6088–91. [PubMed] [Google Scholar]
- [28].van Noesel MM, Versteeg R. Pediatric neuroblastomas: genetic and epigenetic 'danse macabre'. Gene. 2004;325:1–15. doi: 10.1016/j.gene.2003.09.042. [DOI] [PubMed] [Google Scholar]
- [29].Chang CL, et al. Nm23-H1 mutation in neuroblastoma. Nature. 1994;370:335–6. doi: 10.1038/370335a0. [DOI] [PubMed] [Google Scholar]
- [30].Yokoyama A, et al. Evaluation by multivariate analysis of the differentiation inhibitory factor nm23 as a prognostic factor in acute myelogenous leukemia and application to other hematologic malignancies. Blood. 1998;91:1845–51. [PubMed] [Google Scholar]
- [31].Kaetzel DM, McCorkle JR, Novak M, Yang M, Jarrett SG. Potential contributions of antimutator activity to the metastasis suppressor function of NM23-H1. Mol Cell Biochem. 2009;329:161–5. doi: 10.1007/s11010-009-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rochdi MD, et al. Nm23-H2 interacts with a G protein-coupled receptor to regulate its endocytosis through an Rac1-dependent mechanism. J Biol Chem. 2004;279:18981–9. doi: 10.1074/jbc.M312621200. [DOI] [PubMed] [Google Scholar]
- [33].Ishikawa N, Shimada N, Takagi Y, Ishijima Y, Fukuda M, Kimura N. Molecular evolution of nucleoside diphosphate kinase genes: conserved core structures and multiple-layered regulatory regions. J Bioenerg Biomembr. 2003;35:7–18. doi: 10.1023/a:1023433504713. [DOI] [PubMed] [Google Scholar]
- [34].Steeg PS, Horak CE, Miller KD. Clinical-translational approaches to the Nm23-H1 metastasis suppressor. Clin Cancer Res. 2008;14:5006–12. doi: 10.1158/1078-0432.CCR-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marshall JC, Collins J, Marino N, Steeg P. The Nm23-H1 metastasis suppressor as a translational target. Eur J Cancer. 2010;46:1278–82. doi: 10.1016/j.ejca.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem. 1996;271:25107–16. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- [37].Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci U S A. 1997;94:9000–5. doi: 10.1073/pnas.94.17.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–99. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- [39].MacDonald NJ, De la Rosa A, Benedict MA, Freije JM, Krutsch H, Steeg PS. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem. 1993;268:25780–9. [PubMed] [Google Scholar]
- [40].Kim HD, Youn B, Kim TS, Kim SH, Shin HS, Kim J. Regulators affecting the metastasis suppressor activity of Nm23-H1. Mol Cell Biochem. 2009;329:167–73. doi: 10.1007/s11010-009-0109-2. [DOI] [PubMed] [Google Scholar]
- [41].Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- [42].Rumjahn SM, Baldwin KA, Buxton IL. P2y receptor-mediated angiogenesis via vascular endothelial growth factor receptor 2 signaling. Proc West Pharmacol Soc. 2007;50:58–60. [PMC free article] [PubMed] [Google Scholar]
- [43].Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–42. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- [44].Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, Russo ML, Jaju M, Lieberman J. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276:43285–93. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- [45].Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- [46].Goswami SC, Yoon JH, Abramczyk BM, Pfeifer GP, Postel EH. Molecular and functional interactions between Escherichia coli nucleoside-diphosphate kinase and the uracil-DNA glycosylase Ung. J Biol Chem. 2006;281:32131–9. doi: 10.1074/jbc.M604937200. [DOI] [PubMed] [Google Scholar]
- [47].Seong HA, Jung H, Choi HS, Kim KT, Ha H. Regulation of transforming growth factor-beta signaling and PDK1 kinase activity by physical interaction between PDK1 and serine-threonine kinase receptor-associated protein. J Biol Chem. 2005;280:42897–908. doi: 10.1074/jbc.M507539200. [DOI] [PubMed] [Google Scholar]
- [48].Seong HA, Jung H, Ha H. NM23-H1 tumor suppressor physically interacts with serine-threonine kinase receptor-associated protein, a transforming growth factor-beta (TGF-beta) receptor-interacting protein, and negatively regulates TGF-beta signaling. J Biol Chem. 2007;282:12075–96. doi: 10.1074/jbc.M609832200. [DOI] [PubMed] [Google Scholar]
- [49].Jung H, Seong HA, Ha H. NM23-H1 tumor suppressor and its interacting partner STRAP activate p53 function. J Biol Chem. 2007;282:35293–307. doi: 10.1074/jbc.M705181200. [DOI] [PubMed] [Google Scholar]
- [50].Reymond A, Volorio S, Merla G, Al-Maghtheh M, Zuffardi O, Bulfone A, Ballabio A, Zollo M. Evidence for interaction between human PRUNE and nm23-H1 NDPKinase. Oncogene. 1999;18:7244–52. doi: 10.1038/sj.onc.1203140. [DOI] [PubMed] [Google Scholar]
