Abstract
Type 1 narcolepsy, a disorder caused by a lack of hypocretin (orexin), is so strongly associated with human leukocyte antigen (HLA) class II HLA-DQA1∗01:02-DQB1∗06:02 (DQ0602) that very few non-DQ0602 cases have been reported. A known triggering factor for narcolepsy is pandemic 2009 influenza H1N1, suggesting autoimmunity triggered by upper-airway infections. Additional effects of other HLA-DQ alleles have been reported consistently across multiple ethnic groups. Using over 3,000 case and 10,000 control individuals of European and Chinese background, we examined the effects of other HLA loci. After careful matching of HLA-DR and HLA-DQ in case and control individuals, we found strong protective effects of HLA-DPA1∗01:03-DPB1∗04:02 (DP0402; odds ratio [OR] = 0.51 [0.38–0.67], p = 1.01 × 10−6) and HLA-DPA1∗01:03-DPB1∗04:01 (DP0401; OR = 0.61 [0.47–0.80], p = 2.07 × 10−4) and predisposing effects of HLA-DPB1∗05:01 in Asians (OR = 1.76 [1.34–2.31], p = 4.71 × 10−05). Similar effects were found by conditional analysis controlling for HLA-DR and HLA-DQ with DP0402 (OR = 0.45 [0.38–0.55] p = 8.99 × 10−17) and DP0501 (OR = 1.38 [1.18–1.61], p = 7.11 × 10−5). HLA-class-II-independent associations with HLA-A∗11:01 (OR = 1.32 [1.13–1.54], p = 4.92 × 10−4), HLA-B∗35:03 (OR = 1.96 [1.41–2.70], p = 5.14 × 10−5), and HLA-B∗51:01 (OR = 1.49 [1.25–1.78], p = 1.09 × 10−5) were also seen across ethnic groups in the HLA class I region. These effects might reflect modulation of autoimmunity or indirect effects of HLA class I and HLA-DP alleles on response to viral infections such as that of influenza.
Introduction
Type 1 narcolepsy (MIM 161400) is a life-long disorder characterized by sleepiness, cataplexy, and rapid-eye-movement sleep abnormalities. Onset usually occurs in children, adolescents, or young adults. The disease is caused by the loss of hypocretin-producing cells in the lateral hypothalamus.1 Narcolepsy is strongly associated with a specific human leukocyte antigen (HLA) class II molecule, the DQα0102–DQβ0602 heterodimer (abbreviated DQ0602), which is shared by 98% of narcoleptics across ethnic groups and encoded by the HLA-DQA1∗01:02∼DQB1∗06:02 haplotype.2,3 DQ0602 is present in 12%–38% of control individuals across ethnic groups. Genome-wide association studies (GWASs) in narcolepsy have also found associations with loci related to autoimmunity, such as T cell receptor (TCR) loci (TRA [MIM 186880], TRB [MIM 186930]), IL10RB [MIM 123889], IFNAR1 [MIM 107450], CTSH [MIM 116820], P2RY11 [MIM 602697], and ZNF365 [MIM 607818].4–6 These results suggest autoimmune-mediated hypocretin cell destruction that might involve antigen presentation by DQ0602 to CD4+ T cells.
In addition, narcolepsy has a strong environmental component, and most monozygotic twins are discordant.7 In children, where onset is often abrupt and more easily documented, narcolepsy is highly seasonal in that it peaks in the spring or summer.8 Onset follows upper-airway infections, notably of influenza (MIM 614680) or Streptococcus pyogenes (MIM 607395), suggesting triggering effects of infections.8,9
A 4- to 6-fold increase in childhood narcolepsy onset was observed in the spring and summer of 2010, following the 2009 H1N1 swine pandemic flu in China.5,10 Further, vaccination with Pandemrix, an AS03-adjuvanted pandemic H1N1 vaccine approved for use in Europe, was associated with a 3- to 17-fold increased risk of developing childhood narcolepsy in multiple countries, leading to increased incidence in Scandinavia.11–17 For unclear reasons, increased risk of narcolepsy after the use of other H1N1 vaccines has not been reported, and is unlikely to be as strong as that following Pandemrix.18 These findings, together with genetic evidence, suggest that narcolepsy is an autoimmune disease affecting hypocretin neurons and triggered by upper-airway infections.
Because of close physical proximity and a high degree of linkage disequilibrium (LD) observed for the HLA-DRB1 (MIM 142857) and HLA-DQB1 (MIM 604305) loci, it is difficult to assess additional effects of HLA-DR on susceptibility independently of HLA-DQ. In most ethnic groups, DQ0602 is exclusively associated with HLA-DRB1∗15:01, but studies in Chinese and African Americans, two populations where LD between these two alleles is lower, demonstrate that the association is primarily with DQ0602.3,19 Other minor associations have been reported for the DR locus (e.g., for rare HLA-DRB1∗04 subtypes20–23) but have never been confirmed on a large scale and across multiple ethnic groups.
Confirming the importance of HLA-DQ, additional HLA-DQ haplotypes consistently affect narcolepsy susceptibility when observed in trans of the major susceptibility haplotype HLA-DQA1∗01:02∼DQB1∗06:02. Similar trans heterodimer effects have been reported for other autoimmune diseases, such as celiac disease (MIM 212750) and type 1 diabetes (MIM 222100).24–26 In almost all cases, trans haplotypes that affect narcolepsy risk contain HLA-DQ alleles that are similar to HLA-DQA1∗01:02 or HLA-DQB1∗06:02 and, as a result, can cross-heterodimerize with DQ0602. Most notably, HLA-DQA1∗01:01∼DQB1∗05:01, HLA-DQA1∗01:03∼DQB1∗06:03, and HLA-DQA1∗01:03∼DQB1∗06:01 are protective against narcolepsy, whereas DQ0602 homozygosity increases risk in all ethnic groups.2,3,22,26–29 We postulate that this is due to allele competition, a model where risk is proportional to the amount of DQ0602 available and its unique ability to present a putative autoantigen.3,26 The model also predicts that any minor change in the DQ0602 antigen binding groove abolishes predisposition.
In addition to affecting allele competition, HLA-DQB1∗03:01 increases narcolepsy susceptibility when present in trans of DQ0602,3,22,26,27,29 an effect unlikely to be explained by allele competition given that HLA-DQB1∗03:01 does not heterodimerize with HLA-DQA1∗01:02 and thus should not affect DQ0602 dosage.30 Unlike DQ0602 dosage, HLA-DQB1∗03:01 also strongly reduces age of onset,2,5 suggesting that it acts through a different mechanism, for example, development of the TCR repertoire.
The strong and consistent association between narcolepsy and HLA-DQ has obscured studies of other HLA loci, such as HLA class I loci and other class II loci, including HLA-DP. Additional HLA class I effects have been reported in many HLA-class-II-associated diseases, suggesting an involvement of CD8+ T cells. For example, celiac disease and type 1 diabetes have weak HLA class I associations after HLA class II subtypes are controlled for. Type 1 diabetes also shows specific effects of HLA-DRB1∗04 subtypes in the presence of the same HLA-DQ heterodimer in Japan.31 More recently, HLA-DPA1 (MIM 142880) and HLA-DPB1 (MIM 142858) have been associated with several autoimmune diseases primarily associated with HLA-DR or HLA-DQ, such as type 1 diabetes, multiple sclerosis (MIM 126200),32,33 anti-glomerular basement membrane disease (MIM 233450),34 and myasthenia gravis (MIM 254200).35 Of notable interest are associations between HLA-DP and both influenza vaccine responses36 and chronic viral infections, notably of hepatitis B virus.37,38 To address the predisposition of HLA loci other than HLA-DQ in narcolepsy, we performed high-resolution class I and class II typing in HLA-DQ-matched narcoleptics versus control individuals and used imputation to replicate and extend our findings.