- [51].Forus A, et al. Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas-a possible mechanism for altering the nm23-H1 activity. Oncogene. 2001;20:6881–90. doi: 10.1038/sj.onc.1204874. [DOI] [PubMed] [Google Scholar]
- [52].D'Angelo A, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–49. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- [53].Oue N, Yoshida K, Noguchi T, Sentani K, Kikuchi A, Yasui W. Increased expression of h-prune is associated with tumor progression and poor survival in gastric cancer. Cancer Sci. 2007;98:1198–205. doi: 10.1111/j.1349-7006.2007.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Garzia L, et al. Phosphorylation of nm23-H1 by CKI induces its complex formation with h-prune and promotes cell motility. Oncogene. 2008;27:1853–64. doi: 10.1038/sj.onc.1210822. [DOI] [PubMed] [Google Scholar]
- [55].Otsuki Y, Tanaka M, Yoshii S, Kawazoe N, Nakaya K, Sugimura H. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci U S A. 2001;98:4385–90. doi: 10.1073/pnas.071411598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Murakami M, Meneses PI, Knight JS, Lan K, Kaul R, Verma SC, Robertson ES. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int J Cancer. 2008;123:500–10. doi: 10.1002/ijc.23568. [DOI] [PubMed] [Google Scholar]
- [57].Curtis CD, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor alpha alters estrogen-responsive gene expression. Cancer Res. 2007;67:10600–7. doi: 10.1158/0008-5472.CAN-07-0055. [DOI] [PubMed] [Google Scholar]
- [58].Zhu J, Tseng YH, Kantor JD, Rhodes CJ, Zetter BR, Moyers JS, Kahn CR. Interaction of the Ras-related protein associated with diabetes rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc Natl Acad Sci U S A. 1999;96:14911–8. doi: 10.1073/pnas.96.26.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Switzer CH, Cheng RY, Vitek TM, Christensen DJ, Wink DA, Vitek MP. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30:2504–13. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Marino N, Marshall JC, Steeg PS. Protein-protein interactions: a mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch Pharmacol. 2011 doi: 10.1007/s00210-011-0646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Subramanian C, Cotter MA, 2nd, Robertson ES. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med. 2001;7:350–5. doi: 10.1038/85499. [DOI] [PubMed] [Google Scholar]
- [62].Murakami M, Kaul R, Kumar P, Robertson ES. Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus. Mol Cell Biochem. 2009;329:131–9. doi: 10.1007/s11010-009-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saha A, Kaul R, Murakami M, Robertson ES. Tumor viruses and cancer biology: Modulating signaling pathways for therapeutic intervention. Cancer Biol Ther. 2010;10:961–78. doi: 10.4161/cbt.10.10.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, Cai Q, Robertson ES. Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog. 2011;7:e1001275. doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Saha A, Robertson ES. Epstein-Barr Virus-Associated B-cell Lymphomas: Pathogenesis and Clinical Outcomes. Clin Cancer Res. 2011;17:3056–63. doi: 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang Y, Liu X, Xing X, Cui Y, Zhao C, Luo B. Variations of Epstein-Barr virus nuclear antigen 1 gene in gastric carcinomas and nasopharyngeal carcinomas from Northern China. Virus Res. 2010;147:258–64. doi: 10.1016/j.virusres.2009.11.010. [DOI] [PubMed] [Google Scholar]
- [67].Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A. 2006;103:19500–5. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A. 1993;90:9150–4. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lee MA, Diamond ME, Yates JL. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–82. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cohen JI, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A. 1989;86:9558–62. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–25. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Apcher S, Daskalogianni C, Manoury B, Fahraeus R. Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation. PLoS Pathog. 2010;6:e1001151. doi: 10.1371/journal.ppat.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kang MS, et al. Epstein-Barr virus nuclear antigen 1 does not induce lymphoma in transgenic FVB mice. Proc Natl Acad Sci U S A. 2005;102:820–5. doi: 10.1073/pnas.0408774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kang MS, Soni V, Bronson R, Kieff E. Epstein-Barr virus nuclear antigen 1 does not cause lymphoma in C57BL/6J mice. J Virol. 2008;82:4180–3. doi: 10.1128/JVI.02596-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wilson JB, Bell JL, Levine AJ. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 1996;15:3117–26. [PMC free article] [PubMed] [Google Scholar]