Material and Methods
HLA Typing and Selection of Samples
All narcolepsy-affected individuals were HLA-DQB1∗06:02 positive and had clear-cut cataplexy or documented low hypocretin-1 in the cerebrospinal fluid.5,39,40 A subset of samples of Asian and white ethnicity (590 case and 692 control individuals) and sourced from the Stanford Center for Narcolepsy database were typed with deep sequencing (HLA-DRB1, HLA-DQA1, and HLA-DQB1) and IMGT/HLA Database version 3140.41 With this information, a matched set of case and control individuals who shared the same ethnicity, country of origin, and HLA-DQA1 and HLA-DQB1 genotypes were selected, resulting in 322 case and 322 control individuals. For analysis of other loci, we further matched for HLA-DRB1, resulting in 304 case and 304 control individuals. These individuals were then typed for HLA-A (MIM 142800), HLA-B (MIM 142830), HLA-C (MIM 142840), HLA-DPA1, and HLA-DPB1 with the Luminex xMAP Technology at Stanford Medical School Blood Center. This sample constituted the HLA-typed matched set.
Two other cohorts, one white and one Chinese, were also included in the analysis, but in these cases HLA genotypes were imputed from HLA region SNP data. These cohorts did not overlap the 644 HLA-typed samples and constituted the HLA-imputed matched set. The white matched sample was selected among 1,540 case and 10,421 control individuals.39 Samples included previously published subjects sourced from the Stanford Center for Narcolepsy database and worldwide collaborators.39 DNA samples were genotyped on the Illumina ImmunoChip array at the University of Virginia and Stanford University. UCSC Genome Browser hg18 mapping was used as a reference. Illumina manifest file Immuno_BeadChip_1149691_B.bpm was used in the majority of cases. In cases where file Immuno_BeadChip_11419691_A was used, map positions were converted to be consistent with 1149691_B or were omitted from the analysis. Genotypes were called with Illumina GeneExpress (Illumina GenomeStudio GenTrain2.0 algorithm) with extensive additional curation.39 Individuals with a call rate under 0.98 (147 case and 123 control individuals) and samples that were related ( > 0.2) were excluded from further analysis. Data from all sources were merged in forward-strand format. Using the PLINK suite of software,42 we identified 142,054 high-quality SNPs with a call rate above 0.99 (in both case and control individuals separately) and passing Hardy-Weinberg equilibrium (HWE) filtering in control individuals (p > 1 × 10−5). Principal-component analysis for population stratification for this data set is shown in Figure S1.
The Chinese sample included a total of 1,189 narcolepsy subjects, 1,1365 of whom were seen at the sleep laboratory of Peking University People’s Hospital; this unit in the Department of Pulmonary Medicine evaluates patients with sleep disorders and receives referrals from all over China. In addition, 51 Asian samples came from Taiwan (Dr. Huang, National Taiwan University), and two came from Stanford. The individuals had hypocretin deficiency or clear-cut cataplexy and HLA-DQB1∗06:02. Affected subjects were mostly of Han descent (0.87) and from North China (0.85). The majority of the subjects were male (0.67) and children (0.70). Control genotypes from China came from university employees and students (0.41 male). In addition, we had shared control individuals from GWASs underway for colon cancer (MIM 114500) and Sjögren syndrome (MIM 270150). The Chinese data set was genotyped on the Affymetrix Axiom CHB (Han Chinese in Beijing, China) array. Genotypes were called with the Affymetrix Genotyping Console. Individuals who had a call rate < 0.95, were outliers after principal-component analysis (n = 47), or were related (n = 53) were removed, leaving 1,189 case and 1,997 control individuals. For the main association study, we selected SNP variants with a minor allele frequency (MAF) ≥ 0.01, a call rate ≥ 0.90, and a HWE p value ≥ 0.001 in control individuals. Principal-component analysis for population stratification for this data set is shown in Figure S1.
Ethics Statement
Informed consent in accordance with governing institutions was obtained from all subjects. The research protocols were approved by institutional-review-board panels on medical human subjects at Stanford University and the Beijing University People’s Hospital.
HLA Imputation in Samples with GWAS Data
HLA imputation for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1 was performed with the HIBAG package in R version 3.1.1 (July 10, 2014).43 HIBAG is an HLA-imputation tool that uses attribute bootstrap aggregation of several classifiers (SNPs) to select groups of SNPs that predict HLA type.44 For the ImmunoChip cohort, the imputation was performed with the European- and ImmunoChip-specific models from HIBAG. Imputation accuracy was verified by high-resolution typing in 177 individuals, resulting in imputation accuracy of 0.98 in HLA-A, 0.97 in HLA-B, 0.98 in HLA-C, 0.96 in HLA-DRB1, 1.00 in HLA-DQA1, 1.00 in HLA-DQB1, 1.00 in HLA-DPA1, and 0.92 in HLA-DPB1. The lower imputation quality of HLA-DPB1 was due to incorrectly imputed HLA-DPB1∗20:01, HLA-DPB1∗23:01, and HLA-DPB1∗06:01 genotypes, which were rare. Because HIBAG did not have built-in haplotypes for HLA-DPA1, we first built a model for HLA-DPA1 by using the type 1 diabetes consortium sample that had SNP and HLA information for 5,191 individuals from the SNP2HLA package.45,46
For the Chinese cohort, the Affymetrix CHB-specific chip reference panel was used for all loci but HLA-DPA1, for which a reference panel was built with HIBAG and publicly available Singapore Genome Variation Project (SGVP) data. One hundred Han Chinese individuals in SGVP have full 4-digit-level HLA typing and GWAS data available for HLA-DPA1. Imputation was verified for 254 individuals in the HLA class II genes, and the quality was high: 0.95 for HLA-DRB1, 0.94 for HLA-DQA1, and 0.98 for HLA-DQB1. Allele frequencies were within normal ranges according to dbMHC allele frequencies and earlier studies.47
Statistical Analysis
For stratified analysis, all samples were fully matched for country of origin and HLA-DRB1, HLA-DQB1, and HLA-DQA1 genotypes (for analysis of HLA class I and HLA-DP loci) or for country of origin and HLA-DQB1 and HLA-DQA1 genotypes (for analysis of the HLA-DR locus).
The analysis was carried out with carrier frequencies and the chi-square test with package meta.MH in R version 3.1.1 (July 10, 2014).43 Regional association plots were drawn with locus zoom.48 Sub-analyses of HLA loci were carried out with the Mantel-Haenszel test and, in the case of the HLA-DP heterodimer analysis, with a “one-by-one” sequential analysis that removed the effect of the most significant variant. This latter technique is similar to relative predisposition-effect statistics.49 Conditional analyses were performed on the full data sets with PLINK versions 1.7 and 1.9.42 In the conditional analysis, individuals homozygous for HLA-DQB1∗06:02 were removed. Meta-analyses for conditional analysis were performed with GWAMA.50 Nominal p values are reported for associations with p < 0.0005 after a Bonferroni correction for 100 tests. Other significant p < 0.05 associations are shown in Tables S1–S3, S4, S5, S6, S7 and S8, S9, S10–S16, and S17.
Results
HLA Class II Effects in HLA-DQ-Matched Narcolepsy Case and Control Individuals Reveal Strong Effects of HLA-DP
Genotype matching is the most conservative analytical method. The analysis of HLA-DRB1 was done in an HLA-DQ- and country-matched sample composed of 1,221 case and 1,221 control individuals. No residual HLA-DR association with narcolepsy was seen, except for a nominal association with HLA-DRB1∗04:03 (Table S1). Because HLA-DR and HLA-DQ display extremely high LD, all subsequent analyses were performed in a HLA-DRB1-, HLA-DQA1-, and HLA-DQB1-matched sample for a total number of 1,063 case and 1,063 control individuals.
The strongest findings were seen in the HLA class II HLA-DPB1 locus (Table 1), where HLA-DPB1∗04:02 conferred a strong protective effect against narcolepsy. In addition, HLA-DPB1∗05:01 increased the risk in Asians but not in whites (Table 1). Nominally protective effects were seen with HLA-DPB1∗04:01 and HLA-DPB1∗10:01 but not with other DPB1 alleles (Table S2). A nominally significant association was seen with HLA-DPA1∗01:03 (Table S3).
Table 1.
Association between HLA-DPB1 Alleles and Narcolepsy
| HLA-DPB1 Allele |
Asian |
White |
Mantel-Haenszel Test |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | OR (CI) | p | p Heterogeneity Test | |
| 04:02 | 49 (0.112) | 23 (0.052) | 0.44 (0.26–0.74) | 0.0014 | 114 (0.29) | 66 (0.17) | 0.50 (0.35–0.70) | 4.98 × 10−5 | 0.50 (0.38–0.66) | 6.105 × 10−07 | 0.914 |
| 05:01 | 236 (0.538) | 295 (0.67) | 1.76 (1.34–2.32) | 4.71 × 10−5 | 34 (0.09) | 30 (0.08) | 1.29 (0.86–1.92) | 0.221 | 1.48 (1.17–1.88) | 0.001 | 0.106 |
Case and control individuals were matched for HLA-DRB1, HLA-DQA1, and HLA-DQB1 alleles and for country and ethnicity. The p values were calculated with the Mantel-Haenszel test. Abbreviations are as follows: CI, confidence interval; Freq, carrier frequency; and OR, odds ratio.
In order to form functional HLA-DP molecules, the HLA-DPα and HLA-DPβ proteins (encoded by HLA-DPA1 and HLA-DPB1, respectively) need to heterodimerize. Heterodimerization of HLA-DPα and HLA-DPβ can occur in cis (on the same haplotype) or in trans (encoded by different chromosomes), provided that HLA-DPα and HLA-DPβ are biochemically compatible. HLA-DPA1 and HLA-DPB1 encode distinct amino acid motifs in the peptide-binding region, and polymorphisms at these positions determine which peptides can be bound by specific HLA-DP α and β subtypes and how they are presented to T cells (the so-called peptide-binding repertoire). To examine for potential effects in both cis and trans, we next performed stepwise analysis of HLA-DPA1-DPB1 heterodimers.
HLA-DPB1∗04:02 is in high LD with HLA-DPA1∗01:03, whereas HLA-DPB1∗05:01 is in LD with HLA-DPA1∗02:02. However, HLA-DPB1∗05:01 is also seen in cis with HLA-DPA1∗02:01.47,51 We thus tested a stepwise association of all possible heterodimers at the HLA-DP locus with narcolepsy across all samples. In the first pass analysis, HLA-DPA1∗01:03-DPB1∗04:02 (DP0402), followed by protective association with HLA-DPA1∗01:03-DPB1∗04:01 (DP0401), was most significantly associated with narcolepsy (Table 2). In addition, nominally significant associations were seen with HLA-DPA1∗02:02-DPB1∗19:01 and HLA-DPA1∗02:02-DPB1∗05:01 (DP0501) (Table 2).
Table 2.
Association between DPA1-DPB1 Heterodimers and Narcolepsy in Stepwise Analysis
| HLA-DPA1-DPB1 Heterodimer | No. of Control Subjects (Freq) | No. of Case Subjects (Freq) |
Mantel-Haenszel Test |
||
|---|---|---|---|---|---|
| OR (CI) | p | p Heterogeneity Test | |||
| 01:03-04:02 | 160 (0.15) | 88 (0.083) | 0.51 (0.38–0.67) | 1.01 × 10−06 | 0.852 |
| 01:03-04:01 | 516 (0.58) | 459 (0.52) | 0.61 (0.47–0.80) | 2.07 × 10−04 | 0.342 |
| 02:02-19:01 | 7 (0.020) | 0 (0.00) | 0 (0.00–NA) | 0.008 | NA |
| 02:02-05:01 | 193 (0.57) | 218 (0.64) | 1.41 (1.02–1.95) | 0.039 | 0.387 |
The p values were calculated with the chi-square test and Maentel-Haenszel test. The p heterogeneity test is Breslow-Day’s p value. Abbreviations are as follows: CI, confidence interval; Freq, carrier frequency; NA, not available (the exact OR or p value could not be calculated); and OR, odds ratio.
In narcolepsy, the largest risk is seen in individuals homozygous for HLA-DQB1∗06:02 or heterozygous for HLA-DQB1∗03:01 and HLA-DQB1∗06:02. The next largest risk is seen in individuals who are heterozygous but have neutral alleles on the other chromosome, whereas those who carry HLA-DQA1∗01 that is not HLA-DQA1∗01:02 in trans of HLA-DQB1∗06:02 are relatively protected.26 In a final analysis, we tested whether the effect size of HLA-DP was affected by the HLA-DQ risk groups by dividing the sample into groups according to these previously known HLA-DQ risk subgroups.22,26 The effects of HLA-DP did not differ across risk groups (Table S4).
Weak HLA Class I Associations in HLA-Class-II-Matched Narcolepsy Case and Control Individuals
We finally analyzed the effect of HLA-A, HLA-B, and HLA-C loci in HLA-DR- and HLA-DQ-matched subjects (Tables S5, S6, and S7). Nominally significant associations were seen with HLA-A∗02:07 (odds ratio [OR] = 1.66 [1.01–2.74], p = 0.046), HLA-A∗03:01 (OR = 0.79 [0.64–0.97], p = 0.024), HLA-A∗11:01 (OR = 1.43 [1.15–1.78], p = 0.001), HLA-A∗29:02 (OR = 0.50 [0.30–0.85], p = 0.008), HLA-B∗35:03 (OR = 2.30 [1.27–4.18], p = 0.005), HLA-B∗40:02 (OR = 0.54 [0.34–0.87]), HLA-B∗41:02 (OR = 0.14 [0.02–1.15], p = 0.33), HLA-B∗44:03 (OR = 0.55 [0.38–0.81], p = 0.002), HLA-B∗44:05 (OR = 0, p = 0.025), HLA-C∗05:01 (OR = 0.73 [0.54–0.99], p = 0.044), HLA-C∗14:03 (OR = 0.38 [0.15–1.01], p = 0.044), and HLA-C∗16:01 (OR = 0.42 [0.24–0.74]). Similar effects were also found after HLA-DP was matched between case and control individuals for the potential effect of extended haplotypes (Tables S8, S9, and S10).