- [76].Lindner SE, Sugden B. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–68. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- [78].Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–5. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- [79].Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–92. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dheekollu J, Lieberman PM. The replisome pausing factor timeless is required for episomal maintenance of latent epstein-barr virus. J Virol. 2011;85:5853–63. doi: 10.1128/JVI.02425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78:11487–505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hung SC, Kang MS, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci U S A. 2001;98:1865–70. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Norseen J, Johnson FB, Lieberman PM. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol. 2009;83:10336–46. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Habeshaw G, Yao QY, Bell AI, Morton D, Rickinson AB. Epstein-barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt's lymphoma reflect virus strains prevalent in different geographic areas. J Virol. 1999;73:965–75. doi: 10.1128/jvi.73.2.965-975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–8. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- [86].Blake N, et al. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- [87].Kamranvar SA, Gruhne B, Szeles A, Masucci MG. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene. 2007;26:5115–23. doi: 10.1038/sj.onc.1210324. [DOI] [PubMed] [Google Scholar]
- [88].Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc Natl Acad Sci U S A. 2009;106:2313–8. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Owen TJ, et al. Epstein-Barr virus-encoded EBNA1 enhances RNA polymerase III-dependent EBER expression through induction of EBER-associated cellular transcription factors. Mol Cancer. 2010;9:241. doi: 10.1186/1476-4598-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cheng TC, Hsieh SS, Hsu WL, Chen YF, Ho HH, Sheu LF. Expression of Epstein-Barr nuclear antigen 1 in gastric carcinoma cells is associated with enhanced tumorigenicity and reduced cisplatin sensitivity. Int J Oncol. 2010;36:151–60. [PubMed] [Google Scholar]
- [91].Kennedy G, Komano J, Sugden B. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc Natl Acad Sci U S A. 2003;100:14269–74. doi: 10.1073/pnas.2336099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hong M, et al. Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA interference inhibits proliferation of EBV-positive Burkitt's lymphoma cells. J Cancer Res Clin Oncol. 2006;132:1–8. doi: 10.1007/s00432-005-0036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yin Q, Flemington EK. siRNAs against the Epstein Barr virus latency replication factor, EBNA1, inhibit its function and growth of EBV-dependent tumor cells. Virology. 2006;346:385–93. doi: 10.1016/j.virol.2005.11.021. [DOI] [PubMed] [Google Scholar]
- [94].Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010;6:e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Le Roux A, Kerdiles B, Walls D, Dedieu JF, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- [96].Cludts I, Farrell PJ. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J Virol. 1998;72:1862–9. doi: 10.1128/jvi.72.3.1862-1869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J Virol. 1996;70:5909–15. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A. 2011;108:1919–24. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lin J, Johannsen E, Robertson E, Kieff E. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76:232–42. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A. 2005;102:18562–6. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Radkov SA, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday MJ. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–97. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bajaj BG, Murakami M, Cai Q, Verma SC, Lan K, Robertson ES. Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J Virol. 