Conditional Analysis Confirms Independent HLA-DP and Class I Effects
In order to study which HLA-DR and HLA-DQ alleles predispose to narcolepsy in the ImmunoChip and Asian data sets, we first performed stepwise analysis of HLA-DRB1, HLA-DQB1, and HLA-DQA1 loci in whites and Asians. As expected, we saw a strong predisposing effect of the known narcolepsy risk locus HLA-DQB1∗06:02 in whites and Asians (Tables S11 and S12). Similarly strong associations were seen with HLA-DRB1∗15:01, which is in strong LD with HLA-DQB1∗06:02, and with HLA-DQA1∗01:02, which is always present in HLA-DQB1∗06:02 haplotypes but is also found in other haplotypes. Figure 1A also shows GWAS data in the HLA region of whites (from ImmunoChip, see Faraco et al.39) and Asians (from Affymetrix CHB data, see Han et al.5) and a large association with the HLA-DR-DQ region, which obscured all other signals.
Figure 1.
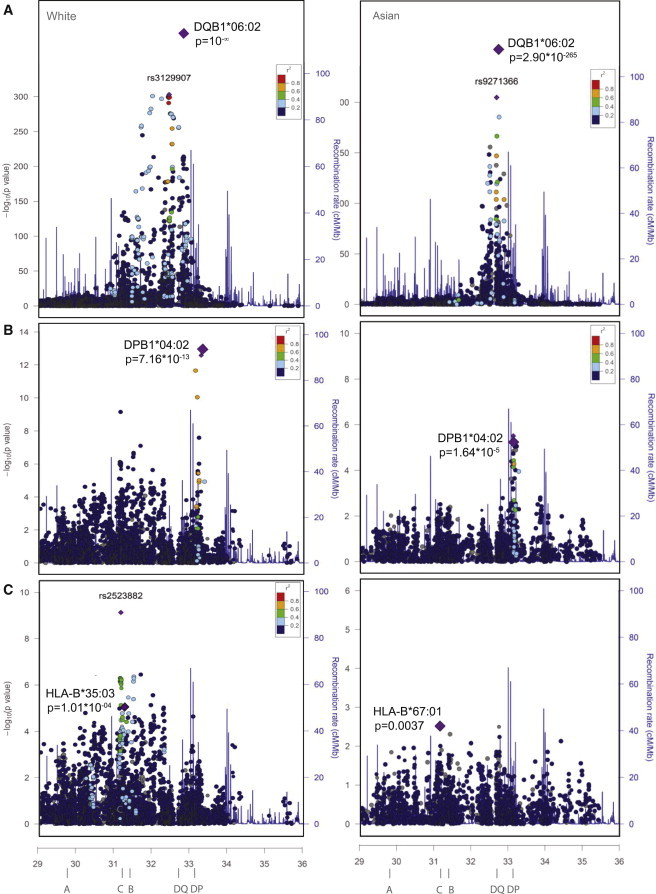
Association between HLA Loci and Narcolepsy
(A) Association of SNPs in the HLA region (Chr6: 29–36 Mb) reveals an overwhelming signal peaking at the level of HLA-DQB1 in white (from Immunochip, see Faraco et al.39) and chinese (from Affymetrix CHB data, see Han et al.5) individuals. Extended LD and association signal within the HLA-DR-DQ region obscure all other signals.
(B) After conditioning for HLA-DRB1, HLA-DQA1, and HLA-DQB1 (significant alleles from the stepwise analysis), a residual association is seen in the HLA-DP region.
(C) After conditioning for all significant HLA class II alleles, a remaining association is seen in the HLA class I region and is most visible proximal to HLA-B. A possible additional peak is seen in white individuals only in the vicinity of PSORS1.
We next performed stepwise conditioning with HLA-DQB1∗06:02 to examine the effects of other HLA-DRB1, HLA-DQA1, and HLA-DQB1 alleles. We did this after excluding subjects homozygous for HLA-DQB1∗06:02. The HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci were analyzed independently. As previously reported in multiple studies,2,3,22,27–29 we detected risk groups of HLA-DRB1, HLA-DQA1, and HLA-DQB1 associations known to act in trans of HLA-DRB1∗1501∼DQA1∗01:02∼DQB1∗06:02: (1) a set of protective alleles (HLA-DRB1∗13:01, HLA-DRB1∗01:01, HLA-DRB1∗08:03, HLA-DQB1∗05:01, HLA-DQB1∗06:03, HLA-DQA1∗01:01, and HLA-DQA1∗01:03) in high LD with similar ORs in both ethnic groups (Tables S11 and S12); (2) additional predisposing effects of HLA-DQA1∗01:02-bearing haplotypes (HLA-DQA1∗01:02 and HLA-DQB1∗05:02 in whites and HLA-DQA1∗01:02 in Asians); and (3) additional predisposing effects of HLA-DQB1∗03:01-bearing haplotypes (Tables S11 and S12). These effects are well established and are consistent with the effect of trans-heterodimerization of DQ1 alleles on DQ0602 and an additional effect of HLA-DQB1∗03:01. In addition, nominally significant effects were seen with HLA-DQB1∗02:02, HLA-DQB1∗04:02, and HLA-DQB1∗03:02 (Tables S11 and S12).
Associations were next conditioned on all significantly associated HLA-DR and HLA-DQ alleles and SNPs. As seen from residual HLA association effects, HLA-DPB1∗04:02 was again highly protective in both whites and Asians (Table 3). Similarly, HLA-DPB1∗05:01 also showed significant predisposing effects in narcolepsy (Table 3). Furthermore, HLA-DPB1∗02:01 was found as an additional association (Table 3; Tables S13 and S14). Figure 1B also shows GWAS data in the HLA region of whites (from ImmunoChip, see Franco et al.39) and Chinese (from Affymetrix CHB data, see Han et al.5) after conditioning for HLA-DR and HLA-DQ; it shows large residual association in the HLA-DP region and a main effect of HLA-DPB1∗04:02.
Table 3.
Association of HLA-DPB1 Alleles after Conditioning for HLA-DRB1, HLA-DQA1, and HLA-DQB1 Effects
|
ImmunoChip |
Chinese GWAS |
Meta-analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-DPB1 Allele | No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | OR | p Meta-analysis | I2 |
| 04:02 | 2,298 (0.22) | 138 (0.090) | 0.47 (0.38–0.58) | 7.15 × 10−13 | 258 (0.13) | 57 (0.048) | 0.38 (0.24–0.59) | 1.64 × 10−05 | 0.45 (0.38–0.55) | 8.99 × 10−17 | 0 |
| 02:01 | 2,527 (0.24) | 347 (0.23) | 1.39 (1.20–1.60) | 1.14 × 10−05 | 776 (0.39) | 640 (0.54) | 1.14 (0.93–1.39) | 0.2037 | 1.30 (1.15–1.46) | 1.74 × 10−05 | 0.584 |
| 05:01 | 400 (0.038) | 113 (0.073) | 1.43 (1.08–1.89) | 0.0123 | 1,187 (0.59) | 776 (0.65) | 1.35 (1.12–1.64) | 0.00186 | 1.38 (1.18–1.61) | 7.11 × 10−05 | 0 |
Abbreviations are as follows: CI, confidence interval; I2, heterogeneity in the meta-analysis as described in Higgins et al.60 (0 means no heterogeneity); and OR, odds ratio.
In a final analysis, we examined HLA class I associations after conditioning on all identified HLA class II (HLA-DR, HLA-DQ, and HLA-DP) effects. Statistically significant predisposing associations were seen with HLA-B∗51:01, HLA-B∗35:03, HLA-B∗18:01, HLA-C∗04:01, and HLA-A∗11:01, whereas HLA-B∗07:02 was protective (Table 4; Tables S15 and S16). Of special interest were associations with HLA-A∗11:01, HLA-B∗51:01, and HLA-B∗35:03 because these were in the same direction across ethnic groups, a finding more suggestive of a direct effect.
Table 4.
Association of HLA Class I Alleles after Conditioning for HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1 Alleles
| HLA Allele |
ImmunoChip |
Chinese GWAS |
Meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | No. of Control Subjects (Freq) | No. of Case Subjects (Freq) | OR (CI) | p | OR (CI) | p Meta-analysis | I2 | |
| HLA-B∗51:01 | 1,114 (0.11) | 189 (0.12) | 1.51 (1.24–1.85) | 5.45 × 10−5 | 248 (0.12) | 171 (0.14) | 1.41 (0.97–2.07) | 0.075 | 1.49 (1.25–1.78) | 1.09 × 10−5 | 0 |
| HLA-B∗35:03 | 158 (0.015) | 65 (0.042) | 1.96 (1.40–2.75) | 1.01 × 10−4 | 32 (0.016) | 20 (0.017) | 1.89 (0.62–5.64) | 0.257 | 1.95 (1.41–2.70) | 5.14 × 10−5 | 0 |
| HLA-B∗07:02 | 2,368 (0.23) | 886 (0.57) | 0.78 (0.68–0.88) | 8.70 × 10−5 | 119 (0.060) | 133 (0.11) | 1.03 (0.69–1.54) | 0.872 | 0.80 (0.71–0.90) | 2.22 × 10−4 | 0.441 |
| HLA-B∗18:01 | 1,047 (0.10) | 251 (0.16) | 1.46 (1.21–1.76) | 8.43 × 10−5 | 26 (0.013) | 15 (0.013) | 0.65 (0.61–1.65) | 0.361 | 1.41 (1.17–1.69) | 2.38 × 10−4 | 0.369 |
| HLA-C∗04:01 | 2,203 (0.21) | 259 (0.17) | 1.42 (1.20–1.69) | 6.89 × 10−5 | 225 (0.11) | 127 (0.11) | 0.92 (0.21–1.38) | 0.688 | 1.33 (1.13–1.56) | 4.45 × 10−4 | 0.732 |
| HLA-A∗11:01 | 1,206 (0.12) | 199 (0.13) | 1.28 (1.05–1.57) | 0.0146 | 654 (0.33) | 460 (0.39) | 1.38 (0.13–1.07) | 0.012 | 1.32 (1.13–1.54) | 4.92 × 10−4 | 0 |
Abbreviations are as follows: CI, confidence interval; Freq, carrier frequency; I2, heterogeneity as described in Higgins et al.60 (0 means no heterogeneity); and OR, odds ratio.
Figure 1C shows GWAS data for whites after conditioning for all class II (HLA-DR, HLA-DQ, and HLA-DP) effects; it shows complex residual association effects in the class I region. A common association is noted in both ethnic groups in the HLA-B region. In addition, a large association, peaking at rs2523882A (OR = 1.41 [1.26–1.57], p = 7.42 × 10−10), is noted in whites in the PSORS1 region. Surprisingly, several CHB panel SNPs with high LD with rs2523882 in Chinese were either weakly (rs2517474G, OR = 0.78 [0.64–0.96], p = 0.016) or not associated (rs3132564, rs62399065, and rs9263475). Because SNP coverage in this region is vastly superior in the ImmunoChip than in the CHB chip, additional fine typing will be needed to extend this observation.
Variation at the Amino Acid Level
In order to study whether amino acid polymorphisms across different HLA subtypes could affect the predisposition to narcolepsy, we imputed all amino acid polymorphisms in HLA alleles encoded by the different HLA-A, HLA-B, HLA-C, HLA-DPA1, and HLA-DPB1 loci and performed association testing in the typed and imputed data sets that had been matched for HLA class II and country of origin.
At HLA-DPB1, no independent amino acid was associated with narcolepsy. At HLA-DPA1, Ala11 and Gln50 were weakly protective (OR = 0.65 [0.47–0.86], p = 0.0029, and OR = 0.68 [0.52–0.88], p = 0.0035, respectively), and these effects recapitulated effects of the protective HLA-DPA1∗01:03 allele. These two HLA-DPA1 amino acids are present together in HLA-DPA1∗01:03, the most frequent HLA-DPA1 allele, which is protective in the context of HLA-DPA1∗01:03-DPB1∗04:02 and HLA-DPB1∗04:01. The lack of strong association with individual HLA-DPB1 amino acids suggests that larger binding motifs underlie the association with narcolepsy.
In the class I region, we found that HLA-A Tyr9 showed the strongest association with narcolepsy (OR = 1.35, [1.13–1.62], p = 0.0012), whereas only weak associations were seen with other amino acids. Interestingly, the predisposing HLA-A∗11:01 allele has this polymorphism, and it is also found in HLA-A∗25:01, which was detected with the conditional analysis.
Finally, we performed stepwise analysis with all class I alleles and HLA-A Tyr9 in the matched data set in order to see which alleles were driving the associations. The associations were nominally significant, and the strongest association was seen with HLA-C∗16:01, followed by HLA-A∗11:01, which explained in the stepwise analysis most of the HLA Tyr 9 association that was not significant after removal of the HLA-A∗11:01 carriers. Similar to the conditioned analysis, nominally significant associations were also seen with HLA-B∗35:03, HLA-B∗41:02, and HLA-B∗51:01 (Table S17).
Discussion
In this study, we discovered HLA risk loci and protective variants for narcolepsy. These effects were independent of the well-established HLA-DQ effects in narcolepsy. The strongest protection was seen with HLA-DPB1∗04:02 across all ethnic groups and data sets. Further, HLA-DPB1∗05:01 predisposed to narcolepsy independently of HLA-DPB1∗04:02 in Chinese individuals, where it is a common allele, confirming a recently published study in Japanese subjects.52 In addition, predisposing HLA class I associations were seen with HLA-A∗11:01, HLA-B∗35:03, and HLA-B∗51:01 across ethnic groups, although these effects were much weaker than HLA-DP effects. Finally, a possible remaining signal not explained by classic HLA gene polymorphisms was found near PSORS1 in the class I region of white subjects.