2008;82:4082–90. doi: 10.1128/JVI.02500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Murakami M, Lan K, Subramanian C, Robertson ES. Epstein-Barr virus nuclear antigen 1 interacts with Nm23-H1 in lymphoblastoid cell lines and inhibits its ability to suppress cell migration. J Virol. 2005;79:1559–68. doi: 10.1128/JVI.79.3.1559-1568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Subramanian C, Robertson ES. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J Virol. 2002;76:8702–9. doi: 10.1128/JVI.76.17.8702-8709.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kaul R, Murakami M, Lan K, Choudhuri T, Robertson ES. EBNA3C can modulate the activities of the transcription factor Necdin in association with metastasis suppressor protein Nm23-H1. J Virol. 2009;83:4871–83. doi: 10.1128/JVI.02286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–12. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- [107].Niinobe M, Koyama K, Yoshikawa K. Cellular and subcellular localization of necdin in fetal and adult mouse brain. Dev Neurosci. 2000;22:310–9. doi: 10.1159/000017455. [DOI] [PubMed] [Google Scholar]
- [108].Matsumoto K, Taniura H, Uetsuki T, Yoshikawa K. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001;272:173–9. doi: 10.1016/s0378-1119(01)00544-3. [DOI] [PubMed] [Google Scholar]
- [109].Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28:8772–84. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Moon HE, Ahn MY, Park JA, Min KJ, Kwon YW, Kim KW. Negative regulation of hypoxia inducible factor-1alpha by necdin. FEBS Lett. 2005;579:3797–801. doi: 10.1016/j.febslet.2005.05.072. [DOI] [PubMed] [Google Scholar]
- [111].Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–82. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- [112].Herschman HR. Prostaglandin synthase 2. Biochim Biophys Acta. 1996;1299:125–40. doi: 10.1016/0005-2760(95)00194-8. [DOI] [PubMed] [Google Scholar]
- [113].Levy GN. Prostaglandin H synthases, nonsteroidal anti-inflammatory drugs, and colon cancer. FASEB J. 1997;11:234–47. [PubMed] [Google Scholar]
- [114].Needleman P, Turk J, Jakschik BA, Morrison AR, Lefkowith JB. Arachidonic acid metabolism. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- [115].Sano H, et al. Expression of cyclooxygenase-1 and -2 in human colorectal cancer. Cancer Res. 1995;55:3785–9. [PubMed] [Google Scholar]
- [116].Wolff H, Saukkonen K, Anttila S, Karjalainen A, Vainio H, Ristimaki A. Expression of cyclooxygenase-2 in human lung carcinoma. Cancer Res. 1998;58:4997–5001. [PubMed] [Google Scholar]
- [117].Hwang D, Scollard D, Byrne J, Levine E. Expression of cyclooxygenase-1 and cyclooxygenase-2 in human breast cancer. J Natl Cancer Inst. 1998;90:455–60. doi: 10.1093/jnci/90.6.455. [DOI] [PubMed] [Google Scholar]
- [118].Ristimaki A, Honkanen N, Jankala H, Sipponen P, Harkonen M. Expression of cyclooxygenase-2 in human gastric carcinoma. Cancer Res. 1997;57:1276–80. [PubMed] [Google Scholar]
- [119].Zimmermann KC, Sarbia M, Weber AA, Borchard F, Gabbert HE, Schror K. Cyclooxygenase-2 expression in human esophageal carcinoma. Cancer Res. 1999;59:198–204. [PubMed] [Google Scholar]
- [120].Molina MA, Sitja-Arnau M, Lemoine MG, Frazier ML, Sinicrope FA. Increased cyclooxygenase-2 expression in human pancreatic carcinomas and cell lines: growth inhibition by nonsteroidal anti-inflammatory drugs. Cancer Res. 1999;59:4356–62. [PubMed] [Google Scholar]
- [121].Peng JP, Chang HC, Hwang CF, Hung WC. Overexpression of cyclooxygenase-2 in nasopharyngeal carcinoma and association with lymph node metastasis. Oral Oncol. 2005;41:903–8. doi: 10.1016/j.oraloncology.2005.05.003. [DOI] [PubMed] [Google Scholar]
- [122].Sawaoka H, Kawano S, Tsuji S, Tsujii M, Gunawan ES, Takei Y, Nagano K, Hori M. Cyclooxygenase-2 inhibitors suppress the growth of gastric cancer xenografts via induction of apoptosis in nude mice. Am J Physiol. 1998;274:G1061–7. doi: 10.1152/ajpgi.1998.274.6.G1061. [DOI] [PubMed] [Google Scholar]