Our strongest findings indicate an independent role for HLA-DP molecules in narcolepsy susceptibility. In narcolepsy, the effect of heterodimerization of HLA-DQA1 and HLA-DQB1 is well established.26 In HLA-DP, there are only three common HLA-DPA1 genes that have very conserved haplotypes with HLA-DPB1. Analysis of possible cis (in the same haplotype) and trans (on the other chromosome) heterodimers revealed that the most protective heterodimer was HLA-DPA1∗01:03-DPB1∗04:02, whereas HLA-DPA1∗02:02-DPB1∗05:01 conferred the largest risk. These haplotypes were observed in cis, and the analysis of trans associations did not improve statistical significance.
The HLA-DP loci are important in the development of autoimmune diseases such as multiple sclerosis (MS),32,53 sarcoidosis (MIM 181000),54 and type 1 diabetes.33 Similar to in our findings, HLA-DPB1∗04:02 has been shown to be protective against type 1 diabetes and sarcoidosis,33,54 whereas HLA-DPB1∗05:01 has been associated with increased risk of MS.53,55 In addition, HLA-DPB1∗05:01 has been associated with non-clearance of viral infections such as that of chronic hepatitis B, whereas similar to in our study, HLA-DPB1∗04:02 is protective against this condition.38
The specific disease mechanisms underlying this new HLA-DP association in narcolepsy remain elusive. Narcolepsy was recently associated with pandemic H1N1 2009 vaccination11–17 and infections.8,10 In addition, streptococcal antibodies were found more frequently in narcoleptics than in matched healthy control individuals.9 These findings suggest that environmental triggers, such as upper-airway winter infections, are strong effectors in the development of narcolepsy. It is thus interesting to speculate that the presence of HLA-DP risk alleles, such as HLA-DPB1∗05:01, results in lower viral clearance or immune response, whereas the opposite might occur with protective alleles, such as HLA-DPB1∗04:02. In this model, a lower clearance of the viral trigger could be critical to the development of autoimmunity. HLA-DPB1∗05:01 has also been shown to be more common in individuals who do not develop seroprotection after hepatitis B vaccination.56
We also observed consistent associations of HLA class I alleles HLA-A∗11:01, HLA-B∗35:03, and HLA-B∗51:01 (predisposing) after correction of all HLA class II effects, suggesting an independent role for these HLA alleles. These findings are similar to those found in other autoimmune diseases, such as MS32,57,58 or type 1 diabetes,59 where the main risk alleles are located in the HLA class II region but residual association is seen in HLA class I. Of notable interest is the fact that in type 1 diabetes, a disease where HLA-DQB1∗06:02 is strongly protective, opposite effects to type 1 diabetes of HLA-A∗11:01 are also seen in narcolepsy. HLA class I effects in these disease might suggest the involvement of CD8+ T or natural killer cells, given that these three alleles are also known killer cell immunoglobulin-like receptor ligands.
To conclude, our findings suggest that the HLA associations in narcolepsy are more complex than previously thought and show that important high-risk variants reside outside the known HLA-DR-DQ risk region, notably in the HLA-DP region, where HLA-DPB1∗04:02 and HLA-DPB1∗05:01 have strong effects. We found additional HLA class I effects, some of which were most compatible with the direct effect of specific HLA alleles, and others will need further confirmation. Our study benefited from the evaluation of two ethnic groups, formal HLA typing, and HLA subtype imputation based on GWAS data. Combining these methods is likely to reveal a more precise picture of the role of the HLA region in autoimmune diseases such as narcolepsy.
Acknowledgments
We thank all the participating subjects, their families, and their physicians. We thank Jing Zhang for technical assistance. We thank collaborators M. Breban, W.M. Chen, P. Concannon, V. Damotte, P. Deloukas, M. Dobrovolná, L. Ehrmann, C. Erhardt, B. Fontaine, P. Geisler, C. Gieger, J. Hallmayer, P.E. Hesla, D. Kemlink, N. Klopp, L. Kolesar, P. Lichtner, S. Nevsimalova, G.T. Nepom, S. Onengut-Gumuscu, F. Poli, S.S. Rich, T.J. Rico, G. Rouleau, K. Sonka, S.D. Thompson, G. Trynka, C. Wijmenga, and J. Winkelmann for genotyping and providing samples for the study. The study was primarily funded by Wake Up Narcolepsy, NIH NS23724, and patient gifts to E.M. Funding to H.M.O. was provided by the Sigrid Juselius Foundation, the Paivikki and Sakari Sohlberg Foundation, and the Orion-Farmos Research Foundation. Funding for the Chinese portion of the study was supported by 973 Program 2015CB856405 and NSFC81420108002 to F.H. We thank the Wellcome Trust (British 1958 Birth Cohort Collection), and KORA (Kooperative Gesundheitsforschung in der Region Augsburg, Germany) for funding control genotypes. The KORA study was initiated and financed by the Helmholtz Zentrum München-German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Ressearch and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences, Ludwig-Maximilians-Universität, as part of LMUinnovativ.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Singapore Genome Variation Project (SGVP), http://www.statgen.nus.edu.sg/∼SGVP/
R project, http://www.r-project.org/
UCSC Human Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
References
- 1.Peyron C., Faraco J., Rogers W., Ripley B., Overeem S., Charnay Y., Nevsimalova S., Aldrich M., Reynolds D., Albin R. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 2000;6:991–997. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 2.Pelin Z., Guilleminault C., Risch N., Grumet F.C., Mignot E., US Modafinil in Narcolepsy Multicenter Study Group HLA-DQB1∗0602 homozygosity increases relative risk for narcolepsy but not disease severity in two ethnic groups. Tissue Antigens. 1998;51:96–100. doi: 10.1111/j.1399-0039.1998.tb02952.x. [DOI] [PubMed] [Google Scholar]
- 3.Han F., Lin L., Li J., Dong S.X., An P., Zhao L., Liu N.Y., Li Q.Y., Yan H., Gao Z.C. HLA-DQ association and allele competition in Chinese narcolepsy. Tissue Antigens. 2012;80:328–335. doi: 10.1111/j.1399-0039.2012.01948.x. [DOI] [PubMed] [Google Scholar]
- 4.Kornum B.R., Kawashima M., Faraco J., Lin L., Rico T.J., Hesselson S., Axtell R.C., Kuipers H., Weiner K., Hamacher A. Common variants in P2RY11 are associated with narcolepsy. Nat. Genet. 2011;43:66–71. doi: 10.1038/ng.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han F., Faraco J., Dong X.S., Ollila H.M., Lin L., Li J., An P., Wang S., Jiang K.W., Gao Z.C. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 2013;9:e1003880. doi: 10.1371/journal.pgen.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallmayer J., Faraco J., Lin L., Hesselson S., Winkelmann J., Kawashima M., Mayer G., Plazzi G., Nevsimalova S., Bourgin P. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat. Genet. 2009;41:708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dauvilliers Y., Maret S., Bassetti C., Carlander B., Billiard M., Touchon J., Tafti M. A monozygotic twin pair discordant for narcolepsy and CSF hypocretin-1. Neurology. 2004;62:2137–2138. doi: 10.1212/wnl.62.11.2137. [DOI] [PubMed] [Google Scholar]
- 8.Han F., Lin L., Warby S.C., Faraco J., Li J., Dong S.X., An P., Zhao L., Wang L.H., Li Q.Y. Narcolepsy onset is seasonal and increased following the 2009 H1N1 pandemic in China. Ann. Neurol. 2011;70:410–417. doi: 10.1002/ana.22587. [DOI] [PubMed] [Google Scholar]
- 9.Aran A., Lin L., Nevsimalova S., Plazzi G., Hong S.C., Weiner K., Zeitzer J., Mignot E. Elevated anti-streptococcal antibodies in patients with recent narcolepsy onset. Sleep. 2009;32:979–983. doi: 10.1093/sleep/32.8.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H., Zhuang J., Stone W.S., Zhang L., Zhao Z., Wang Z., Yang Y., Li X., Zhao X., Zhao Z. Symptoms and occurrences of narcolepsy: a retrospective study of 162 patients during a 10-year period in eastern China. Sleep Med. 2014;15:607–613. doi: 10.1016/j.sleep.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Partinen M., Saarenpää-Heikkilä O., Ilveskoski I., Hublin C., Linna M., Olsén P., Nokelainen P., Alén R., Wallden T., Espo M. Increased incidence and clinical picture of childhood narcolepsy following the 2009 H1N1 pandemic vaccination campaign in Finland. PLoS ONE. 2012;7:e33723. doi: 10.1371/journal.pone.0033723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nohynek H., Jokinen J., Partinen M., Vaarala O., Kirjavainen T., Sundman J., Himanen S.L., Hublin C., Julkunen I., Olsén P. AS03 adjuvanted AH1N1 vaccine associated with an abrupt increase in the incidence of childhood narcolepsy in Finland. PLoS ONE. 2012;7:e33536. doi: 10.1371/journal.pone.0033536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Flanagan D., Barret A.S., Foley M., Cotter S., Bonner C., Crowe C., Lynch B., Sweeney B., Johnson H., McCoy B., Purcell E. Investigation of an association between onset of narcolepsy and vaccination with pandemic influenza vaccine, Ireland April 2009-December 2010. Euro Surveill. 2014;19:15–25. [PubMed] [Google Scholar]
- 14.Persson I., Granath F., Askling J., Ludvigsson J.F., Olsson T., Feltelius N. Risks of neurological and immune-related diseases, including narcolepsy, after vaccination with Pandemrix: a population- and registry-based cohort study with over 2 years of follow-up. J. Intern. Med. 2014;275:172–190. doi: 10.1111/joim.12150. [DOI] [PubMed] [Google Scholar]
- 15.Miller E., Andrews N., Stellitano L., Stowe J., Winstone A.M., Shneerson J., Verity C. Risk of narcolepsy in children and young people receiving AS03 adjuvanted pandemic A/H1N1 2009 influenza vaccine: retrospective analysis. BMJ. 2013;346:f794. doi: 10.1136/bmj.f794. [DOI] [PubMed] [Google Scholar]
- 16.Dauvilliers Y., Arnulf I., Lecendreux M., Monaca Charley C., Franco P., Drouot X., d’Ortho M.P., Launois S., Lignot S., Bourgin P., Narcoflu-VF study group Increased risk of narcolepsy in children and adults after pandemic H1N1 vaccination in France. Brain. 2013;136:2486–2496. doi: 10.1093/brain/awt187. [DOI] [PubMed] [Google Scholar]
- 17.Heier M.S., Gautvik K.M., Wannag E., Bronder K.H., Midtlyng E., Kamaleri Y., Storsaeter J. Incidence of narcolepsy in Norwegian children and adolescents after vaccination against H1N1 influenza A. Sleep Med. 2013;14:867–871. doi: 10.1016/j.sleep.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Montplaisir J., Petit D., Quinn M.-J., Ouakki M., Deceuninck G., Desautels A., Mignot E., De Wals P. Risk of narcolepsy associated with inactivated adjuvanted (AS03) A/H1N1 (2009) pandemic influenza vaccine in Quebec. PLoS ONE. 2014;9:e108489. doi: 10.1371/journal.pone.0108489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki K., Grumet F.C., Lin X., Gelb M., Guilleminault C., Dement W.C., Mignot E. DQ (rather than DR) gene marks susceptibility to narcolepsy. Lancet. 1992;339:1052. doi: 10.1016/0140-6736(92)90571-j. [DOI] [PubMed] [Google Scholar]
- 20.Roh E.Y., Park M.H., Park H., Park D.H., Choi J.B., Kim S.J., Jeong D.U. Association of HLA-DR and -DQ genes with narcolepsy in Koreans: comparison with two control groups, randomly selected subjects and DRB1∗1501-DQB1∗0602—positive subjects. Hum. Immunol. 2006;67:749–755. doi: 10.1016/j.humimm.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 21.Hong S.C., Leen-Kim, Park S.A., Han J.H., Lee S.P., Lin L., Okun M., Nishino S., Mignot E. HLA and hypocretin studies in Korean patients with narcolepsy. Sleep. 2002;25:440–444. [PubMed] [Google Scholar]
- 22.Mignot E., Lin L., Rogers W., Honda Y., Qiu X., Lin X., Okun M., Hohjoh H., Miki T., Hsu S. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am. J. Hum. Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mignot E., Lin L., Li H. HLA allele and microsatellite studies in narcolepsy. In: Hansen J., Dupont B., editors. HLA 2004, Immunobiology of the Human MHC, Proceedings of the 13th International Histocompatibility Workshop and Congress. IHWG Press; Seattle: 2006. pp. 817–823. [Google Scholar]
- 24.Megiorni F., Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J. Biomed. Sci. 2012;19:88. doi: 10.1186/1423-0127-19-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eerligh P., van Lummel M., Zaldumbide A., Moustakas A.K., Duinkerken G., Bondinas G., Koeleman B.P., Papadopoulos G.K., Roep B.O. Functional consequences of HLA-DQ8 homozygosity versus heterozygosity for islet autoimmunity in type 1 diabetes. Genes Immun. 2011;12:415–427. doi: 10.1038/gene.2011.24. [DOI] [PubMed] [Google Scholar]
- 26.Ollila H.M., Fernandez-Vina M., Mignot E. HLA-DQ allele competition in narcolepsy: A comment on Tafti et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014 doi: 10.5665/sleep.4342. Published online October 17, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong S.C., Lin L., Lo B., Jeong J.H., Shin Y.K., Kim S.Y., Kweon Y., Zhang J., Einen M., Smith A. DQB1∗0301 and DQB1∗0601 modulate narcolepsy susceptibility in Koreans. Hum. Immunol. 2007;68:59–68. doi: 10.1016/j.humimm.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Hohjoh H., Terada N., Nakayama T., Kawashima M., Miyagawa T., Honda Y., Tokunaga K. Case-control study with narcoleptic patients and healthy controls who, like the patients, possess both HLA-DRB1∗1501 and -DQB1∗0602. Tissue Antigens. 2001;57:230–235. doi: 10.1034/j.1399-0039.2001.057003230.x. [DOI] [PubMed] [Google Scholar]