- [123].Kaul R, Verma SC, Murakami M, Lan K, Choudhuri T, Robertson ES. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J Virol. 2006;80:1321–31. doi: 10.1128/JVI.80.3.1321-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Murono S, Inoue H, Tanabe T, Joab I, Yoshizaki T, Furukawa M, Pagano JS. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc Natl Acad Sci U S A. 2001;98:6905–10. doi: 10.1073/pnas.121016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ray N, Bisher ME, Enquist LW. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J Virol. 2004;78:12964–74. doi: 10.1128/JVI.78.23.12964-12974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–33. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- [127].Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–8. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- [128].Reddy KB, Krueger JS, Kondapaka SB, Diglio CA. Mitogen-activated protein kinase (MAPK) regulates the expression of progelatinase B (MMP-9) in breast epithelial cells. Int J Cancer. 1999;82:268–73. doi: 10.1002/(sici)1097-0215(19990719)82:2<268::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- [129].Murono S, Yoshizaki T, Sato H, Takeshita H, Furukawa M, Pagano JS. Aspirin inhibits tumor cell invasiveness induced by Epstein-Barr virus latent membrane protein 1 through suppression of matrix metalloproteinase-9 expression. Cancer Res. 2000;60:2555–61. [PubMed] [Google Scholar]
- [130].Gaudin P, et al. TIMP-1/MMP-9 imbalance in an EBV-immortalized B lymphocyte cellular model: evidence for TIMP-1 multifunctional properties. Biochim Biophys Acta. 2000;1499:19–33. doi: 10.1016/s0167-4889(00)00084-7. [DOI] [PubMed] [Google Scholar]
- [131].Trocme C, Gaudin P, Berthier S, Barro C, Zaoui P, Morel F. Human B lymphocytes synthesize the 92-kDa gelatinase, matrix metalloproteinase-9. J Biol Chem. 1998;273:20677–84. doi: 10.1074/jbc.273.32.20677. [DOI] [PubMed] [Google Scholar]
- [132].Takeshita H, Yoshizaki T, Miller WE, Sato H, Furukawa M, Pagano JS, Raab-Traub N. Matrix metalloproteinase 9 expression is induced by Epstein-Barr virus latent membrane protein 1 C-terminal activation regions 1 and 2. J Virol. 1999;73:5548–55. doi: 10.1128/jvi.73.7.5548-5555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Yoshizaki T, Sato H, Furukawa M, Pagano JS. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc Natl Acad Sci U S A. 1998;95:3621–6. doi: 10.1073/pnas.95.7.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kuppers DA, Lan K, Knight JS, Robertson ES. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J Virol. 2005;79:9714–24. doi: 10.1128/JVI.79.15.9714-9724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- [136].Zhang Z, Morla AO, Vuori K, Bauer JS, Juliano RL, Ruoslahti E. The alpha v beta 1 integrin functions as a fibronectin receptor but does not support fibronectin matrix assembly and cell migration on fibronectin. J Cell Biol. 1993;122:235–42. doi: 10.1083/jcb.122.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Denhardt DT, Guo X. Osteopontin: a protein with diverse functions. FASEB J. 1993;7:1475–82. [PubMed] [Google Scholar]
- [138].Busk M, Pytela R, Sheppard D. Characterization of the integrin alpha v beta 6 as a fibronectin-binding protein. J Biol Chem. 1992;267:5790–6. [PubMed] [Google Scholar]
- [139].Charo IF, Nannizzi L, Smith JW, Cheresh DA. The vitronectin receptor alpha v beta 3 binds fibronectin and acts in concert with alpha 5 beta 1 in promoting cellular attachment and spreading on fibronectin. J Cell Biol. 1990;111:2795–800. doi: 10.1083/jcb.111.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Smith JW, Vestal DJ, Irwin SV, Burke TA, Cheresh DA. Purification and functional characterization of integrin alpha v beta 5. An adhesion receptor for vitronectin. J Biol Chem. 1990;265:11008–13. [PubMed] [Google Scholar]
- [141].Brodt P, Fallavollita L, Sawka RJ, Shibata P, Nip J, Kim U, Shibata H. Tumor cell adhesion to frozen lymph node sections--a correlate of lymphatic metastasis in breast carcinoma models of human and rat origin. Breast Cancer Res Treat. 1990;17:109–20. doi: 10.1007/BF01806291. [DOI] [PubMed] [Google Scholar]
- [142].Filardo EJ, Brooks PC, Deming SL, Damsky C, Cheresh DA. Requirement of the NPXY motif in the integrin beta 3 subunit cytoplasmic tail for melanoma cell migration in vitro and in vivo. J Cell Biol. 1995;130:441–50. doi: 10.1083/jcb.130.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Albelda SM, Mette SA, Elder DE, Stewart R, Damjanovich L, Herlyn M, Buck CA. Integrin distribution in malignant melanoma: association of the beta 3 subunit with tumor progression. Cancer Res. 1990;50:6757–64. [PubMed] [Google Scholar]
- [144].Brooks PC, Stromblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin alpha v beta 3. Cell. 1996;85:683–93. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- [145].Huang S, Stupack D, Liu A, Cheresh D, Nemerow GR. Cell growth and matrix invasion of EBV-immortalized human B lymphocytes is regulated by expression of alpha(v) integrins. Oncogene. 2000;19:1915–23. doi: 10.1038/sj.onc.1203509. [DOI] [PubMed] [Google Scholar]