- 29.Tafti M., Hor H., Dauvilliers Y., Lammers G.J., Overeem S., Mayer G., Javidi S., Iranzo A., Santamaria J., Peraita-Adrados R. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwok W.W., Kovats S., Thurtle P., Nepom G.T. HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J. Immunol. 1993;150:2263–2272. [PubMed] [Google Scholar]
- 31.Sugihara S., Ogata T., Kawamura T., Urakami T., Takemoto K., Kikuchi N., Takubo N., Tsubouchi K., Horikawa R., Kobayashi K., Japanese Study Group of Insulin Therapy for Childhood and Adolescent Diabetes (JSGIT) HLA-class II and class I genotypes among Japanese children with Type 1A diabetes and their families. Pediatr. Diabetes. 2012;13:33–44. doi: 10.1111/j.1399-5448.2011.00833.x. [DOI] [PubMed] [Google Scholar]
- 32.Patsopoulos N.A., Barcellos L.F., Hintzen R.Q., Schaefer C., van Duijn C.M., Noble J.A., Raj T., Gourraud P.A., Stranger B.E., Oksenberg J., IMSGC. ANZgene Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013;9:e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varney M.D., Valdes A.M., Carlson J.A., Noble J.A., Tait B.D., Bonella P., Lavant E., Fear A.L., Louey A., Moonsamy P., Type 1 Diabetes Genetics Consortium HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2010;59:2055–2062. doi: 10.2337/db09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo H., Chen M., Cui Z., Yang R., Xu P.C., Zhou X.J., Zhao M.H. The association of HLA-DQB1, -DQA1 and -DPB1 alleles with anti- glomerular basement membrane (GBM) disease in Chinese patients. BMC Nephrol. 2011;12:21. doi: 10.1186/1471-2369-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horiki T., Inoko H., Moriuchi J., Ichikawa Y., Arimori S. Combinations of HLA-DPB1 and HLA-DQB1 alleles determine susceptibility to early-onset myasthenia gravis in Japan. Autoimmunity. 1994;19:49–54. doi: 10.3109/08916939409008008. [DOI] [PubMed] [Google Scholar]
- 36.Moss A.J., Gaughran F.P., Karasu A., Gilbert A.S., Mann A.J., Gelder C.M., Oxford J.S., Stephens H.A., Lambkin-Williams R. Correlation between human leukocyte antigen class II alleles and HAI titers detected post-influenza vaccination. PLoS ONE. 2013;8:e71376. doi: 10.1371/journal.pone.0071376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida N., Sawai H., Kashiwase K., Minami M., Sugiyama M., Seto W.K., Yuen M.F., Posuwan N., Poovorawan Y., Ahn S.H. New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS ONE. 2014;9:e86449. doi: 10.1371/journal.pone.0086449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamatani Y., Wattanapokayakit S., Ochi H., Kawaguchi T., Takahashi A., Hosono N., Kubo M., Tsunoda T., Kamatani N., Kumada H. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 39.Faraco J., Lin L., Kornum B.R., Kenny E.E., Trynka G., Einen M., Rico T.J., Lichtner P., Dauvilliers Y., Arnulf I. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9:e1003270. doi: 10.1371/journal.pgen.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.American Academy of Sleep Medicine . American Academy of Sleep Medicine; Chicago: 2014. The International Classification of Sleep Disorders, Third Edition. [Google Scholar]
- 41.Wang C., Krishnakumar S., Wilhelmy J., Babrzadeh F., Stepanyan L., Su L.F., Levinson D., Fernandez-Viña M.A., Davis R.W., Davis M.M., Mindrinos M. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:8676–8681. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Development Core Team . R Foundation for Statistical Computing; Vienna: 2010. R: A language and environment for statistical computing. [Google Scholar]
- 44.Zheng X., Shen J., Cox C., Wakefield J.C., Ehm M.G., Nelson M.R., Weir B.S. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rich S.S., Concannon P., Erlich H., Julier C., Morahan G., Nerup J., Pociot F., Todd J.A. The Type 1 Diabetes Genetics Consortium. Ann. N Y Acad. Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 47.Hollenbach J.A., Madbouly A., Gragert L., Vierra-Green C., Flesch S., Spellman S., Begovich A., Noreen H., Trachtenberg E., Williams T. A combined DPA1∼DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics. 2012;64:559–569. doi: 10.1007/s00251-012-0615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollenbach J.A., Mack S.J., Thomson G., Gourraud P.A. Analytical methods for disease association studies with immunogenetic data. Methods Mol. Biol. 2012;882:245–266. doi: 10.1007/978-1-61779-842-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mägi R., Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Begovich A.B., Moonsamy P.V., Mack S.J., Barcellos L.F., Steiner L.L., Grams S., Suraj-Baker V., Hollenbach J., Trachtenberg E., Louie L. Genetic variability and linkage disequilibrium within the HLA-DP region: analysis of 15 different populations. Tissue Antigens. 2001;57:424–439. doi: 10.1034/j.1399-0039.2001.057005424.x. [DOI] [PubMed] [Google Scholar]
- 52.Miyagawa T., Toyoda H., Hirataka A., Kanbayashi T., Imanishi A., Sagawa Y., Kotorii N., Kotorii T., Hashizume Y., Ogi K. New susceptibility variants to narcolepsy identified in HLA class II region. Hum. Mol. Genet. 2014 doi: 10.1093/hmg/ddu480. [DOI] [PubMed] [Google Scholar]
- 53.Kira J. Multiple sclerosis in the Japanese population. Lancet Neurol. 2003;2:117–127. doi: 10.1016/s1474-4422(03)00308-9. [DOI] [PubMed] [Google Scholar]
- 54.Wennerström A., Vlachopoulou E., Lahtela L.E., Paakkanen R., Eronen K.T., Seppänen M., Lokki M.L. Diversity of extended HLA-DRB1 haplotypes in the Finnish population. PLoS ONE. 2013;8:e79690. doi: 10.1371/journal.pone.0079690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X.M., Wang C., Zhang K.N., Lin A.Y., Kira J., Hu G.Z., Qu X.H., Xiong Y.Q., Cao W.F., Gong L.Y. Association of susceptibility to multiple sclerosis in Southern Han Chinese with HLA-DRB1, -DPB1 alleles and DRB1-DPB1 haplotypes: distinct from other populations. Mult. Scler. 2009;15:1422–1430. doi: 10.1177/1352458509345905. [DOI] [PubMed] [Google Scholar]
- 56.Wu T.W., Chu C.C., Liao H.W., Lin S.K., Ho T.Y., Lin M., Lin H.H., Wang L.Y. HLA-DPB1 and anti-HBs titer kinetics in hepatitis B booster recipients who completed primary hepatitis B vaccination during infancy. Genes Immun. 2014;15:47–53. doi: 10.1038/gene.2013.62. [DOI] [PubMed] [Google Scholar]
- 57.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Link J., Lorentzen A.R., Kockum I., Duvefelt K., Lie B.A., Celius E.G., Harbo H.F., Hillert J., Brynedal B. Two HLA class I genes independently associated with multiple sclerosis. J. Neuroimmunol. 2010;226:172–176. doi: 10.1016/j.jneuroim.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Nejentsev S., Howson J.M., Walker N.M., Szeszko J., Field S.F., Stevens H.E., Reynolds P., Hardy M., King E., Masters J., Wellcome Trust Case Control Consortium Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