- [146].Choudhuri T, Verma SC, Lan K, Robertson ES. Expression of alpha V integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology. 2006;351:58–72. doi: 10.1016/j.virol.2006.03.031. [DOI] [PubMed] [Google Scholar]
- [147].Klosky JL, Gamble HL, Spunt SL, Randolph ME, Green DM, Hudson MM. Human papillomavirus vaccination in survivors of childhood cancer. Cancer. 2009;115:5627–36. doi: 10.1002/cncr.24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Foresta C, Patassini C, Bertoldo A, Menegazzo M, Francavilla F, Barzon L, Ferlin A. Mechanism of human papillomavirus binding to human spermatozoa and fertilizing ability of infected spermatozoa. PLoS One. 2011;6:e15036. doi: 10.1371/journal.pone.0015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Koutsky L. Epidemiology of genital human papillomavirus infection. Am J Med. 1997;102:3–8. doi: 10.1016/s0002-9343(97)00177-0. [DOI] [PubMed] [Google Scholar]
- [150].Giuliano AR, et al. Epidemiology of human papillomavirus infection in men, cancers other than cervical and benign conditions. Vaccine. 2008;26(Suppl 10):K17–28. doi: 10.1016/j.vaccine.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- [152].Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci U S A. 2000;97:10002–7. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Munger K, Basile JR, Duensing S, Eichten A, Gonzalez SL, Grace M, Zacny VL. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–98. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- [154].Zwerschke W, Jansen-Durr P. Cell transformation by the E7 oncoprotein of human papillomavirus type 16: interactions with nuclear and cytoplasmic target proteins. Adv Cancer Res. 2000;78:1–29. doi: 10.1016/s0065-230x(08)61022-2. [DOI] [PubMed] [Google Scholar]
- [155].Mileo AM, Piombino E, Severino A, Tritarelli A, Paggi MG, Lombardi D. Multiple interference of the human papillomavirus-16 E7 oncoprotein with the functional role of the metastasis suppressor Nm23-H1 protein. J Bioenerg Biomembr. 2006;38:215–25. doi: 10.1007/s10863-006-9037-y. [DOI] [PubMed] [Google Scholar]
- [156].Cai Q, Verma SC, Lu J, Robertson ES. Molecular biology of Kaposi's sarcoma-associated herpesvirus and related oncogenesis. Adv Virus Res. 2010;78:87–142. doi: 10.1016/B978-0-12-385032-4.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Vanni T, Sprinz E, Machado MW, Santana Rde C, Fonseca BA, Schwartsmann G. Systemic treatment of AIDS-related Kaposi sarcoma: current status and perspectives. Cancer Treat Rev. 2006;32:445–55. doi: 10.1016/j.ctrv.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [158].Bonnet F, et al. Malignancy-related causes of death in human immunodeficiency virus-infected patients in the era of highly active antiretroviral therapy. Cancer. 2004;101:317–24. doi: 10.1002/cncr.20354. [DOI] [PubMed] [Google Scholar]
- [159].Engels EA, et al. Cancer risk in people infected with human immunodeficiency virus in the United States. Int J Cancer. 2008;123:187–94. doi: 10.1002/ijc.23487. [DOI] [PubMed] [Google Scholar]
- [160].Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc Natl Acad Sci U S A. 2009;106:11725–30. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Qian LW, Xie J, Ye F, Gao SJ. Kaposi's sarcoma-associated herpesvirus infection promotes invasion of primary human umbilical vein endothelial cells by inducing matrix metalloproteinases. J Virol. 2007;81:7001–10. doi: 10.1128/JVI.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Di Bartolo DL, Cannon M, Liu YF, Renne R, Chadburn A, Boshoff C, Cesarman E. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood. 2008;111:4731–40. doi: 10.1182/blood-2007-09-110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Platt G, Carbone A, Mittnacht S. p16INK4a loss and sensitivity in KSHV associated primary effusion lymphoma. Oncogene. 2002;21:1823–31. doi: 10.1038/sj.onc.1205360. [DOI] [PubMed] [Google Scholar]
- [164].Shamay M, Krithivas A, Zhang J, Hayward SD. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc Natl Acad Sci U S A. 2006;103:14554–9. doi: 10.1073/pnas.0604469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Qin Z, Dai L, Toole B, Robertson E, Parsons C. Regulation of Nm23-H1 and cell invasiveness by Kaposi's sarcoma-associated herpesvirus. J Virol. 2011;85:3596–606. doi: 10.1128/JVI.01596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]